Abstract
Alanine racemase, encoded by the gene alr, is an important enzyme in the synthesis of d-alanine for peptidoglycan biosynthesis. Strains of Mycobacterium smegmatis with a deletion mutation of the alr gene were found to require d-alanine for growth in both rich and minimal media. This indicates that alanine racemase is the only source of d-alanine for cell wall biosynthesis in M. smegmatis and confirms alanine racemase as a viable target gene for antimycobacterial drug development.
Tuberculosis is the world's leading cause of mortality among infectious diseases. Annually nearly 2 million deaths are attributed to tuberculosis, and one-third of the world's population is latently infected with the tuberculosis bacillus (6, 35). Because of its importance to global health, there is great interest in the identification and development of targets for antimycobacterial drug design. One such target of significant historical interest is alanine racemase (5, 14, 22). Alanine racemase is a pyridoxal phosphate-containing homodimeric enzyme that catalyzes the conversion of l-alanine to d-alanine, a key building block in the biosynthesis of the peptidoglycan layer in bacterial cell walls. Alanine racemases are typically absent in eukaryotes but ubiquitous among prokaryotes, which makes this enzyme an attractive target for the development of novel antimicrobials.
Although d-alanine is essential for bacterial cell wall formation, determining which genes are crucial in the d-alanine biosynthesis pathway has proven to be more complicated. Bacteria contain either one or two alanine racemase genes. In species with two genes, one is constitutively expressed and anabolic, while the other is inducible and catabolic (25-27). These genes supply the d-alanine needed for cell wall biosynthesis, and knockout studies with several of these bacteria have established that the alanine racemase enzyme is essential for growth in the absence of exogenous d-alanine (9, 12, 18, 24, 33, 34). In some bacteria, an alternate pathway to d-alanine exists via utilization of d-amino acid transaminase. For example, in Listeria monocytogenes both the dal and dat genes, encoding alanine racemase and d-amino acid transaminase, respectively, must be inactivated in order to produce a d-alanine auxotroph (29). It is also true that for some bacteria that possess a d-amino acid transaminase, such as Bacillus subtilis, alanine racemase has been shown to be essential in both rich and minimal media containing l-alanine (2, 8, 28).
In mycobacteria, the data regarding the essentiality of alanine racemase have been contradictory to date. One study with Mycobacterium smegmatis suggested that even in the absence of d-alanine, alanine racemase was not required for growth (4). In that study the alanine racemase gene was insertionally inactivated. Although the inactivation was not lethal, the mutant displayed a greatly reduced ability to grow and a hypersensitivity to d-cycloserine, an inhibitor of alanine racemase. The authors attributed their unexpected result to the possible presence of an alternate pathway for d-alanine synthesis, possibly involving d-amino acid transaminase. However, the subsequent publication of the M. smegmatis mc2155 genome has failed to show any significant homologies to d-amino acid transaminases (Mycobacterium smegmatis MC2 Genome Page [http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=gms]). In addition, in a landmark study by Sassetti et al. that delineated through genome-wide transposon insertional inactivation all M. tuberculosis genes essential for growth, alanine racemase was clearly identified as essential (21). Because of this unexpected discrepancy between the findings for Mycobacterium tuberculosis and M. smegmatis and because M. smegmatis has shown great utility recently as a surrogate for M. tuberculosis in antimycobacterial drug design (1), we decided to reinvestigate this issue.
Deletion of the alr gene in M. smegmatis results in an obligate growth requirement for d-alanine.
The alr gene was inactivated in M. smegmatis mc2155 (23) by replacement of 99% of its coding sequence with the kanamycin resistance cassette from pUC18K (Table 1) using standard protocols for growth and mutagenesis as previously described (30). All general cloning steps were performed in Escherichia coli DH10B with culturing at 37°C in LB broth at an initial pH of 7.0 or on LB agar plates (20). When needed, sucrose was added to plates at 10% (wt/vol), and kanamycin, hygromycin, and gentamicin were used at final concentrations of 25, 50, and 5 μg/ml, respectively. The flanking regions (1,000 bp) of the alr gene were PCR amplified using the primer pair AlrKO1 and AlrKO2 and the pair AlrKO3 and AlrKO4 (Table 1). The resulting amplicons were then ligated simultaneously with a kanamycin resistance cassette (KpnI-HindIII fragment containing aphA-3 from pUC18K) into the BamHI-XbaI-digested delivery vector pPR23 (19, 30), thus creating pDM2 (Fig. 1). The knockout delivery vector pDM2 was electroporated into the M. smegmatis strain mc2155 (0.2-cm-gap cuvette at 2.5 kV, 1,000 Ω, and 25 μF), and kanamycin-resistant (Kanr) colonies were selected at 28°C to allow replication of the temperature-sensitive vector. The resulting strain (DM5) was grown to exponential phase at 28°C in Luria-Bertani medium supplemented with 0.05% (wt/vol) Tween 80 (Sigma) (LBT) containing 25 μg/ml kanamycin and 5 mM d,l-alanine. Aliquots of the culture were plated on LBT plates containing 10% sucrose, 25 μg/ml kanamycin, and 5 mM d,l-alanine and were incubated at 40°C. Another DM5 culture was prepared and plated in an identical manner, except that d,l-alanine was omitted from all the culture media.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant genotype and/or information | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH10B | F−araD139 Δ(ara leu)7697 ΔlacX74 galU galK rpsL deoR φ80dlacZΔM15 endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | 11 |
| M. smegmatis | ||
| mc2155 | Wild type, ept-1 | 23 |
| DM5 | mc2155 harboring pDM2 (Δalr::aphA-3 Kanr Gmr) | This study |
| DM22 | mc2155 Δalr::aphA-3 Kanr | This study |
| DM30 | DM22 harboring pDM3; Hygr | This study |
| Plasmids | ||
| pPR23 | E. coli-mycobacterial shuttle vector; oriM(Ts) sacB Gmr | 19 |
| pUHA267 | E. coli-mycobacterial shuttle vector; Hygr | AgResearch, Wallaceville, New Zealand |
| pDM2 | pPR23 harboring Δalr::aphA-3; Kanr Gmr | This study |
| pDM3 | pUHA267 harboring the wild-type alr and upstream promoter region; Hygr | This study |
| Primers | ||
| AlrKO1 | 5′-AAATTTGGATCCGACAAGAACATGATCGACAA-3′ | This study |
| AlrKO2 | 5′-AAATTTGGTACCGGTCTGCATCGTCATAATCT-3′ | This study |
| AlrKO3 | 5′-AAATTTAAGCTTGGCAGGACAACAAGATTGAG-3′ | This study |
| AlrKO4 | 5′-AAATTTTCTAGAGCCCTCGACATCCATTGCTT-3′ | This study |
| 5alr2 | 5′-AAATTTCCATGGTGGGGCAGTACTACAACTTC-3′ | This study |
| 3alr2 | 5′-AAATTTAAGCTTGTAGGGGTCTTCCTTGCTCA-3′ | This study |
FIG. 1.
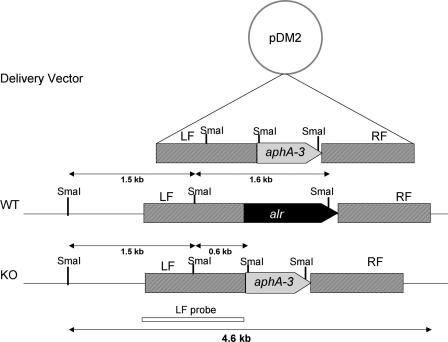
Construction of a Δalr mutant of M. smegmatis mc2155. Replacement of the chromosomal alr gene with the kanamycin cassette (aphA-3 gene) results from crossover events in the right and left flanks. Restriction sites of SmaI and fragments expected in Southern hybridization analysis are indicated for wild-type mc2155 (WT) and for the Δalr mutant (KO). LF, left flank; RF, right flank.
We predicted that deletion of the alr gene would lead to a requirement for d-alanine for growth. As a first test of this hypothesis, selection for the alr deletion was performed in the presence and absence of d,l-alanine. When 5 mM d,l-alanine was included in the initial growth medium and plates, 19 Kanr Sucr colonies appeared on plates at the nonpermissive temperature. Six colonies were picked for characterization, and four out of six analyzed strains (DM22 to DM25) were found to require d-alanine for growth in both LBT and minimal (7H11) media (Table 2). In contrast, the two remaining strains (DM21 and DM26), which did not require d-alanine for growth, remained Gmr, indicating that they still harbored vector sequences and suggesting that spontaneous sacB mutations were responsible for their ability to grow in the presence of sucrose.
TABLE 2.
Antibiotic resistance and growth requirements of six candidate alr deletion mutants (DM21 to DM26) selected in the presence of d-alanine
| Colony/strain | Growth on selective mediuma
|
|||
|---|---|---|---|---|
| Kanamycin | Sucrose | Gentamicin | Without d-Ala | |
| DM21 | + | + | + | + |
| DM22 | + | + | − | − |
| DM23 | + | + | − | − |
| DM24 | + | + | − | − |
| DM25 | + | + | − | − |
| DM26 | + | + | + | + |
M. smegmatis strain DM5, which harbors the Δalr delivery vector pDM2, was grown to exponential phase at 28°C in LBT medium supplemented with 5 mM d,l-alanine. Aliquots of the culture were plated onto LBT-kanamycin-sucrose plates supplemented with 5 mM d,l-alanine. The plates were incubated at 40°C to select for chromosomal integration of the vector.
The genetic selection was repeated in the absence of d,l-alanine supplementation. While 15 Kanr Sucr colonies appeared on selection plates without d,l-alanine supplementation, subsequent testing of colonies for d-alanine auxotrophy showed that all of these strains (six out of six analyzed, strains DM11 to DM16) grew normally in the absence of d-alanine. These colonies could be a result of single recombination events and contain a full-length copy of alr in addition to delivery vector sequences, as indicated by Southern hybridization results described below (Fig. 2). All of these colonies were found to be Gmr, indicating that they still harbored vector sequences, suggesting that spontaneous mutations had occurred in sacB resulting in their ability to grow in the presence of sucrose.
FIG. 2.
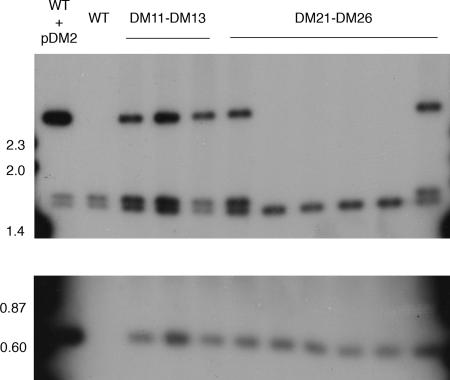
Southern hybridization analysis of candidate Δalr mutants. SmaI digests of genomic DNA were probed with the radiolabeled left flank probe of the deletion construct (Fig. 1). Strains that are wild type at the alr locus are predicted to have doublet bands in the 1.5-kb region, while Δalr strains should have a single band in this region. The lower portion is a longer exposure of the same blot to allow detection of the 0.6-kb band detected in all strains except wild-type mc2155. Molecular masses of size markers are indicated in kilobases. The phenotypes of strains DM21 to DM26 are shown in Table 2.
Confirmation that the alr gene was deleted in the d-alanine auxotrophs was obtained by Southern hybridization analysis (Fig. 2). Genomic DNA was isolated from the parent and mutant strains and digested with SmaI. Hybridization with the left flank of the alr gene (Fig. 1) revealed hybridizing bands of 1.56 kb and 1.64 kb in the wild-type strain mc2155 (Fig. 2, lane 2), while bands of 1.56 kb and 0.6 kb were observed in the Δalr strains (Fig. 2, lanes 7 to 10), as predicted by sequence analysis (Fig. 1). DM5, the wild-type mc2155 strain containing the pDM2 delivery vector (grown at 28°C to allow propagation of the temperature-sensitive vector), shows the presence of an additional plasmid-derived 2.5-kb band (Fig. 2, lane 1), which is not present in the final Δalr mutant strains. All strains selected in the absence of d,l-alanine, as well as the two colonies selected in the presence of d,l-alanine that were not d-alanine auxotrophs, showed the doublet found in the wild-type strain and the 2.5-kb plasmid-derived band, suggesting that these strains were the result of a single crossover event that left both plasmid and wild-type alr sequences in place on the chromosome.
Although the Kanr cassette from pUC18K has been reported to be nonpolar (17), we confirmed that the d-alanine requirement of the Δalr mutant was due only to loss of alanine racemase activity by complementation with a full-length copy of alr supplied in trans. A construct for complementation of the Δalr mutant was cloned into the integrative E. coli/mycobacterial shuttle vector pUHA267 (15). A 1.7-kb PCR product encompassing the alr gene plus approximately 500 bp of upstream DNA was amplified by PCR using primers 5alr2 and 3alr2 (Table 1) and ligated into the NcoI and HindIII sites of pUHA267, creating the plasmid pDM3. pDM3 was electroporated into the Δalr strain DM22 to create the Kanr Hygr strain DM30. Unlike the parental Δalr strain, DM30 was able to grow normally in the absence of d-alanine. This confirms that deletion of the alr gene is necessary and sufficient to create a d-alanine auxotroph and thus also shows that the alr gene is required for survival of M. smegmatis under normal growth conditions.
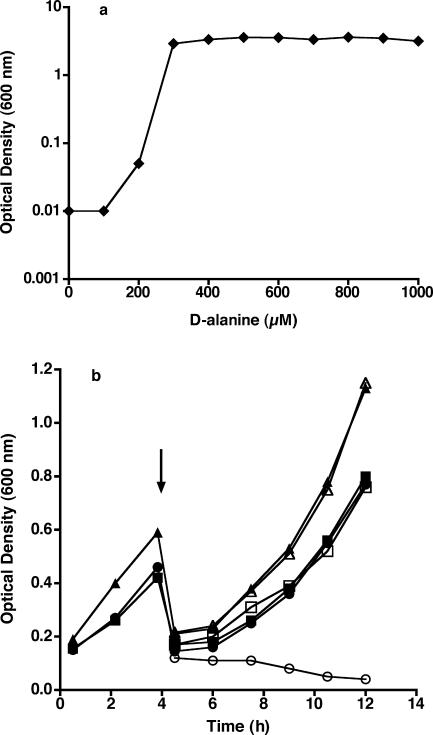
Further characterization of the growth characteristics of the Δalr strain DM22 and the complemented Δalr strain DM30 was carried out in liquid culture (Fig. 3). In order to determine the concentration of d-alanine required for growth of the Δalr strain DM22, LBT medium containing various concentrations of d-alanine was inoculated and incubated overnight at 37°C. As shown in Fig. 3a, growth appeared normal (final optical density at 600 nm, ∼3.5) at all starting d-alanine concentrations of 300 μM or greater, while no growth was seen at or below 200 μM d-alanine. Visual inspection by light microscopy revealed no differences in cell morphology between wild-type M. smegmatis and the Δalr strain DM22 grown in the presence of 300 μM or greater d-alanine. The optical density of DM22 cultures did not increase in the presence of 200 μM d-alanine, and visual inspection by light microscopy of the Δalr strain DM22 in the absence of d-alanine showed on average a doubling or tripling in the length of the bacterial cells. These results suggest that the Δalr strain is unable to generate a critical component of the cell division machinery in the absence of exogenous d-alanine, presumably related to an inability to incorporate d-alanine into peptidoglycan. As shown in Fig. 3b, the growth of DM22 and DM30 in LBT medium supplemented with 1 mM d-alanine was very similar to that of the wild-type strain. When cells were collected by centrifugation and resuspended in LBT medium without d-alanine, growth of the DM30 and wild-type strains continued normally, while growth of the Δalr strain DM22 was arrested (Fig. 3b). The optical density of the DM22 culture without added d-alanine decreased slightly over time.
FIG. 3.
Growth profiles of M. smegmatis mc2155 and mutants. (a) Growth of Δalr strain DM22 in LBT medium as a function of initial d-alanine concentration. (b) Growth of mc2155 (squares), Δalr strain DM22 (circles), and complemented Δalr strain DM30 (triangles) in LBT medium containing 1 mM d-alanine. At the 4-h time point (marked by arrow), cultures were collected by centrifugation, washed with LBT medium, and then split into LBT medium without (open symbols) or with (closed symbols) 1 mM d-alanine.
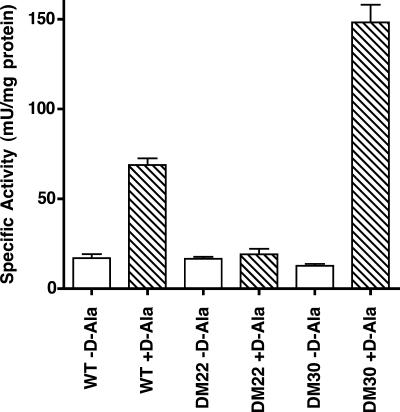
In order to determine the effect of deletion of the alr gene on alanine racemase activity, cleared cell lysates were prepared from cultures of wild-type M. smegmatis, the Δalr strain DM22, and the complemented Δalr strain DM30 and assayed for enzyme activity (Fig. 4). M. smegmatis lysates were prepared from cultures grown in LBT supplemented with 1 mM d-alanine. Cells were collected by centrifugation and then washed twice and concentrated 25-fold in 50 mM Tris-HCl (pH 8). Cells were disrupted by three passages through a French press at 18,000 lb/in2, and lysates were cleared of cell debris by centrifugation at 20,000 × g for 15 min. Cell membranes were separated from the cytoplasmic fraction by centrifugation at 140,000 × g for 45 min at 4°C, and the supernatant (soluble fraction containing alanine racemase) was passed through a 0.22-μm-pore-size filter. Alanine racemase activity was measured spectrophotometrically at 23°C in a coupled assay in the d- to l-alanine direction in which l-alanine is formed by alanine racemase found in the lysate and then deaminated to pyruvate by the NAD+-dependent l-alanine dehydrogenase from B. subtilis, and the formation of NADH is measured at 340 nm (7). Aliquots of the soluble fraction were added to 50 mM Tricine, pH 8.5, 10 mM NAD+, and 0.2 U/ml l-alanine dehydrogenase, so that baseline reduction of NAD+ could be determined in the absence of l-alanine. d-Alanine was then added to an initial concentration of 5 mM to start the reaction. Protein concentrations of the extracts were determined by the Bradford method (3). As shown in Fig. 4, crude extracts of the Δalr strain DM22 contained no detectable alanine racemase activity above the background levels detected in the absence of the d-alanine substrate. In contrast, wild-type mc2155 contained levels of alanine racemase activity significantly higher than background, and the complemented Δalr strain DM30 contained approximately double the alanine racemase activity of the wild type. The cause of the higher levels of racemase activity in DM30 is unknown but could be related to multiple insertions at the integration site of the complementing vector.
FIG. 4.
Alanine racemase activity in cell lysates. Cell lysates of wild-type mc2155 (WT), the Δalr strain DM22, and the complemented Δalr strain DM30 were prepared, and alanine racemase activity was determined. Rates are expressed in mU/mg protein where 1 unit corresponds to 1 μmol l-alanine produced per minute. Background rates in the absence of d-alanine substrate (white bars) and rates in the presence of d-alanine (striped bars) are the means of triplicate measurements, and the error bars denote standard deviations.
The results of this study are congruent with studies that demonstrate that the deletion of the alr gene in other bacteria has been found to lead to d-alanine auxotrophy. In some bacteria, such as Escherichia coli and Salmonella enterica serovar Typhimurium, there are two genes encoding alanine racemase activity (16, 33), and both of these genes must be deleted before a requirement for d-alanine is observed (27, 32). Other bacterial species, such as Lactobacillus plantarum and Lactococcus lactis, have a single gene for alanine racemase, the deletion of which leads to a requirement for d-alanine for growth (18, 24). Still other species, such as Listeria monocytogenes, express a d-amino acid transaminase that must be deleted along with the alr gene to yield d-alanine auxotrophy (29). Our searches of the M. smegmatis genome sequence (Mycobacterium smegmatis MC2 Genome Page [http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=gms]) for an additional alanine racemase gene or for a potential d-amino acid transaminase gene were unsuccessful. Together with the results of Sassetti et al., which identified the alanine racemase gene as an essential gene in M. tuberculosis (21), this strongly suggests that alr is essential for M. smegmatis. Our results resolve a discrepancy in the literature and show that M. smegmatis, like L. plantarum and L. lactis, has a single gene for alanine racemase, the deletion of which leads to a requirement for d-alanine for growth.
The results of Chacon et al. differ from our own and from those of Sassetti et al. in that they found that insertional inactivation of the alr gene in M. smegmatis did not lead to d-alanine auxotrophy (4, 21). It is known that insertional inactivation can sometimes still allow for the production of active protein (13, 31), which is why gene inactivation by deletion is a more definitive technique. It is possible, therefore, that their insertional disruption of the alr gene did not abolish all alanine racemase activity. Notably, cell lysates from their mutant were assayed and displayed no measurable alanine racemase activity. They report, however, that there is a 4% background in their alanine racemase assay, which could mask a low residual racemase activity in their extracts. Interestingly, it has been reported that proteolysis of alanine racemase from Salmonella enterica serovar Typhimurium results in the production of protein fragments that associate and retain 3% of their wild-type activity (10). Such a truncated species would not have been detected in the assay, but it could have retained enough activity to account for the growth which they reported. It is known that the alanine racemase from M. tuberculosis has a Vmax that is orders of magnitude (>100-fold) less than that of the enzyme from Pseudomonas aeruginosa (26, 27); thus, mycobacteria can survive on very low levels of alanine racemase activity. Chacon et al. report significantly altered growth characteristics associated with their mutant, which would be consistent with a low level of alanine racemase activity. In this report, in which 99% of the alr gene was deleted, we can state unequivocally that the possibility for the production of active alanine racemase has been removed. Therefore, we can report with confidence that the alanine racemase gene is essential for growth of M. smegmatis in the absence of d-alanine and that the development of alanine racemase inhibitors as antimycobacterial agents warrants further investigation.
Acknowledgments
This work was supported by funding from the University of Otago, the Robert A. Welch Foundation, the National Institutes of Health, the Thrash Foundation, and the Foundation for the Centers for Molecular Research in Infectious Diseases.
Footnotes
Published ahead of print on 7 September 2007.
We dedicate this article to the late John Curtis Thrash, Jr., a noted philanthropist and businessman from Houston, Texas.
REFERENCES
- 1.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 2.Berberich, R., M. Kaback, and E. Freese. 1968. d-Amino acids as inducers of l-alanine dehydrogenase in Bacillus subtilis. J. Biol. Chem. 243:1006-1011. [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Chacon, O., Z. Feng, N. B. Harris, N. E. Caceres, L. G. Adams, and R. G. Barletta. 2002. Mycobacterium smegmatis d-alanine racemase mutants are not dependent on d-alanine for growth. Antimicrob. Agents Chemother. 46:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copie, V., W. S. Faraci, C. T. Walsh, and R. G. Griffin. 1988. Inhibition of alanine racemase by alanine phosphonate: detection of an imine linkage to pyridoxal 5′-phosphate in the enzyme inhibitor complex by solid-state 15N nuclear magnetic resonance. Biochemistry 27:4966-4970. [DOI] [PubMed] [Google Scholar]
- 6.Dye, C., G. P. Garnett, K. Sleeman, and B. G. Williams. 1998. Prospects for worldwide tuberculosis control under the WHO DOTS strategy. Directly observed short-course therapy. Lancet 352:1886-1891. [DOI] [PubMed] [Google Scholar]
- 7.Esaki, N., and C. T. Walsh. 1986. Biosynthetic alanine racemase of Salmonella typhimurium: purification and characterization of the enzyme encoded by the alr gene. Biochemistry 25:3261-3267. [DOI] [PubMed] [Google Scholar]
- 8.Ferrari, E., D. J. Henner, and M. Y. Yang. 1985. Isolation of an alanine racemase gene from Bacillus subtilis and its use for plasmid maintenance in B. subtilis. Nat. Biotechnol. 3:1003-1007. [Google Scholar]
- 9.Franklin, F. C., and W. A. Venables. 1976. Biochemical, genetic, and regulatory studies of alanine catabolism in Escherichia coli K12. Mol. Gen. Genet. 149:229-237. [DOI] [PubMed] [Google Scholar]
- 10.Galakatos, N. G., and C. T. Walsh. 1987. Specific proteolysis of native alanine racemases from Salmonella typhimurium: identification of the cleavage site and characterization of the clipped two-domain proteins. Biochemistry 26:8475-8480. [DOI] [PubMed] [Google Scholar]
- 11.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hols, P., C. Defrenne, T. Ferain, S. Derzelle, B. Delplace, and J. Delcour. 1997. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J. Bacteriol. 179:3804-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iredale, J. P. 1999. Demystified gene knockouts. Mol. Pathol. 52:111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambert, M. P., and F. C. Neuhaus. 1972. Mechanism of d-cycloserine action: alanine racemase from Escherichia coli W. J. Bacteriol. 110:978-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobocka, M., J. Hennig, J. Wild, and T. Klopotowski. 1994. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J. Bacteriol. 176:1500-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palumbo, E., C. F. Favier, M. Deghorain, P. S. Cocconcelli, C. Grangette, A. Mercenier, E. E. Vaughan, and P. Hols. 2004. Knockout of the alanine racemase gene in Lactobacillus plantarum results in septation defects and cell wall perforation. FEMS Microbiol. Lett. 233:131-138. [DOI] [PubMed] [Google Scholar]
- 19.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 22.Silverman, R. B. 1988. The potential use of mechanism-based enzyme inactivators in medicine. J. Enzyme Inhib. 2:73-90. [DOI] [PubMed] [Google Scholar]
- 23.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 24.Steen, A., E. Palumbo, M. Deghorain, P. S. Cocconcelli, J. Delcour, O. P. Kuipers, J. Kok, G. Buist, and P. Hols. 2005. Autolysis of Lactococcus lactis is increased upon d-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 187:114-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strych, U., M. Davlieva, J. P. Longtin, E. L. Murphy, H. Im, M. J. Benedik, and K. L. Krause. 2007. Purification and preliminary crystallization of alanine racemase from Streptococcus pneumoniae. BMC Microbiol. 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strych, U., H. C. Huang, K. L. Krause, and M. J. Benedik. 2000. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 41:290-294. [DOI] [PubMed] [Google Scholar]
- 27.Strych, U., R. L. Penland, M. Jimenez, K. L. Krause, and M. J. Benedik. 2001. Characterization of the alanine racemases from two mycobacteria. FEMS Microbiol. Lett. 196:93-98. [DOI] [PubMed] [Google Scholar]
- 28.Tanizawa, K., S. Asano, Y. Masu, S. Kuramitsu, H. Kagamiyama, H. Tanaka, and K. Soda. 1989. The primary structure of thermostable d-amino acid aminotransferase from a thermophilic Bacillus species and its correlation with l-amino acid aminotransferases. J. Biol. Chem. 264:2450-2454. [PubMed] [Google Scholar]
- 29.Thompson, R. J., H. G. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tran, S. L., and G. M. Cook. 2005. The F1Fo-ATP synthase of Mycobacterium smegmatis is essential for growth. J. Bacteriol. 187:5023-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanhaesebroeck, B., J. L. Rohn, and M. D. Waterfield. 2004. Gene targeting: attention to detail. Cell 118:274-276. [DOI] [PubMed] [Google Scholar]
- 32.Walsh, C. T. 1989. Enzymes in the d-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem. 264:2393-2396. [PubMed] [Google Scholar]
- 33.Wasserman, S. A., C. T. Walsh, and D. Botstein. 1983. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 153:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wijsman, H. J. 1972. The characterization of an alanine racemase mutant of Escherichia coli. Genet. Res. 20:269-277. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 2007. WHO fact sheet: tuberculosis. World Health Organization, Geneva, Switzerland. http://www.who.int/mediacentre/factsheets/fs104/en/.