Abstract
Streptococcus gordonii is a primary colonizer of the multispecies biofilm on tooth surfaces forming dental plaque and a potential agent of endocarditis. The recent completion of the genome sequence of the naturally competent strain Challis allowed the design of a spotted oligonucleotide microarray to examine a genome-wide response of this organism to environmental stimuli such as signal peptides. Based on temporal responses to synthetic competence signaling peptide (CSP) as indicated by transformation frequencies, the S. gordonii transcriptome was analyzed at various time points after CSP exposure. Microarray analysis identified 35 candidate early genes and 127 candidate late genes that were up-regulated at 5 and 15 min, respectively; these genes were often grouped in clusters. Results supported published findings on S. gordonii competence, showing up-regulation of 12 of 16 genes that have been reported to affect transformation frequencies in this species. Comparison of CSP-induced S. gordonii transcriptomes to results published for Streptococcus pneumoniae strains identified both conserved and species-specific genes. Putative intergenic regulatory sites, such as the conserved combox sequence thought to be a binding site for competence sigma factor, were found preceding S. gordonii late responsive genes. In contrast, S. gordonii early CSP-responsive genes were not preceded by the direct repeats found in S. pneumoniae. These studies provide the first insights into a genome-wide transcriptional response of an oral commensal organism. They offer an extensive analysis of transcriptional changes that accompany competence in S. gordonii and form a basis for future intra- and interspecies comparative analyses of this ecologically important phenotype.
Many oral streptococci, including Streptococcus gordonii, are naturally transformable, being able to take up exogenous DNA from the environment and incorporate it into their genome. These commensal streptococci are the primary colonizers of multispecies biofilms on the tooth surface that form dental plaque. The close proximity of cohabitants in this niche makes oral streptococci potential recipients of horizontally transferred DNA and potential reservoirs for biologically important genetic determinants. Thus, their mechanisms for DNA uptake have broad ecological significance. Although a genome-wide genetic study of competence in S. gordonii has not yet been undertaken, current experimental evidence suggests that mechanisms involved in competence development and transformation in S. gordonii are similar to those in the well-characterized system of Streptococcus pneumoniae (10, 20). The comC product is produced as the precursor of competence signaling peptide (CSP) and then processed and secreted by an ABC transporter and accessory protein, encoded by comA and comB, respectively. Extracellular CSP binds to sensor histidine kinase, encoded by comD, which, with the response regulator, encoded by comE, forms a two-component signal transduction system involved in competence regulation (12).
Unlike the stationary-phase competence response of Bacillus subtilis, the competence response of S. pneumoniae occurs in mid-log growth phase in at least two temporal stages (reviewed in reference 4). Global genome analyses of two different strains of S. pneumoniae using microarrays (7, 29) have extended genetic and biochemical studies and confirmed that CSP induces gene expression as early as 5 min after exposure to the peptide; these genes have been designated early-response genes. S. pneumoniae genes with increased expression 15 min after CSP exposure have been designated late-response competence genes. In general, expression of CSP-induced S. pneumoniae genes returns to basal levels after approximately 35 min. Although early genes are primarily involved in coordination of the CSP response, late genes involve expression of genes encoding peptides such as bacteriocins, genes involved in DNA uptake and repair, and stress responses (29). S. pneumoniae transcriptome studies (7, 29) indicate that stress response genes are also involved in a delayed response, increasing expression levels consistently over a 40-min time period. Conversely, a subset of genes is repressed in response to CSP.
Although S. gordonii is more closely related taxonomically to S. pneumoniae based on 16S rRNA sequences (16), these commensal plaque bacteria occupy the same ecological niche on the tooth surface as the cariogenic oral species Streptococcus mutans. The competence response of S. pneumoniae differs from that of S. mutans in terms of temporal stages and characteristics of peptide transporters used (reviewed in reference 26). Although the responses of individual S. mutans gene clusters to CSP have been studied, the transcriptome response has not yet been described. The completion of the S. gordonii strain Challis CH1 genome sequence provides an opportunity to construct a genome-wide microarray to provide an extensive analysis of transcriptional changes that accompany competence in this species. The present studies document both similarities and differences of the S. gordonii competence response compared to other streptococcal competence systems. The occurrence of species-specific components within the shared bacterial competence phenotype highlights the necessity of investigating individual species to fully understand how competence and transformation shape their evolution and fitness for colonization and survival.
MATERIALS AND METHODS
Transformation frequencies in response to synthetic CSP.
To establish the effective level of CSP to be used to study the transcriptome response, strain Challis CH1 (33) cells were grown in Todd-Hewitt broth (Becton Dickinson and Company) with 8 mM HCl to inhibit natural competence (6). Spectrophotometric readings and viable counts confirmed that the additional HCl had no effect on the log-phase growth curve of strain CH1 compared to the growth curve in nonacidified Todd-Hewitt medium. In order to determine effects of various concentrations of CSP, synthetic peptide (H-DVRSNKIRLWWENIFFNKK-OH; synthesized by Mimotopes Pty, Ltd.) based upon the peptide described by Havarstein et al. (12) and confirmed by the sequence of the S. gordonii Challis CH1 comC locus (SGO_2147) was added to aliquots of cells grown to an optical density at 600 nm (OD600) of 0.4 to achieve CSP concentrations of 25, 50, 100 and 200 ng/ml. At 5, 15, and 40 min thereafter, ca. 1 μg/ml of streptococcal plasmid pVA749 (25) or 10 μg/ml of chromosomal DNA from the streptomycin-resistant Challis strain CH1S (34) was added to the CSP-induced cells. After incubation for 1 h at 36°C, the cell mixtures were serially diluted and plated on Todd-Hewitt agar plates with either 10 μg/ml erythromycin or 1,000 μg/ml streptomycin, respectively, to select for transformants. Parallel control cells for all experiments had no CSP added. Plates were incubated for 48 h at 36°C in 5% CO2. Transformation frequencies were calculated as the number of antibioitic-resistant transformants per ml divided by the number of viable CFU per ml.
A temporal curve of the competence response was generated by treating parallel cultures of CH1 cells (OD600 = 0.4.) with 100 ng/ml CSP and then adding pVA749 DNA at 2- or 5-min intervals over a 110-min time period. The cell mixutures were treated with 2 U of DNase 9 min after addition of the plasmid in order to degrade any DNA that had not entered the cells yet still allow transformation with the 5.2-kb plasmid. All cell mixtures were incubated at 36°C for 1 h and then serially diluted and plated as described above to determine transformation frequencies. Negative control cultures that were not CSP induced were included in all experiments.
Isolation of Streptococcus gordonii total RNA.
Ten-milliliter culture aliquots were removed at time zero or at 5, 15, and 40 min after CSP exposure and immediately transferred to an equal volume of hot acid phenol (pH 4.3, ∼100°C), vortexed for 20 s to mix, and incubated for an additional 10 min at 100°C. After cooling on ice, the lysate was extracted first with acid phenol-chloroform and then with chloroform. Total RNA was recovered from the extracted lysate by addition of sodium acetate (pH 5.2) to 0.3 M and an equal volume of isopropanol. Precipitated RNA was recovered by centrifugation, resuspended in RNase-free water (Ambion, Austin, TX), and treated with RNase-free DNase (Ambion) to remove remaining genomic DNA. Remaining contaminants were removed by purification of RNA on QIAGEN RNeasy columns (QIAGEN, Valencia, CA). RNA purity was determined on an Agilent BioAnaylzer. Only RNA preparations with RNA integrity numbers of >7.0 were used in these experiments. Purified RNA was stored at −80°C prior to labeling for use in microarray hybridizations.
Streptococcus gordonii DNA microarray.
A 70-mer oligonucleotide DNA microarray was used for these studies. The initial automated annotation of 2,195 open reading frames (ORFs) from the S. gordonii CH1 genome completed at The Institute for Genomic Research was used in the OligonucleotidePicker oligonucleotide finding program (http://pga.mgh.harvard.edu/oligopicker) to identify unique and nonoverlapping 70-mer oligonucleotides corresponding to each ORF. Probes representing each of these ORFs were included in the microarray. Subsequent manual curation of the automated annotation eliminated 44 of the original ORFs (indicated in this article as ORFX) and resulted in a final tally of 2,151 ORFs or loci (indicated in this article as SGOX), which have been deposited in GenBank. All of the original ORFs that were not updated as loci in the final genome annotation were maintained and analyzed in our microarray experiments and are labeled ORFX because they provide insights into the transcription of intergenic regions. Hybridization of cDNA to the unique 70-mer probes in the microarrays indicates that the corresponding genomic region was transcribed but does not ensure that the region of the transcript encodes a functional translated protein corresponding to the designated ORF. The 70-mer oligonucleotides were synthesized by Invitrogen (Carlsbad, CA), diluted to a final concentration of 25 μM in Corning microarray spotting buffer, and spotted in replicates of six onto UltraGap microarray slides (Corning Life Sciences, Acton, MA) using an Intelligent Automation Systems spotting robot (Brooks Automation, Chelmsford, MA). After being printed, the slides were cross-linked and then stored in a desiccator at room temperature.
Microarray target preparation.
S. gordonii RNA was used for indirect labeling as previously described (28) with either Cy5 or Cy3 dye (GE Healthcare Bio-Sciences Corp., Piscataway, NJ), leading to the synthesis of 3 μg of cDNA with 150 to 200 pmol of dye molecule incorporated per microgram of cDNA produced. Briefly, RNA was first converted to amino-allyl cDNA by reverse transcription with Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) in a deoxynucleotide triphosphate mixture containing amino-allyl dUTP. The RNA template was removed by alkaline hydrolysis, and amino-allyl cDNA was recovered by purification on QIAGEN QIAquick PCR columns (QIAGEN). The cDNA was dried in a SpeedVac, labeled by coupling to either Cy3 or Cy5 NHS-Cy dye (GE Healthcare Bio-Sciences Corp.), and purified on QIAGEN QIAquick PCR columns (QIAGEN). The quality of the probes and their specific activity were confirmed by a spectrophotometric scan from 200 to 700 nm. Only targets with >200 pmol of dye incorporation per sample and a ratio of less than 50 nucleotides/dye molecules were used for hybridizations.
Hybridization of Streptococcus gordonii spotted oligonucleotide microarray.
Prior to hybridization, the slides were blocked by prehybridization at 42°C in blocking buffer (1% bovine serum albumin, 5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate). The slides were washed with water and isopropanol and dried prior to hybridization. The dried targets were resuspended in 60 μl of hybridization buffer (50% formamide, 5× SSC, 0.1% SDS, 0.5 mg salmon sperm DNA), heated at 95°C for 10 min, rapidly cooled, and added to a slide under a lifterslip (Erie Scientific Company, Portsmouth, NH). Each slide was placed at 42°C for 16 h in a sealed hybridization chamber. The arrays were then washed successively in 1× SSC and 0.1× SSC and dried.
Microarray analysis.
Images of the hybridized arrays were obtained with a GenePix 4000A microarray scanner (Axon Instruments, Foster City, CA) and analyzed using TIGR Spotfinder (www.tigr.org/software) to measure fluorescence signals. The data set was normalized by applying the LOWESS algorithm (block mode; smooth parameter, 0.33) using MIDAS (www.tigr.org/software). Replicate spots that had Cy3-to-Cy5 ratios beyond 1.5 standard deviations from the average ratio were discarded, and values for the remaining replicates were then averaged to determine the final ratio reported for each ORF. For each time point comparison, at least one flip-dye experiment per RNA batch was performed. Two independent RNA batches were used per time point. Results from all experiments are included in the data presented.
qRT-PCR analysis.
cDNA templates were synthesized from sample and control RNAs using the Bio-Rad iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative reverse transcriptase PCRs (qRT-PCRs) were carried out using the iCycler reverse transcriptase PCR system (Bio-Rad) using the iTaq SYBR green supermix with ROX (Bio-Rad) and primers designed to amplify PCR products of ∼100 bp in length. A standard curve was prepared using serial 10-fold dilutions of a cDNA template of known concentration. Relative RNA abundance measurements at individual time points after induction of competence were made by comparing PCR-formation profiles to each other.
GenBank accession number.
The genome sequence of S. gordonii strain Challis CH1 was deposited in GenBank with accession number s_gordonii_challis_n CP000725.
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE8758.
RESULTS AND DISCUSSION
S. gordonii transformation frequencies in response to CSP.
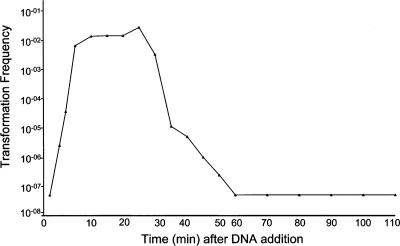
As suggested by previous studies with strains of S. gordonii (10), increasing levels of synthetic CSP over the range of 25, 50, 100, or 200 ng/ml did not appear to affect the competence response. Each of these CSP concentrations resulted in transformation frequencies with chromosomal DNA ranging from 5 × 10−3 to 1 × 10−2, comparable to levels seen when cells were made competent by serum (34). These results with S. gordonii contrasted with those reported for planktonic cells of S. mutans, which respond to CSP in a dose-dependent manner over this concentration range (17). Most importantly, no transformants were obtained for any of the parallel control cultures without CSP, indicating that the peptide was necessary and responsible for the transformability of the CH1 cells. Based on these data, 100 ng/ml CSP was used for further characterization of the competence response of strain CH1. A time course study (Fig. 1) verified that the cells needed more than 2 min of exposure to CSP in order to take up exogenous DNA and that this response began to decline after 30 min. This temporal response was more similar to that of S. pneumoniae (29) than to that of S. mutans; the latter species has been reported to require up to 2 h to attain an optimum competence response (17).
FIG. 1.
Transformability of strain CH1 cells in response to CSP. At time0 100 ng/ml CSP was added to aliquots of log phase cells (OD600 = 0.4). Replicative plasmid DNA carrying an erythromycin resistance determinant was added to individual aliquots of cells at various time points (abscissa). Two units of DNase were added 9 min after the addition of DNA. All procedures were carried out at 36°C. Cell mixtures were incubated for 1 h and plated on Todd-Hewitt agar containing 10 μg/ml erythromycin. Transformation frequencies for CSP-induced cells at each time point, calculated as the number of erythromycin-resistant colonies/ml divided by the number of viable CFU/ml on antibiotic-free Todd-Hewitt agar, are shown on the ordinate. Limits of detection were <5 × 10−8. No erythromycin-resistant colonies were detected in any parallel control cultures without CSP at any time point. Results shown are representative of three independent experiments.
Microarrays identify a temporal transcriptional response to CSP.
Based upon the transformation frequency time course (Fig. 1), transcriptome analysis of the competence response was examined over a 40-min time period. The temporal criteria previously used for S. pneumoniae microarray data (29) were used for initial classification of S. gordonii CSP-responsive loci. Candidate genes were identified if they showed an average of greater than twofold increase in expression after exposure to CSP. Genes were classified as “early” if they showed up-regulation at 5 min after CSP exposure and classified as “late” if up-regulation did not occur until the 15-min time point. In a few cases there were minor variations within gene clusters, where one gene within a putatively cotranscribed region began to show up-regulation at 5 min whereas the remaining genes in the region did not show up-regulation until 15 min. Careful examination of S. pneumoniae competence transcriptome data shows similar variations (29). Table S1 in the supplemental material shows numerical values for all the genes up-regulated (LOWESS normalized values of >2-fold) in response to CSP and their coordinates with respect to the assigned origin of replication of the S. gordonii genome. Since the microarray was constructed before the final annotation of the S. gordonii genome sequence was completed, some of the probes detect transcripts in intergenic regions that were initially annotated as putative ORFs. Thus, the extent of the transcripts through intergenic regions can be visualized. In such cases, we have included the original ORF names to clarify this. Selected gene regions are expanded and shown in a colorimetric display (Fig. 2) of relative expression levels of selected up-regulated genes at 5, 15, and 40 min after CSP exposure. Based upon criteria of greater than twofold increase in expression at 5 min, 35 candidate early genes were identified. These genes fell into nine clusters consisting of two or more adjacent genes that in each case were transcribed in the same direction with each other and nine individual genes that were flanked by genes that were not up-regulated. The results are generally consistent with findings for S. pneumoniae that the early genes were generally transporters and regulators, often occurring in clusters (29).
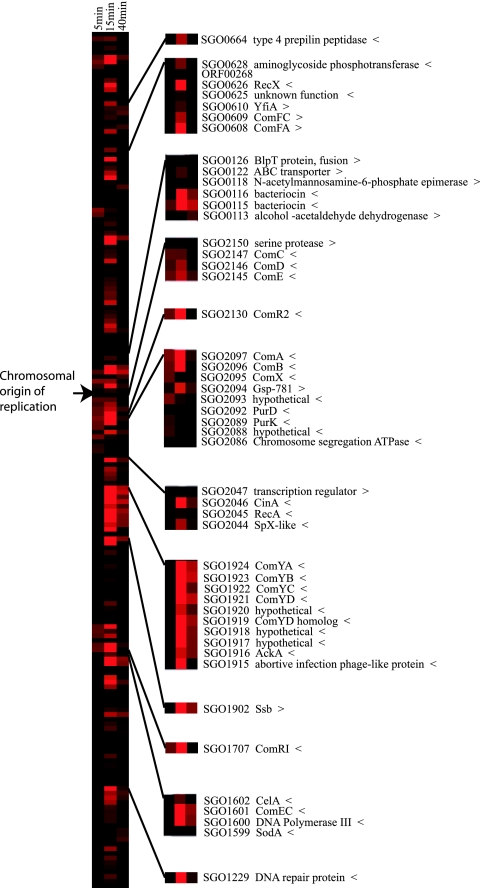
FIG. 2.
Colorimetric representation of relative expression levels of CSP-induced S. gordonii genes. Transcript levels as measured by cDNA hybridized to a sixfold redundant S. gordonii microarray and averaged for two replicate hybridizations are shown at 5, 15, and 40 min after CSP induction. Results for 185 up-regulated gene loci (expression ratio, >2; induced/uninduced) were ordered vertically based upon their position in the chromosome (9). Temporally invariant genes (expression levels averaging ≤2 over the course of the experiment) were omitted. Color codes represent cDNA levels at each time point as indicated at the top of the columns, over the range from ≤2 (black) to 30.4 (bright red). Enlarged side panels show expression patterns for representative genes. For numerical values for all up-regulated genes, see Table S1 in the supplemental material. The “>” (5′-3′) and “<” (3′-5′) symbols following the name of each genetic locus indicate the direction of transcription with reference to the chromosomal origin of replication.
Based upon the same criteria at 15 min after CSP exposure, 127 candidate late genes were initially identified in S. gordonii. Most of these are arranged in approximately 21 clusters of 2 or more genes. These include 32 ORFs encoding hypothetical or conserved hypothetical proteins as well as S. gordonii homologs of other identified streptococcal late competence genes (see Table S1 in the supplemental material).
Overall, the up-regulated genes tend to be organized in clusters (Fig. 2). From an evolutionary standpoint, the clustering of the CSP-responsive genes has intriguing implications for the evolutionary acquisition of these genes by horizontal transfer. However, shared functional relationships of the clustered genes cannot be assumed, and positional effects must be considered when analyzing the competence response. It is possible that dispensable genes are up-regulated as a result of their position adjacent to essential genes. Up-regulation of downstream genes may result from read-through from strongly induced upstream promoters, especially in cases of weak intergenic transcriptional terminators. It has also been suggested that in S. pneumoniae, convergent downstream genes are expressed from competence-induced promoters in a form of antisense transcriptional regulation (30). The pattern of gene expression in S. gordonii (Fig. 2) supports these possibilities and provides a basis for future studies outside the scope of this initial analysis to characterize underlying genetic regulatory mechanisms.
Microarrays confirm CSP-induced expression of S. gordonii genes known to be involved in competence.
Products of early responsive genes included the competence secretion apparatus encoded by the first two genes in the comABX cluster, comA and comB. The S. gordonii comX gene is not related to the similarly designated S. pneumoniae comX alternative sigma factor gene; however, this region of DNA, which may encode a small peptide, is essential for S. gordonii competence (21). Like the identical comX1 and comX2 genes found in S. pneumoniae, the S. gordonii identical comR1 and comR2 competence sigma factor genes (13) were up-regulated at 5 min. The genes immediately flanking the comR determinants did not show CSP induction at any of the time points in this study. The strain Challis comCDE genes were up-regulated at 5 min, confirming the start of the competence cascade. This up-regulation was verified by qRT-PCR (Table 1). As in S. pneumoniae, the S. gordonii comCDE genes are located near the chromosomal origin of replication. The adjacent divergent serine protease HtrA/DegP (SGO_2150), which did not show a 5- or 15-min CSP response, is located next to the chromosome segregation protein (SGO_2151), which is followed in the circular chromosome by the chromosome replication initiator protein, DnaA (SGO_0001). It has been hypothesized that the localization of the com gene cluster near the origin of replication could provide advantages for synchronizing the log-phase competence response with replication initiation to control the copy number of the comCDE genes in response to environmental influences that affect cell division (5). The conserved chromosomal location in both S. gordonii and S. pneumoniae supports this hypothesis.
TABLE 1.
Quantitative real-time PCR data for selected S. gordonii genesa
| Locus | Gene | T0 Qt. avg (SD) | T5 Qt. avg (SD) | T15 Qt. avg (SD) | Gene expression ratiob
|
|
|---|---|---|---|---|---|---|
| T5/T0 | T15/T0 | |||||
| SGO_0773 | ccpA | 21,191,149.0 (1,204,396.6) | 14,221,354.0 (340,341.8) | 5,190,967.0 (153,186.9) | 0.7 | 0.2 |
| SGO_0478 | cwp | 532,721.6 (76,538.1) | 852,681.7 (63,916.4) | 890,846.9 (50,984.0) | 1.6 | 1.7 |
| SGO_0298 | vanR | 1,666,434.7 (18,397.2) | 1,362,722.6 (100,448.8) | 779,398.3 (36,221.8) | 0.8 | 0.5 |
| SGO_2145 | comD | 1,084,301.1 (59,522.4) | 353,052,608.0 (4,219,968.0) | 162,309,768.0 (14,092,434.5) | 325.6 | 149.7 |
| SGO_2144 | comE | 412,183.1 (11,049.2) | 169,425,200.0 (12,508,074.1) | 78,864,680.0 (1,677,777.7) | 411.0 | 191.3 |
| SGO_2105 | abpA | 222,849,064.0 (749,159.8) | 134,761,056.0 (21,550,148.3) | 60,919,676.0 (381,679.3) | 0.6 | 0.3 |
| SGO_2001 | rpsB | 57,243,782.0 (2,931,305.5) | 37,882,854.0 (5,400,878.8) | 19,134,599.0 (80,962.3) | 0.7 | 0.3 |
| SGO_1816 | scaR | 3,514,630.9 (246,373.9) | 3,717,323.8 (140,567.8) | 1,704,124.5 (140,567.8) | 1.1 | 0.5 |
| SGO_1197 | topA | 276,780.4 (5,055.5) | 373,193.5 (46,398.2) | 10,370,308.5 (693,210.0) | 1.3 | 37.5 |
cDNA templates were synthesized from sample and control RNAs using the Bio-Rad iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), and reactions were carried out using the iTaq SYBR green supermix with ROX (Bio-Rad) and primers designed to amplify ca. 100-bp PCR products. Relative RNA abundance measurements at individual time points after induction of competence were made by comparing PCR-formation profiles to each other. Values are expressed as absolute quantification (Qt.) for each duplicate reaction at time zero (T0), 5 min (T5), or 15 min (T15). Dilutions of 16S rRNA genes were used as calibrators. The endogenous control gene was gyrA.
Ratios indicating a <2-fold increase in gene expression compared with the value for time zero correspond to genes that did not show CSP induction (see Table S1 in the supplemental material).
The comYA-YD gene cluster, shown to be essential for DNA transformation (22), showed maximum expression at 15 min after CSP induction (Fig. 2). SGO_0115 and SGO_0116, which encode peptides involved in bacteriocin and hemolysin activity in S. gordonii strain DL1 (14), were also strongly up-regulated at 15 min (Fig. 2). The recA gene, which has been shown to be necessary for efficient DNA transformation (34), is located within a cluster of genes downstream of a cinA homolog that is strongly up-regulated at 15 min (Fig. 2; see Table S1 in the supplemental material). The present results support the genetic studies and indicate that these genes are directly induced by CSP.
In contrast, it has been previously reported that the genes in the S. gordonii adcCBA operon (SGO_1936, SGO_1937, and SGO_1938, respectively) (19, 27), which are essential for manganese homeostasis and in vitro biofilm formation, are involved in competence, since strains defective in these genes showed lower transformation frequencies. However, the present microarray analysis suggests that this may not be a direct effect of these genes in response to CSP, since neither the adcCBA gene cluster nor those genes adjacent to them were up-regulated at any of the time points in our studies. Similarly, previous studies (15) have shown that disruption of hppA (SGO_1328), encoding a lipoprotein involved in hexa- and heptapeptides, resulted in decreased transformation frequency compared to that of parental S. gordonii strain Challis DL1 cells. In contrast, disruption of the functionally related adjacent genes hppG (SGO_1327) and hppH (SGO_1325), both encoding oligopeptide binding lipoproteins, did not affect transformation frequencies (15). In the present microarray analyses, none of these gene loci, nor those near them, were up-regulated in response to CSP. It is of note that hppA, which did affect competence when disrupted (15), is located within a putative polycistronic operon located approximately 4.7 kbp from comR1 (SGO_1333). Although the extent of the hpp transcript is not known, disruption of hppA may have had cis effects on comR1 expression, thereby influencing the observed transformation frequencies. Alternatively, hppA may be involved in transport of an as yet unidentified essential peptide(s) or factor(s) necessary for efficient transformation. There are many possible explanations for discrepancies between our microarray results and those of previous genetic studies with S. gordonii. Differences in gene expression may be due to strain differences, medium components, or other environmental effects. Alternatively, the previously identified genes may be involved in aspects of the competence response that are not directly affected by the CSP peptide within the time frame examined in the present studies. Finally, it is possible that the sensitivities of the microarray probes for these genes are not sufficient to detect subtle changes in expression of these genes. Nevertheless, the microarray results do provide genome-wide insights into gene expression that are consistent with our present limited knowledge of competence in S. gordonii.
Examination of DNA regions preceding the early and late CSP-induced S. gordonii genes.
In S. pneumoniae, early genes are often preceded by imperfect direct repeats with a consensus sequence of aCAnTTcaG-12(n)-aCAgTTgaC (uppercase letters correspond to the most conserved bases [29]); this region is thought to be the ComE binding site (35). Putative imperfect direct repeats that did not have this S. pneumoniae consensus sequence were found preceding the comR alleles of S. gordonii, but disruption of at least one of these repeats did not affect transformation frequencies (13). Indeed, genome-wide computer-assisted searches for DNA conserved repeat patterns preceding ORFs (32) did not identify this consensus preceding any of the S. gordonii candidate early genes. Although functional studies are outside the range of the present paper, these microarray studies provide an essential basis for designing studies to define the ComE binding site in S. gordonii.
In S. pneumoniae, a consensus octomer preceded by a run of T's, designated the combox or cinbox (1), is present upstream of some but not all of the late competence genes. This DNA element is thought to act as a promoter for alternative sigma factor comX-directed polymerase (24). This consensus sequence was found upstream of 12 CSP-induced S. gordonii genes or gene clusters (Fig. 3), including comYA, cinArecA, and sthA and sthB, all of which have experimental data supporting their role in the competence response (14, 22, 34). Nine of these identified S. gordonii genes or gene clusters have homologs in both S. pneumoniae sequenced strains TIGR4 and R6. For example, DNA topoisomerase I (SGO_1197), which showed up-regulation in response to CSP by both microarray (see Table S1 in the supplemental material) and qRT-PCR (Table 1) analyses, is located within a cluster of genes with putative DNA processing and acid tolerance functions, similar to those found in S. pneumoniae.
FIG. 3.
S. gordonii gene clusters up-regulated in response to CSP at 15 min and preceded by a putative combox regulatory site (1). Putative proteins encoded by each gene are shown. (A) Late gene consensus sequences. Genetic loci and proteins encoded by the first up-regulated ORF downstream of the combox sequence are listed. Comparison to the consensus combox sequence and the distances to the putative start codon of the ORF immediately downstream of the consensus sequence are shown. In the case of the combox upstream of the bacteriocin SthA, the distance to SGO_0117, encoding a hypothetical protein (HP) that was not expressed in any of the experiments, as well as the distance to SGO_0116, encoding the SthA bacteriocin, are both shown. S. pneumoniae homologs in strains TIGR4 (gene loci SP) and R6 (gene loci SPR) for the S. gordonii loci are shown. All S. pneumoniae homologs except SPR2021 are preceded by a similar combox sequence (2) and have been shown to be up-regulated in response to CSP in both strains R6 and Rx (7, 29). SGO_2094 does not have a significant homolog in S. pneumoniae strain TIGR4, whereas strain R6 does carry a GSP781 homolog (SPR2021); the gene encoding this protein has not been implicated in the S. pneumoniae competence response (7). (B) S. gordonii gene clusters with identified comboxes. Clusters of genes up-regulated at 15 min after CSP exposure and positioned downstream of the combox consensus sequence (C) are shown. Gene loci are designated by the SGO number above each arrow; putative encoded proteins are shown within the arrows. Directions of the arrows indicate the 5′→3′ direction of each ORF in relation to the chromosomal origin of replication. Arrow sizes are not to scale. Arrows with dashed lines do not represent 15-min CSP-responsive ORFs and either were classified as representing early-response competence genes (encoding ComA, ComB, and ComX) or were not expressed in the microarray under any of the conditions tested (HP encoded by SGO_0117).
A combox sequence was also found upstream of the S. gordonii locus SGO_2094, encoding a putative general stress response protein, Gsp781. S. pneumoniae strain TIGR4 does not possess a homolog for this gene; strain R6 does have a homolog, but it was not found to be up-regulated in a CSP response transcriptome study (7). Expression of SGO_2094, located immediately adjacent to the convergently transcribed comABX, is up-regulated ca. threefold at 5 min (Fig. 2; also see Table S1 in the supplemental material). However, at 15 min, when the intergenically located comX signal has decreased to less than 2-fold, the gsp781 response continued to increase to >8-fold, suggesting that gsp781 is a late-response gene, thereby supporting the functional role of the identified combox sequence (Fig. 3). Interestingly, the S. gordonii genome also has another Gsp781 homolog (SGO_2017) convergently located approximately 14 kbp closer to the chromosomal origin of replication. This homolog, which is not preceded by a combox consensus sequence, is 49.7% identical to SGO_2094 at the nucleotide level and was not up-regulated in the microarrays, further supporting the importance of the putative combox rather than the sequence of the gene or encoded protein in the response to CSP.
Each of the two bacteriocin peptides encoded by SGO_0115 and SGO_0116 (14), which do not have homologs in S. pneumoniae, is preceded by its own combox sequence (Fig. 3). SGO_0116 is separated from its combox by the previously identified (14) hypothetical protein SGO_0117, placing the start site 347 bp downstream from the combox. Although SGO_0117 is more favorably positioned, with a putative start codon 108 bp downstream from the combox, cDNA targets did not hybridize to the SGO_0117 probe in any of the microarrays of CSP-induced or noninduced cultures, suggesting that the chromosomal region encoding this hypothetical protein is not transcribed under the conditions in this study.
S. gordonii genes homologous to S. pneumoniae genes involved in competence.
Although relatively few genes have been experimentally confirmed to be involved in S. gordonii competence, the present genome-wide analysis has verified that a number of homologs to genes involved in the competence responses of other streptococcal species are also up-regulated in S. gordonii. Since the competence response of S. pneumoniae has been the subject of extensive genetic studies (18) and has been analyzed at the transcriptome level in both strains Rx (29) and R6 (7), this species provides an excellent initial comparison for characterizing the S. gordonii CSP response. Not surprisingly, many of the S. pneumoniae homologs are up-regulated in S. gordonii in response to CSP. A complete list of S. pneumoniae competence genes and the response of the closest homolog based on amino acid similarity in S. gordonii is shown in Table S2 in the supplemental material. The studies investigating the strain Rx competence response used microarray probes from the sequenced S. pneumoniae strain TIGR4 (29), so the up-regulated genes in this strain are designated with the TIGR4 gene loci (SP); the strain R6 competence response (7) was determined by using a strain R6 microarray (gene loci SPR). It is important to note that the S. gordonii homologs for this comparison were based upon the similarities of the entire amino acid sequences of the S. pneumoniae and S. gordonii ORFs as compared by BLAST analysis. Some of the S. pneumoniae genes did not have highly similar homologs in S. gordonii, as is indicated by a low E value (see Table S2 in the supplemental material); nevertheless, the most-similar ORFs were examined to screen for expression similarities. The results highlighted species-specific differences in CSP-induced gene expression.
Of the 124 CSP-inducible genes identified in S. pneumoniae, 67 were individually dispensable for DNA transformation; only 23 were required for transformation (29). Of the early CSP-responsive S. pneumoniae genes, only eight were required: comA, comB, comC, comD, comE, sigma factors comX1 and comX2, and comW. All except comW have highly similar homologs in S. gordonii (see similarity values in Table S2 of the supplemental material) that were up-regulated 5 min after CSP induction (see Table S1 in the supplemental material). In S. pneumoniae, the early CSP response gene comW encodes an essential competence cofactor (23). The S. gordonii ORF most similar to comW, with a low E-value of 5.5e−03 (see Table S2 in the supplemental material), is SGO_1502, encoding a putative 24-kDa conserved hypothetical protein. SGO_1502 is located immediately downstream of loci encoding a putative DNA response regulator, a histidine kinase, and a second conserved hypothetical protein. Neither SGO_1502 nor any of its immediately flanking genes were up-regulated in response to CSP (see Table S1 in the supplemental material). Furthermore, a derivative of S. gordonii strain CH1 was constructed by allelic replacement (unpublished data) in which the 645 bp of SGO_1502 and 35 bp of its upstream intergenic region were replaced with a polar spectinomycin resistance determinant; this strain had the same transformation frequencies as the parental strain, CH1, further indicating that SGO_1502 was not required for competence. Further studies will be needed to determine if an as yet unidentified factor, playing a role similar to that of comW, is involved in the S. gordonii competence response.
In addition to the genes similar to those in S. pneumoniae, additional species-specific candidate early CSP-responsive genes were identified in S. gordonii, such as SGO_1721, encoding an FeS assembly protein that showed consistent up-regulation of more than fourfold at 5 min after CSP exposure; this gene does not have a CSP-induced homolog in S. pneumoniae. Additional early and late CSP-induced genes in S. gordonii either alone or in gene clusters appear to be involved in metabolism or transport (see Table S1 in the supplemental material). Results such as these emphasize the importance of studying competence in multiple streptococcal species to clarify the role of essential and nonessential genes and to gain insights into underlying molecular mechanisms.
Although S. gordonii and S. pneumoniae share many common patterns in competence response related to the classes of genes that are up-regulated, amino acid sequence similarities alone were not positive predictors of CSP responsiveness (see Table S2 in the supplemental material). The S. gordonii genome does not have highly similar homologs for all the S. pneumonia CSP-responsive genes (see Table S2 in the supplemental material). It has been suggested (29) that some of the S. pneumoniae CSP-responsive genes may not be directly involved in the competence response but instead are up-regulated due to their location within specific gene clusters. S. gordonii up-regulated gene patterns (Fig. 2) support the suggestion that some genes may be CSP induced as the result of their chromosomal position near essential competence genes rather than their functional gene product. It is possible that genes within competence clusters that are not essential for transformation may play other biological roles in the competence response, such as killing of surrounding cells or involvement in DNA acquisition (3). Comparison of the S. pneumoniae transcriptome response to that of S. gordonii provides a basis for future assessment of candidate genes for functional analysis to clarify roles of essential or dispensable genes.
Two-component systems.
Although both S. gordonii and S. mutans occupy the same environment in the oral cavity, the present results support recent studies suggesting that the genetic bases for their competence responses may differ. Competence in S. pneumoniae and S. gordonii appears to be due to a ComDE two-component signal transduction system (reviewed in reference 26). In contrast, a Blp-like two-component system, which is involved in bacteriocin-like peptide transport, appears to be responsible for the competence cascade in S. mutans (11, 17). Although S. pneumoniae has determinants for both ComDE and BlpRH two-component systems, the former is responsible for competence in this species (8). Similarly, although the S. gordonii genome contains a BlpR DNA response homolog (SGO_0374) and a BlpH histidine kinase homolog (SGO_0375), neither of these genes was up-regulated in response to CSP. Future studies designed to dissect the S. gordonii competence cascade may also provide evolutionary insights into the competence response.
Delayed-response genes.
In S. pneumoniae, genes whose expression continued to increase and peak after early- and late-response genes were designated delayed-response genes (29). In general, these genes encoded chaperones and proteases involved in stress response, such as GroES, GroEL, and DnaK. The delayed response was not as evident in S. gordonii. The homologs for the groES, groEL, and dnaK genes (see Table S1 in the supplemental material) were not up-regulated in S. gordonii up to 40 min after CSP exposure when expression of the early and late genes had generally returned to basal levels. Similarly, the S. gordonii serine protease HtrA/DegP (SGO_2150), another classic heat shock protein which has been implicated in mediating S. pneumoniae competence (31) and was reported among the delayed-response S. pneumoniae genes (29), is similarly located near the chromosomal origin of replication in S. gordonii but was not differentially expressed in the present transcriptome analyses (see Tables S1 and S2 in the supplemental material). Although these findings do not rule out an indirect role of stress response genes in the S. gordonii competence response or that the differential expression occurred at a time point that was not tested in these studies, the delayed response in S. gordonii was not as evident based upon the S. pneumoniae criteria.
Although they did not show a delayed-response temporal pattern, two S. gordonii stress-associated genes did appear to be synchronous with the other late genes responsive to CSP at 15 min. The putative acid tolerance gene satD (SGO_1196), which is located within a gene cluster flanked by the gene encoding the DNA-processing Smf protein (Fig. 2), as well as the gene encoding the general stress protein Gsp781 (SGO_2094), are both within late gene clusters downstream of combox sequences (Fig. 3). Additional studies will be needed to characterize the S. gordonii delayed response and the role of stress response genes in the competence response of this species.
Down-regulated genes.
Genes repressed in response to CSP are less well characterized in streptococcal species. Like S. pneumoniae (7), S. gordonii had a two- to fivefold decrease in expression of several ribosomal protein genes and genes involved in metabolism (see Table S3 in the supplemental material), including the gene encoding catabolite control protein A (ccpA). Down-regulation of ccpA was supported by qRT-PCR data that showed the ratio of amplicons at 15 min to the initial value at time zero was only 0.2, indicating a fivefold decrease (Table 1). S. pneumoniae competence response transcriptome studies have shown strain-specific results for down-regulated genes. For example, S. pneumoniae alcohol dehydrogenase was shown to be down-regulated in strain TIGR4, while its homolog was up-regulated in strain R6 (7). Such variations in up- and down-regulated genes have been speculated to be due to different growth conditions (7). Indeed, in the present study, it is difficult to interpret if minimal changes in gene expression at 40 min are directly or indirectly due to CSP response or due to changes accompanying growth of the culture. Growth-related expression changes may be more evident in S. gordonii as the time post-CSP approaches the ca. 55-min generation time of Todd-Hewitt-grown cells. Future studies will be aimed at delineating temporal responses to verify the specificity of the various classes of genes affected by CSP.
Concluding remarks.
The present studies describe the first genome-wide response of the oral bacterium S. gordonii and the first functional insights into the complete genome of a commensal oral bacterial species. The results are consistent with known genetic data for S. gordonii and related streptococcal species, demonstrating the usefulness of the constructed 70-mer microarray for monitoring biological responses in S. gordonii. Competence is an ideal subject for these initial studies, since the response of a bacterial culture to CSP synchronizes the cells, thereby facilitating interpretation of the cells’ expression responses. The completion of these studies offers an extensive analysis of transcriptional changes that accompany competence in S. gordonii and highlights similarities and differences between the responses of this species and those of other streptococcal competence systems. The S. gordonii CSP response was more similar to that of S. pneumoniae than that reported for S. mutans, even though S. gordonii occupies an ecological niche in a multispecies biofilm on the tooth surface that is more similar to that of S. mutans. Species-specific differences in both CSP-induced genes and putative DNA regulatory regions were found that provide a basis for future studies that may identify patterns of essential genes in different streptococcal species that express a competence phenotype. Although microarrays have inherent limitations and a functional analysis of each of the putative CSP-responsive genes is beyond the scope of this initial study, these data do provide an essential basis for future analyses of comparative genomic competence responses and will serve as a powerful tool for monitoring S. gordonii global responses to environmental stimuli and signaling factors.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant DE11090 from the National Institute of Dental and Craniofacial Research.
We thank D. A. Morrison, University of Illinois at Chicago, for helpful advice and discussions.
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Campbell, E. A., S. Y. Choi, and H. R. Masure. 1998. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol. Microbiol. 27:929-939. [DOI] [PubMed] [Google Scholar]
- 2.Claverys, J. P., and B. Martin. 1998. Competence regulons, genomics and streptococci. Mol. Microbiol. 29:1126-1127. [DOI] [PubMed] [Google Scholar]
- 3.Claverys, J. P., B. Martin, and L. S. Havarstein. 2007. Competence-induced fratricide in streptococci. Mol. Microbiol. 64:1423-1433. [DOI] [PubMed] [Google Scholar]
- 4.Claverys, J. P., M. Prudhomme, and B. Martin. 2006. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 60:451-475. [DOI] [PubMed] [Google Scholar]
- 5.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35:251-259. [DOI] [PubMed] [Google Scholar]
- 6.Colman, G. 1969. Transformation of viridans-like streptococci. J. Gen. Microbiol. 28:275-286. [DOI] [PubMed] [Google Scholar]
- 7.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 8.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaustad, P., and D. A. Morrison. 1998. Induction of transformation in streptococci by synthetic competence stimulating peptides. Methods Cell Sci. 20:65-70. [Google Scholar]
- 11.Hale, J. D. F., N. C. K. Heng, R. W. Jack, and J. R. Tagg. 2005. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J. Bacteriol. 187:5036-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 13.Heng, N. C., J. R. Tagg, and G. R. Tompkins. 2006. Identification and characterization of the loci encoding the competence-associated alternative sigma factor of Streptococcus gordonii. FEMS Microbiol. Lett. 259:27-34. [DOI] [PubMed] [Google Scholar]
- 14.Heng, N. C., J. R. Tagg, and G. R. Tompkins. 2007. Competence-dependent bacteriocin production by Streptococcus gordonii DL1 (Challis). J. Bacteriol. 189:1468-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkinson, H. F., R. A. Baker, and G. W. Tannock. 1996. A binding-lipoprotein-dependent oligopeptide transport system in Streptococcus gordonii essential for uptake of hexa- and heptapeptides. J. Bacteriol. 178:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawamura, Y., X. G. Hou, F. Sultana, H. Miura, and T. Ezaki. 1995. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int. J. Syst. Bacteriol. 45:406-408. [DOI] [PubMed] [Google Scholar]
- 17.Kreth, J., J. Merritt, W. Shi, and F. Qi. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57:392-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, M. S., B. A. Dougherty, A. C. Madeo, and D. A. Morrison. 1999. Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes. Appl. Environ. Microbiol. 65:1883-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loo, C. Y., K. Mitrakul, I. B. Voss, C. V. Hughes, and N. Ganeshkumar. 2003. Involvement of the adc operon and manganese homeostasis in Streptococcus gordonii biofilm formation. J. Bacteriol. 185:2887-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lunsford, R. D. 1998. Streptococcal transformation: essential features and applications of a natural gene exchange system. Plasmid 39:10-20. [DOI] [PubMed] [Google Scholar]
- 21.Lunsford, R. D., and J. London. 1996. Natural genetic transformation in Streptococcus gordonii: comX imparts spontaneous competence on strain wicky. J. Bacteriol. 178:5831-5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunsford, R. D., and A. G. Roble. 1997. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J. Bacteriol. 179:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, P., H. Li, and D. A. Morrison. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 54:172-183. [DOI] [PubMed] [Google Scholar]
- 24.Luo, P., and D. A. Morrison. 2003. Transient association of an alternative sigma factor, ComX, with RNA polymerase during the period of competence for genetic transformation in Streptococcus pneumoniae. J. Bacteriol. 185:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macrina, F. L., J. A. Tobian, K. R. Jones, and R. P. Evans. 1981. Molecular cloning in the streptococci, p. 195-210. In A. Hollaender, R. DeMoss, S. Kaplan, J. Konisky, D. Savage, and R. Wolfe (ed.), Genetic engineering of microorganisms for chemicals. Plenum Publishing Corp., New York, NY.
- 26.Martin, B., Y. Quentin, G. Fichant, and J. P. Claverys. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14:339-345. [DOI] [PubMed] [Google Scholar]
- 27.Mitrakul, K., C. Y. Loo, C. Gyurko, C. V. Hughes, and N. Ganeshkumar. 2005. Mutational analysis of the adcCBA genes in Streptococcus gordonii biofilm formation. Oral Microbiol. Immunol. 20:122-127. [DOI] [PubMed] [Google Scholar]
- 28.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 30.Rimini, R., B. Jansson, G. Feger, T. C. Roberts, M. de Francesco, A. Gozzi, F. Faggioni, E. Domenici, D. M. Wallace, N. Frandsen, and A. Polissi. 2000. Global analysis of transcription kinetics during competence development in Streptococcus pneumoniae using high density DNA arrays. Mol. Microbiol. 36:1279-1292. [DOI] [PubMed] [Google Scholar]
- 31.Sebert, M. E., K. P. Patel, M. Plotnick, and J. N. Weiser. 2005. Pneumococcal HtrA protease mediates inhibition of competence by the CiaRH two-component signaling system. J. Bacteriol. 187:3969-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stothard, P. 2000. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28:1102-1104. [DOI] [PubMed] [Google Scholar]
- 33.Sulavik, M. C., G. Tardif, and D. B. Clewell. 1992. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J. Bacteriol. 174:3577-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vickerman, M. M., D. G. Heath, and D. B. Clewell. 1993. Construction of recombination-deficient strains of Streptococcus gordonii by disruption of the recA gene. J. Bacteriol. 175:6354-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ween, O., P. Gaustad, and L. S. Havarstein. 1999. Identification of DNA binding sites for ComE, a key regulator of natural competence in Streptococcus pneumoniae. Mol. Microbiol. 33:817-827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.