Abstract
Bacterial peptidoglycan hydrolases are considered to have destructive potential, which in the presence of inhibitory concentrations of cell wall synthesis inhibitors is involved in cell lysis. Therefore, the expression and activity of autolytic enzymes must be tightly regulated in growing cells. We describe here a series of experiments undertaken to examine further the coordination between cell wall synthesis and degradation. Cell growth in the presence of subinhibitory concentrations of β-lactam antibiotics was used to determine the effects of the partial inhibition of cell wall synthesis on the status of the autolytic system in Staphylococcus aureus. Our results revealed that, despite increased in vitro hydrolysis of cell walls by autolytic enzymes due to hypo-cross-linked peptidoglycans, cells grown in the presence of β-lactams were dramatically less prone to autolysis as a result of decreased transcription and enzymatic activities of several major autolytic enzymes. Similar repression of autolytic enzymatic activity and transcription was also observed when cell wall synthesis was disturbed by lowering the level of transcription of pbpB, the gene encoding the major transpeptidase in S. aureus. Our data show that the perturbation of cell wall synthesis in growing cells of S. aureus induces strong repression of the autolytic system and provide evidence for transcriptional regulation between cell wall synthetic and hydrolytic enzymes.
Peptidoglycan (PG), a heteropolymer composed of glycan strands interconnected by oligopeptides, is the major component of the cell walls of gram-positive bacteria. PG is a dynamic structure that undergoes constant and simultaneous synthesis and degradation during cell growth. Bacteria produce several hydrolases that specifically cleave various covalent bonds in the PG, including N-acetylmuramyl-l-alanine amidases, N-acetylglucosaminidases, N-acetylmuramidases, endopeptidases, and transglycosylases. The physiological functions of these enzymes remain largely unknown; however, it has been proposed that in various bacterial species they play important roles in a variety of cellular processes, such as cell wall growth, cell wall turnover, cell separation, the recycling of muropeptides, lysis induced by cell wall synthesis inhibitors, the establishment of competence for genetic transformation, flagellum formation, sporulation, and bacterial pathogenicity processes (for a review, see references 1, 16, 30, 38, and 39).
More than 20 bacteriolytic bands in Staphylococcus aureus can be detected using zymography, suggesting that S. aureus produces several PG hydrolases (42). Up to 13 genes encoding known or putative PG hydrolases in the genome of S. aureus strain NCTC 8325 have been described previously, but only three genes (atl, sle1/aaa, and lytM) and their products have been characterized. The major autolytic enzyme Atl is a bifunctional autolysin initially produced as a 138-kDa protein that subsequently undergoes proteolytic processing to generate the two major autolytic enzymes, a 62-kDa N-acetylmuramyl-l-alanine amidase and a 51-kDa N-acetylglucosaminidase (21, 28). Atl is involved in the separation of daughter cells after cell division, in cell wall turnover, and in antibiotic-induced lysis (7, 29, 44-46, 52). Sle1 (or Aaa) is a 32-kDa protein with N-acetylmuramyl-l-alanine amidase activity (14, 20). A sle1 mutant forms clusters, suggesting the involvement of Sle1 in the separation of daughter cells during cell division. LytM is a 32-kDa protein with glycylglycine endopeptidase activity and is distributed uniformly on the cell surface, suggesting that LytM plays a role in cell growth (34, 35). LytH, LytA, and LytN (33, 50, and 46 kDa, respectively) are PG hydrolases with N-acetylmuramyl-l-alanine amidase activity (10, 19, 43, 50). However, none of the genes or gene products listed above are essential for bacterial growth, since mutants have been described previously; even a sle1 and atl double mutant is viable, but its growth is significantly impaired (20).
Because of their potential to compromise cell wall integrity, the expression and activities of autolytic enzymes must be tightly regulated. At the posttranscriptional level, the activities of autolytic enzymes in S. aureus have been shown to be modulated by heat shock, NaCl, lipoteichoic acids, and the activities of proteases (4, 12, 26, 27, 32, 36, 47, 51). In addition, the expression of some autolytic enzymes is regulated at the transcriptional level. The expression of atl is stimulated by low temperatures and by the presence of NaCl (7). Several two-component signal transduction systems and global regulators are involved in the regulation of the autolytic activity in S. aureus. MgrA (or Rat) (17, 18, 25), ArlRS (8, 24), LytSR (2, 3, 13), and SarA (9, 27) negatively modulate autolytic activity. In contrast, agr (9) and the cidABC operon (37) function as positive regulators of autolysis.
In this study, we examined the consequences of cell wall synthesis disturbance on the autolytic activity in exponentially growing cells of S. aureus, either by growing cells in the presence of subinhibitory concentrations (half the MICs) of β-lactams or by downregulating the expression of the pbpB gene encoding the major transpeptidase penicillin-binding protein 2 (PBP2). We found that the perturbation of cell wall synthesis triggers the repression of the autolytic system in S. aureus, which provides evidence for close regulation between cell wall synthetic and hydrolytic enzymes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Characteristics of S. aureus strains used in this study are described in Table 1. Bacterial cultures were grown in tryptic soy broth (TSB; Difco Laboratories) or on tryptic soy agar (Difco Laboratories) at 37°C with aeration.
TABLE 1.
S. aureus strains used in this study
| Strain | Relevant characteristics | MIC (μg/ml) of:
|
Source or reference | ||
|---|---|---|---|---|---|
| Ceftizoxime | Oxacillin | Cefotaxime | |||
| 27s | Derivative of RN450; antibiotic sensitive | 0.8 | 0.1 | 0.8 | R. Novick |
| COL | Methicillin-resistant clinical isolate; mecA | 800 | 400 | 800 | RU collectiona |
| RUSAL9 | COL atl::Tn551 mutant | 400 | 100 | 100 | 29 |
| 27sspac::pbpB | 27s derivative in which pbpB is under the control of the spac promoter | 23 | |||
| COLspac::pbpB | COL derivative in which pbpB is under the control of the spac promoter | 31 | |||
RU, The Rockefeller University.
Population analysis profiles.
The susceptibilities of S. aureus strains to antibiotics were determined by population analysis as previously described (Table 1) (49). Oxacillin and cefotaxime were purchased from Sigma-Aldrich, and ceftizoxime was purchased from USPC, Inc.
Analysis of PG composition.
PG was purified from 1-liter cultures of bacteria grown at 37°C to mid-exponential phase, as previously described (5). Purified PG was digested with mutanolysin (Sigma-Aldrich), and muropeptides were reduced with sodium borohydride (Sigma-Aldrich) and separated on a Hypersil ODS column (Thermo Electron Corporation).
TX-100-stimulated whole-cell autolysis.
Cells were grown to mid-exponential phase, chilled in an ice-ethanol bath, harvested, and washed with ice-cold water. Cells were then suspended to an optical density (OD) at 620 nm of 1.0 in 50 mM glycine buffer, pH 8.0, containing 0.01% Triton X-100 (TX-100) as previously described (6). The autolytic rate at 37°C was measured for 3 h as a decrease of OD at 620 nm.
Preparation of autolytic enzyme extracts.
Cells were grown to mid-exponential phase in 500 ml of TSB at 37°C with aeration, chilled in an ice-ethanol bath, harvested, and washed with ice-cold 50 mM Tris-HCl (pH 7.5)-150 mM NaCl. Cells were extracted with 500 μl of 4% sodium dodecyl sulfate (SDS) for 30 min at room temperature with stirring or with 500 μl of 3 M LiCl-0.1% TX-100 for 30 min at 4°C with stirring, as previously described (42). Supernatants were used as a source of enzymes. Protein concentrations were determined with the modified Lowry protein assay kit (Pierce) with bovine serum albumin as a standard.
Enzymatic hydrolysis of crude cell walls in vitro.
Crude cell walls were isolated from cells grown to mid-exponential phase by boiling in 4% SDS, and then preparations were extensively washed with hot water to remove SDS. Crude cell walls were suspended in 50 mM Tris-HCl, pH 7.5, to an OD at 620 nm of 0.5, and LiCl cell extracts (10 μg of proteins/ml) were added. The hydrolysis at 37°C was measured as a decrease of OD at 620 nm over 4 h.
Zymographic analysis.
SDS cell extracts (10 μg of proteins) were separated by SDS-polyacrylamide gel electrophoresis on a 10% resolving gel containing crude cell walls (0.1%) (42). Gels were washed three times with water for 15 min and once with 50 mM Tris-HCl (pH 7.5)-0.1% TX-100-10 mM CaCl2-10 mM MgCl2 for 30 min at room temperature, and then the gels were incubated at 37°C in the buffer described above (40).
RNA isolation and Northern blot analysis.
Total RNA was extracted from cultures grown up to an OD at 620 nm of 0.7. RNA (5 μg) was resolved by electrophoresis on 1.2% agarose-0.66 M formaldehyde gels in morpholinepropanesulfonic acid running buffer. RNA was blotted onto Hybond-N+ membranes (GE Healthcare) using a turbo blotter alkaline transfer system (Schleicher and Schuell) with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). PCR-amplified DNA probes were labeled with [α-32P]dCTP (GE Healthcare) by using a Ready-To-Go labeling kit (GE Healthcare) and hybridized under high-stringency conditions. The blots were subsequently washed and autoradiographed. The transcription of the housekeeping gene pta or the intensity of 16S rRNA bands in the gel was used as an internal control.
RESULTS
Autolysis of whole cells grown in the presence of subinhibitory concentrations of β-lactams.
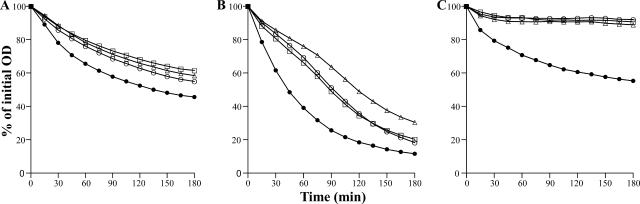
A β-lactam-susceptible strain, 27s, a methicillin-resistant clinical isolate, COL, and the atl mutant of COL, RUSAL9, were used in this study (Table 1). Cells were grown in the presence of subinhibitory concentrations (half the MICs) of various β-lactam antibiotics, and the rates of TX-100-stimulated autolysis of whole cells were compared with the rates for untreated control cultures. Cells from cultures grown in the presence of cefotaxime and ceftizoxime, which are specific for PBP2, or oxacillin showed a decreased rate of autolysis compared with those from untreated control cultures for all the strains tested (Fig. 1). A similar decrease of the autolytic rate was also observed when cells were grown in the presence of subinhibitory concentrations of additional PBP-specific inhibitors, such as cloxacillin (PBP1), cephradine (PBP3), and cefoxitin (PBP4), moenomycin (an inhibitor of transglycosylase activity), and early-stage cell wall synthesis inhibitors, such as fosfomycin, d-cycloserine, and bacitracin (data not shown). However, growing cells in the presence of subinhibitory concentrations of DNA synthesis inhibitors (novobiocin and ciprofloxacin) had no detectable effect on the autolytic rate (data not shown).
FIG. 1.
TX-100-stimulated autolysis of whole cells grown in the presence of subinhibitory concentrations of β-lactams. Cells of strains 27s (A), COL (B), and RUSAL9 (C) grown in the absence (•) or in the presence of subinhibitory concentrations (half the MICs) of ceftizoxime (○), oxacillin (▵), and cefotaxime (□) were suspended in 50 mM glycine buffer, pH 8.0, containing 0.01% TX-100. The rates of autolysis were monitored as a decrease of OD at 620 nm.
In vitro susceptibilities of cell walls and activities and amounts of autolytic enzymes in cells grown in the presence of subinhibitory concentrations of β-lactams.
To determine whether the decreased rates of autolysis of cells grown in the presence of β-lactams were due to alterations in the activities and/or the amounts of autolytic enzymes or to modifications of the substrate (cell walls), autolytic enzymes and cell walls were isolated from cells grown in the absence and in the presence of subinhibitory concentrations (half the MICs) of β-lactams.
The susceptibilities of cell walls to degradation by autolytic enzymes were compared. Crude cell walls isolated from cells grown in the absence and in the presence of antibiotics were suspended in 50 mM Tris-HCl, pH 7.5, and subjected to hydrolysis by the same autolytic enzyme extract obtained from COL. In the absence of an antibiotic, 27s cell walls were more susceptible than COL and RUSAL9 cell walls (Fig. 2A). We also determined the muropeptide compositions of the PG components of these crude cell walls and observed a good correlation between the cell wall susceptibility to autolytic enzymes and the degree of PG cross-linking (Table 2): the more cross-linked the PG was, the less susceptible the cell wall was to hydrolysis by the autolytic enzyme extracts. Growth in the presence of β-lactams, which reduced the PG cross-linking (Table 2), increased the susceptibility of cell walls to hydrolysis by autolytic enzymes in vitro (Fig. 2A). This enhanced hydrolysis of cell walls isolated from cells grown in the presence of β-lactams was also observed with autolytic enzymes extracted from 27s (data not shown).
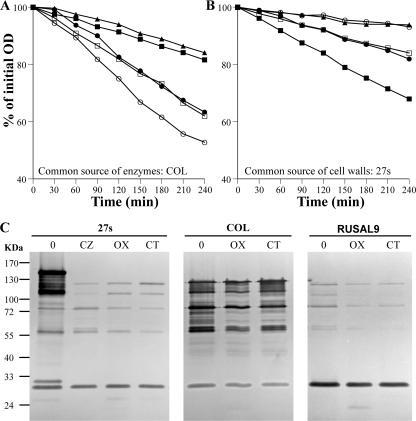
FIG. 2.
In vitro susceptibility of cell walls and activity of autolytic enzymes in cells grown in the presence of subinhibitory concentrations of β-lactams. (A) COL LiCl cell extract (10 μg of proteins/ml) was used to test the susceptibility of crude cell walls isolated from cells grown in the absence and in the presence of β-lactams (at half the MICs) to degradation by autolytic enzymes. Results for crude cell walls of 27s (•), 27s grown in the presence of ceftizoxime (○), COL (▪), COL grown in the presence of oxacillin (□), and RUSAL9 (▴) are shown. (B) 27s crude cell walls were subjected to hydrolysis by LiCl cell extracts (10 μg of proteins/ml) prepared from cells grown in the absence and in the presence of β-lactams (at half the MICs). Results for LiCl cell extracts from 27s (•), 27s grown in the presence of ceftizoxime (○), COL (▪), COL grown in the presence of oxacillin (□), and RUSAL9 (▴) are shown. (C) Zymographic analysis of autolytic enzyme extracts. SDS cell extracts (10 μg of proteins) prepared from cells grown in the absence (0) and in the presence of ceftizoxime (CZ), oxacillin (OX), and cefotaxime (CT) at half the MICs were separated on a 10% resolving gel containing 27s crude cell walls. After renaturation and incubation at 37°C, bacteriolytic bands were observed as clear zones in the opaque gel that appeared as dark bands against a dark background.
TABLE 2.
Muropeptide compositions of PG purified from cells grown in the absence and in the presence of subinhibitory concentrations (half the MICs) of β-lactams
| Strain | Antibiotic (concn, μg/ml) | % Of total muropeptides in the form of:
|
|||
|---|---|---|---|---|---|
| Monomers | Dimers | Tri-, tetra-, or pentamers | Higher oligomers | ||
| 27s | None | 6.4 | 15.2 | 29.5 | 46.3 |
| Ceftizoxime (0.4) | 16.2 | 21.8 | 32.4 | 28.9 | |
| COL | None | 6.6 | 12.2 | 26.5 | 52.2 |
| Oxacillin (200) | 29.7 | 27.1 | 28.4 | 14 | |
| RUSAL9 | None | 7.6 | 13.1 | 25.7 | 52.5 |
These results suggested that the decreased rates of autolysis in cells grown in the presence of β-lactams were most likely related to alterations in the amounts and/or activities of autolytic enzymes. In order to test this suggestion, another set of experiments was designed in which a common cell wall substrate isolated from 27s was subjected to hydrolysis by autolytic enzymes extracted from cells grown in the absence or presence of β-lactams. In the absence of an antibiotic, the levels of activity of autolytic enzymes from strain COL were higher than those of autolytic enzymes from strain 27s, whereas there was hardly any lysis in the case of RUSAL9 autolytic enzymes (Fig. 2B). Autolytic enzymes extracted from cells grown in the presence of antibiotics degraded cell walls at a lower rate than those from controls grown without an antibiotic (Fig. 2B). These differences in quantitative hydrolytic activities prompted us to analyze the zymographic profile of SDS extracts from cells grown in the absence and in the presence of subinhibitory concentrations of β-lactams (Fig. 2C). Only a few weak bands and one strong band of low molecular mass that may correspond to Sle1 and/or LytM (20, 34) were detected in the extract from the atl mutant, indicating that the majority of the bacteriolytic bands observed in the other extracts (138-, 115-, 85-, 62-, and 51-kDa bands) corresponded to Atl and its processed intermediates. Compared with the bands from extracts prepared from untreated controls, decreased intensity and even the disappearance of some bands from the extracts prepared from cells grown in the presence of β-lactams were observed. Similar patterns of autolytic enzymes were observed regardless of the source of the cell walls used (data not shown). The activities of autolytic enzymes assayed by quantitative and zymographic analyses were in good agreement with the rates of TX-100-induced autolysis, suggesting that the reduced rates of whole-cell autolysis in the presence of subinhibitory concentrations of β-lactams were the result of decreased activities and/or amounts of autolytic enzymes.
Expression levels of genes encoding autolytic enzymes in cells grown in the presence of subinhibitory concentrations of β-lactams.
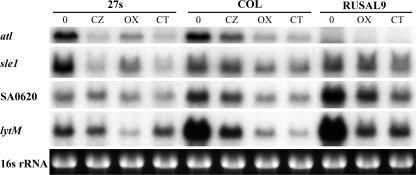
To further determine whether the reduced activities and/or amounts of autolytic enzymes in the presence of subinhibitory concentrations (half the MICs) of β-lactams were the result of changes in the transcription profiles of these enzymes, we compared the levels of expression of genes encoding autolytic enzymes by Northern blot analysis (Fig. 3). Several genes (atl, sle1, and lytM) encoding enzymes with previously documented bacteriolytic activities and also SA0620, encoding a putative autolysin, were tested. The exposure of COL cells to subinhibitory concentrations of β-lactams decreased the expression of atl, sle1, SA0620, and lytM. Similar changes in levels of gene expression in the COL atl mutant were observed. The transcription of atl and sle1 in 27s was also strongly repressed, whereas no changes in SA0620 transcription were observed. The level of expression of lytM in 27s was very low compared to that in COL and was only slightly reduced in the presence of β-lactams.
FIG. 3.
Levels of expression of genes encoding autolytic enzymes in cells grown in the presence of subinhibitory concentrations of β-lactams. RNA was purified from cultures of 27s, COL, and RUSAL9 strains grown to the mid-exponential phase in the absence (0) and in the presence of ceftizoxime (CZ), oxacillin (OX), and cefotaxime (CT) at half the MICs. RNA (5 μg) was resolved by electrophoresis on agarose-formaldehyde gels. After transfer, membranes were hybridyzed with 32P-labeled atl, sle1, SA0620, and lytM DNA probes.
Effects of pbpB transcription on whole-cell autolysis.
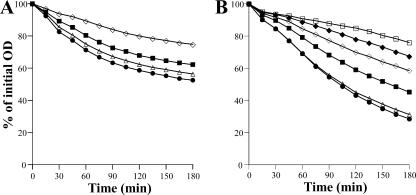
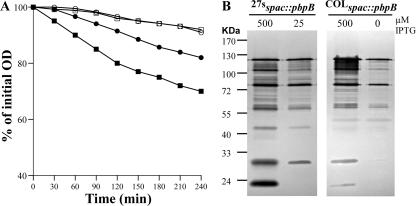
Since the perturbation of cell wall synthesis and structure by the partial inhibition of the normal functioning of PBPs by β-lactam antibiotics led to the repression of autolysis, we investigated the effects of the perturbation of cell wall synthesis by lowering the level of transcription of pbpB and hence the amount of PBP2, the protein considered to be the major transpeptidase in S. aureus (23, 31). We used two conditional mutants, 27sspac::pbpB and COLspac::pbpB, in which the pbpB gene is under the control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter (23, 31). The conditional mutants were grown in the presence of an optimal concentration of IPTG (500 μM) and suboptimal concentrations of IPTG. The conditional mutant in the 27s background was unable to grow in the presence of IPTG concentrations below 25 μM, whereas the conditional mutant in the COL background, in which PBP2a is present, could grow in the absence of IPTG. The rates of TX-100-stimulated whole-cell autolysis were compared. In parallel with the decreasing concentration of IPTG (and hence pbpB transcription and PBP2 amount), the rate of autolysis progressively decreased (Fig. 4).
FIG. 4.
Effects of pbpB transcription on TX-100-stimulated whole-cell autolysis. Cells were suspended in 50 mM glycine buffer, pH 8.0, containing 0.01% TX-100, and the rates of autolysis were monitored as a decrease of OD at 620 nm. 27sspac::pbpB (A) and COLspac::pbpB (B) were grown in TSB supplemented with decreasing concentrations of IPTG, 500 μM (•), 100 μM (▵), 50 μM (▪), 25 μM (⋄), and 12.5 μM (⧫), and in the absence of IPTG (□).
Effects of pbpB transcription on the activities and amounts of autolytic enzymes.
The activities and amounts of autolytic enzymes in the conditional mutants 27sspac::pbpB and COLspac::pbpB grown in the presence of optimal and suboptimal concentrations of IPTG and in the absence of IPTG were compared. Autolytic enzyme extracts were tested against a common cell wall substrate (27s cell walls) in 50 mM Tris-HCl, pH 7.5 (Fig. 5A), or analyzed by zymography (Fig. 5B). In the presence of a low concentration of IPTG or in the absence of IPTG, the hydrolysis of cell walls was slowed down and the pattern of the bacteriolytic bands changed: several bands were either absent or less intense.
FIG. 5.
Effects of pbpB transcription on the in vitro activity of autolytic enzymes. (A) 27s crude cell walls were suspended in 50 mM Tris-HCl, pH 7.5, and subjected to hydrolysis by LiCl cell extracts (10 μg of proteins/ml) prepared from strain 27sspac::pbpB grown in the presence of 500 μM (•) and 25 μM (○) IPTG and from strain COLspac::pbpB grown in the presence of 500 μM IPTG (▪) and in the absence of IPTG (□). (B) Zymographic analysis of autolytic enzyme extracts. SDS cell extracts (10 μg of proteins) prepared from strains 27sspac::pbpB and COLspac::pbpB grown in the presence of different concentrations of IPTG were separated on a 10% resolving gel containing 27s crude cell walls. After renaturation and incubation at 37°C, bacteriolytic bands were observed as clear zones in the opaque gel that appeared as dark bands against a dark background.
Effects of pbpB transcription on levels of expression of genes encoding autolytic enzymes.
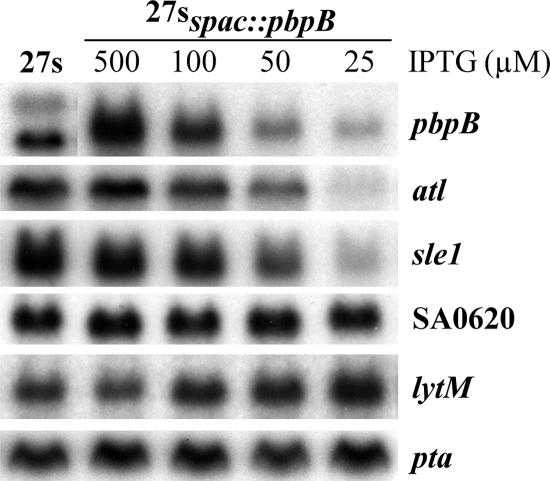
We compared the levels of expression of genes encoding autolytic enzymes by Northern blot analysis when the conditional mutant 27sspac::pbpB was grown in the presence of decreasing concentrations of IPTG (Fig. 6). In parallel with the reduced transcription of pbpB, the expression of atl and sle1 was progressively downregulated whereas that of the lytM gene was slightly upregulated. We did not observe any changes in the transcription of SA0620.
FIG. 6.
Effects of pbpB transcription on the levels of expression of genes encoding autolytic enzymes. RNA (5 μg) was purified from cultures of 27sspac::pbpB grown to the mid-exponential phase in the presence of decreasing concentrations of IPTG and resolved by electrophoresis on agarose-formaldehyde gels. After transfer, membranes were hybridized with 32P-labeled pbpB, atl, sle1, SA0620, lytM, and pta DNA probes. The wild-type strain 27s was used as a control.
Effects of removal and readdition of IPTG on autolysis of the conditional mutant 27sspac::pbpB.
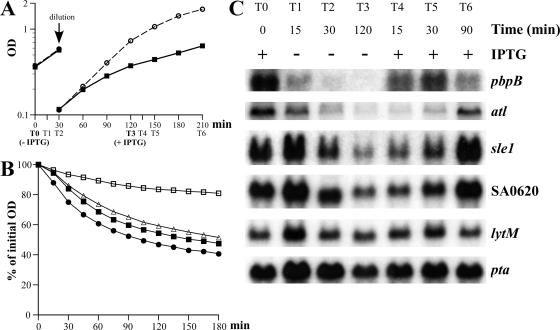
The S. aureus conditional mutant 27sspac::pbpB was grown in the presence of an optimal concentration of IPTG up to an OD of 0.3 (time zero [T0]). The culture was centrifuged and washed twice, and cells were resuspended in medium without IPTG (Fig. 7A). Samples were removed 15 min (T1) and 30 min (T2) later. At this time, dilution of the culture was performed to maintain cells in exponential growth phase, and the culture was further incubated without IPTG for 90 min (T3). After 2 h without IPTG, an optimal concentration of IPTG was added back and samples were removed after 15 min (T4), 30 min (T5), and 90 min (T6). The susceptibility of whole cells to autolysis was assayed at T0, T2, T3, and T6 (Fig. 7B). After the removal of IPTG, the rate of autolysis gradually decreased (T2 and T3). This effect was reversible since the rate of autolysis returned to close to normal 90 min after the readdition of IPTG to the culture (T6). In parallel, the levels of expression of genes encoding autolytic enzymes from T0 through T6 were determined (Fig. 7C). As soon as 15 min after the removal of IPTG, the transcription of pbpB decreased extensively, and no transcript was detected after 30 and 120 min. When IPTG was added back, pbpB transcripts were again detected after 15 min. The transcription of atl, sle1, and SA0620 progressively decreased and increased after the removal and the readdition of IPTG, respectively; however, these effects were slightly delayed compared to the rapid changes in pbpB transcription. No changes in the level of expression of lytM were observed. The transcription of autolytic enzymes was downregulated as soon as 30 min after the removal of IPTG, while no differences in the growth rates between the IPTG-depleted and control cultures were yet detected (Fig. 7), suggesting that the transcription of cell wall synthetic enzymes and that of hydrolytic enzymes are unlikely to be linked through growth rate.
FIG. 7.
Effects of removal and readdition of IPTG on autolysis of the conditional mutant 27sspac::pbpB. (A) Growth curve of a 27sspac::pbpB conditional mutant (▪) grown in the absence of IPTG (− IPTG) for 120 min (T0 to T3) and then in the presence of IPTG (+ IPTG) for 90 min (T3 to T6), as described in the text. In parallel, a control culture of 27sspac::pbpB (○) was grown in the presence of an optimal concentration of IPTG. (B) Cell susceptibility to autolysis was assayed at T0 (•) and at 30 min (T2; ▵) and 120 min (T3; □) after the removal of IPTG and 90 min (T6; ▪) after the readdition of IPTG. Cells were suspended in 50 mM glycine buffer, pH 8.0, containing 0.01% TX-100, and the rates of autolysis were monitored as a decrease of OD at 620 nm. (C) RNA (5 μg) was purified at T0 through T6 and resolved by electrophoresis on agarose-formaldehyde gels. After transfer, membranes were hybridized with 32P-labeled pbpB, atl, sle1, SA0620, lytM, and pta DNA probes.
DISCUSSION
Numerous reports in the literature have suggested that the inhibition of cell wall synthesis by inhibitory concentrations of antibiotics affecting cell wall synthesis induces cell wall degradation and cell lysis in many bacteria by a process that involves autolytic enzymes, since mutants defective in autolysin show reduced rates of cell wall turnover and the absence of lysis in the presence of cell wall synthesis inhibitors (29, 48). Moreover, it is well known that the treatment of S. aureus cells with subinhibitory concentrations of β-lactam antibiotics results in the production of PG with drastically reduced cross-linking. This form of hypo-cross-linked PG was also shown to be more prone to in vitro enzymatic degradation by autolysins (33). Therefore, one may expect that growing bacteria in the presence of subinhibitory concentrations of β-lactams would weaken the cell wall integrity, resulting in an increase in the proneness of these bacteria to autolysis.
However, the data presented in this communication show the opposite. While S. aureus cells grown in the presence of subinhibitory concentrations of oxacillin, ceftizoxime, or cefotaxime did produce a hypo-cross-linked PG that was indeed more susceptible to in vitro enzymatic degradation by autolytic enzymes, the cells exhibited dramatically reduced susceptibility to autolysis. This decreased rate of autolysis was paralleled by substantial decreases in the amounts and/or activities of autolytic enzymes as shown by zymographic analysis and quantitative enzymatic hydrolysis of cell walls. Most striking was the drastically reduced transcription of at least four determinants of autolytic enzymes (atl, sle1, lytM, and SA0620) in β-lactam-treated bacteria.
When the perturbation of cell wall synthesis was provoked not by β-lactam inhibition of PBPs but by reducing the transcription of the pbpB gene encoding one of the major S. aureus transpeptidases, PBP2, the same observations were still noted: decreased susceptibility of cells to autolysis, decreased amounts and/or activities of autolytic enzymes, and strikingly reduced transcription of the major autolytic enzyme genes, atl and sle1. Moreover, the inhibition of pbpB transcription in the conditional mutant 27sspac::pbpB was quickly followed by an inhibition of transcription of the autolytic enzyme genes, which was reversible after the readdition of IPTG to the cultures.
While the doubling time of cultures grown in the presence of subinhibitory concentrations of cell wall synthesis inhibitors (and other classes of antibiotics) increased (for example, from 22 to 32, 29, and 37 min for 27s in the presence of ceftizoxime, oxacillin, and cefotaxime, respectively), the perturbation of cell wall synthesis and the repression of autolysis are unlikely to be coupled through reduced growth rate. (i) The decreased autolytic rates observed for the cells grown in the presence of antibiotics were specific for cell wall synthesis inhibitors, and no effect on autolysis was observed in the presence of DNA synthesis inhibitors. (ii) Growing cells at low temperatures decreased the growth rate but inversely resulted in an increased autolytic rate (data not shown). (iii) IPTG-depleted and control cultures of the conditional mutant 27sspac::pbpB grew at identical rates for almost 60 min, whereas whole-cell autolysis and the transcription of autolytic enzymes were already repressed 30 min after the removal of IPTG (Fig. 7).
Our observations allow several conclusions. (i) Growing bacteria in the presence of subinhibitory concentrations of cell wall synthesis inhibitors provoked the repression of the potentially destructive autolytic system in S. aureus, which may be considered a defense mechanism of the bacterium to prevent any damage to the cell wall. A balance between the building and breaking of PG covalent bonds during cell wall synthesis has been suggested previously by several authors (for a review, see reference 15). Our data provide evidence that such tight regulation between the activities of cell wall synthetic enzymes and cell wall hydrolytic enzymes does indeed exist. (ii) The strikingly reduced transcription of autolytic enzyme genes indicates that the decreased autolytic activity is unlikely to be due to altered translocation or enhanced release into the culture supernatant but rather to the decreased production of the enzymes. (iii) The expression of several determinants of autolytic enzymes was altered, suggesting that at least one global regulator or two-component signal transduction system may be involved in their regulation. (iv) A reduced rate of transcription of the pbpB gene encoding the major cell wall synthetic enzyme PBP2 brought about the same—reversible—response in the cell susceptibility to autolysis and in the activities and transcription of autolytic enzymes, suggesting the existence of some transcriptional regulation between cell wall synthetic and hydrolytic enzymes. Using a DNA microarray assay, Sobral et al. recently identified extensive and genome-wide changes in the transcription profile of S. aureus in response to a reduced rate of transcription of the cell wall precursor synthetic gene murF. In agreement with our data, similar repression of autolytic activity was observed in response to a disturbance in cell wall synthesis (41).
Several two-component signal transduction systems (LytSR, Agr, and ArlRS) and global regulators (SarA and MgrA) are involved in the regulation of the autolytic activity in S. aureus (2, 3, 8, 9, 18, 24, 25, 27). Moreover, the two-component sensory regulatory system VraSR is capable of sensing the perturbation of cell wall synthesis after the exposure of S. aureus cells to cell wall synthesis inhibitors or a decrease in the amount of PBP2 (11, 22). Which of these regulatory systems may sense the perturbations of cell wall synthesis and negatively control autolytic activity in S. aureus is under investigation.
Acknowledgments
This work was supported by a grant from the U.S. Public Health Service, 5 RO1 AI045738.
Footnotes
Published ahead of print on 7 September 2007.
REFERENCES
- 1.Archibald, A. R., I. C. Hancock, and C. R. Harwood. 1993. Cell wall structure, synthesis, and turnover, p. 381-410. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria. American Society for Microbiology, Washington, DC.
- 2.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cleveland, R. F., J. V. Holtje, A. J. Wicken, A. Tomasz, L. Daneo-Moore, and G. D. Shockman. 1975. Inhibition of bacterial wall lysins by lipoteichoic acids and related compounds. Biochem. Biophys. Res. Commun. 67:1128-1135. [DOI] [PubMed] [Google Scholar]
- 5.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 6.de Jonge, B. L., H. de Lencastre, and A. Tomasz. 1991. Suppression of autolysis and cell wall turnover in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J. Bacteriol. 173:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325/4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujimoto, D. F., and K. W. Bayles. 1998. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J. Bacteriol. 180:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimura, T., and K. Murakami. 1997. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzymes. J. Bacteriol. 179:6294-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardete, S., S. W. Wu, S. Gill, and A. Tomasz. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob. Agents Chemother. 50:3424-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilpin, R. W., A. N. Chatterjee, and F. E. Young. 1972. Autolysis of microbial cells: salt activation of autolytic enzymes in a mutant of Staphylococcus aureus. J. Bacteriol. 111:272-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heilmann, C., J. Hartleib, M. S. Hussain, and G. Peters. 2005. The multifunctional Staphylococcus aureus autolysin Aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73:4793-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holtje, J. V. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol Rev. 62:181-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holtje, J. V., and E. I. Tuomanen. 1991. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J. Gen. Microbiol. 137:441-454. [DOI] [PubMed] [Google Scholar]
- 17.Ingavale, S., W. van Wamel, T. T. Luong, C. Y. Lee, and A. L. Cheung. 2005. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect. Immun. 73:1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 19.Jayaswal, R. K., Y. I. Lee, and B. J. Wilkinson. 1990. Cloning and expression of a Staphylococcus aureus gene encoding a peptidoglycan hydrolase activity. J. Bacteriol. 172:5783-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kajimura, J., T. Fujiwara, S. Yamada, Y. Suzawa, T. Nishida, Y. Oyamada, I. Hayashi, J. Yamagishi, H. Komatsuzawa, and M. Sugai. 2005. Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58:1087-1101. [DOI] [PubMed] [Google Scholar]
- 21.Komatsuzawa, H., M. Sugai, S. Nakashima, S. Yamada, A. Matsumoto, T. Oshida, and H. Suginaka. 1997. Subcellular localization of the major autolysin, ATL and its processed proteins in Staphylococcus aureus. Microbiol. Immunol. 41:469-479. [DOI] [PubMed] [Google Scholar]
- 22.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 23.Leski, T. A., and A. Tomasz. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 187:1815-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, X., L. Zheng, C. Landwehr, D. Lunsford, D. Holmes, and Y. Ji. 2005. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187:5486-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madiraju, M. V., D. P. Brunner, and B. J. Wilkinson. 1987. Effects of temperature, NaCl, and methicillin on penicillin-binding proteins, growth, peptidoglycan synthesis, and autolysis in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1727-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manna, A. C., S. S. Ingavale, M. Maloney, W. van Wamel, and A. L. Cheung. 2004. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J. Bacteriol. 186:5267-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshida, T., M. Sugai, H. Komatsuzawa, Y. M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshida, T., and A. Tomasz. 1992. Isolation and characterization of a Tn551-autolysis mutant of Staphylococcus aureus. J. Bacteriol. 174:4952-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perkins, H. R. 1980. The bacterial autolysins, p. 437-456. In H. J. Rogers, H. R. Perkins, and J. B. Ward (ed.), Microbial cell walls. Chapman and Hall, London, United Kingdom.
- 31.Pinho, M. G., S. R. Filipe, H. de Lencastre, and A. Tomasz. 2001. Complementation of the essential peptidoglycan transpeptidase function of penicillin-binding protein 2 (PBP2) by the drug resistance protein PBP2A in Staphylococcus aureus. J. Bacteriol. 183:6525-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qoronfleh, M. W., J. E. Gustafson, and B. J. Wilkinson. 1998. Conditions that induce Staphylococcus aureus heat shock proteins also inhibit autolysis. FEMS Microbiol. Lett. 166:103-107. [DOI] [PubMed] [Google Scholar]
- 33.Qoronfleh, M. W., and B. J. Wilkinson. 1986. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of β-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob. Agents Chemother. 29:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramadurai, L., and R. K. Jayaswal. 1997. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J. Bacteriol. 179:3625-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramadurai, L., K. J. Lockwood, M. J. Nadakavukaren, and R. K. Jayaswal. 1999. Characterization of a chromosomally encoded glycylglycine endopeptidase of Staphylococcus aureus. Microbiology 145:801-808. [DOI] [PubMed] [Google Scholar]
- 36.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, K. C., B. A. Firek, J. B. Nelson, S. J. Yang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shockman, G. D., and J. F. Barrett. 1983. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu. Rev. Microbiol. 37:501-527. [DOI] [PubMed] [Google Scholar]
- 39.Shockman, G. D., and J. V. Holtje. 1994. Microbial peptidoglycan (murein) hydrolases, p. 131-166. In J. M. Ghuysen and R. Hackenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science B.V., Amsterdam, The Netherlands. [Google Scholar]
- 40.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobral, R. G., A. E. Jones, S. G. Des Etages, T. J. Dougherty, R. M. Peitzsch, T. Gaasterland, A. M. Ludovice, H. de Lencastre, and A. Tomasz. 2007. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189:2376-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugai, M., T. Akiyama, H. Komatsuzawa, Y. Miyake, and H. Suginaka. 1990. Characterization of sodium dodecyl sulfate-stable Staphylococcus aureus bacteriolytic enzymes by polyacrylamide gel electrophoresis. J. Bacteriol. 172:6494-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugai, M., T. Fujiwara, H. Komatsuzawa, and H. Suginaka. 1998. Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. Gene 224:67-75. [DOI] [PubMed] [Google Scholar]
- 44.Sugai, M., H. Komatsuzawa, T. Akiyama, Y. M. Hong, T. Oshida, Y. Miyake, T. Yamaguchi, and H. Suginaka. 1995. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J. Bacteriol. 177:1491-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugai, M., S. Yamada, S. Nakashima, H. Komatsuzawa, A. Matsumoto, T. Oshida, and H. Suginaka. 1997. Localized perforation of the cell wall by a major autolysin: atl gene products and the onset of penicillin-induced lysis of Staphylococcus aureus. J. Bacteriol. 179:2958-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi, J., H. Komatsuzawa, S. Yamada, T. Nishida, H. Labischinski, T. Fujiwara, M. Ohara, J. Yamagishi, and M. Sugai. 2002. Molecular characterization of an atl null mutant of Staphylococcus aureus. Microbiol. Immunol. 46:601-612. [DOI] [PubMed] [Google Scholar]
- 47.Tobin, P. J., N. Mani, and R. K. Jayaswal. 1994. Effect of physiological conditions on the autolysis of Staphylococcus aureus strains. Antonie Leeuwenhoek 65:71-78. [DOI] [PubMed] [Google Scholar]
- 48.Tomasz, A., P. Moreillon, and G. Pozzi. 1988. Insertional inactivation of the major autolysin gene of Streptococcus pneumoniae. J. Bacteriol. 170:5931-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomasz, A., S. Nachman, and H. Leaf. 1991. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob. Agents Chemother. 35:124-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., N. Mani, P. A. Pattee, B. J. Wilkinson, and R. K. Jayaswal. 1992. Analysis of a peptidoglycan hydrolase gene from Staphylococcus aureus NCTC 8325. J. Bacteriol. 174:6303-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, W., A. N. Chatterjee, and F. E. Young. 1978. Regulation of bacterial cell walls: correlation between autolytic activity and cell wall turnover in Staphylococcus aureus. J. Bacteriol. 134:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]