Abstract
Molecular signals, including Nod factors and succinoglycan, are necessary for the establishment of nitrogen-fixing nodules (Fix+) in Medicago truncatula-Sinorhizobium meliloti symbiosis. This report shows that M. truncatula-S. meliloti interactions involve ecotype-strain specificity, as S. meliloti Rm41 and NRG247 are Fix+ (compatible) on M. truncatula A20 and Fix− (incompatible) on M. truncatula A17, the Fix phenotypes are reversed with S. meliloti NRG185 and NRG34, and there is a correlation between the host specificity and succinoglycan oligosaccharide structure. S. meliloti NRG185 produces oligosaccharides that are almost fully succinylated, with two succinate groups per subunit, whereas the oligosaccharides produced by S. meliloti Rm41 include many nonsuccinylated subunits, as well as subunits with a single succinate group and others with malate. The results of this study demonstrated the following: (i) incompatibility is not a consequence of an avirulence factor or lack of Nod factor activity; (ii) the Fix+ phenotypes are succinoglycan dependent; (iii) there is structural variability in the succinoglycan oligosaccharide populations between S. meliloti strains; (iv) the structural nature of the succinoglycan oligosaccharides is correlated to compatibility; most importantly, (v) an S. meliloti Rm41 derivative, carrying exo genes from an M. truncatula A17-compatible strain, produced a modified population of succinoglycan oligosaccharides (similar to the donor strain) and was Fix+ on A17.
Sinorhizobium meliloti and Medicago sativa (alfalfa) enter into a nitrogen-fixing symbiosis, and the initiation of a functional symbiosis involves an exchange of signal molecules. Plant-derived flavonoids activate the transcription of bacterial nodulation (nod) genes, resulting in the biosynthesis of N-acylated chito-oligosaccharides (Nod factors). The Nod factors then elicit root hair curling and nodule initiation in the plant (11, 14). The subsequent infection of the nodule by the microsymbiont results in the development of nitrogen-fixing nodules (Fix+) containing physiologically distinct bacteroids. Sinorhizobium spp. lipopolysaccharides, capsular polysaccharides (K antigens), and exopolysaccharides (succinoglycan or galactoglucan) are involved in the infection process, and specific polysaccharide mutants yield nonfunctional “pseudonodules” and a Fix− phenotype (9, 26, 29, 36).
Succinoglycan (exo) mutants of S. meliloti Rm1021 elicit Fix− nodules due to a lack of infection thread development (10, 31, 38). Succinoglycan is composed of octasaccharide repeat units (one galactosyl and seven glucosyl residues) substituted with pyruvyl (1-carboxyethylidene), acetyl, and succinyl groups (2, 23, 24, 43). The functional oligosaccharin, an oligosaccharide with signal activity (1), consists of three repeats (5, 53), and the presence of the succinyl groups is essential for activity (30). S. meliloti also produces K antigens (46), and the KR5 antigen of S. meliloti Rm41 exoB functionally replaces succinoglycan in the infection of alfalfa (8, 41, 42, 45, 48, 54). Unlike exopolysaccharides, however, K antigen composition and structure varies significantly between Sinorhizobium spp. strains (15-17, 37, 45-47, 49), and not all K antigens can promote Fix+ symbiosis on alfalfa (6).
In addition to succinoglycan, S. meliloti Rm1021 expR101 also produces a second exopolysaccharide, galactoglucan, consisting of acetylated, pyruvylated disaccharide repeating units (one galactosyl and one glucosyl residue), which can promote infection thread development on alfalfa (18, 22, 32, 56). Purified galactoglucan added to the inoculum of S. meliloti Rm1021 exopolysaccharide mutants was shown to promote infection at levels as low as 7 × 10−12 M (21). In contrast to alfalfa, galactoglucan and K antigens cannot functionally replace succinoglycan in the S. meliloti-M. truncatula symbiosis (18; this report), and although all three bacterial polysaccharides may function in the nodulation of alfalfa, there are differences in the plant response to each (38).
Succinoglycan, galactoglucan, and K antigen production are genetically distinct: the majority of the succinoglycan biosynthetic genes (exo/exs) are located in a 27-kb cluster on pSYMb (20, 44), a megaplasmid that carries many of the genes involved in symbiosis, whereas galactoglucan production is directed by the exp genes located in a distinct 23-kb cluster on pSYMb (7). Capsule expression in S. meliloti involves, at least, three separate gene regions genes located on pSYMb and the chromosome (25, 27, 28, 40).
M. truncatula is a model legume that is closely related to alfalfa (3), and although there is no evidence for host specificity in S. meliloti-M. sativa symbiosis, host specificity in S. meliloti-M. truncatula interactions has been demonstrated (52). This report shows that wild-type (WT) S. meliloti strains Rm41 and NRG247 are Fix+ (compatible) with M. truncatula A20 but are Fix− (incompatible) with M. truncatula A17, that the phenotypes are reversed with S. meliloti strains NRG185 and NRG34, and that the incompatible phenotype on M. truncatula is similar to that of an exopolysaccharide/K antigen mutant on alfalfa. This study examined the biochemical basis for this ecotype-strain specificity (compatibility versus incompatibility) in M. truncatula-S. meliloti interactions.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and production of succinoglycan.
The S. meliloti strains used in this study (Table 1) were grown under standard conditions (19) for both the plant assays and succinoglycan analyses. For polysaccharide analysis, cells were centrifuged at 20,000 × g for 30 min, and the supernatant, which contains the succinoglycan, was concentrated by roto-evaporation and exhaustively dialyzed against distilled H2O (dH2O) at 4°C. The preparation was then concentrated, as before, and freeze-dried. For crude determination of succinoglycan production, cells were plated on medium containing calcofluor (31).
TABLE 1.
Strains, plasmids, and compatibility phenotypes on M. truncatula A17 and A20
| Strain or plasmid | Characteristics or genotype (source)a | Compatibility
|
|
|---|---|---|---|
| A17 | A20 | ||
| S. meliloti strains | |||
| NRG185 | WTb from M. sativa; North America (37) | +c | −d |
| NRG34 | WT from M. sativa; North America (37) | + | − |
| Rm41 | WT from M. sativa; Europe (42) | − | + |
| NRG247 | WT from M. sativa; North America (37) | − | + |
| Rm1021 | Derivative of S. meliloti SU47 (35) | +/−e | +/− |
| GRC01 | NRG185 exoB::Tn5 (EPS−) (this work) | − | − |
| AK631 | Rm41 exoB631 (EPS−) (42) | − | − |
| PP674 | AK631 rkpA::Tn5 (EPS− KPS−) (40) | − | − |
| PP699 | Rm41 rkpA::Tn5 (KPS−) (40) | − | + |
| TOR33 | NRG185 rkpA::Tn5 (KPS−) (this work) | + | − |
| TJR41 | Rm41 pEX312 (this work) | +/− | +/− |
| TJR42 | Rm41 pD56 (this work) | − | + |
| NRG185 + Rm41 | Coinoculation on M. truncatula A17 | + | NTf |
| Plasmids | |||
| pEX312 | Tcr pLAFR1 with Rm1021 exo subclone (33) | ||
| pD56 | Tcr pLAFR1 with Rm1021 exo subclone (33) | ||
| pPP428::Tn5-674 | Tcr pLAFR1 rkp-1 (pPP428 rkpA::Tn5) (42) | ||
| pRK602 | pRK600′ΩTn5 (exoB::Tn5) (31) | ||
EPS, exopolysaccharide; KPS, K-antigen polysaccharide.
WT recovered from alfalfa nodules.
+, compatible (Fix+); numerous (25 to 45) functional nodules and healthy dark green plants.
−, incompatible (Fix−); no functional nodules, only pseudonodules, and unhealthy light green to yellow plants.
+/−, compatible (Fix+) but low efficiency; few functional nodules and light green plants.
NT, not tested.
Strain construction.
The K antigen mutants of S. meliloti were constructed by marker exchange (19), using pPP428::Tn5-674 (42), containing the rkp-1 region with a Tn5 insertion in rkpA (Peter Putnoky, University of Pécs, Hungary). The succinoglycan mutants were constructed by marker exchange, using pRK602 (31), a plasmid containing a Tn5 insertion in exoB (G. C. Walker, MIT). The mobilization of pEX312 and pD56 (exo subclones from Rm1021) into S. meliloti Rm41 was in done in triparental matings (13). S. meliloti Rm1021 exo subclones were used because they were readily available, and no other strains of S. meliloti have had the exo region sequenced and cloned.
Plant nodulation assays.
M. truncatula Jemalong seeds were obtained from Burkiss Seeds (Armidale, NSW, Australia), and the M. truncatula Jemalong A17 and M. truncatula A20 seeds were provided by Doug Cook (Department of Plant Pathology, University of California, Davis). Surface-sterilized seeds were sown into plastic pots on sterile potting soil and covered with 1 cm of vermiculite. As the cotyledons appeared, the plantlets were inoculated with S. meliloti by flooding the base of the stem with 200 to 300 μl of cells (107 to 108 CFU/ml) in phosphate-buffered saline. The plants were maintained in a controlled growth chamber, with 14 h of light (26°C) and 10 h of dark (20°C) at 60% humidity, and watered daily with dH2O and weekly with Jensen's N-free nutrient solution. The plants were checked periodically for general health and nodule morphology. The presence of numerous elongated, pink nodules and healthy green plants at 3 to 4 weeks postinoculation was diagnostic for the compatible phenotype, and the presence of only pseudonodules (small, white, and round) and yellow, stunted plants was diagnostic for the incompatible phenotype. No pink nodules were ever found on any incompatible host plants at the day 28 postinoculation time point. The initial assays were performed twice, in flats, with 300 to 400 plants, and the follow-up nodulation studies with all the WT strains and mutants were repeated three times (30 to 45 plants/experiment), with 6 to 8 plants/pot. All assays that involved mutants included compatible and incompatible WT strains as controls. Nodules were removed and tested for nitrogen fixation functionality by the acetylene reduction method (50). This verified that those nodules that appeared elongated and pink were functional in nitrogen reduction and those that were round and white were not. For the plant assays with the S. meliloti Rm41 derivatives, carrying pEX312 and pD56, bacterial cells were removed from the pink nodules and streaked onto plates containing tetracycline.
In some cases, the above-ground parts of the plants were harvested and weighed; chlorophyll was extracted with dimethyl sulfoxide, and the chlorophyll b content was measured (4). From the results, the micrograms of chlorophyll b/g of shoot tissue (wet weight) content was calculated.
For pouch experiments, sterilized seeds were placed in sterile water and incubated at 4°C for 36 h, after which time they were transferred to room temperature. After 24 h, the seeds with emerging radicals were transferred to the paper fold at the top of a growth pouch (Cyg; Mega International, MN) wrapped in aluminum foil, and the seedlings were inoculated when the roots were about 1 cm long. The plants were watered daily and fertilized twice weekly with Jensen's N-free nutrient solution with Fe-EDTA complex as iron source (34, 51).
Size exclusion chromatography.
The concentrated samples were separated according to size, using a 1.5- by 100-cm column packed with Bio-Gel of A-5m (Bio-Rad, Richmond, CA) and a pH-neutral eluent (50 mM Na2HPO4, 100 mM NaCl, pH 7). Fractions (1.6 ml) were collected, and aliquots were analyzed by the phenol-sulfuric acid assay (12); the fractionated succinoglycan was pooled according to size, dialyzed against dH2O, and lyophilized.
Carbohydrate analysis.
Carbohydrate composition analysis was performed by the gas-liquid chromatography analysis of the trimethylsilyl methyl glycosides (55), using a 30-m DB-5 capillary column (J&W Scientific, Folsum, CA), and by gas-liquid chromatography-(electron impact) mass spectrometry analysis of the partially methylated alditol acetates, using a 30-m SP-2330 capillary column (Supelco-Sigma-Aldrich, St. Louis, MO). For the latter, the oligosaccharides were permethylated (Hakomori method) prior to reduction and acetylation, as described previously (55). These analyses showed that the preparations contained only succinoglycan, and the absence of galactoglucan was verified by polyacrylamide gel electrophoresis analysis (21) and nuclear magnetic resonance (NMR) analyses, which showed no trace of the diagnostic galactoglucan α-anomeric resonance (data not shown).
MALDI-TOF MS.
Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) analysis was performed with an Applied Biosystems (Framingham, MA) Voyager DE PRO mass spectrometer in the positive mode. The instrument was operated at an accelerating voltage of 25 kV and a grid voltage at 85%. The sample was ionized with a nitrogen laser (λ, 337 nm) with a pulse width of 3 ns and 4 to 7.5 μJ. The acquisition mass range for this study was 500 to 10,000 atomic mass units (amu), with 150 laser shots/spectrum. The samples were dissolved in H2O and mixed with a matrix of 100 mM 2,5-dihydroxybenzoic acid in 75% ethanol, and the matrix-sample mixture (1 to 2 μl) was applied to the probe and dried. The sample was serially diluted with matrix to obtain the optimal concentration. The mass ions represent M + Na + 1.
FAB-MS.
Fast atom bombardment-mass spectrometry (FAB-MS) was performed on a ZAB-SE instrument (VG, Manchester, England) in the negative mode, with an ionizing voltage of 70 eV and an accelerating voltage of 10 kV, operating at low resolution (ca. 1:800). The samples were dissolved in ultrapure H2O, and 1 μl was added to a matrix of thioglycerol (2 μl). The scan range was 300 to 3,000 amu. The superior resolution yielded the expected succinoglycan monomeric oligosaccharide (SMO) mass ion at M + Na − 1, with an additional ∼0.6 amu due to the natural abundance of 13C.
Organic acid analysis.
The esterified organic acids were released from the succinoglycan by base treatment and analyzed by high-performance liquid chromatography (HPLC), using a Rezex column (Phenomenex, Torrance, CA) on a Dionex BioLC apparatus (Dionex Corp., Sunnyvale, CA) and 0.05 N sulfuric acid as the eluent. The refractive index of the eluent was monitored with a RID-10A detector (Shimadzu Corp., Kyoto, Japan). The relative elution points of succinate, acetate, and malate were established with authentic standards (pyruvate was not released).
Proton (1H) NMR spectroscopy.
1H NMR analyses of the succinoglycan preparations were performed with a Varian UNITY INOVA 300-MHz NMR spectrometer (Varian Inc., Palo Alto, CA). The samples were exchanged twice with D2O and then dissolved in 0.6 ml D2O, and 1H NMR spectra were obtained at 25°C. Acetate and pyruvate each contain a methyl group (-CH3) and succinate contains two methylene groups (-CH2-CH2-), and as all evidence has shown that succinoglycan is always fully pyruvylated (53), the integration value for the acetyl group resonance was assigned a value of 3.0, to represent the area that results from a methyl group. Using this scheme, fully pyruvylated, acetylated, and doubly succinylated repeats would show integration values of 3, 3, and 8, respectively.
RESULTS
S. meliloti-M. truncatula symbiosis shows strain-ecotype specificity.
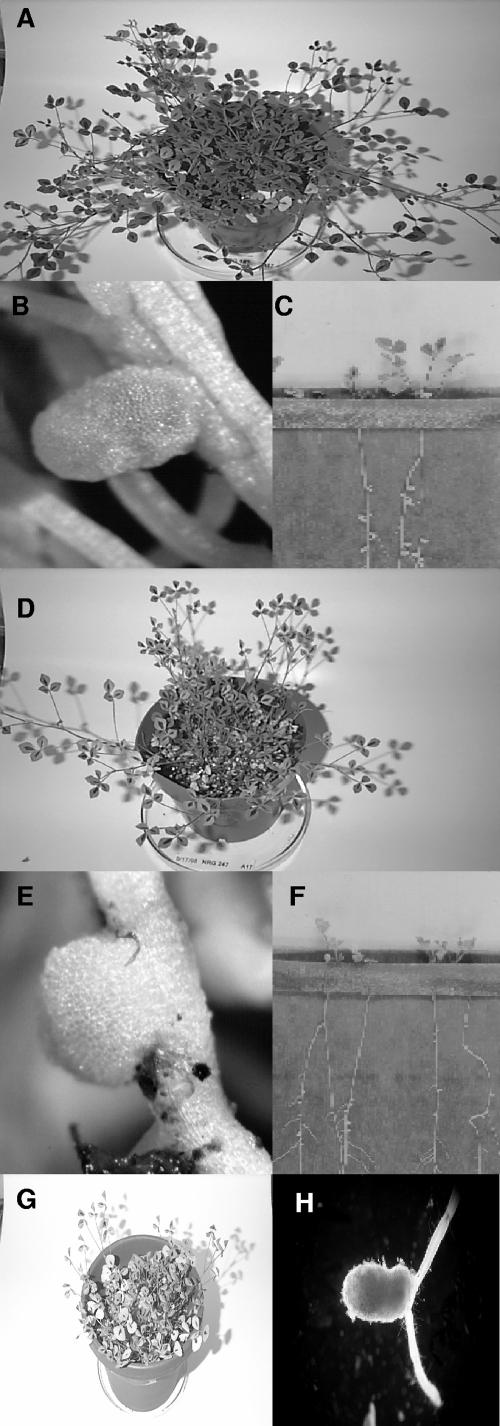
M. truncatula Jemalong or Jemalong A17 (identical in all experiments) and A20, which is genetically distinct from A17 (39), were used for all plant assays. The results of the experiments are given in Table 1. M. truncatula Jemalong or Jemalong A17 plants inoculated with S. meliloti NRG185 and S. meliloti NRG34 were dark green and exhibited robust growth after 4 weeks, showing that those strains were able to effectively nodulate M. truncatula Jemalong A17 (Fig. 1A; Fix+, compatible interaction). Examination of the roots showed the presence of numerous (25 to 45/plant) elongated, pink nodules (Fig. 1B). In contrast, the Jemalong A17 plants inoculated with S. meliloti NRG247 or S. meliloti Rm41 were yellow, stunted, and produced runners (Fig. 1D; Fix−, incompatible interaction), and only small, white, round pseudonodules were present on the roots (Fig. 1E). The round, white nodules and elongated, pink nodules from several plants were removed and tested for acetylene reduction, and only the latter were able to reduce acetylene; thus, the observable elongated, pink phenotype is indicative of Fix+ nodules. When inoculated onto M. truncatula A20, the compatibility phenotypes of the four strains were reversed (Table 1), showing a mutually exclusive pattern of compatibility. None of the strains ever elicited a functional nodule at the day 28 postinoculation time point on an incompatible host plant in these assays, which included hundreds of plants over the course of this study.
FIG. 1.
(A) M. truncatula Jemalong plants inoculated with S. meliloti NRG185 at 28 days postinoculation. (B) Root nodule from plants in panel A. (C) M. truncatula Jemalong plants in a growth pouch inoculated at 10 days after inoculation with S. meliloti NRG185. (D) M. truncatula Jemalong plants inoculated with S. meliloti NRG247 at 28 days postinoculation. (E) Root nodule from plants in panel D. (F) M. truncatula Jemalong plants in a growth pouch inoculated 10 days after inoculation with S. meliloti NRG247. (G) M. truncatula Jemalong A17 plants inoculated with S. meliloti NRG185 exoB::Tn5 (succinoglycan mutant) at 28 days postinoculation; the nodules were identical to those in panel E. (H) Root nodule from M. truncatula Jemalong A17 plants inoculated with S. meliloti TJR41 (Rm41, pEX312), which carries part of the S. meliloti Rm1021 exo (succinoglycan) gene region.
The plant assays were repeated in growth pouches (Fig. 1C and F), and the results were the same as the experiments using pots, although the incompatible hosts exhibited extremely poor health within 10 days. The numerous Fix+ nodules are clearly shown in Fig. 1C. The pouch experiments also clarified the kinetics of rapid nodulation, i.e., it was observed that S. meliloti NRG185 actually elicited the first noticeably pink nodules in about 6 or 7 days.
One exception to the host-specific pattern of compatibility was S. meliloti Rm1021. Although S. meliloti Rm1021 was Fix+ on both M. truncatula A17 and A20, it was much less efficient on both host plants than the other compatible strains, e.g., the number of nitrogen-fixing nodules was consistently ≤10% of what is typically found on A17 plants inoculated with S. meliloti NRG185, and the development of those nodules was delayed up to 3 days. However, the formation of Fix+ nodules on all A17 and A20 plants tested demonstrated that S. meliloti Rm1021 is compatible on both hosts.
Due to the yellowing demonstrated by the incompatible hosts, the results of the nodulation assays were quantified by chlorophyll b content analyses. An average of 670 (± 56 [standard error]) μg/g of chlorophyll b was extracted from the tissue of the compatible plants (not including those inoculated with S. meliloti Rm1021) compared to an average of 241 (± 21) μg/g from the incompatible plants. The compatibility, but relative inefficiency, of Rm1021 on both host plants was clearly demonstrated in these assays, with an average of 397 (± 17) μg/g of chlorophyll b extracted from M. truncatula A17 and A20 leaves; this was ∼36% the level of chlorophyll b extracted from the compatible plants inoculated with the other strains, using the incompatible average as a baseline.
The compatible (Fix+) phenotype requires succinoglycan production.
Nodulation assays were then performed with M. truncatula A17 and A20 and the S. meliloti polysaccharide mutants (Table 1). All plants, including the compatible hosts, inoculated with succinoglycan mutants (exoB) were yellow and stunted (M. truncatula A17 plants inoculated with S. meliloti NRG185 exoB::Tn5 are shown in Fig. 1G), and only pseudonodules were found on the roots; thus, the Fix+ phenotype requires succinoglycan production. The Fix− phenotype of the exoB mutants also showed that the K antigens cannot functionally replace succinoglycan on M. truncatula, as neither strain carries a mutation in the rkp genes and both constitutively produce K antigens of the size range required for symbiosis on alfalfa (46, 48).
The K antigen mutants (rkpA), which are unaffected in succinoglycan production, were Fix+ on the compatible host plants and Fix− on the incompatible host plants; and so the K antigens are not required for compatibility and do not act as strain-specific avirulence factors on incompatible hosts. Also, the succinoglycan-K antigen double mutant (exoB rkpA) was unable to infect any plants, showing that there is no combined effect, positive or negative, of the two polysaccharides. Finally, nodulation developed normally in M. truncatula A17 plants inoculated with both S. meliloti NRG185 and S. meliloti Rm41, showing that the incompatible strain does not inhibit the Fix+ phenotype.
The introduction of pEX312, a plasmid containing a fragment of the S. meliloti Rm1021 exo gene region, into S. meliloti Rm41 confers a Fix+ phenotype on M. truncatula A17.
Two S. meliloti Rm41 derivatives were constructed using pEX312 and pD56 (33), which contain subclones of the upstream and downstream segments, respectively, of the S. meliloti Rm1021 exo gene region. S. meliloti TJR42(pD56) and S. meliloti TJR41(pEX312) were then applied to the nodulation assays on M. truncatula A17. There was no change in the compatibility phenotype for S. meliloti TJR42, relative to WT Rm41. However, S. meliloti TJR41 was Fix+ on M. truncatula A17, and the efficiency and kinetics of nodulation (few functional nodules that were delayed in development) were similar to the donor strain, Rm1021 (Table 1). A nitrogen-fixing nodule (verified by acetylene reduction) elicited and occupied by S. meliloti TJR41 is shown in Fig. 1H. The bacteria were recovered from the infected nodules and shown to have retained pEX312 by growth on plates containing tetracycline.
Host specificity is correlated to succinoglycan structure.
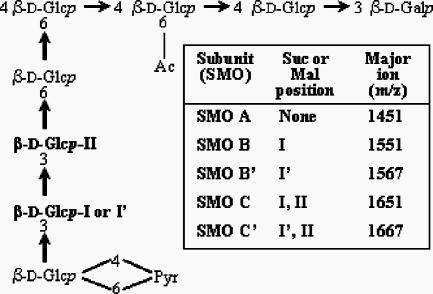
As the incompatible phenotype of the WT strains used in this study is similar to the Fix− phenotype associated with exo (succinoglycan) mutants, an analysis of the succinoglycan oligosaccharides from incompatible and compatible strains was undertaken. The succinoglycan oligosaccharides from S. meliloti NRG185, S. meliloti Rm41, and S. meliloti TJR41 were isolated and analyzed for comparison to the S. meliloti Rm1021 succinoglycan, which has been extensively studied by others (2, 5, 23, 24, 43, 53). The potential octasaccharide repeats, described previously (38), are shown in Fig. 2 as SMOs, including SMO A, SMO B, and SMO C, each of which differs in the degree of succinylation. Although poorly resolved at the high-mass range, MALDI-TOF MS analyses verified that the low-molecular-weight (LMW) fraction included the SMO, dimeric (SDO), and trimeric (STO) oligosaccharides (data not shown), with no evidence for the presence of tetrameric or larger oligosaccharides; this pattern of polymerization has been demonstrated by others (53). Glycosyl residue analyses confirmed that the preparations contained succinoglycan: linkage analysis yielded the expected partially methylated alditol acetates for both preparations, and composition analyses showed the presence of the two hexosyl residues at the expected ∼7:1 ratio.
FIG. 2.
Potential SMOs, as reported by Wang et al. (53). The potential succinylation sites are indicated by I and II. B′ and C′ represent replacement of succinate with malate in some oligosaccharides at I′ (see text). The major ions are mass + Na + 1 from MALDI-MS in the positive mode. Abbreviations: Glc, glucose; Gal, galactose; Ac, acetate; Pyr, pyruvate; Suc, succinate; Mal, malate.
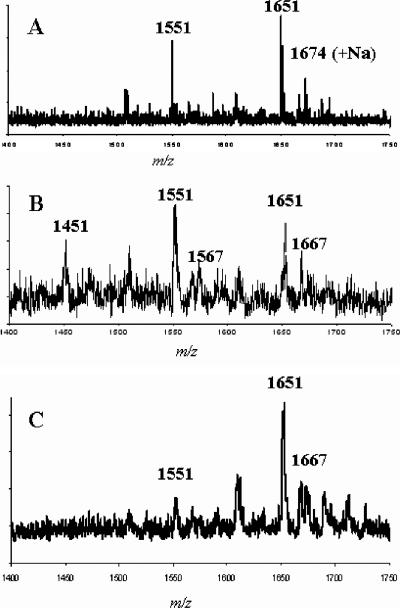
The major ions detected for the positive-mode MALDI-TOF MS analyses were Na adducts (mass + Na + 1) (Fig. 2). The MALDI-TOF MS analyses of the SMOs from S. meliloti NRG185 (compatible with M. truncatula A17) yielded primarily the ions for SMO C (m/z 1,651) and additional Na adducts (Fig. 3A). Ions corresponding to SMO B (m/z 1,551) and additional Na adducts were detected at low intensities, but there was no evidence for the presence of SMO A. The analyses of the SMOs from S. meliloti Rm41 (compatible with M. truncatula A20) yielded a spectrum (Fig. 3B) that was much more complex and significantly different from the S. meliloti NRG185 spectrum. The results showed that SMO B was present in greater abundance than SMO C, as shown by the ion at m/z 1,551, and a significant ion corresponding to SMO A was detected at m/z 1,451.
FIG. 3.
Positive-mode MALDI-TOF MS analyses of SMOs from S. meliloti NRG185 (A), S. meliloti Rm41 (B), and S. meliloti TJR41 (C). The major ions are mass + Na + 1; additional salt adducts are also present.
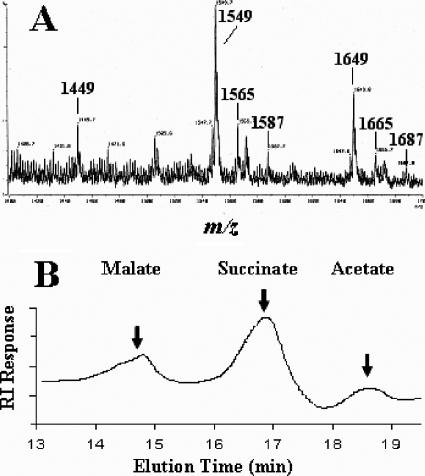
An unexpected result of the MALDI-MS analyses of the S. meliloti Rm41 preparation were ions at m/z 1,567 and m/z 1,667 (Fig. 3B), which represent a modified SMO with an additional oxygen atom (i.e., 1,551 + 16 and 1,651 + 16). Due to the poor resolution of the MALDI-MS spectra, the preparation was analyzed by FAB-MS in the negative mode (major ions are mass + Na − 1), which provided much better resolution. The FAB-MS clearly resolved the ions associated with the unknown (Fig. 4A), with significant ions at m/z 1,565 and m/z 1,665 (M + Na − 1). The presence of additional Na adduct ions, at m/z 1,587 and m/z 1,687, showed that they were not fragments, artifacts, or noise. As there were no ions detected at m/z 1,465 or m/z 1,487, the modification did not involve the pyruvyl or acetyl substituents, such as the substitution of glycolate for acetate, and so the only plausible explanation for the observation was that malate replaced succinate in some oligosaccharides. The LMW succinoglycan preparation from S. meliloti Rm41 was base treated to release the O-esterified substituents (succinate, acetate, and malate), and the solubilized organic acids were analyzed by HPLC (Fig. 4B). The results consistently showed a peak at the elution point of a malate standard. There were no FAB-MS ions, indicating the presence of two malate substituents on one SMO, and so, based on the biosynthesis hypothesis of Wang et al. (16), present evidence suggests that only the penultimate branch glycosyl residue may be malylated (SMO B′ and SMO C′ in Fig. 2).
FIG. 4.
(A) Negative-mode FAB-MS analysis of SMOs from S. meliloti Rm41. Major ions are mass + Na − 1. (B) HPLC analysis of the base-released substituents (succinate, acetate, and malate) from the S. meliloti Rm41 succinoglycan oligosaccharides; the arrows indicate the elution points of the indicated standards.
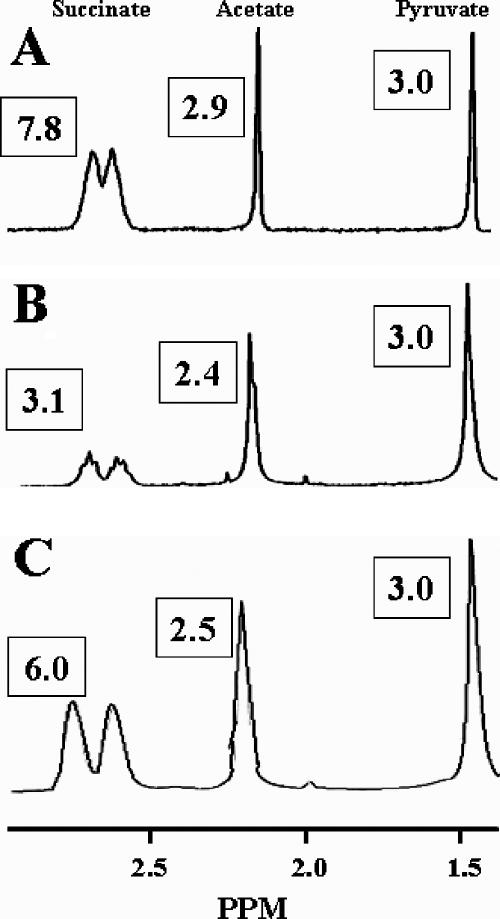
The LMW succinoglycan preparations were then analyzed by 1H NMR (Fig. 5), in order to establish the relative abundance of the substituents in each oligosaccharide population. In contrast to the MS analyses, the NMR analyses yielded the aggregate spectra for the SMOs, SDOs, and STOs present in the preparations. The integration values are given in Fig. 5, and the relative abundances of the substituents are given in Table 2. The presence of malate could not be detected in these analyses because the malate resonances were not resolved from the major succinoglycan resonances. The 1H NMR analysis of the S. meliloti NRG185 preparation (Fig. 5A) yielded results similar to the MS analysis of the SMOs, i.e., the integration value of the succinate resonances indicated that nearly all of the potential succinylation sites are occupied in the LMW succinoglycan (I and II) (Fig. 2), with a molar ratio of 1.95 succinyl groups/repeat. In contrast, the integration values for the 1H NMR analyses of the S. meliloti Rm41 preparation showed a 0.78 molar ratio of succinyl groups/repeat, and there is less than molar substitution with acetate (0.80).
FIG. 5.
1H NMR analyses of low-molecular-weight succinoglycan oligosaccharides from S. meliloti NRG185 (A), S. meliloti Rm41 (B), and S. meliloti TJR41 (C). The spectra are aggregates of the SMO, SDO, and STO components. The diagnostic resonances are labeled at the top of the figure, and the integration values of each are given in boxes. The value for pyruvate was set to 3 for comparative purposes (see Materials and Methods).
TABLE 2.
Substitution patterns in the LMW succinoglycan from various S. meliloti strains
| S. meliloti strain | Molar ratio (substituent/repeat) or presence of substituent
|
|||
|---|---|---|---|---|
| Pyruvatea | Acetatea | Succinatea | Malateb | |
| Rm1021 | 1.00 | 0.70 | 1.40 | − |
| NRG185 | 1.00 | 0.97 | 1.95 | − |
| Rm41 | 1.00 | 0.80 | 0.78 | + |
| TJR41 | 1.00 | 0.84 | 1.50 | + |
The molar ratio of succinate in the LMW succinoglycan produced by S. meliloti TJR41 (Rm41, pEX312) is similar to S. meliloti Rm1021.
The MALDI-MS (Fig. 3C) and 1H NMR (Fig. 5C) analyses of the succinoglycan oligosaccharide preparation from S. meliloti TJR41 showed that it was very different from the parental strain, although malate-containing SMOs were present. Importantly, the molar ratios of succinate and acetate were similar to S. meliloti Rm1021 (Table 2), the donor strain. Although, S. meliloti TJR41 produces an abundance of SMO C, like S. meliloti NRG185 (Fig. 3), the lower overall molar ratio of succinate (Table 2) indicates that there must be much less C in the SDO and STO populations produced by TJR41 compared to NRG185.
DISCUSSION
The results of this study show that M. truncatula-S. meliloti interactions may involve ecotype-strain specificity, as S. meliloti strains Rm41 and NRG247 are Fix+ (compatible) with M. truncatula A20 and Fix− (incompatible) with M. truncatula A17, and the Fix phenotypes are reversed with S. meliloti strains NRG185 and NRG34. Although it may not be universal in M. truncatula-S. meliloti interactions, this pattern of host specificity appears to be common: in addition to the strains and ecotypes reported here, M. truncatula A25 and A29 were recently tested, and they had compatibility phenotypes that were identical to A20 and A17, respectively, and four other WT strains of S. meliloti have also been assayed and each demonstrated host specificity with M. truncatula (B. L. Reuhs, unpublished data).
The presence of pseudonodules on the incompatible host plants demonstrated that each strain produced functional Nod factors and that nodule development, not initiation, determines compatibility. It was also shown that the compatible interactions are succinoglycan dependent, as exoB mutants were Fix− in all plant assays, and that structural differences in the succinoglycan oligosaccharides are correlated to host specificity. MALDI-TOF MS (Fig. 3) and NMR (Fig. 5) analyses showed that S. meliloti NRG185 produces almost exclusively oligosaccharides that are fully succinylated, with two succinyl groups/repeat (i.e., C, C-C, and C-C-C) (Fig. 2), whereas S. meliloti Rm41 also produces many nonsuccinylated repeats (A) as well as repeats with a single succinate group (B) and those with malate (B′ and C′), which could yield a variety of dimeric and trimeric oligosaccharides.
Furthermore, the introduction of pEX312, which contains an upstream fragment of the S. meliloti Rm1021 exo gene region, into S. meliloti Rm41 resulted in a modified production of succinoglycan oligosaccharides by the derivative strain, S. meliloti TJR41, and conferred to it a Fix+ phenotype on M. truncatula A17. Although S. meliloti Rm1021 is relatively inefficient with M. truncatula A17, pEX312 and pD56 were used in this study because the exo genes from other S. meliloti strains have not been sequenced and cloned. The overall degrees of succinylation of the oligosaccharides produced by S. meliloti Rm1021 and S. meliloti TJR41 are similar (but not identical, which is likely due to the fact that TJR41 is merodiploid) and very different from the parent strain, and this may explain why S. meliloti TJR41 is much less efficient on A20 than S. meliloti Rm41. The exo subclone in pEX312 includes exoH, which encodes a succinyltransferase, and this alone may explain the difference in succinylation between S. meliloti TJR41 and S. meliloti Rm41; however, the nonrandom distribution of succinate between the SMO population and the SDO and STO populations from S. meliloti TJR41 suggests that complex regulatory and polymerization systems are probably involved.
The results of this study suggest a direct role for succinoglycan in host specificity, and it appears that the different ecotypes of M. truncatula may require distinct succinoglycan oligosaccharides for infection and Fix+ symbiosis. However, direct causality has not been firmly established, only a correlation, and some other mechanism, or combination of mechanisms, may be responsible for this phenomenon. If, however, the succinoglycan oligosaccharides are the host specificity determinants, they probably have a signal function, and the active oligosaccharide required for compatibility on M. truncatula A17 is likely to be C-C-C, as the molar ratio of succinate for the oligosaccharides from S. meliloti NRG185 is 1.95 (Table 2). In contrast, S. meliloti Rm41 produces all three primary subunits, as well as the malate-containing oligosaccharides, and the molar ratio of succinate for the oligosaccharides is 0.78. Thus, an oligosaccharide required for compatibility on M. truncatula A20 would likely be very different and include, or consist exclusively of, nonsuccinylated subunits (e.g., A-A-A) or subunits with malate. Such a mechanism would explain the host specificities of S. meliloti Rm41 and S. meliloti NRG185, as the former is unlikely to produce significant quantities of C-C-C and the latter would almost certainly not produce any A-A-A.
The identification of ecotype-strain specificities in M. truncatula-S. meliloti interactions adds a new dimension to the study of plant-microbe interactions, and the fact that M. truncatula is used as a model legume enhances the possibility of studying host specificity in the plant. The additional signal step, if that is what determines compatibility, allows for a further compartmentalization of symbiotic events in the plant cell, as the compatible and incompatible interactions may be compared and contrasted, independent of Nod factor activity. In addition to host specificity, this report also brought to light the fact that there is variability in the succinoglycan oligosaccharide structure among S. meliloti strains. Although the backbone of succinoglycan may be conserved, the variation in substitution patterns among strains could yield strain-specific differences in symbiosis, even with alfalfa.
Acknowledgments
This work was supported by grant MCB-9728564 from the National Science Foundation (B.L.R.) and the Purdue Research Foundation.
We thank Gordon Campbell for advice and help on strain construction, Karl Wood for advice on mass spectrometry, and Ron Turco for a critical review of the manuscript.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Albersheim, P., and A. G. Darvill. 1985. Oligosaccharins. Sci. Am. 253:58-64.3906895 [Google Scholar]
- 2.Åman, P., M. McNeil, L.-E. Franzén, A. G. Darvill, and P. Albersheim. 1981. Structural elucidation, using HPLC-MS and GLC-MS, of the acidic polysaccharide secreted by Rhizobium meliloti strain 1021. Carbohydr. Res. 95:263-282. [Google Scholar]
- 3.Barker, D. G., S. Bianchi, F. Blondon, Y. Dattée, G. Duc, S. Essad, P. Flament, P. Gallusci, G. Génier, P. Guy, X. Muel, J. Tourneur, J. Dénarié, and T. Huguet. 1990. Medicago truncatula, a model plant system for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 8:40-49. [Google Scholar]
- 4.Barnes, J. D., L. Balaguer, E. Manrique, E. Elvira, and A. W. Davison. 1992. A reappraisal of the use of DMSO for the extraction and determination of chlorophylls a and b in lichens and higher plants. Environ. Exp. Botany 32:85-100. [Google Scholar]
- 5.Battisti, L., J. C. Lara, and J. A. Leigh. 1992. Specific oligosaccharide form of the Rhizobium meliloti exopolysaccharide promotes nodule invasion in alfalfa. Proc. Natl. Acad. Sci. USA 89:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker, A., N. Fraysse, and L. Sharypova. 2005. Recent advances in studies on structure and symbiosis-related function of rhizobial K-antigens and lipopolysaccharides. Mol. Plant-Microbe Interact. 18:899-905. [DOI] [PubMed] [Google Scholar]
- 7.Becker, A., S. Ruberg, H. Kuster, A. A. Roxlau, M. Keller, T. Ivashina, H. P. Cheng, G. C. Walker, and A. Puhler. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179:1375-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becquart-de Kozak, I., B. L. Reuhs, D. Buffard, C. Breda, J. S. Kim, R. Esnault, and A. Kondorosi. 1997. Role of the K-antigen subgroup of capsular polysaccharides in the early recognition process between Rhizobium meliloti and alfalfa leaves. Mol. Plant-Microbe Interact. 10:114-123. [Google Scholar]
- 9.Carlson, R. W., B. L. Reuhs, L. S. Forsberg, and E. L. Kannenberg. 1999. Rhizobial cell surface carbohydrates, p. 53-90. In J. B. Goldberg (ed.), Genetics of bacterial polysaccharides. CRC Press, Boca Raton, FL.
- 10.Cheng, H.-P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denarie, J., and P. Roche. 1992. Rhizobium nodulation signals, p. 295-324. In D. P. S. Verma (ed.), Molecular signals in plant-microbe communications. CRC Press, Boca Raton, FL.
- 12.Dubois, M., K. Gilles, J. Hamilton, P. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 20:350-356. [Google Scholar]
- 13.Finan, T. M., B. Kunkel, G. F. Devos, and E. R. Signer. 1986. A second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, R. F., and S. R. Long. 1992. Rhizobium-plant signal exchange. Nature 357:655-660. [DOI] [PubMed] [Google Scholar]
- 15.Forsberg, L. S., and B. L. Reuhs. 1997. Structural characterization of the K antigens from Rhizobium fredii USDA257: evidence for a common structural motif, with strain-specific variation, in the capsular polysaccharides of Rhizobium spp. J. Bacteriol. 179:5366-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraysse, N., B. Lindner, Z. Kaczynski, L. Sharypova, O. Holst, K. Niehaus, and V. Poinsot. 2005. Sinorhizobium meliloti strain 1021 produces a low-molecular-mass capsular polysaccharide that is a homopolymer of 3-deoxy-D-manno-oct-2-ulosonic acid harboring a phospholipid anchor. Glycobiology 15:101-108. [DOI] [PubMed] [Google Scholar]
- 17.Gil-Serrano, A. M., M. A. Rodriguez-Carvajal, P. Tejero-Mateo, J. L. Espartero, M. Menendez, J. Corzo, J. E. Ruiz-Sainz, and A. M. Buendia-Claveria. 1999. Structural determination of a 5-acetamido-3,5,7,9-tetradeoxy-7(3-hydroxybutyramido)-l-glycero-l-manno-nonulosonic acid-containing homopolysaccharide isolated from Sinorhizobium fredii HH103. Biochem. J. 342:527-535. [PMC free article] [PubMed] [Google Scholar]
- 18.Glazebrook, J., and G. C. Walker. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661-672. [DOI] [PubMed] [Google Scholar]
- 19.Glazebrook, J., and G. C. Walker. 1991. Genetic techniques in Rhizobium meliloti. Methods Enzymol. 204:398-418. [DOI] [PubMed] [Google Scholar]
- 20.Glucksmann, M. A., T. L. Reuber, and G. C. Walker. 1993. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti—a model for succinoglycan biosynthesis. J. Bacteriol. 175:7045-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez, J. E., B. L. Reuhs, and G. C. Walker. 1996. Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa. Proc. Natl. Acad. Sci. USA 93:8636-8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Her, G.-R., J. Glazebrook, G. C. Walker, and V. N. Reinhold. 1990. Structural studies of a novel exopolysaccharide produced by a mutant of Rhizobium meliloti strain Rm1021. Carbohydr. Res. 198:305-312. [DOI] [PubMed] [Google Scholar]
- 23.Hisamatsu, M., J. Abe, A. Amemura, and T. Harada. 1980. Structural elucidation of succinoglycan and related polyscaccharides from Agrobacterium and Rhizobium by fragmentation with two special β-D-glucanase and methylation analysis. Agric. Biol. Chem. 44:1049-1055. [Google Scholar]
- 24.Jansson, P. E., L. Kenne, B. Lindberg, H. Ljunggren, J. Lonngren, U. Ruden, and S. Svensson. 1977. Demonstration of an octasaccharide repeating unit in extracellular polysaccharide of Rhizobium meliloti by sequential degradation. J. Am. Chem. Soc. 99:3812-3815. [DOI] [PubMed] [Google Scholar]
- 25.Kereszt, A., E. Kiss, B. L. Reuhs, R. W. Carlson, A. Kondorosi, and P. Putnoky. 1998. Novel rkp gene clusters of Sinorhizobium meliloti involved in capsular polysaccharide production and the invasion of the symbiotic nodule: rkpK gene encodes for a UDP-glucose dehydrogenase. J. Bacteriol. 180:5426-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kijne, J. W. 1992. The Rhizobium infection process, p. 349-398. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, NY.
- 27.Kiss, E., A. Kereszt, F. Barta, S. Stephens, B. L. Reuhs, A. Kondorosi, and P. Putnoky. 2001. The rkp-3 gene region of Sinorhizobium meliloti Rm41 contains strain-specific genes that determine K antigen structure. Mol. Plant-Microbe Interact. 14:1395-1403. [DOI] [PubMed] [Google Scholar]
- 28.Kiss, E., B. L. Reuhs, J. S. Kim, A. Kereszt, G. Petrovis, P. Putnoky, I. Dusha, R. W. Carlson, and A. Kondorosi. 1997. The rkpGHI and -J genes are involved in capsular polysaccharide production by Rhizobium meliloti. J. Bacteriol. 179:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leigh, J. A., and D. L. Coplin. 1992. Exopolysaccharides in plant-bacterial interactions. Annu. Rev. Microbiol. 46:307-346. [DOI] [PubMed] [Google Scholar]
- 30.Leigh, J. A., J. W. Reed, J. F. Hanks, A. M. Hirsch, and G. C. Walker. 1987. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell 51:579-587. [DOI] [PubMed] [Google Scholar]
- 31.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levery, S. B., H. Zahn, C. C. Lee, J. A. Leigh, and S. Hakomori. 1991. Structural analyses of a second acidic exopolysaccharide of Rhizobium meliloti that can function in alfalfa root nodule invasion. Carbohydr. Res. 210:339-348. [DOI] [PubMed] [Google Scholar]
- 33.Long, S., J. W. Reed, J. Himawan, and G. C. Walker. 1988. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J. Bacteriol. 170:4239-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lullien, V., D. G. Barker, P. De Lajudie, and T. Huguet. 1987. Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol. Biol. 9:469-478. [DOI] [PubMed] [Google Scholar]
- 35.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noel, K. D. 1992. Rhizobial polysaccharides required in symbioses with legumes, p. 341-357. In D. P. S. Verma (ed.), Molecular signals in plant-microbe communications. CRC Press, Boca Raton, FL.
- 37.Olsen, P., S. Wright, M. Collins, and W. Rice. 1994. Patterns of reactivity between a panel of monoclonal antibodies and forage Rhizobium strains. Appl. Environ. Microbiol. 60:654-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellock, B. J., H.-P. Cheng, and G. C. Walker. 2000. Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penmetsa, R. V., and D. R. Cook. 2000. Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol. 123:1387-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrovics, G., P. Putnoky, B. L. Reuhs, J. S. Kim, T. A. Thorp, D. Noel, R. W. Carlson, and A. Kondorosi. 1993. The presence of a novel type of surface polysaccharide in Rhizobium meliloti requires a new fatty acid synthase-like gene cluster involved in symbiotic nodule development. Mol. Microbiol. 8:1093-1094. [DOI] [PubMed] [Google Scholar]
- 41.Putnoky, P., E. Grosskopf, D. T. C. Ha, G. B. Kiss, and A. Kondorosi. 1988. Rhizobium fix genes mediate at least two communication steps in symbiotic nodule development. J. Cell Biol. 106:597-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putnoky, P., G. Petrovics, A. Kereszt, E. Grosskopf, D. T. C. Ha, Z. Banfalvi, and A. Kondorosi. 1990. Rhizobium meliloti lipopolysaccharide and exopolysaccharide can have the same function in the plant-bacterium interaction. J. Bacteriol. 172:5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinhold, B. B., S. Y. Chan, T. L. Reuber, A. Marra, G. C. Walker, and V. N. Reinhold. 1994. Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J. Bacteriol. 176:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 45.Reuhs, B. L., R. W. Carlson, and J. S. Kim. 1993. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reuhs, B. L., D. P. Geller, J. S. Kim, J. E. Fox, V. S. Kumar Kolli, and S. G. Pueppke. 1998. Sinorhizobium fredii and S. meliloti produce structurally conserved lipopolysaccharides and strain-specific K antigens. Appl. Environ. Microbiol. 64:4930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reuhs, B. L., S. B. Stephens, D. P. Geller, J. S. Kim, J. Glenn, J. Przytycki, and T. Ojanen-Reuhs. 1999. Epitope identification for a panel of anti-Sinorhizobium meliloti monoclonal antibodies and application to the analysis of K antigens and lipopolysaccharides from bacteroids. Appl. Environ. Microbiol. 65:5186-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reuhs, B. L., M. N. V. Williams, J. S. Kim, R. W. Carlson, and F. Cote. 1995. Suppression of the Fix− phenotype of Rhizobium meliloti exoB by lpsZ is correlated to a modified expression of the K polysaccharide. J. Bacteriol. 177:4289-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez-Carvajal, M. A., J. A. Rodrigues, M. E. Soria-Diaz, P. Tejero-Mateo, A. Buendia-Claveria, R. Gutierrez, J. E. Ruiz-Sainz, J. Thomas-Oates, and A. M. Gil-Serrano. 2005. Structural analysis of the capsular polysaccharide from Sinorhizobium fredii HWG35. Biomacromolecules 6:1448-1456. [DOI] [PubMed] [Google Scholar]
- 50.Soberon, M., H. D. Williams, R. K. Poole, and E. Escamilla. 1989. Isolation of a Rhizobium-Phaseoli cytochrome mutant with enhanced respiration and symbiotic nitrogen fixation. J. Bacteriol. 171:465-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somerville, C. R., and W. L. Ogren. 1982. Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem. J. 202:373-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tirichine, L., F. De Billy, and T. Huguet. 2001. mtsym6, a gene conditioning Sinorhizobium strain-specific nitrogen fixation in Medicago truncatula. Plant Physiol. 123:845-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, L.-X., Y. Wang, B. J. Pellock, and G. C. Walker. 1999. Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J. Bacteriol. 181:6788-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, M. N. V., R. I. Hollingsworth, S. Klein, and E. R. Signer. 1990. The symbiotic defect of Rhizobium meliloti exopolysaccharide mutants is suppressed by lpsZ+, a gene involved in lipopolysaccharide biosynthesis. J. Bacteriol. 172:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.York, W. S., A. G. Darvill, M. McNeil, T. T. Stevenson, and P. Albersheim. 1985. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118:3-40. [Google Scholar]
- 56.Zhan, H., S. B. Levery, C. C. Lee, and J. A. Leigh. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. USA 86:3055-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]