Abstract
Mycobacterium avium subsp. paratuberculosis causes an enteric infection in cattle, with a great impact on the dairy industry in the United States and worldwide. Characterizing the gene expression profile of M. avium subsp. paratuberculosis exposed to different stress conditions, or shed in cow feces, could improve our understanding of the pathogenesis of M. avium subsp. paratuberculosis. In this report, the stress response of M. avium subsp. paratuberculosis on a genome-wide level (stressome) was defined for the first time using DNA microarrays. Expression data analysis revealed unique gene groups of M. avium subsp. paratuberculosis that were regulated under in vitro stressors while additional groups were regulated in the cow samples. Interestingly, acidic pH induced the regulation of a large number of genes (n = 597), suggesting the high sensitivity of M. avium subsp. paratuberculosis to acidic environments. Generally, responses to heat shock, acidity, and oxidative stress were similar in M. avium subsp. paratuberculosis and Mycobacterium tuberculosis, suggesting common pathways for mycobacterial defense against stressors. Several sigma factors (e.g., sigH and sigE) were differentially coregulated with a large number of genes depending on the type of each stressor. Subsequently, we analyzed the virulence of six M. avium subsp. paratuberculosis mutants with inactivation of differentially regulated genes using a murine model of paratuberculosis. Both bacterial and histopathological examinations indicated the attenuation of all gene mutants, especially those selected based on their expression in the cow samples (e.g., lipN). Overall, the employed approach profiled mycobacterial genetic networks triggered by variable stressors and identified a novel set of putative virulence genes. A similar approach could be applied to analyze other intracellular pathogens.
Mycobacterium avium subsp. paratuberculosis is the causative agent of Johne's disease in cattle (10) and might be implicated in Crohn's disease in humans (27, 28). Infection with M. avium subsp. paratuberculosis results in a chronic infection of cows characterized by granulomatous enteritis, chronic diarrhea, and eventually death. The disease is prevalent in the United States and worldwide, causing an estimated annual loss of more than $200 million for the dairy industry in the United States alone (21), and is considered one of the most serious infectious diseases in dairy cattle worldwide (8, 12). Both clinically ill and healthy but infected animals can shed M. avium subsp. paratuberculosis bacilli in their feces intermittently, exposing bacteria to variable adverse conditions such as low nutrients and variable temperatures. Contamination of the environment by the M. avium subsp. paratuberculosis-containing feces becomes the most common source of infection, especially for young calves who can be infected through the fecal-oral route (9). Clinically infected cows can shed 106 to 108 CFU/g of fecal material that can easily spread the infection to new calves, for which the infectious dose is 103 CFU/animal (51). Following the complete sequencing of the M. avium subsp. paratuberculosis genome (20), large-scale analysis of the genome contents of both clinical and environmental isolates of M. avium subsp. paratuberculosis identified several novel diagnostic targets (2, 29, 52). In this report, we test the hypothesis that profiling the gene expression patterns of M. avium subsp. paratuberculosis isolates exposed to different stress conditions or directly isolated from infected cows will provide insights into the genetic basis of virulence in M. avium subsp. paratuberculosis. Using a similar approach for stool samples of cholera patients, Larocque et al. (19) gained more insights into the pathogenesis of Vibrio cholerae.
Once reaching the intestinal lumen, virulent strains of M. avium subsp. paratuberculosis can invade intestinal tissue effectively within hours (53) and establish a persistent infection in the intestinal mucosa (26) and mesenteric lymph nodes, which can last for years (51). Earlier reports showed that M. avium subsp. paratuberculosis bacilli can survive and proliferate inside the phagosomes of infected macrophages by using mechanisms that are not completely understood (49). Preventing phagosome acidification is one of the possible scenarios by which M. avium subsp. paratuberculosis avoids macrophage killing (38). However, the genetic basis of such a mechanism of survival remains elusive. Recently, the expression profiles of the infected bovine macrophages were characterized to reveal different patterns of gene expression between cows clinically or subclinically infected with M. avium subsp. paratuberculosis (11). In this report, we profiled the mycobacterial response to several stress inducers (stressors) such as oxidative stress, heat shock, and acidic pH to mimic microenvironments that M. avium subsp. paratuberculosis bacilli might face during survival inside macrophages. We also took advantage of a relatively simple protocol to isolate a large number of M. avium subsp. paratuberculosis bacilli from clinically infected cows to define the transcriptional profile of M. avium subsp. paratuberculosis continuously shed in the feces. The latter analysis could uncover the transcriptional machinery of bacilli that are most likely to transmit infections to naïve animals. In general, our analysis identified the “stressome,” the bacterial stress responses on a genome-wide level, employed by M. avium subsp. paratuberculosis to survive in hostile microenvironments. In addition, the contribution of a selected list of stress-responsive genes to M. avium subsp. paratuberculosis survival was examined in a mouse model of paratuberculosis, which identified a set of novel virulence factors including several lipases involved in lipid degradation in M. avium subsp. paratuberculosis.
MATERIALS AND METHODS
Bacteria.
M. avium subsp. paratuberculosis ATCC 19698 was used for all in vitro transcriptional profiling. M. avium subsp. paratuberculosis K-10 was used only for generating the lipN mutant and for infecting a control group of mice. For all cultures, M. avium subsp. paratuberculosis ATCC 19698 and M. avium subsp. paratuberculosis K-10 were grown in Middlebrook 7H9 broth (Difco, Sparks, MD) supplemented with 0.5% glycerol, 2 μg/ml mycobactin J (Allied Monitor, Fayette, MO), 0.05% Tween 80, and 10% ADC (2% glucose, 5% bovine serum albumin fraction V, and 0.85% NaCl) at 37°C with shaking at 100 rpm/min. For animal inoculation, bacterial cultures of an optical density at 600 nm of 1.0 were harvested, washed once in phosphate-buffered saline (PBS), and resuspended in an equal volume of PBS. When needed, serial dilutions of M. avium subsp. paratuberculosis cultures were plated on 7H10 agar supplemented with 0.5% glycerol, 2 μg/ml mycobactin J, and 10% ADC. Agar plates for organ colony counting from animal tissues were also supplemented with 5 mg/ml vancomycin, 30 mg/ml amphotericin B, and 10 mg/ml nalidixic acid to reduce bacterial and fungal contamination, especially when intestines were plated.
RNA extraction from bacterial cultures.
To profile the mycobacterial stress response to variable conditions, M. avium subsp. paratuberculosis ATCC 19698 cultures were allowed to grow to mid-log phase (optical density at 600 nm, 0.5) and aliquots were subjected to one of the following stressors: (i) shift to 45°C, (ii) addition of H2O2 to a final concentration of 10 mM, (iii) low pH obtained by adding HCl (pH 5.5), or (iv) treatment with hexadecylpyridinium chloride (HPC; Sigma, St. Louis, MO) to a final concentration of 1%. Measuring the pH of 1% HPC solution indicated its acidity to be pH 5.0. All cultures were exposed to each treatment for 3 h before bacterial pellets were harvested by centrifugation at 3,200 × g for 20 min. Total RNA from mycobacterial cultures was extracted using protocols that we established before with a few modifications (44, 45). Briefly, bacterial pellets (107 to 108 CFU) were resuspended in 4 ml TRIzol reagent (Invitrogen, Carlsbad, CA), split into four 2-ml screw-cap tubes each with 3.0 g of 0.1 mm zirconia/silica beads (BioSpect Products, Inc.), and disrupted in a Mini-BeadBeater-8 (BioSpect Products, Inc.) at top speed four times for 30 s each with 30-s intervals on ice. Following a 10-min incubation at room temperature, the supernatant was transferred to RNase-free tubes and centrifuged at 11,000 rpm for 15 min. RNA was isolated from the supernatant with chloroform and isopropanol treatments, washed with 75% ethanol, air dried, and resuspended in RNase-free H2O as described before (45). To remove contaminating DNA, RNA samples were treated with DNase I (Invitrogen) (10 U/μg) at 37°C for 30 min. The quality and quantity of the extracted RNA were examined with agarose gel electrophoresis (see Fig. S1 in the supplemental material) and an Ultrospec 3100 pro UV/Visible spectrophotometer (GE Healthcare, Piscataway, NJ).
Cow fecal samples.
Feces from Holstein cows with a documented history of Johne's disease were collected by the Johne's Testing Center, University of Wisconsin—Madison. Before mycobacterial culturing or direct isolation of RNA, 3 g of fecal samples was treated with 30 ml of 1% HPC for 16 h to eliminate fungal and nonmycobacterial contaminants. This procedure was used before to decontaminate M. avium subsp. paratuberculosis-contaminated samples (16, 31). The upper liquid layer was carefully transferred and centrifuged at 3,200 × g for 20 min to harvest mycobacterial pellets from infected animals. These bacterial pellets (107 CFU) were directly used for RNA extraction without further enrichment or addition of antibiotic. At least 10 bacterial pellets were collected from each cow to obtain enough bacterial RNA for DNA microarray analysis. RNA was extracted from bacterial pellets as described above. To confirm the identity of the M. avium subsp. paratuberculosis bacilli isolated from the fecal samples, we used PCR to amplify IS900 sequences from all bacterial pellets recovered from the feces (data not shown) before proceeding to RNA extraction. Additionally, DNase I-treated RNA isolated from cow samples was subjected to standard reverse transcription (46) and PCR amplification using specific primers for tlyA, lipM, and lipN genes (Table 1). These amplicons were further sequenced to confirm the identity of the amplified transcripts.
TABLE 1.
PCR primers used in this study
| Primer | Gene and direction | Purpose | Sequence |
|---|---|---|---|
| AMT137 | accD5, forward | qRT-PCR | CTTCAACATCCCGATCATCA |
| AMT138 | accD5, reverse | qRT-PCR | AGCCCATCACGCAGTAGG |
| AMT145 | ptrBa, forward | qRT-PCR | CGGGTCTACGACATCGACTT |
| AMT146 | ptrBa, reverse | qRT-PCR | CCGTAGCCGTAAATCAGTGC |
| AMT147 | fabG3, forward | qRT-PCR | GTTACACCGCAACCAAATTC |
| AMT148 | fabG3, reverse | qRT-PCR | TACACCACCAGGTTGGACAC |
| AMT817 | tlyA, forward | qRT-PCR | CTGTCGTTCATCTCGCTGTG |
| AMT818 | tlyA, reverse | qRT-PCR | CGCAGGAAGTATTCGACGTT |
| AMT819 | lipM, forward | qRT-PCR | CTGGGTCAAACAACACATCG |
| AMT820 | lipM, reverse | qRT-PCR | GATCGACTTGATCAGCAGTCC |
| AMT823 | lipN, forward | qRT-PCR | TGGTTCGAATCGCAGTACCT |
| AMT824 | lipN, reverse | qRT-PCR | CGAGCTGGAACAGGTTGG |
| AMT924 | MAP4287c, forward | qRT-PCR | ATTGCACGCCTCCGATCT |
| AMT925 | MAP4287c, reverse | qRT-PCR | CAGCACCGTCAGCGTCTC |
| AMT278 | lipN upstream, forward | Gene knockout | ATATATACTAGTACCTTGGCGATGTACTTGC |
| AMT279 | lipN upstream, reverse | Gene knockout | ATATATAAGCTTTCTGATCCATGCGACGGG |
| AMT280 | lipN downstream, forward | Gene knockout | ATATATTCTAGAGGGTGCGGTGTCGGTCAG |
| AMT281 | lipN downstream, reverse | Gene knockout | ATATATGGTACCGCCGGCCAGTGAATCAGG |
| AMT125 | Hygromycin resistance gene, forward | Mutant confirmation | GGGAAGACCTCGGAATGG |
| AMT126 | Hygromycin resistance gene, reverse | Mutant confirmation | CTGCGGAACGACCAGGAAT |
| AMT222 | lipN_104, forward | Mutant confirmation | CGAACCCAGGTACCGCAG |
| AMT223 | lipN_919, reverse | Mutant confirmation | ATGCTGTCCGGTCTGCGC |
Microarray sample preparation and hybridization.
For all microarray experiments, tiled-oligonucleotide DNA microarrays designed from the genome sequence of M. avium subsp. paratuberculosis K-10 were constructed by NimbleGen Systems Inc. (Madison, WI) (37, 52). Attempts were made to design 14 unique pairs of 24-mer oligonucleotides covering 100% of the predicted open reading frames (ORFs) in the M. avium subsp. paratuberculosis genome (20). However, because of the large number of sequence repeats in the M. avium subsp. paratuberculosis genome, only 91.2% of M. avium subsp. paratuberculosis predicted ORFs were used to construct the M. avium subsp. paratuberculosis DNA microarrays. For every perfect match probe of the ORF sequence, there is a corresponding mismatch probe with mutations at the 6th and 12th positions. For each hybridization, total RNA samples were reverse transcribed to cDNA and fluorescently labeled according to a protocol established earlier by our group and NimbleGen (37, 52). Briefly, RNA (5 μg) was incubated with 0.5 μg of random nonamer, 0.01 mM of dithiothreitol, 1 mM of deoxynucleoside triphosphates, and 300 U of SuperScript II reverse transcriptase (Invitrogen) at 42°C for 90 min. The remaining RNA was removed by adding NaOH, and the pH was equilibrated by adding HEPES. The generated cDNA was randomly fragmented with 0.1 U of DNase I at 37°C for 5 to 10 min and purified with Millipore YM-10 microfilters (Billerica, MA). The fragmented cDNA was labeled with biotin using terminal deoxynucleotide transferase (Promega, Madison, WI) in the presence of 1 μM of biotin-N6-ddATP (Perkin-Elmer Life Sciences Inc., Boston, MA) at 37°C for 1 h. Hybridizations were conducted using NimbleGen hybridization chambers as described before (52) by adding biotin-labeled samples heated to 95°C for 5 min. After 12 to 16 h of hybridization, the slides were washed in nonstringent (6× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA (pH 7.7)] and 0.01% Tween 20) and stringent (100 mM morpholineethanesulfonic acid, 0.1 M NaCl, and 0.01% Tween 20) buffers for 5 min each followed by adding Cy3-streptavidin (GE Healthcare). An Axon GenePix 4000B laser scanner (Axon Instrument, Union City, CA) was used to scan the slides at 5-μm resolution.
Data analysis.
Hybridization signals were extracted from the scanned images using the NimbleScan software (NimbleGen). The signal intensity of each ORF was represented by the mean of the 14 probe signals calculated by subtracting mismatch signals from perfect match signals to eliminate background signals. Two hybridizations from two biological replicates of each examined condition were analyzed with a total of 28 data points collected for each examined ORF. Raw hybridization signals were normalized by scaling the mean signal for each array to 1,000. Signal intensities of <5 were transformed to 5 to accommodate genes with undetectable transcripts in one treatment but not the others. The normalized linear signals were loaded to the R program (http://www.r-project.org/) with the EBarrays package, which employs empirical Bayes statistics to identify differentially expressed genes between two conditions by calculating the posterior probability of differential expression using the Lognormal-Normal model (17). Differential gene expression was considered significant only if genes have a probability of differential expression of >0.5 and a change of >±2-fold. Normalized signal intensities were also analyzed using hierarchical clustering algorithms implemented in TIGR Multi-Experiment Viewer software 4.0b (http://www.tigr.org/software/microarray.shtml).
Quantitative PCR.
For a selected list of genes (Table 1), we performed a SYBR green-based, quantitative real-time PCR (qRT-PCR) to evaluate the performance of the DNA microarrays. To confirm the absence of genomic DNA in the RNA sample used for qRT-PCR, IS900-specific primers were used to amplify RNA samples. Only IS900 amplicon-negative RNA samples were allowed for subsequent qRT-PCR analysis. The templates for qRT-PCR (cDNA) were prepared as described above for DNA microarray analysis. All primers used here (Table 1) were designed with web-based tools, Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), and analyzed using the BLAST algorithm against the whole GenBank database to confirm their specific binding to the target sequences (E scores were <0.05 for primers that were >18-mers). PCRs were prepared using Bio-Rad (Hercules, CA) iQ-SYBR Green Supermix and run using the 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA). The threshold cycle (CT) of each gene was normalized to the CT of the 16S rRNA from the same cDNA sample. The expression changes (n-fold) were calculated by comparing the normalized CT of treated samples to that of the control sample as previously detailed (48).
Construction of lipN-knockout mutant.
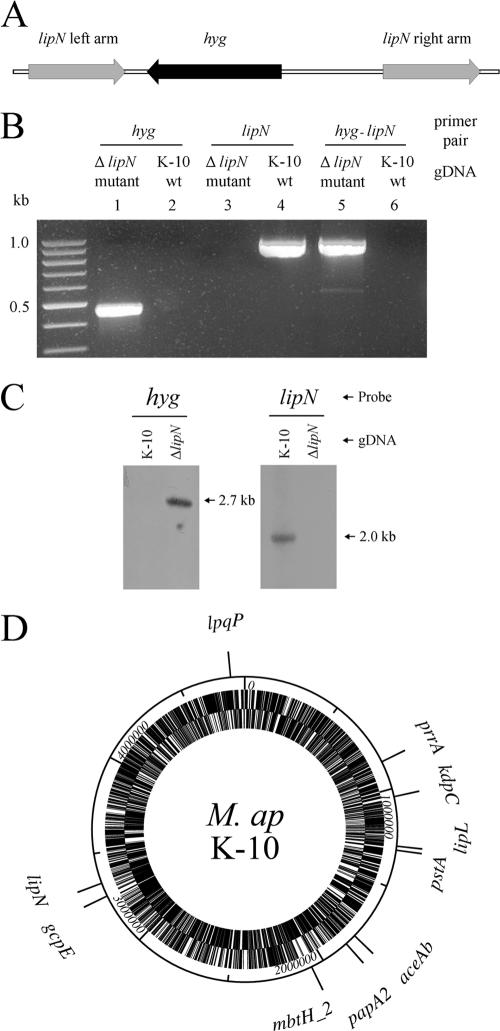
The specialized transduction protocol used earlier for Mycobacterium tuberculosis (4) was employed to generate an in-frame deletion of 1.1 kb of the lipN gene from M. avium subsp. paratuberculosis strain K-10. Briefly, the lipN allelic-exchange substrate was generated using the pYUB854 shuttle vector. Fragments of ∼700 bp upstream and downstream of the lipN coding region were amplified with primers (Table 1) that introduced restriction sites compatible with the cloning sites in pYUB854. Restriction enzyme-digested amplicons representing both the left and right flanking regions of lipN were gel purified using the Wizard Gel Clean-Up System (Promega) and subsequently cloned into pYUB854 to generate the pYUB854::lipN construct. The DNA of the temperature-sensitive phasmid phAE87 (3) was self-ligated to generate concatemers and subsequently digested with PacI. DNA of the pYUB854::lipN construct was also digested with PacI and ligated with digested phAE87 concatemers to replace the pYUB328 cosmid sequence using an in vitro lambda-packaging system (GIGAPackII; Stratagene, La Jolla, CA). The packaged phage particles were transduced into Escherichia coli HB101, and the shuttle phasmid DNA was extracted from the mixture of hygromycin-resistant colonies. Mycobacterium smegmatis cells were transformed with the purified phasmid DNA by electroporation, and the transformants were allowed to produce plaques at 30°C. The recombinant mycobacteriophage in the lysate was subsequently propagated and titrated in M. smegmatis. High-titer phage constructs were further transduced to M. avium subsp. paratuberculosis K-10 at the nonpermissive temperature (37°C) with a multiplicity of infection of 10. The genotype of lipN-deletion mutants growing on hygromycin-containing plates was confirmed with PCR and Southern blotting analyses as outlined before (36, 47).
Virulence assay of M. avium subsp. paratuberculosis mutants in mice.
BALB/c mice were purchased from Harlan (Indianapolis, IN) at 3 weeks of age and kept in a pathogen-free environment according to our approved protocol from the Institutional Animal Care and Use Committee, University of Wisconsin—Madison. Groups of mice (n = 10 to 20 each) were inoculated with either M. avium subsp. paratuberculosis K-10 or one of its isogenic mutants generated by homologous recombination (ΔlipN strain) or insertional mutagenesis (ΔlipL, ΔaceAB, ΔmbtH2, ΔprrA, and ΔlpqP strains) selected from a previous analysis of an M. avium subsp. paratuberculosis mutant library (36). All insertional mutants were constructed in M. avium subsp. paratuberculosis ATCC 19698 as described before (36). PBS-washed and resuspended bacteria were dispersed using a cup-horned sonicator (Fisher Scientific, Pittsburgh, PA) and injected into the mice intraperitoneally. Inocula were adjusted to yield a dose of 107 to 108 CFU/mouse. Mouse groups (n = 3 to 6) were sacrificed at 3, 6, and 12 weeks postinfection (WPI), and samples from liver and intestine were collected for bacterial and histopathological examinations as previously described (36).
RESULTS
M. avium subsp. paratuberculosis responses to variable stress conditions.
Following oral infection, M. avium subsp. paratuberculosis bacilli face variable microenvironments during translocation across the alimentary tract to reach their final destination in the macrophages (53). To gain insights into bacterial responses upon exposure to stressors, we examined the transcriptional profile of M. avium subsp. paratuberculosis cultures following treatments with defined stimuli such as acidic pH, heat shock, or oxidative stress using tiled-oligonucleotide DNA microarrays. Heat treatment at 45°C was chosen to identify the general stress response of M. avium subsp. paratuberculosis, while acidic pH and oxidative conditions were chosen to profile M. avium subsp. paratuberculosis exposure to the environment of the abomasums or within macrophages, respectively. The 3-h exposure time was selected based on the rapid change in gene transcripts in bacteria (30, 54) and the complex nature of the mycobacterial cell wall that controls the flow of stressors. A similar time frame was employed before for M. tuberculosis cultures (34, 50). Preliminary analysis of the hybridization replicates revealed a high level of correlation among biological replicates (r > 0.8) (Fig. 1A) with detectable levels of hybridization in >70% of all the ORFs carried in the M. avium subsp. paratuberculosis K-10 genome (see Table S1 in the supplemental material). Considering all of the ORFs with detectable hybridization signals (above background level) in at least one of the examined samples indicated the presence of transcripts in all ORFs examined by DNA microarrays (91.2% of the included ORFs). It is noteworthy that not all detected transcripts were significantly regulated (see below). The detection of such a high percentage of transcripts confirms the gene predictions used for the M. avium subsp. paratuberculosis genome (20). qRT-PCR analysis of a selected list of genes further confirmed the regulation of 85% of the examined genes, consistent with their regulation estimated by DNA microarrays (see Table S2 in the supplemental material).
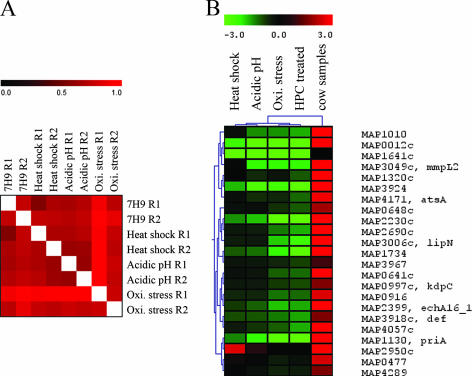
FIG. 1.
Microarray analysis of M. avium subsp. paratuberculosis cultures exposed to variable stressors. (A) A scatter plot analysis displaying a high reproducibility between biological replicates (r > 0.9) of M. avium subsp. paratuberculosis cultures following exposure to oxidative stressors. (B) Venn diagram of genes that significantly changed expression levels under various in vitro stress conditions.
Of the three profiles examined with DNA microarrays, the low-pH-treated samples had the largest number of genes with significantly altered expression levels (n = 597) compared to untreated samples. Heat shock and oxidative stress gave lower numbers of significantly regulated genes (31 and 155, respectively) (see Tables S3, S4, and S5 in the supplemental material). As expected, the shift in culture temperature to 45°C induced a set of genes such as dnaK, dnaJ, htpX, groEL1, groEL2, and hsp18. Heat shock of M. tuberculosis induced responses in orthologous genes similar to those in the previously mentioned list (42, 43), further validating the DNA microarrays used for gene expression profiling. Genes encoding several regulatory factors (based on sequence annotation) were also up-regulated (e.g., MAP3575, MAP0661c, and MAP3331), suggesting the activation of an array of heat shock-responsive genes. Similarly, transcriptional levels of several other regulatory genes were altered significantly during exposure to oxidative and low-pH treatments. Specifically, one of the extracytoplasmic sigma factors, sigH, was up-regulated by the oxidative stress, consistent with an earlier finding in M. tuberculosis (32). In addition, 35kd_ag (MAP2855c) and a gene encoding a regulatory protein, whiB2, were among the significantly induced genes under oxidative stress. Previously, whiB2 and 35kd_ag were up-regulated during nutrient starvation (5) and heat shock (43) in M. tuberculosis, respectively, suggesting that these two genes could play a role in relief of the oxidative stress in M. avium subsp. paratuberculosis. Conversely, another sigma factor, sigD, was down-regulated when acidic-pH samples were examined. In fact, the majority of the significantly regulated genes were down-regulated (n = 402), while only 195 genes were up-regulated at acidic pH. This profile was similar to the one generated by M. tuberculosis following exposure to low pH (14). Among the up-regulated genes are those involved in mycobactin metabolism (mbtC) and general stress-responsive genes such as htpX, clpX, and relA. The high number of regulated genes following exposure to low pH, in addition to the type of activated gene (i.e., stress-responsive genes), emphasized the importance of change in pH to the pathogenesis of M. avium subsp. paratuberculosis.
Further analysis of genes with a significant change in transcription levels under all examined stressors (Fig. 1B) identified a list of seven genes that were shared among all treatments. Only one gene in this list, MAP3430 (pmmB), was down-regulated, while the other six genes were up-regulated including the heat shock-responsive genes (htpX, dnaJ, and groEL2) (43) and the essential genes infB and pmmA in M. tuberculosis (33). Genes with unknown functions (MAP2720c and MAP0156) were also included in this list. Earlier microarray analysis (43) of M. tuberculosis cultures exposed to high temperature implicated four of these genes (htpX, dnaJ, groEL2, and infB) in the heat shock response. This profile suggests a common pathway involving these genes in response to stressors in both M. avium subsp. paratuberculosis and M. tuberculosis.
Transcriptional profiling of M. avium subsp. paratuberculosis isolated from naturally infected cows.
Because fecal-oral transmission is the expected route of M. avium subsp. paratuberculosis infection in cattle, it is of great importance to understand the gene expression pattern of M. avium subsp. paratuberculosis bacilli shed in feces. We hypothesized that genes responsible for the persistence of M. avium subsp. paratuberculosis in feces could contribute to M. avium subsp. paratuberculosis virulence during infection. The whole-genome microarray approach was used to analyze in vitro cultures and bacterial pellets collected from fecal materials of infected cows. Both clinical signs and continuous shedding of M. avium subsp. paratuberculosis by the sampled cows confirmed their late stage of Johne's disease. During the recovery of M. avium subsp. paratuberculosis from the fecal samples, HPC treatment was used to eliminate nonmycobacterial contaminants (16, 31). Culturing of the decontaminated fecal samples indicated an M. avium subsp. paratuberculosis load of 107 CFU/gram of fecal sample, suggesting a good possibility for directly isolating M. avium subsp. paratuberculosis and purifying bacterial RNA for DNA microarrays. Nonetheless, a few bacterial colonies were isolated from fecal pellets when LB medium was used to culture fecal samples. To confirm the identity of transcripts isolated from fecal cow samples, amplicons were sequenced from purified RNA samples (see Fig. S1 in the supplemental material) following reverse transcription. In all examined genes (n = 3), BLAST analysis indicated their identity to be M. avium subsp. paratuberculosis or M. avium subsp. avium and not any other bacterial genes (Table 2). Because we examined only a small number of genes, we cannot confirm that all transcripts identified in the fecal samples were generated from M. avium subsp. paratuberculosis. Previously, decontamination with HPC was shown to greatly reduce bacterial and fungal contaminants but with little or no effect on the recovery of viable M. avium subsp. paratuberculosis (16, 31). As a control, the transcriptional profile of HPC-treated cultures was also analyzed to delineate the impact of using HPC on M. avium subsp. paratuberculosis transcripts during our protocol for sample decontamination.
TABLE 2.
List of gene transcripts examined by reverse transcriptase PCR and sequence analysis
| Gene name | Amplicon size (bp) | E value
|
||
|---|---|---|---|---|
| M. avium subsp. paratuberculosis | M. avium | Other bacteria | ||
| MAP1401, tlyA | 263 | 2.0E-83 | 4.0E-80 | No hit |
| MAP2041, lipM | 247 | 3.0E-55 | 3.0E-50 | No hit |
| MAP3006c, lipN | 238 | 4.0E-59 | 9.0E-56 | No hit |
To our surprise, microarray analysis of HPC-treated samples identified significant change in a large number of gene transcripts (n = 649), where most of the general stress-responsive genes observed in acidic pH or heat shock were also induced (e.g., mbtC, relA, htpX, and hsp18) (see Table S6 in the supplemental material). Accordingly, all of the data generated from the cow samples were compared to HPC-treated cultures to neutralize the effect of HPC treatment on M. avium subsp. paratuberculosis. Interestingly, transcriptional analysis of cow samples identified the largest number of genes (n = 1,082) that were significantly different from the HPC-treated samples (see Table S7 in the supplemental material). Among the highly activated genes are those involved in the survival of M. tuberculosis during chronic infection (e.g., ung and icl encoding isocitrate lyase) (25, 48) or those involved in nitrate reduction (narH) (40). Other genes included those involved in lipid metabolism such as lipN and lipI, suggesting a unique nutrient requirement for M. avium subsp. paratuberculosis isolated from fecal samples. Genes involved in variable DNA repair mechanisms (e.g., lexA and uvrD2) or heat shock (htpG) were among the group of up-regulated genes. However, other general stress-responsive genes were among the group of repressed genes such as relA, hsp18, and clpB and the transcriptional regulator whiB3, indicating the unique nature of the M. avium subsp. paratuberculosis strains isolated from cow samples. Finally, two sigma factors (sigI and sigG) were among the significantly induced genes in cow samples. In M. tuberculosis, only sigI was induced following exposure to mild cold (22), suggesting an adaptive response of M. avium subsp. paratuberculosis in feces to a lower temperature than that of the host (normal body temperature for a cow is 39°C). Overall, both the large number of genes regulated in M. avium subsp. paratuberculosis isolated from the cow samples and the type of activated/repressed genes portrayed bacilli that shared common features with in vitro-treated cultures but maintained a group of unique genes that were regulated only in the cow samples.
Grouping the stress-responsive genes.
Analysis of significantly regulated genes of M. avium subsp. paratuberculosis in samples collected from cows or exposed to defined stressors identified a significantly large number of genes involved in mycobacterial stress response to variable stimuli (almost 25% of the encoded genes). Further analysis of genes based on their transcriptional patterns could identify gene groups coregulated to perform a similar function. In this analysis, all genes with detectable levels of transcription (genes with hybridization signals higher than background level) were included and displayed a high level of correlation among all conditions examined (Fig. 2A). Consequently, we applied a hierarchical clustering algorithm (13) to identify unique transcriptional patterns of M. avium subsp. paratuberculosis genes during exposure to individual stressors. Based on the overall clustering of the transcripts, the profile of the cow samples was different from all in vitro cultures examined. The profile of the acidic-pH samples was closely related to that of the HPC-treated samples, and both profiles occupied a cluster node related to that of the cow fecal samples (Fig. 2B). On the other hand, samples exposed to either oxidative stress or heat shock were in a separate cluster node, suggesting a different profile for these conditions compared to the cow samples. Interestingly, one of the recently identified virulence factors in M. avium subsp. paratuberculosis (36), the kdpC gene, was clustered among a group of genes activated only in the cow samples. Other genes in this cluster included a fatty acid degradation lipase/esterase (lipN) and an orthologue to an M. tuberculosis virulence gene, mmpl2 (7). It is possible that this cluster of genes is also involved in M. avium subsp. paratuberculosis virulence.
FIG. 2.
Hierarchical cluster analysis of the gene expression levels collected from M. avium subsp. paratuberculosis cultures exposed to variable stressors. (A) A heat map displaying the overall correlation among replicates of all examined stressors. (B) An example of cluster analysis showing genes activated only in the cow samples. Note the dendrogram displayed at the top of the image reflecting the overall relationship among examined samples. A color bar is presented at the top of each panel with a range from 0 to 1 (black to red) for panel A or from −3 to 3 (green to red) for panel B.
At the gene level, the employed clustering algorithm identified groups of coregulated genes that were shared among stress conditions, such as low pH and HPC treatment (see Table S8 in the supplemental material). In both samples, genes involved in mycobactin biosynthesis (mbtH2), mycolic acid biosynthesis (cmaA2), and a protein kinase (pknB) were induced, suggesting a potential role for their encoded proteins in the acidic response. Other genes that were shared among acidity, HPC treatment, and the cow samples included mce1_1 and mce4 (both involved in cell entry), atpC, and a transcriptional regulator gene, kdpE, suggesting a role of this group of genes in response to acidic pH during infection. Following exposure to low pH, 63% of the orthologous genes that were regulated by acidity in M. avium subsp. paratuberculosis and M. tuberculosis (n = 36) were also repressed (14) in both organisms, suggesting similar responses to acidity. Examples of the repressed genes include fadE12, murA, and aspB. Additionally, we compared the oxidative response of M. tuberculosis (34) to the one obtained in M. avium subsp. paratuberculosis. Among the gene groups that were regulated by H2O2 in M. tuberculosis (32), 25 to 38% of M. avium subsp. paratuberculosis gene orthologues were also induced or repressed, respectively, by the addition of H2O2. Examples of the shared activated genes include sigH, sigB, icl, trxB2, and katG (see Table S8 in the supplemental material). Taken together, the shared profiles indicated conserved responses to acidity and oxidative radicals usually present in mycobacterial environments, such as within macrophages (39).
To gain insights into the regulation of genetic networks in the stressome, gene clusters were inspected for genes coregulated with sigma factors, the global transcriptional regulators. A total of 19 sigma factors are predicted in the genome of M. avium subsp. paratuberculosis, where only sigA and sigB are considered essential sigma factors, while the rest are considered extracytoplasmic (20). Seven extracytoplasmic sigma factors, sigD, sigF1, sigG, sigH, sigI, sigL, and ECF-6, were significantly regulated >±2-fold under any particular stressor, confirming their participation in regulating several gene groups. For example, cluster analysis identified a group of 71 M. avium subsp. paratuberculosis genes that were coexpressed with sigD while another group of 77 genes were coexpressed with sigG. Both gene groups were activated only in cow samples compared to the rest of the examined samples. Interestingly, both sigma factors were shown to be involved in M. tuberculosis virulence during chronic infection (sigD) (6) or survival in the macrophage (sigG) (15). Overall, cluster analysis portrayed both unique and common features of M. avium subsp. paratuberculosis responding to variable stimuli. The identified profiles also suggested the involvement of several highly regulated gene groups that could contribute to M. avium subsp. paratuberculosis virulence and pathogenesis under control of a set of sigma factors such as sigD, sigE, and sigH.
Stress-responsive genes are important for M. avium subsp. paratuberculosis survival in animals.
As suggested by differential gene expression and hierarchical clustering, several genes displayed unique patterns of expression depending on the examined stressor. To test the hypothesis that stress-regulated genes contribute to M. avium subsp. paratuberculosis survival, we employed a strategy based on selecting mutants with inactivation/deletion of genes induced under stress conditions. These genes were tested for survival in a murine model of paratuberculosis. The lipN gene, which was up-regulated in the cow samples, was targeted for deletion mutagenesis (4) as a representative of genes involved in lipid degradation. In M. tuberculosis, a mutant of the lipF gene that is a homolog to M. avium subsp. paratuberculosis lipN had an important effect on mycobacterial persistence in mice (18). Using homologous recombination, a 1.1-kb DNA fragment of the lipN coding region was replaced with a hygromycin-resistant gene cassette using M. avium subsp. paratuberculosis K-10 (4). The deletion of lipN was verified by both PCR and Southern blot analysis (Fig. 3), confirming the ability of the specialized transduction system developed for M. tuberculosis to knock out genes in M. avium subsp. paratuberculosis. To test the virulence of the generated mutant, we compared the colonization levels and histopathology of mice infected with the ΔlipN mutant to the wild-type strain of M. avium subsp. paratuberculosis K-10. Additionally, we used the same murine model to assess the virulence of a selected list of M. avium subsp. paratuberculosis mutants generated in M. avium subsp. paratuberculosis ATCC 19698 and identified during a large-scale screening of M. avium subsp. paratuberculosis transposon mutants (36). All of the examined mutants have insertions in genes that were up-regulated in vivo (lipL and lpqP) or in acidic pH (prrA, aceAB, and mbtH2) to variable degrees (Table 3). Figure 3D displays the location of disrupted genes in the examined mutants.
FIG. 3.
Gene deletion in M. avium subsp. paratuberculosis. (A) The design of lipN-knockout allelic-exchange substrates using the pYUB854 cloning vector (4). (B) PCR confirmation of the lipN-knockout mutant using genomic DNA (gDNA) from the wild type (wt) and the ΔlipN mutant and primer pairs designed for the hygromycin resistance gene, lipN, or the recombinant region after allelic exchange. A 2% agarose gel showed amplicons from the hygromycin resistance gene only when the mutant genomic DNA was used (lane 1), whereas the lipN sequence was amplified only from the wild-type genomic DNA (lane 4). (C) Southern blot analysis of the lipN-knockout mutant. Genomic DNA was digested with XhoI and Acc65I (for hygromycin detection) or XhoI and ScaI (for lipN detection) and detected with hygromycin or lipN probes. The lipN sequence was absent from the lipN-knockout mutant genomic DNA. (D) A genomic map showing the distribution of the 10 genes inactivated by transposon mutagenesis or homologous recombination examined in this and previous studies (36, 53).
TABLE 3.
Changes in expression of the list of genes selected for further screening of their mutants in micea
| Gene name | Low-pH group
|
Cow samples
|
||
|---|---|---|---|---|
| Fold change | PDE | Fold change | PDE | |
| MAP0834c, prrA | 3.55 | 0.07 | −35.71 | 0.64 |
| MAP1228, lipL | −1.91 | 0.11 | 34.34 | 0.52 |
| MAP3006c, lipN | −3.36 | 0.10 | 33.79 | 0.55 |
| MAP1643, aceAB | 282.3 | 0.85 | −32.87 | 0.31 |
| MAP1872c, mbtH2 | 5.26 | 0.10 | −3.97 | 0.09 |
| MAP4288, lpqP | −4.28 | 0.11 | 4.94 | 0.13 |
Change (n-fold) with posterior differential of expression (PDE) is listed based on Bayesian statistical analysis. Bold indicates positive values for change (n-fold).
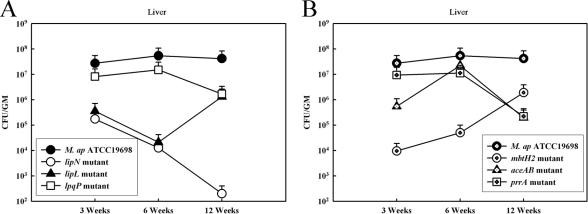
Interestingly, the colonization levels of all examined mutants (n = 6) were highly reduced compared to the wild-type strains (Fig. 4), at least at one time point of the infection. The ΔlipN mutant was consistently cleared from livers and intestines of mice, especially by 12 WPI compared to its parent strain, M. avium subsp. paratuberculosis K-10. Other mutants regained some of the colonization levels after 6 WPI (e.g., mbtH2 and lipL), suggesting the contribution of the disrupted genes to the initial stages of infection. It is also noteworthy that colonization levels of both M. avium subsp. paratuberculosis ATCC 19698 and K-10 were similar at 3 and 6 WPI but not at 12 WPI. Histopathological analysis of liver sections of infected animals showed a lower level of granuloma formation compared to the result from mice infected with the wild-type strain (see Fig. S2 in the supplemental material), verifying the attenuated phenotype indicated by the colonization data. In addition, more granulomas were observed in mutants with higher bacterial counts in the organs. Overall, screening of the M. avium subsp. paratuberculosis mutants in the murine model of paratuberculosis suggested the participation of lipL and mbtH2 in the initial stage of infection. However, the rest of the genes (lipN, lpqP, aceAB, and prrA) could participate in tissue colonization throughout early and late infection. Previously, M. avium subsp. paratuberculosis mutants with insertional mutations in pstA, kdpC, gcpE, and papA2 genes (Fig. 3D) were attenuated in the murine model of paratuberculosis (36). Interestingly, the former three genes were induced by 67.1-, 3.2-, and 88.2-fold in the cow samples compared to the in vitro samples, respectively, while papA2 was induced by 4.6-fold at low pH. When tested in a calf model of intestinal invasion, the ΔpstA mutant was also attenuated (53). Overall, mutants with disruption/deletion of genes highly induced in cow feces or acidic-pH samples were attenuated in the murine model of paratuberculosis, suggesting a key role for these two classes of genes in M. avium subsp. paratuberculosis survival during infection.
FIG. 4.
Screening of M. avium subsp. paratuberculosis mutants in the murine model of paratuberculosis. Mouse groups were intraperitoneally inoculated with 108 CFU/mouse for the M. avium subsp. paratuberculosis ATCC 19698 wild-type strain or its isogenic mutant while groups inoculated with M. avium subsp. paratuberculosis K-10 or its ΔlipN isogenic mutant were inoculated with 107 CFU/mouse. Liver and intestine tissues were collected at 3, 6, and 12 WPI. Only data for the liver are shown here. (A) Colonization levels of three mutants with disruption of genes activated in cow samples compared to levels obtained from mouse groups infected with either M. avium subsp. paratuberculosis ATCC 19698 or M. avium subsp. paratuberculosis K-10. (B) Colonization levels of three mutants with disruption of genes activated in acidic pH compared to levels obtained from a mouse group infected with the wild-type M. avium subsp. paratuberculosis ATCC 19698. Error bars represent standard errors (±) of colony counts from different samples at each time (n = 3 to 6).
DISCUSSION
For a long time, the molecular basis of the pathogenesis of M. avium subsp. paratuberculosis infection has not been completely understood, despite the great economic impact of Johne's disease on the dairy industry (21) and the potential involvement of M. avium subsp. paratuberculosis in Crohn's disease in humans (27). Fortunately, with the advance of genomic analysis of microbial pathogens, the genome sequence of M. avium subsp. paratuberculosis K-10 became available (20). This has sparked investigation of the genomic variations among isolates of M. avium subsp. paratuberculosis and created novel diagnostic targets (2, 35, 52). Nonetheless, the regulation of expressed genes responsible for establishing infection on a genome-wide scale remained poorly understood. Here, we report the first effort to profile gene transcripts of M. avium subsp. paratuberculosis exposed to several stressors or isolated directly from infected cows. Although unique profiles emerged for M. avium subsp. paratuberculosis following exposure to stressors, there was a set of common genes that shared transcriptional levels among all examined stressors. This strongly indicates their involvement in the universal mycobacterial response to stress conditions including the family of heat shock proteins (e.g., hsp and htpX).
Surprisingly, a large number of genes were induced during exposure to acidic pH compared to other in vitro conditions, suggesting the importance of the change in pH to the survival of M. avium subsp. paratuberculosis. However, the largest number of regulated genes was identified when the transcriptional profile of M. avium subsp. paratuberculosis isolated from cow samples was analyzed, indicating the unique challenge for M. avium subsp. paratuberculosis in order to survive in the cow excreta. Such analysis is complicated by the potential presence of other bacterial transcripts despite the decontamination protocol that we used here. Also, variations among transcription profiles of clinical isolates and the standard strain (ATCC 19698) used for all in vitro stressors could further complicate the analysis of the cow samples. Finally, we realize that analyzing M. avium subsp. paratuberculosis isolated from fecal samples would not necessarily mimic the host microenvironments (i.e., cow intestine). However, feces- or host-adapted mycobacteria are the most efficient forms for transmitting infections to naïve animals as shown by both historical (1) and experimental (41) evidence. In fact, inactivation of the three genes that were induced in the fecal samples was detrimental to the survival of M. avium subsp. paratuberculosis in mice, which was similar to the results obtained from the inactivation of genes up-regulated by low pH. Some mutants (e.g., ΔlipL and ΔmbtH2 mutants) partially regained their ability to colonize livers by 12 WPI, suggesting the importance of these genes to initiating a successful infection. On the other hand, other mutants (e.g., ΔlipN and ΔprrA mutants) were not efficient in colonizing livers throughout the examined times, suggesting a potential role for these genes in establishing chronic infection. Further analysis is needed to verify the attenuation associated with the examined genes (e.g., complementation analysis) and to characterize the mechanisms of attenuation exhibited by each mutant in a better model for Johne's disease such as the calf model.
Throughout this report, we analyzed the transcriptional profiles of M. avium subsp. paratuberculosis in comparison to those previously reported for M. tuberculosis (14, 24, 43, 48). At this point, few experimental data are available for the transcriptional responses of M. avium subsp. paratuberculosis, which stifled our comparative analysis. Nonetheless, several common responses were found between M. avium subsp. paratuberculosis and M. tuberculosis at the transcriptional level (e.g., heat shock and oxidative responses), reflecting the conserved evolutionary relationship between the two types of mycobacteria. Also, it indicates the similarity between the two types of infections at the cellular level (e.g., both organisms survive the macrophage microenvironment). However, variations among transcriptional profiles were also found. In M. tuberculosis, only a small number of genes (n = 81) (14) compared to a large number of M. avium subsp. paratuberculosis genes (n = 597) were regulated under acidic pH. In contrast, a higher number of genes significantly responded to oxidative stress in M. tuberculosis (n = 761) (34) compared to only 155 genes in M. avium subsp. paratuberculosis. Such a difference between the two pathogens could reflect the disparity between the microarray platforms or protocols used by each laboratory. Such disparity in responses could also reflect a genuine difference between the microenvironments of the alveolar and enteric macrophages for M. tuberculosis and M. avium subsp. paratuberculosis, respectively.
In addition to common and unique M. avium subsp. paratuberculosis genetic circuits identified for each examined stressor, the employed transcriptional analysis confirmed the gene predictions of a large number of ORFs (91.2% of the genome). As previously suggested (23), environmental stressors usually induce different sigma factors depending on the nature of stimuli. Our microarray analysis suggested the involvement of sigH in the M. avium subsp. paratuberculosis response to oxidative and heat shock stressors as shown previously for M. tuberculosis (32). It is also possible that sigD and sigG participate in the persistent infection with M. avium subsp. paratuberculosis while sigI is involved in adaptation to cold shock, as suggested for M. tuberculosis (22). The rest of the sigma factors may not play a critical role under the examined stress conditions. More analysis is needed to further analyze the role of stress-induced genes in M. avium subsp. paratuberculosis survival during natural infection and to better understand the mechanisms used by M. avium subsp. paratuberculosis to survive in hostile microenvironments. Such knowledge will improve the ability to control M. avium subsp. paratuberculosis infections. Our strategy for transcriptional profiling and subsequent assessment of specific mutants in animal models of infection could be applied to other intracellular pathogens to elucidate their mechanisms of virulence.
Supplementary Material
Acknowledgments
We acknowledge Elizabeth A. Vu, Bassam Abomoelak, and Charles Czuprynski for reading the manuscript. We thank Becky Manning and Gail Thomas for providing fecal materials from infected cows and Sarah K. Ward for the bioinformatic analysis.
Research reported here is supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (AGRICCREE 2003-02230) and the Animal Formula Fund (WIS01093) as well as the Johne's Disease Integrated Program (2004-35605-14243).
Footnotes
Published ahead of print on 10 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ayele, W. Y., M. Macháèková, and I. Pavlík. 2001. The transmission and impact of paratuberculosis infection in domestic and wild ruminants. Vet. Med. 6-7:205-224. [Google Scholar]
- 2.Bannantine, J. P., E. Baechler, Q. Zhang, L. L. Li, and V. Kapur. 2002. Genome-scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardarov, S., J. Kriakov, C. Carriere, S. W. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bardarov, S., M. S. Pavelka, V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 5.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 6.Calamita, H., C. Ko, S. Tyagi, T. Yoshimatsu, N. E. Morrison, and W. R. Bishai. 2005. The Mycobacterium tuberculosis SigD sigma factor controls the expression of ribosome-associated gene products in stationary phase and is required for full virulence. Cell. Microbiol. 7:233-244. [DOI] [PubMed] [Google Scholar]
- 7.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 8.Cetinkaya, B., K. Egan, D. A. Harbour, and K. L. Morgan. 1996. An abattoir-based study of the prevalence of subclinical Johne's disease in adult cattle in south west England. Epidemiol. Infect. 116:373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, D. M., A. J. Radford, G. W. De Lisle, and H. Billman-Jacobe. 1994. Diagnosis and epidemiology of bovine tuberculosis using molecular biological approaches. Vet. Microbiol. 94:83-89. [DOI] [PubMed] [Google Scholar]
- 10.Collins, M. T., D. C. Sockett, W. J. Goodger, T. A. Conrad, C. B. Thomas, and D. J. Carr. 1994. Herd prevalence and geographic distribution of, and risk factors for, bovine paratuberculosis in Wisconsin. J. Am. Vet. Med. Assoc. 204:636-641. [PubMed] [Google Scholar]
- 11.Coussens, P. M., C. J. Colvin, K. Wiersma, A. Abouzied, and S. Sipkovsky. 2002. Gene expression profiling of peripheral blood mononuclear cells from cattle infected with Mycobacterium paratuberculosis. Infect. Immun. 70:5494-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Lisle, G. W., G. F. Yates, and R. H. Montgomery. 2003. The emergence of Mycobacterium paratuberculosis in farmed deer in New Zealand—a review of 619 cases. N. Z. Vet. J. 51:58-62. [DOI] [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant, I. R., E. I. Hitchings, A. McCartney, F. Ferguson, and M. T. Rowe. 2002. Effect of commercial-scale high-temperature, short-time pasteurization on the viability of Mycobacterium paratuberculosis in naturally infected cows' milk. Appl. Environ. Microbiol. 68:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kendziorski, C. M., M. A. Newtone, H. Lan, and M. N. Goululd. 2003. On parameteric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 22:3899-3914. [DOI] [PubMed] [Google Scholar]
- 18.Lamichhane, G., M. Zignol, N. J. Blades, D. E. Geiman, A. Dougherty, J. Grosset, K. W. Broman, and W. R. Bishai. 2003. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 100:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larocque, R. C., J. B. Harris, M. Dziejman, X. Li, A. I. Khan, A. S. Faruque, S. M. Faruque, G. B. Nair, E. T. Ryan, F. Qadri, J. J. Mekalanos, and S. B. Calderwood. 2005. Transcriptional profiling of Vibrio cholerae recovered directly from patient specimens during early and late stages of human infection. Infect. Immun. 73:4488-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linnabary, R. D., G. L. Meerdink, M. T. Collins, J. R. Stabel, R. W. Sweeney, M. K. Washington, and S. J. Wells. 2001. Johne's disease in cattle. Counc. Agric. Sci. Technol. 17:1-10. [Google Scholar]
- 22.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 23.Manganelli, R., R. Proveddi, S. Rodrigue, J. Beaucher, L. Gaudreau, and I. Smith. 2004. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J. Bacteriol. 186:895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 25.McKinney, J. D., K. Höner zu Bentrup, E. J. Muñoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 26.Momotani, E., D. L. Whipple, A. B. Thiermann, and N. F. Cheville. 1988. Role of M cells and macrophages in the entrance of Mycobacterium paratuberculosis into domes of ileal Peyer's patches in calves. Vet. Pathol. 25:131-137. [DOI] [PubMed] [Google Scholar]
- 27.Naser, S. A., G. Ghobrial, C. Romero, and J. F. Valentine. 2004. Culture of Mycobacterium avium subspecies paratuberculosis from the blood of patients with Crohn's disease. Lancet 364:1039-1044. [DOI] [PubMed] [Google Scholar]
- 28.Naser, S. A., I. Shafran, D. Schwartz, F. El Zaatari, and J. Biggerstaff. 2002. In situ identification of mycobacteria in Crohn's disease patient tissue using confocal scanning laser microscopy. Mol. Cell. Probes 16:41-48. [DOI] [PubMed] [Google Scholar]
- 29.Paustian, M. L., A. Amonsin, V. Kapur, and J. P. Bannantine. 2004. Characterization of novel coding sequences specific to Mycobacterium avium subsp. paratuberculosis: implications for diagnosis of Johne's disease. J. Clin. Microbiol. 42:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips, B. J., and W. Kaplan. 1976. Effect of cetylpyridinium chloride on pathogenic fungi and Nocardia asteroides in sputum. J. Clin. Microbiol. 3:272-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raman, S., T. S. Song, X. L. Puyang, S. Bardarov, W. R. Jacobs, and R. N. Husson. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sassetti, C. M., and E. J. Rubin. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin, S. J., C.-W. Wu, H. Steinberg, and A. M. Talaat. 2006. Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect. Immun. 7:3825-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh-Gasson, S., R. D. Green, Y. J. Yue, C. Nelson, F. Blattner, M. R. Sussman, and F. Cerrina. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17:974-978. [DOI] [PubMed] [Google Scholar]
- 38.Souza, C. D., O. A. Evanson, and D. J. Weiss. 2006. Mitogen activated protein kinase p38 pathway is an important component of the anti-inflammatory response in Mycobacterium avium subsp. paratuberculosis-infected bovine monocytes. Microb. Pathog. 41:59-66. [DOI] [PubMed] [Google Scholar]
- 39.Springer, B., S. Master, P. Sander, T. Zahrt, M. McFalone, J. Song, K. G. Papavinasasundaram, M. J. Colston, E. Boettger, and V. Deretic. 2001. Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC in virulent and avirulent strains and effect of ahpC inactivation. Infect. Immun. 69:5967-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stermann, M., L. Sedlacek, S. Maass, and F. C. Bange. 2004. A promoter mutation causes differential nitrate reductase activity of Mycobacterium tuberculosis and Mycobacterium bovis. J. Bacteriol. 186:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart, D. J., J. A. Vaughan, P. L. Stiles, P. J. Noske, M. L. V. Tizard, S. J. Prowse, W. P. Michalski, K. L. Butler, and S. L. Jones. 2006. A long-term study in Angora goats experimentally infected with Mycobacterium avium subsp paratuberculosis: clinical disease, faecal culture and immunological studies. Vet. Microbiol. 113:13-24. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, G. R., B. D. Robertson, and D. B. Young. 2004. Analysis of the function of mycobacterial DnaJ proteins by overexpression and microarray profiling. Tuberculosis 84:180-187. [DOI] [PubMed] [Google Scholar]
- 43.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Myobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 44.Talaat, A. M., S. T. Howard, I. W. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talaat, A. M., P. Hunter, and S. A. Johnston. 2000. Genome-directed primers for selective labeling of bacterial transcripts for DNA microarray analysis. Nat. Biotechnol. 18:679-682. [DOI] [PubMed] [Google Scholar]
- 46.Talaat, A. M., R. Lyons, S. T. Howard, and S. A. Johnston. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl. Acad. Sci. USA 101:4602-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talaat, A. M., and M. Trucksis. 2000. Transformation and transposition of the genome of Mycobacterium marinum. Am. J. Vet. Res. 61:125-128. [DOI] [PubMed] [Google Scholar]
- 48.Talaat, A. M., S. K. Ward, C.-W. Wu, E. Rondon, C. Tavano, J. P. Bannantine, R. Lyons, and S. A. Johnston. 2007. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J. Bacteriol. 189:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tooker, B. C., J. L. Burton, and P. M. Coussens. 2002. Survival tactics of M. paratuberculosis in bovine macrophage cells. Vet. Immun. Immunopathol. 87:429-437. [DOI] [PubMed] [Google Scholar]
- 50.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittington, R. J., and E. S. G. Sergeant. 2001. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust. Vet. J. 79:267-278. [DOI] [PubMed] [Google Scholar]
- 52.Wu, C.-W., J. Glasner, M. T. Collins, S. Naser, and A. M. Talaat. 2006. Whole-genome plasticity among Mycobacterium avium subspecies: insights from comparative genomic hybridizations. J. Bacteriol. 188:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, C. W., M. Livesey, S. K. Schmoller, E. J. B. Manning, H. Steinberg, W. C. Davis, M. J. Hamilton, and A. M. Talaat. 2007. Invasion and persistence of Mycobacterium paratuberculosis during early stages of Johne's disease in calves. Infect. Immun. 75:2110-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaslaver, A., A. E. Mayo, R. Rosenberg, P. Bashkin, H. Sberro, M. Tsalyuk, M. G. Surette, and U. Alon. 2004. Just-in-time transcription program in metabolic pathways. Nat. Genet. 36:486-491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.