Abstract
The filamentous cyanobacterium Anabaena sp. strain PCC 7120 forms heterocysts in a semiregular pattern when it is grown on N2 as the sole nitrogen source. The transition from vegetative cells to heterocysts requires marked metabolic and morphological changes. We show that a trimeric pore-forming outer membrane β-barrel protein belonging to the TolC family, Alr2887, is up-regulated in developing heterocysts and is essential for diazotrophic growth. Mutants defective in Alr2887 did not form the specific glycolipid layer of the heterocyst cell wall, which is necessary to protect nitrogenase from external oxygen. Comparison of the glycolipid contents of wild-type and mutant cells indicated that the protein is not involved in the synthesis of glycolipids but might instead serve as an exporter for the glycolipid moieties or enzymes involved in glycolipid attachment. We propose that Alr2887, together with an ABC transporter like DevBCA, is part of a protein export system essential for assembly of the heterocyst glycolipid layer. We designate the alr2887 gene hgdD (heterocyst glycolipid deposition protein).
Gram-negative bacteria use a type I export system to transfer proteins or other molecules, like siderophores or fatty acids, to the cell surface (37, 50, 55). The proteinaceous substrates contain a C-terminal secretion signal essential and sufficient for export. The information for targeting and translocation, however, seems to be presented in the form of a secondary structure element rather than in the primary sequence as no clear amino acid motif has been identified. The machinery of type I export systems bridges the periplasm, allowing transfer of their substrates beyond the outer membrane. The export system is composed of a translocase spanning the plasma membrane with an α-helical, pore-forming domain and a cytosolic domain energizing the translocation process. Additionally, an adaptor protein associates with the translocase and, after substrate association, induces an interaction with an outer membrane TolC-like exit tunnel to conduct the secretion. Whereas TolC-like proteins from gram-negative proteobacteria have been amply investigated (37, 50, 54, 55), nothing is known about these systems in cyanobacteria. Based on sequence similarity (40) and proteomic analysis of the cell wall fraction (42), we have recently proposed the existence of a protein of this type, Alr2887, in Anabaena sp. strain PCC 7120, for which a function in lipid transfer has previously been proposed (40).
Anabaena sp. strain PCC 7120 is a filamentous cyanobacterium which starts a program of cell differentiation upon nitrogen starvation that results in the appearance of nitrogen-fixing cells. These cells are called heterocysts and are arranged in a semiregular pattern along the filament (63). Three mechanisms protect the oxygen-labile nitrogenase from irreversible inactivation by O2 produced by oxygenic photosynthesis: (i) formation of extra envelope layers outside the gram-negative cell wall, (ii) degradation of photosystem II antennae and a decrease in photosystem II activity, and (iii) an increase in O2-consuming respiration. The process of cell differentiation takes about one generation time (20 h). Heterocysts provide the vegetative cells with their products (combined nitrogen) and obtain from the vegetative cells metabolites that serve as reductants for N2 fixation and C skeletons for nitrogen assimilation (1, 20, 63).
Formation of the extra envelope of the cell wall occurs in several steps. First, a homogeneous layer of polysaccharides (heterocyst envelope polysaccharides [HEPs]) is formed by deposition of the material outside the gram-negative cell wall (10, 26). Subsequently, specific glycolipids (heterocyst envelope glycolipids [HGLs]) are made and deposited between this protective layer and the outer membrane, forming the so-called laminated layer (26, 31). This layer is a barrier for entry of gases (31, 59, 61). HGLs are polyhydroxy alcohols (with 26 to 28 C atoms) that are glycosidically linked at C-1 to glucose (reference 25 and references therein). Fatty acid synthases, polyketide synthases, ketoreductases, dehydrases, acyl transferases, or thioesterases are likely to be involved in the synthesis of the glycolipids (19, 32). How transport and deposition of the HGLs beyond the cell wall occur is still unknown.
Several genes that are involved in the formation of heterocyst-specific cell wall layers have been identified. hepA, hepB, and hepC encode proteins with similarity to ABC-type transporters, glycosyltransferases, and UDP-galactose lipid carrier transferases, respectively (40, 64). Furthermore, alr2825, alr2827, alr2831, alr2833, alr2837, alr2839, and alr2841 are localized on a well-defined specific “HEP island” in the chromosome encoding enzymes with putative functions in the synthesis of the HEP layer (33). Additionally, the hglB, hglC, hglD, and hglE genes encode enzymes necessary for the synthesis of heterocyst glycolipids (4, 9). In turn, the gene product of hglK is needed for formation of the glycolipid layer (5). Finally, the devBCA operon codes for an exporter belonging to the ABC-type transport family essential for HGL layer formation (21). A TolC-like outer membrane protein like Alr2887 (40, 42) would be the missing part of the export system that includes DevBCA, whose substrate is not yet known.
In this work, we analyzed the relationship of Alr2887 to proteins belonging to the TolC family and investigated its possible function. Alr2887 is required for deposition of the glycolipid layer of the heterocyst envelope, and it could have a role in the secretion of proteinaceous substrates involved in this process. We therefore designated the alr2887 gene hgdD (heterocyst glycolipid deposition protein).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
This study was carried out with the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120, also known as Nostoc sp. strain 7120, and several mutant derivatives described in Table 1. Wild-type Anabaena sp. strain PCC 7120 was grown photoautotrophically at 30°C in liquid BG110 medium (51) or A&A medium at a 1:4 dilution (2). All Fox− strains were grown in medium containing 5 mM NH4NO3 and 5 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-NaOH (pH 8). Cultures of mutant strains NMP-alr2887-GFP, NME-alr2887-GFP, and NMΔ-alr2887 contained 2 μg of streptomycin and 2 μg of spectinomycin ml−1; cultures of DR181 contained 50 μg neomycin ml−1 (21, 47). Undiluted medium was solidified with 1.5% agar (Difco, Heidelberg, Germany). Heterocyst formation was induced in liquid cultures by washing the cells three times with combined nitrogen-free medium and incubating them in BG110 medium or A&A medium at a 1:4 dilution under growth conditions. The growth on agar-solidified medium was determined, without antibiotics, using standard BG11 medium, BG110 medium, or BG110 medium supplemented with 4 mM NH4Cl and 8 mM TES-NaOH (pH 8). The amount of biomass in a culture was estimated by determining the concentration of chlorophyll a (Chl) in methanolic extracts.
TABLE 1.
Anabaena strains used in this study
| Strain | Resistance | Genotype | Relevant properties |
|---|---|---|---|
| PCC 7120 | Wild type | ||
| DR181 | Nmr | alr2887::C.K3 | Fox− Het+ Hgl+ Hen− |
| NMP-alr2887-GFP | Spr Smr | alr2887::gfp | C-terminal GFP-protein fusion |
| NME-alr2887-GFP | Spr Smr | Palr2887-gfp in nucA region | N-terminal GFP fusion |
| NMΔ-alr2887 | Spr Smr | alr2887::pCSV3 | Fox− Het+ Hgl+ Hen− |
Genetic procedures.
Transformation of Escherichia coli strains and isolation and manipulation of plasmid DNA were performed by standard methods (53). PCRs were done with the Triple master PCR system (Eppendorf, Hamburg, Germany). Total DNA of Anabaena sp. strain PCC 7120 was isolated as described previously (8).
Mutant DR181 was generated by transfer of pIM181 (Table 2) from E. coli to Anabaena sp. strain PCC 7120 by triparental mating (62). Plasmid pIM181 was constructed as follows. The entire open reading frame (ORF) of alr2887 was amplified from the chromosomal DNA of Anabaena sp. strain PCC 7120 by PCR using primers Oligo 309 and Oligo 310 containing XhoI restriction sites (Table 3). The resulting ca. 2,300-bp fragment was cloned into pGEMT, and the C.K3 cassette, encoding neomycin phosphotransferase (14), was inserted with XbaI ends between the two NheI sites of pIM125, deleting a 513-bp internal fragment of alr2887. The mutagenized gene was cut from this construct (pIM136) with endonuclease XhoI and ligated into the unique XhoI site of the positive selection vector pRL271 (6), resulting in pIM181. After conjugal transfer, putative double recombinants were obtained by the positive selection method described by Cai and Wolk (8). Double recombination and complete segregation of the mutated gene were confirmed by PCR using primers Oligo 309 and Oligo 310.
TABLE 2.
Plasmids used in this study
| Plasmid | Marker(s) | Properties | Reference or source |
|---|---|---|---|
| pCSEL21 | Apr | pIC20R with gene-GFP insertion | 48 |
| pCSV3 | Spr Smr | pRL500 with substituted Apr gene | 49 |
| pCSEL24 | Apr Spr Smr | pBR322 containing Anabaena sp. 2-kb nucA-nuiA fragment and C.S3 cassette | E. Flores, CSIC |
| pGEMT | Apr | A/T cloning vector | Promega Corp. |
| pIM125 | Apr | pGEMT with PCR fragment of alr2887 (2,300-bp insert) | This study |
| pIM136 | Apr Kmr | pIM125 with C.K3 | This study |
| pIM181 | Cmr Emr Kmr | pRL271 with insert of pIM136 | This study |
| pNMP-alr2887-GFP | Spr Smr | pCSV3 with fragment of alr2887 | This study |
| pNME-alr2887-GFP | (Apr) Spr Smr | pCSEL24 with alr2887 promoter-gfp fusion | This study |
| pNMΔ-alr2887 | Spr Smr | pCSV3 with fragment of alr2887 | This study |
| pRL271 | Cmr Emr | Positive selection vector with sacB | 5 |
TABLE 3.
Primers used for cloning
| Primer | Oligonucleotide sequence |
|---|---|
| NME-alr2887-GFP-F | 5′-ATCGATCGATACAGGTACAGGTAAAACCCTGTTA-3′ |
| NME-alr2887-GFP-R | 5′-ATCGGATATCATAGAATAAGTGTTGTCCTTTCACCG-3′ |
| NMP-alr2887-GFP-F | 5′-ATCGATCGATTTAGCAGGGCTGTGGAACCAAT-3′ |
| NMP-alr2887-GFP-R | 5′-ATCGGATATCCTGACTACTAATTAATGCTCTAGAAGT-3′ |
| NMΔ-alr2887-F | 5′-ATCGGGATCCCCAGCAGATACTCAGTCACCAA-3′ |
| NMΔ-alr2887-R | 5′-ATCGGGATCCAGCAATACGGACTTGTTCATCTGC-3′ |
| Oligo 309 | 5′-CTCGAGGGTATCGATAGAGAAC-3′ |
| Oligo 310 | 5′-CTCGAGGCCATCATCTCGGTAATTG-3′ |
| GFP1 | 5′-CCTCTCCACTGACAGAGAATTTTT-3′ |
| GFP2 | 5′-GGGTAAGTTTTCCGTATGTTGCAT-3′ |
| alr2887-P1 | 5′-AACCTGATTCCCAATGCCAATCC-3′ |
| alr2887-P4 | 5′-AAATCTCCTTCGCAGTCCTCAATTAA-3′ |
An internal 600-bp fragment of the alr2887 coding region was amplified by PCR using genomic DNA of strain PCC 7120 and oligonucleotides containing BamHI restriction sites (NMΔ-alr2887-F and NMΔ-alr2887-R [Table 3]). The restricted PCR product was cloned into pCSV3 (Table 1) containing an Spr Smr gene cassette, resulting in plasmid pNMΔ-alr2887. This plasmid was amplified through transformation into E. coli DH5α, and the sequence was confirmed by conventional sequencing. Transformation of Anabaena sp. strain PCC 7120 by conjugal transfer of pNMΔ-alr2887 was performed as described previously (15), generating plasmid integration mutants by single recombination (strain NMΔ-alr2887). Segregation of the mutants was confirmed by Southern blotting of the genomic DNA performed by the standard procedure (53), using digoxigenin-labeled DNA generated by PCR using primers NMΔ-alr2887-F and NMΔ-alr2887-R (Table 3) as probes (Roche, Mannheim, Germany).
A 500-bp fragment of the alr2887 ORF encoding the C terminus of the protein was amplified by PCR using genomic DNA of strain PCC 7120 and primers with ClaI/EcoRV restriction sites (NMP-alr2887-GFP-F and NMP-alr2887-GFP-R [Table 3]). The restricted PCR product was cloned into pCSEL21 (48) to generate an in-frame product with the gene coding for green fluorescent protein (GFP). The plasmid was amplified through transformation into E. coli DH5α, and the sequence was confirmed by conventional sequencing. Subsequently, the fusion construct was excised by restriction with EcoRI. The resulting fragment was cloned into the pCSV3 cargo plasmid, generating pNMP-alr2887-GFP, in which the gfp ORF was added to the last coding triplet of the alr2887 gene. Conjugation with Anabaena sp. strain PCC 7120 was performed as described above, resulting in a single recombinant (strain NMP-alr2887-GFP), whose genetic structure was confirmed by PCR using primers GFP1 or GFP2 (confirming insertion) and alr2887-P4 (testing for the wild type) in combination with alr2887-P1.
The 800-bp upstream region of alr2887, including the first eight codons of the coding sequence, was amplified by PCR using genomic DNA of Anabaena sp. strain PCC 7120 and primers with ClaI/EcoRV restriction sites (NME-alr2887-GFP-F and NME-alr2887-GFP-R [Table 3]) The restricted PCR product was cloned into pCSEL21 in front of the gfp ORF. The fusion fragment was excised by digestion with PstI/EcoRI and ligated into the cargo vector pCSEL24, resulting in pNME-alr2887-GFP. Conjugation to Anabaena sp. strain PCC 7120 was performed as described above, resulting in a single recombinant (strain NME-alr2887-GFP), whose genomic structure was confirmed by PCR using primer GFP2 in combination with primer NME-alr2887-GFP-F.
RNA isolation and analysis.
Total RNA was isolated from cells in 100-ml cultures starved for nitrogen for different time intervals as previously described (56). Electrophoretic separation in denaturing agarose gels and blotting onto Hybond+ membranes (GE-Healthcare, Freiburg, Germany) were performed as previously described (44). RNA size markers from Roche were visualized in the gel by staining with ethidium bromide. After blotting with a vacuum, the membrane was hybridized with a 32P-labeled probe. The probe was a 2,300-bp PCR fragment containing the alr2887 ORF amplified using DNA of plasmid pIM125 as the template and Oligo 309 and Oligo 310 as the primers. Hybridization and visualization were performed as described previously (44).
Microscopic visualization.
Filaments of Anabaena sp. strain PCC 7120 were visualized with a standard reverse light microscope (DM1000; Leica, Germany). Heterocyst-containing cultures were stained with 0.5% Alcian blue in a 50% ethanol solution prior to microscopy.
Fluorescence imaging of the strains expressing N-terminal and C-terminal protein-GFP fusions was performed with a TCS SP2 or TCS PS5 Leica confocal microscope (Wetzlar, Germany) with an HCX PLAN-APO 63× 1.4-numerical-aperture oil immersion objective. All images were obtained using the same microscope setting in order to compare intensities. GFP was excited at 488 nm supplied by an argon ion laser. GFP fluorescence was analyzed by collection through a window from 500 to 570 nm, and Anabaena autofluorescence was monitored by collection through a window from 630 to 700 nm (45).
The promoter activity was further determined by measuring the GFP fluorescence of strain NME-alr2887-GFP and comparing it to the wild-type fluorescence after excitation at 480 nm and recording the emission in a window between 500 and 570 nm (LS55; Perkin Elmer, Germany). The integral of each spectrum was determined and corrected for the background fluorescence obtained for the wild-type strain. The results for three independent measurements and three independent clones carrying the NME-alr2887-GFP construct are presented below.
Cell fractionation and treatment.
Cells were grown in 3 liters of BG11 medium supplemented with 10 mM NaHCO3 (and 2 μg of streptomycin ml−1 and 2 μg of spectinomycin ml−1 for the mutants) and bubbled with a mixture of air and CO2 (1%, vol/vol) to a concentration of about 5 μg Chl ml−1. The cells were washed twice in BG110 medium, reinoculated into 1.5 liters of of BG110 medium, once again supplemented with 10 mM NaHCO3 (and 2 μg of streptomycin ml−1 and 2 μg of spectinomycin ml−1 for the mutants), and incubated under growth conditions for 2 or 3 days. Heterocysts of the wild type and the mutants were isolated essentially as described previously (28). Fractionation was performed as described previously (42, 43).
Cell wall fractions (60 μl) were washed by incubation with 8 M urea, 0.1 M sodium carbonate, or 1 M sodium chloride for 30 min on ice, followed by sedimentation of the membrane fraction by centrifugation at 256,000 × g and 4°C for 10 min. Pelleted membranes and the remaining supernatant were subjected to polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and immunodecorated by antibodies against the outer membrane protein Alr2269 (Omp85) or against GFP (rabbit anti-GFP; immunoglobulin G; Invitrogen, Oregon).
Secreted proteins were isolated from 1-liter cultures that had been grown in BG11 medium to a concentration of about 5 μg Chl ml−1 and incubated in the absence of combined nitrogen for 9 h. The protease inhibitor phenylmethylsulfonyl fluoride and a protease inhibitor cocktail (Complete; 1 tablet/500 ml; Roche, Mannheim, Germany) were added to the suspension, and the cells were pelleted by centrifugation for 5 min at 4,000 × g at room temperature. The supernatant was cleared by two rounds of centrifugation at 9,000 × g for 10 min at 4°C. Ammonium sulfate was added to the supernatant to a final concentration of 60% (wt/vol), and the resulting solution was incubated for 30 min. The precipitate was collected by centrifugation at 30,000 × g for 10 min at 4°C. The clear pellet was dialyzed overnight against 3 M urea-20 mM Tris (pH 7.4), and subsequently one-half of the sample was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) analysis.
Electron microscopy.
Preparation of the samples for transmission electron microscopy was performed as described previously (21, 22) except that Epon was used for embedding. In short, the procedure involved fixation with glutaraldehyde and KMnO4, dehydration with increasing concentrations of ethanol, and poststaining with uranyl acetate and lead citrate. The samples were examined with a Zeiss EM10C microscope at 80 kV.
Lipid analysis.
Lipid analysis was performed as previously described (21). In brief, lipids were extracted from filaments, isolated heterocysts, and cell wall fractions by addition of methanol-chloroform (1:2). The organic solvent was evaporated in a stream of nitrogen. Lipids were dissolved in 200 μl of chloroform and chromatographed on thin-layer plates of silica gel (Kieselgel 60 F254; Merck, Darmstadt, Germany) using a running medium containing 170 ml chloroform, 30 ml methanol, 20 ml acetic acid, and 7.4 ml distilled water. Lipids were visualized by sprinkling the plate with 25% sulfuric acid and exposing it to 220°C for 30 s.
Nitrogenase activity.
Anabaena sp. strains (Table 1) were grown photoautotrophically in a shaker in BG11 medium to a density of 3 to 5 μg Chl ml−1 and induced by incubation in BG110 medium for 48 h. Nitrogenase activity was determined by the acetylene reduction assay under light (150 microeinsteins m−2 s−1) in an atmosphere containing 14% acetylene in air or under anoxic conditions (24, 57). For anoxic conditions, 10 μM 3-(3,4-dichlorophenol)-1,1-dimethylurea was added to the cell suspension, and the flask containing the cells was sealed with a rubber stopper, bubbled with argon for 3 min, and incubated under culture conditions for 1 h before the assay was started by addition of acetylene. Ethylene production was found to be linear under these conditions for at least 1 h.
Homology modeling.
According to the PHYRE server (http://www.sbg.bio.ic.ac.uk/∼phyre/), TolC of E. coli (PDB:1EK9) (36) is the best template for homology modeling of Alr2887. The suggested alignment was manually adjusted, and a homology model was constructed with Modeler v8.2 using a multiple-template approach (23, 52). The situation with the length of the two extracellular loops in Alr2887 is the reverse of that in the crystal structure of TolC (1EK9). In order to avoid de novo modeling of long loops, we used the long loop of 1EK9 (amino acids 257 to 278) as a template for the long loop in Alr2887 (see Fig. S1 in the supplemental material). Coiled-coil prediction was performed by the REPPER server (30).
Phylogenetic analysis.
Sequences of TolC-like proteins were collected (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein) and filtered for a maximal sequence identity of 95% with CD-HIT (38), revealing 178 sequences. Sequences with uncertain annotation were controlled by InterProScan (65). A multiple alignment was produced with MAFFT v5.861 (35), and IQPNNI v3.0.1 (41, 58) was used to reconstruct a maximum likelihood phylogeny, assuming the WAG model (60) and constant rates across sites.
RESULTS
Expression of hgdD.
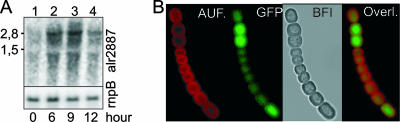
The hgdD (alr2887) gene is up-regulated during nitrogen step-down (13, 40). Transcripts about 2.8 to 3 kb long were detected with a labeled hgdD probe in RNA isolated after different nitrogen starvation times. A clear increase in the message after 6 and 9 h and a relative decline after 12 h were observed (Fig. 1A). Consistently, the activity of the hgdD promoter, tested with an N-terminal HgdD-GFP fusion (present in strain NME-alr2887-GFP), was highest in developing heterocysts, which were frequently found as doubles (Fig. 1B).
FIG. 1.
HgdD is up-regulated in proheterocysts upon nitrogen step-down. (A) RNA was isolated from NH4+-grown cells (lane 1) and from cells that had been starved for combined nitrogen for 6, 9, or 12 h (lanes 2 to 4). Samples contained 30 μg RNA. The blots were successively hybridized with 32P-labeled probes for hgdD (upper panel) and rnpB (lower panel), which were used as loading and transfer controls, respectively. The sizes of RNA standards are indicated on the left. (B) N-terminal HgdD-GFP translational fusion in strain NME-alr2887-GFP as visualized by confocal microscopy. The cyanobacterial autofluorescence (AUF.), the GFP fluorescence (GFP), the overlay of the two signals (Overl.), and a bright-field image (BFI) of the NME-alr2887-GFP strain 16 h after nitrogen step-down are shown.
HgdD is a TolC-like outer membrane protein.
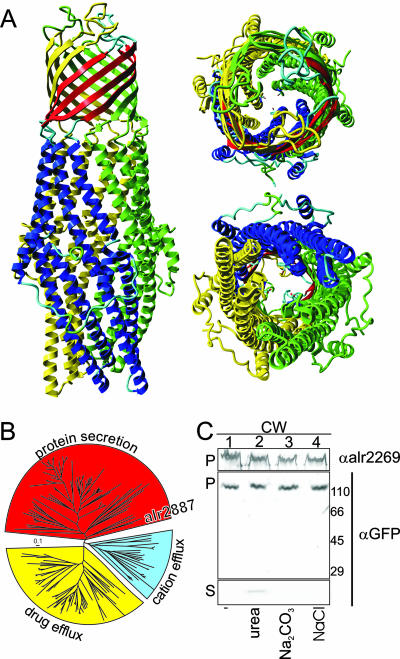
A relationship between HgdD (the Alr2887 protein) and proteins belonging to the TolC family can be proposed based on amino acid similarity (40, 42) or homology modeling (Fig. 2A). We identified HgdD as a TolC-like protein on the basis of the analysis performed by the PHYRE server. In accordance with TolC structures, a homotrimer is proposed, in which each monomer contributes four transmembrane β-strands to the channel. The trimeric structure of the channel assembled by HgdD corroborates the results obtained by blue native PAGE analysis (42). Following this prediction, a model for HgdD containing large α-helical regions, which extend the channel deep into the periplasm towards the cytoplasmic membrane, was created (Fig. 2A). Construction of a phylogenetic tree for TolC-like proteins (Fig. 2B) revealed that HgdD clustered with the group of the TolC-like proteins involved in secretion (3), especially members of the CyaE group secreting cyclolysin (27).
FIG. 2.
Sequence of HgdD is related to those of TolC proteins. (A) Model of HgdD generated as described in the text, showing structural homology to the outer membrane protein TolC. Side, top, and bottom views are shown. Further details are available in the supplemental material. The coordinates are available upon request. (B) Tree of 178 sequences of members of the TolC family (for details, see Fig. S2 in the supplemental material), representing a maximum likelihood phylogeny, assuming the WAG model and constant rates across sites. The tree is based on a multiple alignment. Bar = 0.1 amino acid substitution. The functional categories were assigned as described by Andersen et al. (3). (C) Isolated cell walls (CW) from lysed heterocysts of strain NMP-alr2887-GFP (lane 1) were treated with 8 M urea (lane 2), 0.1 M sodium carbonate (lane 3), or 1 M NaCl (lane 4). The pelleted membrane fraction (P) or the supernatant (S) was probed with antibodies against the outer membrane protein Alr2269 (αalr2269), which was used as a control, or GFP (αGFP). The migration positions of some molecular mass markers (in kDa) are indicated on the right for the pellet fraction probed with anti-GFP.
To test the proposed outer membrane localization (42, 43), we analyzed a C-terminal HgdD-GFP translational fusion that is expressed from the native hgdD promoter of the Anabaena sp. chromosome (present in strain NMP-alr2887-GFP). Using microscopic analysis, we did not observe localization at the periphery of the cells as expected for an outer membrane protein, perhaps because of a low fluorescence signal of the exported protein (not shown). However, using isolated cell wall fractions from NMP-alr2887-GFP, the outer membrane localization of HgdD-GFP was confirmed by immunodecoration with anti-GFP antibodies (Fig. 2C, lane 1). The antibodies did not recognize any protein in Anabaena sp. not containing a GFP tag (not shown). To confirm the membrane insertion of the fusion protein, the cell wall fraction was washed with 8 M urea (Fig. 2C, lane 2), 0.1 M sodium carbonate (lane 3), or 1 M sodium chloride (lane 4). The outer membrane protein Alr2269 (Omp85-like protein [7, 18]) was detected in the membrane pellet independent of the treatment used (Fig. 2C, lanes 1 to 4). The same was true for HgdD-GFP, which was detected only in minor amounts in the supernatant after incubation with 8 M urea. The migration of the HgdD-GFP fusion protein is consistent with its calculated size, about 110 kDa. Therefore, the HgdD-GFP fusion was present in the cell wall and HgdD appeared to be membrane inserted.
hgdD mutants do not form the glycolipid layer of heterocysts.
Having assigned HgdD to the TolC protein family, we aimed to explore the function of this protein. Two independent approaches for site-directed mutagenesis of hgdD were used. Strain DR181 resulted from partial deletion of the gene and insertion of cassette C.K3 into the gene by double recombination with the chromosome. In strain NMΔ-alr2887, the pCSV3 plasmid was inserted into the chromosome at the hgdD locus by single homologous recombination. None of these insertions compromised the expression of the downstream ORF alr2888 as determined by Northern blot analysis (not shown). These insertions resulted in a clear Fox− phenotype (not shown), which confirms the findings for an alr2887 transposon mutant (17, 40). However, one of the mutants, NMΔ-alr2887, was tested and found to develop nitrogenase activity when it was incubated under an anoxic atmosphere. Under oxic conditions, the NMΔ-alr2887 mutant showed a nitrogenase activity of about 0.07 nmol of ethylene μg Chl−1 h−1, compared to 5.32 nmol of ethylene μg Chl−1 h−1 for the wild-type Anabaena sp. strain. However, when the organisms were incubated in an anoxic atmosphere in the presence of 3-(3,4-dichlorophenol)-1,1-dimethylurea to inhibit oxygen-evolving photosynthesis, the activity of the mutant increased fivefold to about 0.34 nmol of ethylene μg Chl−1 h−1, while the wild-type produced 6.42 nmol of ethylene μg Chl−1 h−1. In prolonged anaerobic incubations in the presence of acetylene (1 to 3 days), the mutant released substantial amounts of ethylene that were close to the levels produced by the wild type. This suggests that new nitrogenase was synthesized and accumulated during the anaerobic incubation.
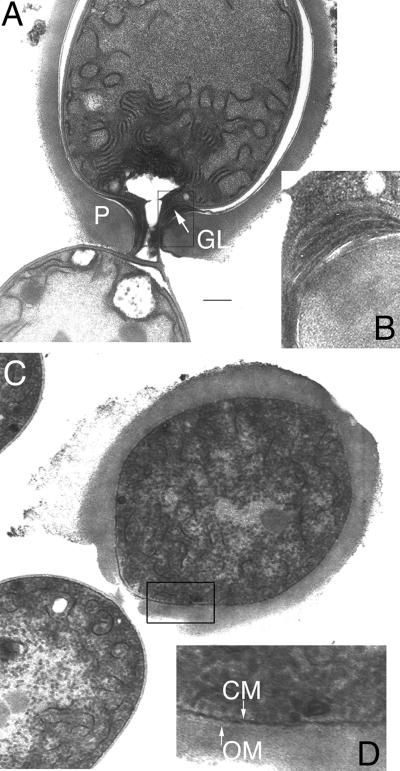
The morphology of heterocysts of the mutant strains grown without combined nitrogen for 2 days was analyzed with respect to the presence of heterocyst-specific envelope layers (Fig. 3). In ultrathin sections, the homogeneous layer was clearly visible and encompassed the enlarged immature heterocyst. The laminated layer, consisting of glycolipids, was deposited on top of the outer membrane in the wild type but could not be detected in any of the mutants (Fig. 3C and D; data not shown for strain NMΔ-alr2887). Rearrangement of the inner thylakoid membranes and formation of the so-called honeycomb membranes did not occur in the mutants. Instead, the membranes dissolved and had an irregular short confluent appearance. The number of glycogen granules increased substantially in both vegetative cells and heterocysts, which is an indication that combined nitrogen was depleted (16). Polar structures like those in the heterocysts of the wild type were not made. The lack of the HGL layer led to early arrest of heterocyst differentiation of the hgdD mutants similar to that observed in Hgl− mutant M7, which is defective in devA (21, 22).
FIG. 3.
Ultrastructure of heterocysts of hgdD mutants: transmission electron micrographs of ultrathin sections of a connection of a heterocyst and a vegetative cell of the wild-type Anabaena sp. strain (A and B) and mutant DR181 (C and D). Magnifications of the cell wall and heterocyst envelope, indicated by squares in panels A and C, are shown for the wild-type strain and DR181 in panels B and D, respectively. In panel A, the white space surrounding the cell wall resulted from dehydration of the protoplast during sample preparation. Bar = 1 μm for panels A and C. GL, laminated layer; P, homogeneous layer; CM, cytoplasmic membrane; OM, outer membrane.
HgdD functions in the formation of the laminated layer.
HgdD could act on the formation of the laminated layer by different mechanisms. It could be involved in export of the HGLs, lipid moieties, or enzymes needed for assembly of the constituents of the laminated layer. It could also influence the HGL synthesis, or the phenotype obtained could be pleiotropic because HgdD might be important for the biogenesis of the outer membrane per se. We did not find any evidence for the latter possibility. No significant changes in the outer membrane proteome occurred as analyzed by SDS-PAGE (not shown), and the whole-cell functions analyzed were not altered. For instance, we could not obtain a significant change in the uptake of amino acids like arginine, phenylalanine, glutamine, aspartic acid, or glutamic acid (49) in the deletion mutant NMΔ-alr2887 (not shown).
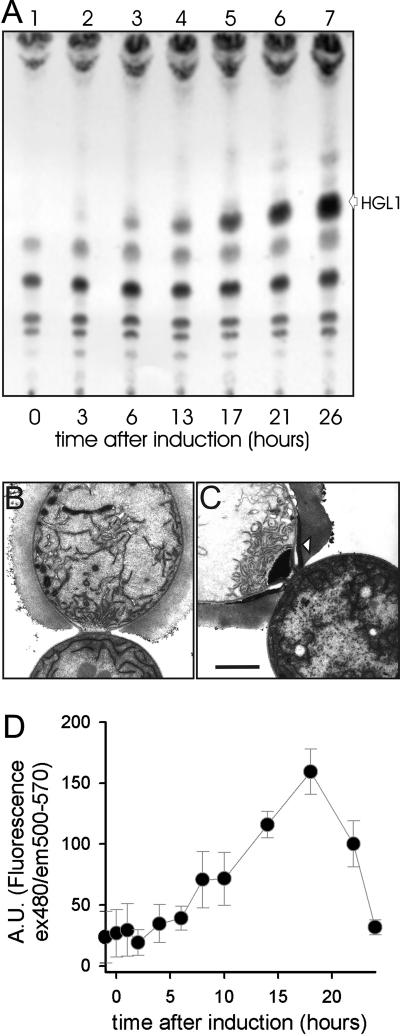
In wild-type filaments, heterocyst-specific glycolipids accumulated to a detectable level 6 h after induction of heterocyst formation, and the amount increased with time (Fig. 4A). The synthesis of HGLs preceded the formation of the laminated layer, because 14 h after nitrogen step-down this layer was not observed yet (Fig. 4B). The first evidence for the HGL layer in electron micrographs was found 17 h after nitrogen step-down (not shown), and the layer was fully developed at 20 h (Fig. 4C). Interestingly, rather than following the presence of HGLs in lipid extracts, the HGL layer appeared after the production of HgdD, as determined by the GFP intensity in N-terminal HgdD-GFP fusion strain NME-alr2887-GFP (Fig. 4D). This observation is consistent with the hypothesis that HgdD is involved in HGL layer formation rather than in HGL production.
FIG. 4.
Progress of HGL synthesis and HGL layer formation. (A) Thin-layer chromatography of lipids of the wild-type Anabaena sp. strain before (lane 1) or at indicated times after (lanes 2 to 7) transfer to BG110 medium. (B and C) Representative transmission electron micrographs of ultrathin sections of filaments of the wild-type Anabaena sp. strain 14 h (B) or 20 h (C) after transfer to BG110 medium. The arrowhead in panel C indicates the HGL layer after 20 h. Bar = 1 μm. (D) The fluorescence of the NME-alr2887-GFP strain was determined, and the difference from the wild-type strain background for three independent measurements and three independent NME-alr2887-GFP clones is shown for the indicated times after nitrogen step-down. A.U., arbitrary units; ex, excitation; em, emission.
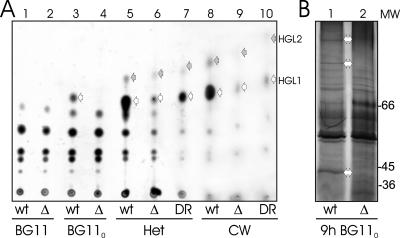
To further test this observation, we compared the lipid distributions of wild-type strain PCC 7120 and mutant strains DR181 and NMΔ-alr2887. All these strains produced the major heterocyst-specific glycolipid (heterocyst-specific glycolipid 1 [HGL1]) of Anabaena sp. strain PCC 7120 (25, 61) when they were incubated in combined nitrogen-free medium for 3 days (Fig. 5A), even though the amount of glycolipids in the mutant strains was reduced and glycolipids were detectable only in isolated heterocysts (Fig. 5A, compare lane 4 with lanes 6 and 7). Heterocyst-specific glycolipid 2 (HGL2) could also be detected in isolated heterocysts of the hgdD mutants (lanes 6 and 7). Because cell wall lipids are synthesized at the plasma membrane (12), the reduction in the level of glycolipids could be interpreted as a defect in synthesis or in correct deposition into the cell wall. Discriminating between these two possibilities can be done by analysis of the lipid content of isolated cell walls. If the mutation causes a defect in synthesis, the amounts of glycolipids detectable in heterocysts and cell walls should be comparable. If there is a defect in the deposition of glycolipids as a special envelope layer of the heterocysts, the lipids might accumulate in the plasma membrane after synthesis, but not in the cell wall. Isolated cell walls from heterocysts of the wild-type Anabaena sp. strain had a high HGL1 content and also produced the HGL2 spot, indicating that the laminated layer of the heterocyst envelope copurified with our cell wall preparations. Analysis of the lipid content of the cell walls isolated from heterocysts of the mutants showed that the level of glycolipids was drastically reduced in them (Fig. 5A, lanes 9 and 10). Therefore, the heterocyst-specific glycolipids were produced but did not appear in the cell wall and did not assemble as a laminated layer.
FIG. 5.
Heterocyst-specific glycolipids and secreted proteins in hgdD mutants. (A) Thin-layer chromatography of lipids of the wild-type Anabaena sp. strain (wt) (lanes 1, 3, 5, and 8), mutant NMΔ-alr2887 (Δ) (lanes 2, 4, 6, and 9), and mutant DR181 (DR) (lanes 7 and 10). Lipids were extracted from filaments grown in BG11 medium (lanes 1 and 2), from filaments 3 days after transfer to BG110 medium (lanes 3 and 4), from isolated heterocysts (Het) (lanes 5, 6, and 7), or from isolated heterocyst cell walls (CW) (lanes 8, 9, and 10). The lipids of the cell wall fractions show a slightly altered migration behavior due to edge effects of the thin-layer chromatography plate. The HGL lipids are indicated by arrows (open arrows, HGL1; gray arrows, HGL2). In lanes 9 and 10 the area where HGL2 would be expected is also indicated. (B) Proteins secreted from the wild type or NMΔ-alr2887 mutant 9 h after nitrogen step-down were concentrated and subjected to SDS-PAGE, followed by silver staining. The positions of molecular weight standards (MW) are indicated on the right, and proteins not secreted from the mutant are indicated by arrows.
Altered protein secretion.
The phylogenetic analysis suggested that HgdD is a protein-secreting TolC-like protein (Fig. 2). To test this possibility, we compared the distributions of the proteins secreted from the wild-type Anabaena sp. strain and mutant strain NMΔ-alr2887 after 9 h of nitrogen deprivation (Fig. 5B). At this stage of development, HGLs are synthesized but the laminated layer is not yet deposited (Fig. 4). At least three high-molecular-weight proteins, which were secreted from the wild type, were not observed in supernatants of the mutant (Fig. 5B). This observation, which was corroborated using samples from later time points (not shown), is consistent with the hypothesis that HgdD is involved in the secretion of specific proteins.
DISCUSSION
HgdD (Alr2887) was previously found in a proteomic analysis of cell walls from vegetative cells (42) and heterocysts (43) of Anabaena sp. strain PCC 7120, suggesting that this protein has a role in both types of cells. However, the expression of hgdD (alr2887) is up-regulated during nitrogen step-down (Fig. 1A and 4D) (13, 40), especially in proheterocysts (Fig. 1B), suggesting a further or specific function in heterocyst differentiation. It has been reported that transposition of Tn5-1063 into this gene abolishes diazotrophic growth (17, 40). Confirming the phenotype of the previously isolated transposon mutant, site-directed hgdD insertional mutants (strains DR181 and NMΔ-alr2887) had a Fox− phenotype with no growth on N2 and subsequent filament fragmentation (not shown).
In the Anabaena chromosome, the hgdD gene is part of a cluster of genes encoding proteins of unknown function (alr2887, alr2888, alr2889, and alr2890) that are located immediately downstream of the kaiA, kaiB, and kaiC genes (34). With an hgdD probe, an approximately 2.8- to 3-kb transcript is detected that can cover hgdD, its 5′ promoter region, and alr2888 (Fig. 1A). A transcript that was a similar size was detected with an alr2888 probe (results not shown). The latter probe also detected an approximately 1-kb transcript that can cover alr2888 and alr2889. Whereas the 2.8- to 3-kb transcript was not observed with RNA isolated from the DR181 and NMΔ-alr2887 mutants, the 1-kb transcript was present in these RNA preparations. Therefore, although alr2888 is cotranscribed with hgdD in the wild-type Anabaena sp. strain, hgdD-independent transcription of alr2888 that is not impaired in the mutants also takes place. This makes it unlikely that the Fox− phenotype of the hgdD mutants results from a polar effect of the insertions in hgdD, although it is still possible that Alr2888 and Alr2889 contribute to the same function as HgdD.
Growth on nitrate or ammonia is not affected in the hgdD mutants, suggesting that HgdD does not have an essential role in vegetative cells under standard laboratory conditions (not shown). An hgdD mutant exhibits a Fox− Fix+ phenotype, suggesting that this type of mutant is unable to provide the microoxic environment necessary for nitrogenase activity. This phenotype has also been described for mutants with an aberrant heterocyst envelope (17, 46) or mutants whose respiration is affected, like the cox2 cox3 double mutants defective in heterocyst-specific terminal respiratory oxidases (57). Even though the hgdD mutants show normal synthesis of the HGLs, they cannot form the laminated HGL layer (Fig. 3 and 5). This phenotype could be explained by a reduction in the incorporation of the HGLs into the heterocyst-specific envelope that is external to the gram-negative cell wall. The maximal expression of HgdD fits between two developmental stages, accomplished synthesis of the glycolipids and appearance of the laminated layer (Fig. 4). As predicted by protein modeling and phylogenetic clustering, HgdD belongs to the TolC family of outer membrane proteins (Fig. 2). TolC forms a trimeric 12-stranded β-barrel in the outer membrane and spans the periplasmic space by forming an α-barrel, and our results are consistent with a cell wall localization for HgdD (Fig. 2C). The TolC-like proteins are commonly involved in the export from the cells of gram-negative bacteria of various molecules, like drugs and proteins (37, 50, 54, 55), and HgdD specifically associates in a phylogenetic analysis with the protein secretion TolC-like proteins (Fig. 2B). We observed changes in the pool of secreted proteins of an hgdD mutant (Fig. 5), which is consistent with the hypothesis that HgdD belongs to the group containing the protein-secreting TolC-like proteins. The proteins that are not secreted in the hgdD mutant remain to be identified, and the possibility that they are not secreted because of an indirect effect of the mutation on development should also be explored.
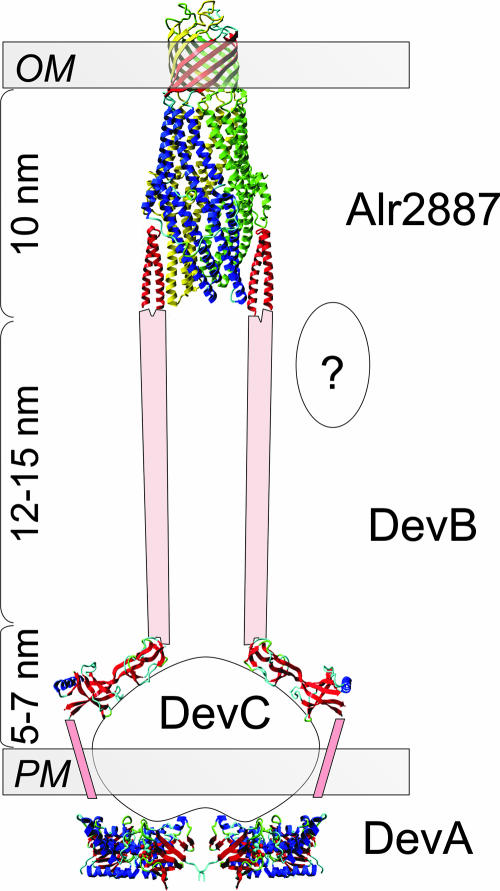
Outer membrane efflux proteins like TolC function together with a traffic ATPase, also known as an ABC transporter (37, 50, 54, 55). The phenotype of the hgdD mutants is remarkably similar to that of the mutants defective in the DevBCA traffic ATPase (21, 22). Mutants with mutations in devA (alr3712), devB (alr3710), or devC (alr3711) do not depose the laminated layer, even though HGL synthesis is not impaired in them. DevA represents an ATP binding cassette, and DevC represents a membrane channel of an ABC transporter. These proteins are similar to the plasma membrane-localized protein translocase HlyB (21, 22, 37, 39), which combines these two domains (see Fig. S1 in the supplemental material). Therefore, a 1:1 stoichiometry between DevC and DevA is expected (Fig. 6). DevC contains an additional domain (amino acid 40 to amino acid 254), which is similar to the periplasmic exposed region of AcrB from E. coli (amino acid 590 to amino acid 869) (11), which is not found in HlyB. On the basis of similarity, this domain could present a contact site for the periplasmic membrane fusion protein DevB. This protein shows similarity to HlyD (37) in the proposed N-terminal transmembrane domain, which is not present in AcrA, and its HlyB docking region (Fig. 6; see Fig. S1 in the supplemental material). One difference between DevB and HlyD is remarkable: DevB contains an extension of the coiled-coil domain with a periodicity of 18 as predicted by the REPPER server. According to the nomenclature of coiled-coil domains, this domain might be a hemagglutinin-like domain (29). The estimated length of this additional coiled-coil system is in the range from 12 to 15 nm, assuming it is a dimeric antiparallel coiled-coil domain (Fig. 6). Hence, DevB and HgdD together would be able to bridge a distance of about 30 nm, which corresponds to the estimated distance between the outer and plasma membranes of Anabaena sp. Therefore, HgdD is a good candidate for a component of a complex together with DevBCA, which bridges the two membranes in order to export molecules essential for formation of the laminated layer of the heterocyst envelope.
FIG. 6.
Proposed functional arrangement of HgdD (Alr2887). The structural composition of the HgdD/DevABC complex is shown, as discussed in the text. HgdD was modeled as shown in Fig. 2A, DevA was modeled using MJ0796, a bacterial ATP binding cassette (PDB:1L2T), as the template, and sections of DevB were modeled using MexA (PDB:1VF7) as the template. The pink boxes represent an extension of the two-helix coiled coil of DevB bridging the periplasm and attaching to HgdD (Alr2887). The ellipse with a question mark indicates that further elements stabilizing the complex might exist. OM, outer membrane; PM, cytoplasmic membrane.
Homologues of devB (hgdB, all5347) and devC (hgdC, all5346) that are involved in temporal and spatial aspects of HGL deposition have been described previously (19). However, the cell walls are different from the cell wall described here, since the laminated layer is found even though it is aberrantly distributed (19). HglK, a protein with an unknown function and putative membrane localization, is directly or indirectly involved in deposition of the HGLs (5). Mutants with mutations in hglK develop thylakoid distensions that could accumulate nonexported glycolipids. Therefore, the phenotypes of the hgdD mutants described here resemble the phenotypes of mutants with mutations in the DevBCA exporter rather than other hgl or hgd genes.
To summarize, the outer membrane-localized TolC-like protein HgdD may have a nonessential constitutive function, but it is essential during nitrogen starvation. The mutant phenotype and sequence homology support the hypothesis that HgdD has a TolC-like function in export of enzymes involved in the assembly of the laminated layer outside the gram-negative outer membrane in the heterocyst envelope. In addition to the three proteins observed to be absent in the secretome of the mutant (Fig. 5), All2736, a protein of unknown function, might also be a substrate for HgdD. The all2736 gene is induced like hgdD (13), and All2736 has previously been found to comigrate in blue native PAGE with HgdD (42).
Supplementary Material
Acknowledgments
We thank Serena Schmidt von Braun for help with fluorescence microscopy and Anna Scherzinger for some RNA preparations.
Financial support from the Deutsche Forschungsgemeinschaft (grant SFBTR1-C7) and the Volkswagenstiftung to E.S., from the Boehringer Ingelheim Fonds to S.M., from the Deutsche Forschungsgemeinschaft (grant Ma1359/2-3) to I.M., and from the Ministerio de Educación y Ciencia, Spain, to E.F. (grant BFU2005-07672) is acknowledged.
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, D. G., and P. S. Duggan. 1999. Heterocyst and akinete differentiation in cyanobacteria. New Phytol. 144:3-33. [Google Scholar]
- 2.Allen, M. B., and D. I. Arnon. 1955. Studies on nitrogen-fixing blue-green algae. I. Growth and nitrogen fixation by Anabaena cylindrica Lemm. Plant Physiol. 30:366-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen, C., C. Hughes, and V. Koronakis. 2000. Chunnel vision. Export and efflux through bacterial channel-tunnels. EMBO Rep. 1:313-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, C. C., K. S. Ramaswamy, S. Endley, L. A. Scappino, J. W. Golden, and R. Haselkorn. 1997. Suppression of heterocyst differentiation in Anabaena PCC 7120 by a cosmid carrying wild-type genes encoding enzymes for fatty acid synthesis. FEMS Microbiol. Lett. 151:23-30. [DOI] [PubMed] [Google Scholar]
- 5.Black, K., W. J. Buikema, and R. Haselkorn. 1995. The hglK gene is required for localization of heterocyst-specific glycolipids in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:6440-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black, T. A., Y. Cai, and C. P. Wolk. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77-84. [DOI] [PubMed] [Google Scholar]
- 7.Bredemeier, R., T. Schlegel, F. Ertel, A. Vojta, L. Borissenko, M. T. Bohnsack, M. Groll, A. von Haeseler, and E. Schleiff. 2007. Functional and phylogenetic properties of the pore-forming beta-barrel transporters of the Omp85 family. J. Biol. Chem. 282:1882-1890. [DOI] [PubMed] [Google Scholar]
- 8.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, E. L., M. F. Cohen, and J. C. Meeks. 1997. A polyketide-synthase-like gene is involved in the synthesis of heterocyst glycolipids in Nostoc punctiforme strain ATCC 29133. Arch. Microbiol. 167:251-258. [DOI] [PubMed] [Google Scholar]
- 10.Cardemil, L., and C. P. Wolk. 1976. The polysaccharides from heterocyst and spore envelopes of a blue-green alga. Methylation analysis and structure of the backbones. J. Biol. Chem. 251:2967-2975. [PubMed] [Google Scholar]
- 11.Das, D., Q. S. Xu, J. Y. Lee, I. Ankoudinova, C. Huang, Y. Lou, A. Degiovanni, R. Kim, and S. H. Kim. 2007. Crystal structure of the multidrug efflux transporter AcrB at 3.1Å resolution reveals the N-terminal region with conserved amino acids. J. Struct. Biol. 158:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doerrler, W. T. 2006. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol. Microbiol. 60:542-552. [DOI] [PubMed] [Google Scholar]
- 13.Ehira, S., and M. Ohmori. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 14.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119-138. [DOI] [PubMed] [Google Scholar]
- 15.Elhai, J., and C. P. Wolk. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747-754. [DOI] [PubMed] [Google Scholar]
- 16.Ernst, A., H. Kirschenlohr, J. Diez, and P. Böger. 1984. Glycogen content and nitrogenase activity in Anabaena variabilis. Arch. Microbiol. 140:120-125. [Google Scholar]
- 17.Ernst, A., T. Black, Y. Cai, J. M. Panoff, D. N. Tiwari, and C. P. Wolk. 1992. Synthesis of nitrogenase in mutants of the cyanobacterium Anabaena sp. strain PCC 7120 affected in heterocyst development or metabolism. J. Bacteriol. 174:6025-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ertel, F., O. Mirus, R. Bredemeier, S. Moslavac, T. Becker, and E. Schleiff. 2005. The evolutionarily related beta-barrel polypeptide transporters from Pisum sativum and Nostoc PCC7120 contain two distinct functional domains. J. Biol. Chem. 280:28281-28289. [DOI] [PubMed] [Google Scholar]
- 19.Fan, Q., G. Huang, S. Lechno-Yossef, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for synthesis and deposition of envelope glycolipids in Anabaena sp. strain PCC 7120. Mol. Microbiol. 58:227-243. [DOI] [PubMed] [Google Scholar]
- 20.Fay, P. 1992. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol. Rev. 56:340-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1998. The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27:1193-1202. [DOI] [PubMed] [Google Scholar]
- 22.Fiedler, G., M. Arnold, S. Hannus, and I. Maldener. 1999. An ABC exporter is essential for the localisation of envelope material in heterocysts of cyanobacteria, p. 529-537. In G. A. Peschek, W. Loeffelhardt, and G. Schmetterer (ed.), The phototrophic prokaryotes. Plenum Publishing Corporation, New York, NY.
- 23.Fiser, A., R. K. Do, and A. Sali. 2000. Modeling of loops in protein structures. Protein Sci. 9:1753-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flores, E., R. Pernil, A. M. Muro-Pastor, V. Mariscal, I. Maldener, S. Lechno-Yossef, Q. Fan, C. P. Wolk, and A. Herrero. 2007. Septum-localized protein required for filament integrity and diazotrophy in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:3884-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gambacorta, A., I. Romano, G. Sodano, and A. Trincone. 1998. Heterocyst glycolipids from nitrogen fixing cyanobacteria other than Nostocaceae. Phytochemistry 48:801-805. [Google Scholar]
- 26.Giddings, T. H., Jr., and L. A. Staehelin. 1979. Changes in thylakoid structure associated with the differentiation of heterocysts in the cyanobacterium, Anabaena cylindrica. Biochim. Biophys. Acta 546:373-382. [DOI] [PubMed] [Google Scholar]
- 27.Glaser, P., H. Sakamoto, J. Bellalou, A. Ullmann, and A. Danchin. 1988. Secretion of cyclolysin, the calmodulin-sensitive adenylate cyclase-haemolysin bifunctional protein of Bordetella pertussis. EMBO J. 7:3997-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden, J. W., L. L. Whorff, and D. R. Wiest. 1991. Independent regulation of nifHDK operon transcription and DNA rearrangement during heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 173:7098-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruber, M., and A. N. Lupas. 2003. Historical review: another 50th anniversary—new periodicities in coiled coils. Trends Biochem. Sci. 28:679-685. [DOI] [PubMed] [Google Scholar]
- 30.Gruber, M., J. Soding, and A. N. Lupas. 2005. REPPER—repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 33:W239-W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haury, J. F., and C. P. Wolk. 1978. Classes of Anabaena variabilis mutants with oxygen-sensitive nitrogenase activity. J. Bacteriol. 136:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hopwood, D. A., and D. H. Sherman. 1990. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu. Rev. Genet. 24:37-66. [DOI] [PubMed] [Google Scholar]
- 33.Huang, G., Q. Fan, S. Lechno-Yossef, E. Wojciuch, C. P. Wolk, T. Kaneko, and S. Tabata. 2005. Clustered genes required for the synthesis of heterocyst envelope polysaccharide in Anabaena sp. strain PCC 7120. J. Bacteriol. 187:1114-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213, 227-253. [DOI] [PubMed] [Google Scholar]
- 35.Katoh, K., K. Misawa, K. Kuma, and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 37.Koronakis, V., J. Eswaran, and C. Hughes. 2004. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 73:467-489. [DOI] [PubMed] [Google Scholar]
- 38.Li, W., and A. Godzik. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658-1659. [DOI] [PubMed] [Google Scholar]
- 39.Maldener, I., G. Fiedler, A. Ernst, F. Fernandez-Pinas, and C. P. Wolk. 1994. Characterization of devA, a gene required for the maturation of proheterocysts in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 176:7543-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maldener, I., S. Hannus, and M. Kammerer. 2003. Description of five mutants of the cyanobacterium Anabaena sp strain PCC 7120 affected in heterocyst differentiation and identification of the transposon-tagged genes. FEMS Microbiol. Lett. 224:205-213. [DOI] [PubMed] [Google Scholar]
- 41.Minh, B. Q., L. S. Vinh, A. von Haeseler, and H. A. Schmidt. 2005. pIQPNNI: parallel reconstruction of large maximum likelihood phylogenies. Bioinformatics 21:3794-3796. [DOI] [PubMed] [Google Scholar]
- 42.Moslavac, S., R. Bredemeier, O. Mirus, B. Granvogl, L. A. Eichacker, and E. Schleiff. 2005. Proteomic analysis of the outer membrane of Anabaena sp. strain PCC 7120. J. Proteome Res. 4:1330-1338. [DOI] [PubMed] [Google Scholar]
- 43.Moslavac, S., V. Reisinger, M. Berg, O. Mirus, O. Vosyka, M. Plöscher, E. Flores, L. A. Eichacker, and E. Schleiff. 2007. The proteome of the heterocyst cell wall in Anabaena sp. PCC 7120. Biol. Chem. 388:823-829. [DOI] [PubMed] [Google Scholar]
- 44.Muro-Pastor, A. M., A. Valladares, E. Flores, and A. Herrero. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377-1385. [DOI] [PubMed] [Google Scholar]
- 45.Muro-Pastor, A. M., E. Olmedo-Verd, and E. Flores. 2006. All4312, an NtcA-regulated two-component response regulator in Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 256:171-177. [DOI] [PubMed] [Google Scholar]
- 46.Murry, M. A., and C. P. Wolk. 1989. Evidence that the barrier to the penetration of oxygen into heterocysts depends upon two layers of the cell envelope. Arch. Microbiol. 151:469-474. [Google Scholar]
- 47.Olmedo-Verd, E., E. Flores, A. Herrero, and A. M. Muro-Pastor. 2005. HetR-dependent and -independent expression of heterocyst-related genes in an Anabaena strain overproducing the NtcA transcription factor. J. Bacteriol. 187:1985-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olmedo-Verd, E., A. M. Muro-Pastor, E. Flores, and A. Herrero. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picossi, S., M. L. Montesinos, R. Pernil, C. Lichtle, A. Herrero, and E. Flores. 2005. ABC-type neutral amino acid permease N-I is required for optimal diazotrophic growth and is repressed in the heterocysts of Anabaena sp. strain PCC 7120. Mol. Microbiol. 57:1582-1592. [DOI] [PubMed] [Google Scholar]
- 50.Poole, K. 2001. Multidrug resistance in Gram-negative bacteria. Curr. Opin. Microbiol. 4:500-508. [DOI] [PubMed] [Google Scholar]
- 51.Rippka, R., J. Dereules, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain stories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 52.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 53.Sambrook, J. F., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 54.Sharff, A., C. Fanutti, J. Shi, C. Calladine, and B. Luisi. 2001. The role of the TolC family in protein transport and multidrug efflux. From stereochemical certainty to mechanistic hypothesis. Eur. J. Biochem. 268:5011-5026. [DOI] [PubMed] [Google Scholar]
- 55.Thanassi, D. G., and S. J. Hultgren. 2000. Multiple pathways allow protein secretion across the bacterial outer membrane. Curr. Opin. Cell Biol. 12:420-430. [DOI] [PubMed] [Google Scholar]
- 56.Valladares, A., A. M. Muro-Pastor, M. F. Fillat, A. Herrero, and E. Flores. 1999. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 449:159-164. [DOI] [PubMed] [Google Scholar]
- 57.Valladares, A., A. Herrero, D. Pils, G. Schmetterer, and E. Flores. 2003. Cytochrome c oxidase genes required for nitrogenase activity and diazotrophic growth in Anabaena sp. PCC 7120. Mol. Microbiol. 47:1239-1249. [DOI] [PubMed] [Google Scholar]
- 58.Vinh, L. S., and A. von Haeseler. 2004. IQPNNI: moving fast through tree space and stopping in time. Mol. Biol. Evol. 21:1565-1571. [DOI] [PubMed] [Google Scholar]
- 59.Walsby, A. E. 1985. The permeability of heterocysts to the gases nitrogen and oxygen. Proc. R. Soc. Lond. B 226:345-366. [Google Scholar]
- 60.Whelan, S., and N. Goldman. 2001. A general model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691-699. [DOI] [PubMed] [Google Scholar]
- 61.Winkenbach, F., C. P. Wolk, and M. Jost. 1972. Lipids of membranes and in the cell envelope in heterocysts of a blue-green alga. Planta 107:69-80. [DOI] [PubMed] [Google Scholar]
- 62.Wolk, C. P., A. Vonshak, P. Kehoe, and J. Elhai. 1984. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc. Natl. Acad. Sci. USA 81:1561-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 64.Wolk, C. P. 2000. Heterocyst formation in Anabaena. ASM Press, Washington, DC.
- 65.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.