Abstract
In the cyanobacterium Synechococcus sp. strain PCC 7942, a circadian clock-related gene, pex, was identified as the gene prolonging the period of the clock. A PadR domain, which is a newly classified transcription factor domain, and the X-ray crystal structure of the Pex protein suggest a role for Pex in transcriptional regulation in the circadian system. However, the regulatory target of the Pex protein is unknown. To determine the role of Pex, we monitored bioluminescence rhythms that reported the expression activity of the kaiA gene or the kaiBC operon in pex deficiency, pex constitutive expression, and the wild-type genotype. The expression of kaiA in the pex-deficient or constitutive expression genotype was 7 or 1/7 times that of the wild type, respectively, suggesting that kaiA is the target of negative regulation by Pex. In contrast, the expression of the kaiBC gene in the two pex-related genotypes was the same as that in the wild type, suggesting that Pex specifically regulates kaiA expression. We used primer extension analysis to map the transcription start site for the kaiA gene 66 bp upstream of the translation start codon. Mapping with deletion and base pair substitution of the kaiA upstream region revealed that a 5-bp sequence in this region was essential for the regulation of kaiA. The repression or constitutive expression of the kaiA transgene caused the prolongation or shortening of the circadian period, respectively, suggesting that the Pex protein changes the period via the negative regulation of kaiA.
The circadian clock, which drives many biological phenomena with a period of about 24 h, including gene expression, has been observed in prokaryotic cyanobacteria and in eukaryotic cells (5). Cyanobacteria exhibit robust circadian rhythms related to many of their biological activities (4, 7, 15, 19, 27, 29, 30). Molecular genetic techniques, e.g., efficient transformation and gene targeting by precise homologous recombination, have been used for studying the cyanobacterial Synechococcus sp. strain PCC 7942 (3, 6, 24). By integrating molecular genetic properties and a monitoring system for the in vivo gene expression of bioluminescence, we previously isolated several types of clock mutants that exhibited altered bioluminescence rhythms, including short- or long-period rhythms and even arrhythmias (16). We then cloned the causative gene cluster, composed of kaiA and kaiBC, for all the circadian mutants and named it kaiABC (9, 16). Positive and negative transcriptional autoregulation by the Kai proteins with a period of about 24 h was found, as observed in eukaryotic circadian clocks, but the Kai proteins seem to function robustly to sustain the transcription rhythm of the kaiBC operon over that of the genome, rather than to maintain the period of the clock (11, 18, 20). However, little is known about the regulation of this gene cluster.
A circadian clock-related gene, pex (for period-extender) (17), was originally isolated as a genomic DNA region through which the C22a mutant harboring the kaiC1 mutation in the kaiC locus (circadian period of 22 h) was suppressed (9, 16, 17). Cells with constitutive expression of pex showed a period prolongation phenotype (28 h). In contrast, the lack of pex shortened the period by about 1 h, suggesting its physiological function in regulating the clock (17). Database analyses of the domain structure classified Pex as a PadR family (pfam03551) protein. PadR in Lactobacillus binds to a promoter of its target gene (padA) and regulates the metabolism of an environmental toxin (2, 8). The pex-deficient strain of Synechococcus showed an abnormal phase angle of the clock (the acrophase of the kaiBC gene expression rhythm was about 3 h earlier than that of the wild type) under diurnal light-dark cycles, and pex mRNA and Pex protein increased in the dark period (31). X-ray crystal analysis of the Pex protein in Synechococcus showed a winged-helix protein, a structure commonly found in DNA-binding transcription factors, such as the multiple antibiotic resistance repressor (MarR) family proteins (1, 22). In addition, Pex specifically bound to an upstream DNA fragment of kaiA in vitro (1).
Here, we demonstrate that kaiA expression is significantly more abundant in pex-deficient cells than in wild-type cells, suggesting that Pex functions to repress kaiA expression. The bioluminescence reporter for kaiA was also used to find cis elements for Pex in the upstream region of kaiA. A 5-bp element (AGAGA) downstream from the kaiA transcription start site was essential to the negative regulation of kaiA by Pex. We were able to reproduce the period alteration that occurs in pex-related mutants by the exogenous up- or down-regulation of kaiA expression.
MATERIALS AND METHODS
Bacterial strains, medium, and cultures.
We used wild-type Synechococcus sp. strain PCC 7942 and bioluminescence reporter strains for the psbAI, kaiA, and kaiBC genes (9, 15). In addition, the psbAI reporter strain lacking or constitutively expressing the pex gene (17) was used to obtain kaiAp::luxAB and kaiBCp::luxAB reporter strains harboring the pex-related genotype. A kaiBC reporter strain in which kaiA was inactivated by a nonsense mutation at the fourth codon (9) was used in a repression experiment for kaiA expression. The Synechococcus cultures for RNA analysis were grown in BG-11 liquid medium (26) under the standard light conditions previously described (17).
Monitoring of gene expression as bioluminescence.
We used an automated photon-counting apparatus (9) with a photomultiplier tube to monitor the expression of kaiA or kaiBC gene bioluminescence. Synechococcus cells were grown for 3 to 4 days on solid BG-11 agar in 40-mm plastic dishes under standard conditions to give 30 to 60 colonies (0.2 mm in diameter). After a 12-h dark treatment for entrainment of the clock, the dishes were placed in the photon-counting apparatus. The bioluminescence intensity was normalized to the number of colonies. A representative rhythm among three to six replicates was found for each reporter strain.
DNA sequencing and sequence analysis.
DNA sequencing was carried out using a Taq DyeDeoxy terminator cycle sequencing kit and a model 373A DNA sequencing system (Applied Biosystems, Foster City, CA).
Northern blotting.
Cultivation conditions and total RNA isolation were performed as described previously (9, 17). After sampling and RNA extraction, 5 μg of total RNA was subjected to electrophoresis on 1.0% agarose gel containing 1.0% formaldehyde, blotted onto positively charged nylon membranes, and hybridized with a digoxigenin (DIG)-labeled kaiA probe, as described previously (9, 17). Chemiluminescence images and the hybridization signal were obtained and quantified using a Fluor-S MultiImager (Bio-Rad, Hercules, CA).
Primer extension analysis.
A primer extension experiment was performed using standard procedures (17, 28). The oligonucleotide used as a primer was 5′-CCGTCGATTCCACCCAAATGC-3′, which corresponds to nucleotides +89 to +109 of the kaiA gene. To make sequencing ladders, we carried out a sequencing reaction using the same primer. The labeling efficiency of the primer with [γ-32P]ATP was >5,000 Ci/mmol.
Construction of plasmids for repression or induction of kaiA gene expression.
To make control cells for the repression experiment with the kaiA gene, we constructed a pTS2kaiA plasmid. We amplified a 1-kb DNA segment that contained the promoter and open reading frame of kaiA from the Synechococcus genome by using PCR primers with a BglII linker sequence, i.e., 5′-CGAGATCTAAACAACAGCCCTCTATCATCTCAG-3′ (−92 to −68 of the kaiA gene; the BglII restriction site is underlined) and 5′-GAAGATCTAACAGGATAAAGAG-3′ (+958 to +971; the BglII restriction site is underlined), and digested this with the BglII restriction enzyme. The BglII digest was inserted into a unique BamHI site in neutral site II (NSII) (GenBank/EMBL/DDBJ database accession no. U44761) in the targeting vector pTS2K (M. Ishiura and S. Kutsuna, unpublished data). The amplified 1-kb kaiA fragment was also inserted into a unique BamHI site downstream of the trc promoter (trcp) in another NSII targeting vector, pTS2Ktrcp. We then selected the plasmid inserted into the kaiA fragment inverting to trcp. The obtained plasmid, pTS2kaiA or pTS2kaiA::trcp, was used for the transformation of a kaiA-inactivated strain of the kaiBC reporter.
For the constitutive expression of kaiA, we constructed plasmid pTS2trcp::kaiA-GTG. A 0.9-kb section of the open reading frame of kaiA was amplified using the primer sequences 5′-ATAGATCTTAAGACTCAGTCCTGACAGGAGCGACTGCG-3′ (+41 to +67; the BglII restriction site is underlined) and 5′-GAAGATCTAACAGGATAAAGAG-3′ (+958 to +971; the BglII restriction site is underlined) and digested with BglII. The digest was inserted into the BamHI site downstream of trcp in the pTS2Ktrcp plasmid. The obtained plasmid was introduced into a wild-type kaiBC reporter strain.
Deletion mapping of the negative element in the kaiA upstream region.
From the 0.8-kb SmaI-XhoI segment, which carried the promoter of kaiA, we constructed a series of deletion derivatives carrying upstream regions of various lengths (Table 1). To make a deletion derivative (nucleotides −92 to +402), we used a DraI-XhoI restriction fragment of the kaiA region. Appropriate segments for the other derivatives were synthesized by PCR using the following oligonucleotides: upper primers 5′-GAAGGCCTAACTTTTGAGAACTGT-3′ (−66 to −51 of kaiA; the StuI restriction site is underlined here and in the following sequences), 5′-GAAGGCCTGTGGACAAAGCGATC-3′ (−44 to −30), 5′-GAAGGCCTTGAGCTGCAGTGCTA-3′ (−20 to −6), 5′-GAAGGCCTAATTTTTCCTTTGTCC-3′ (+6 to +21), and 5′-GAAGGCCTATCTGTCTGCAGACT-3′ (+30 to +44) and the lower primer (5′-GTGGTTGGCCCCCATCAGCAT-3′ (+481 to +501), which corresponded to a sequence downstream of the XhoI site in the kaiA gene. The PCR products were digested with StuI and XhoI. The digests were inserted between the unique sites EcoRV and XhoI upstream of luxAB in the pTS2Slux plasmid (17). Oligonucleotides 5′-CGGGAGCTCTACAGTAATCGACTCC-3′, which corresponded to a sequence upstream of the SmaI site, and 5′-AAACGCTCGAGACGCAGTCGCTCCTGT-3′ (+53 to +68; the XhoI site is underlined) were used in the PCR. The amplified DNA was digested with SmaI and XhoI and inserted between the unique sites in pTS2Slux. By PCR using the SK primer as the upper one (5′-TCTAGAACTAGTGGATC-3′ [Toyobo, Osaka, Japan]), which corresponded to the plasmid sequence upstream of the SmaI site of the kaiA promoter region fused to luxAB in the kaiA expression reporter construct with the genomic segment NSII, and using six primers as lower ones (5′-CAGGCCTGACTGAGTCTGCAGAC-3′ [+34 to +49], 5′-CAGGCCTGACTGAGTCTGCAGAC-3′, 5′-TAGGCCTAGATTAATCTCTGGAC-3′ [+18 to +33], 5′-TAGGCCTCTGGACAAAGGAAAAA-3′ [+8 to +23], 5′-CAGGCCTAAAATTTAATTTAGCC-3′ [−5 to +11], and 5′-AAGGCCTGCTCATGAGGCCGCG-3′ [−30 to −16]; StuI sites are underlined), the downstream deletion fragments of the kaiA upstream region with a vector portion were amplified from the kaiA expression reporter construct for NSII. These products were digested with BamHI and StuI and inserted between the unique sites BamHI and EcoRV in pTS2Slux. The deletion of the kaiA upstream region was checked by sequencing with the M13 reverse primer (Toyobo, Osaka, Japan). We introduced the obtained derivatives into wild-type Synechococcus and measured the bioluminescence quantitatively after the selection of the transformed cells with spectinomycin (40 μg ml−1), as described previously (9).
TABLE 1.
Relative bioluminescence of deletion derivatives of kaiA reportersa
| Gene or reporter | Upstream region | % Bioluminescence ± SD | No. of samples |
|---|---|---|---|
| kaiA | −457 to +402 | 100 ± 9 | 3 |
| D1 | −92 to +402 | 28 ± 5 | 4 |
| D2 | −66 to +402 | 45 ± 6 | 4 |
| D3 | −44 to +402 | 53 ± 3 | 4 |
| D4 | −20 to +402 | 1 | 3 |
| D5 | +6 to +402 | 1 | 3 |
| D6 | +25 to +402 | 2 | 3 |
| D7 | −457 to +68 | 59 ± 8 | 4 |
| D8 | −457 to +49 | 68 ± 8 | 4 |
| D9 | −457 to +33 | 102 ± 21 | 4 |
| D10 | −457 to +23 | 385 ± 25 | 4 |
| D11 | −457 to +11 | 441 ± 18 | 4 |
| D12 | −457 to −16 | 11 ± 1 | 4 |
The reporters were recombined in the NSII genomic region.
For the preparation of deletion derivatives of the kaiA reporter in the targeting site NSI, we inserted the deletion fragments of the kaiA upstream region into the upstream region of luxAB in pTS1CluxΔ. The obtained plasmids were targeted into NSI, which has psbAp::luxAB reporter DNA in the genome of the bioluminescence reporter strain AMC149 (15). The selection of the transformation was performed on BG-11 agar with a chloramphenicol concentration of 7.5 μg ml−1.
Mutation analysis of the negative element in the promoter region of kaiA.
We used PCR to line up mutated D9 reporters with base pair substitutions. The SK primer (Toyobo), which corresponds to the upstream part of kaiA in the plasmid pTS1CluxΔ, and mutant primers with the StuI linker were used to amplify the mutation fragments, as performed for the upper primer. The sequences of the lower mutation primers were as follows: for M1, 5′-TAGGCCTAGATTAATCTCTCCTCAAAGGAAAAATTTAATT-3′; for M2, 5′-TAGGCCTAGTAATTTCTCTGGACAAAGGAAAAA-3′; and for M3, (5′-TAGGCCTAGATTAATCACAGGACAAAGGAAAAAT-3′ (mutations are underlined). The PCR products were digested with BamHI and StuI and then cloned into the BamHI-SmaI multicloning sequence of the plasmid.
RESULTS
Pex as a negative regulator of a circadian clock gene, kaiA.
Previously, using a bioluminescence reporter for the photosynthesis gene psbAI, we determined that the free-running period of the cells of pex-deficient (Δpex) or pex constitutive expression (trcp::pex, with the inducer 1 mM isopropyl-β-d-thiogalactopyranoside [IPTG]) genotype was about 1 h shorter or about 3 h longer, respectively, than that of the wild-type pex locus (pex+) (17), suggesting that changes in pex expression may affect certain aspects of the clock gene cluster kaiABC. To find this type of abnormality in the period mutants, we also used the bioluminescence reporter genes (luxAB). Approximately 0.8-kb upstream regions of kaiA, including its open reading frame (∼0.3 kb), or kaiBC (∼0.9 kb) were fused to luxAB. The obtained constructs were introduced into the cells with Δpex or trcp::pex. We then examined the cells obtained for bioluminescence. Compared with bioluminescence rhythms in pex+, we confirmed that the timing of the rhythms in Δpex and trcp::pex was advanced or delayed in every cycle, leading to shortened or lengthened circadian periods, respectively (Fig. 1A). The effects on the period were the same as that of a clock-regulated bioluminescence reporter for the psbAI gene (17). The bioluminescence level for kaiA expression in Δpex or trcp::pex was 7 times (mean relative level ± standard deviations = 731.6% ± 37.8%; n = 6) or <1/7 times (15.3% ± 2.7%; n = 5), respectively, that in pex+ (100 ± 9.2%; n = 6) over the time course of the rhythm (Fig. 1A). However, the bioluminescence of the kaiBC expression level was the same in the three genotypes (pex+, 157.8% ± 8.5%, n = 6; Δpex, 138.4% ± 10.5%, n = 6; trcp::pex, 162.2% ± 8.5%, n = 4) (Fig. 1A). These results suggest that the period mutants have abnormal accumulations of kaiA mRNA.
FIG. 1.
Expression of kaiABC in the circadian period mutants. The expression of kaiA and kaiBC was analyzed in strains carrying the pex+, Δpex, and trcp::pex genotypes. (A) Panels represent the bioluminescence rhythms of the expression activity of the upstream region of kaiA or kaiBC (kaiA::luxAB or kaiBC::luxAB, respectively). The ordinate shows the percentage of relative bioluminescence; the abscissa shows the hours in continuous light after 12 h of darkness. The reporter gene fusions were recombined in the genomic region NSI. To activate trcp::pex gene fusion in a trcp::pex strain, we used the inducer 1 mM IPTG. (B) The upper panel shows results of Northern blotting of kaiA mRNA in the three genotypes after 6 h in constant light (kaiA mRNA) and used total RNA (rRNA). Five micrograms of total RNA was applied. The fluorescence image of the gel stained with ethidium bromide confirmed that an equal amount of the total RNA existed in each sample by referring the 23S and 16S ribosomal RNAs among the genotypes. For the lower panel, the relative level was calculated by comparing the amount of kaiA mRNA in each genotype to the total value of the three genotypes. The mean ± standard error (error bars) (n = 3) of the calculated relative level of each genotype is shown in the graph. kaiA mRNA was detected with a DIG-labeled kaiA probe by using PCR-based DIG-dUTP (DIG DNA labeling mix; Roche Diagnostics, Mannheim, Germany) incorporated into the PCR product of the kaiA coding region. Synechococcus cultures were grown under standard light conditions until the optical density at 730 nm reached 0.2. Three independent sets of experiments were conducted. LL, continuous light condition of 50 μmol m−2 s−1.
Northern blotting confirmed the level of kaiA mRNA accumulation in the three genotypes of the cells (Fig. 1B, upper panel). The cells of pex+, Δpex, and trcp::pex were examined under standard light conditions following 12 h of darkness to reset the clock. After 6 h in continuous light, we collected the cultures and examined kaiA mRNA. The accumulation of kaiA mRNA in Δpex or trcp::pex cells was significantly (about two times) higher or lower, respectively, than that in pex+ (Fig. 1B, lower panel). Thus, the deficiency or constitutive expression of pex caused an increase or decrease in the accumulation of kaiA mRNA, respectively, suggesting that Pex negatively regulates kaiA expression through the 0.8-kb kaiA upstream region.
Transcription start site of kaiA.
We analyzed the 5′ terminus of kaiA mRNA by using the primer extension method (Fig. 2). There were three 5′ termini, at nucleotides 64, 65, and 66 upstream of the translation initiation codon of kaiA. We mapped the 5′ terminus of kaiA mRNA at nucleotide 66, the potentially stable one of kaiA. Thus, it is probably the start site for the transcription of the kaiA gene. Hereafter, the position of nucleotide 66 is referred to as +1.
FIG. 2.
Transcription start site of kaiA determined using the primer extension method. Thirty micrograms of total RNA was hybridized to a primer with the 5′ end labeled with γ-32P and reverse transcribed with reverse transcriptase. The product was analyzed on a sequencing gel (lane P). A sequencing ladder (lanes G, A, T, and C) was obtained by sequencing reactions in which the same primer was used as a sequencing primer. The arrowhead indicates the 5′ termini of kaiA mRNA; the arrow indicates the direction of transcription. To clear the bands obtained in lane P, the area is shown at a lower threshold than the sequence ladder.
Deletion analysis of the upstream region of the kaiA gene.
To find the postulated regulatory element(s) in the kaiA upstream region, we constructed deletion derivatives of the kaiA reporter and introduced them into wild-type cells. These reporter genes were recombined into a genome region, NSII. We then compared the bioluminescence rhythms of the colonies obtained. The peak bioluminescence on the second day was applied to calculate the expression activity. The deletions from −457 to −44 (reporters D1, D2, and D3) showed expression activity (Table 1), but further deletions (reporters D4, D5, and D6) diminished the activity. Therefore, sequences in the region of −44 and lower are essential to the promoter per se, consistent with the putative transcription start site. In addition, the −457 to −44 region might function to positively regulate the promoter. Although D1, D2, and D3 reporter strains showed expression activity, the deletion of a downstream region (+49 to +402 [D7 and D8]) decreased the activity. In contrast, the deletions from +11 to +49 (D9, D10, and D11) tended to increase the activity, whereas further deletions (+11 to −16 [D12]) decreased the activity significantly. This result implies that a negative regulatory element(s) occurs in the DNA (+11 to +33). Therefore, the upper region (−457 to −44) and lower region (+11 to +49) were roughly mapped as positive and negative regulatory regions, respectively. We deduced that the D3-D11 overlapping regions (−44 to +11) are the minimum promoter regions.
Analysis of the negative regulatory element upstream of kaiA using pex-deficient mutants.
We examined whether the predicted negative regulatory element was related to the postulated function of Pex. We measured the bioluminescence of the deletion derivatives in pex+ or Δpex cells (Table 2). Compared to pex+, the expression activities of D3, D8, and D9 in Δpex cells were 2.6, 3.6, and 4.7 times that of their expression in pex+ cells. The activities of further deletions, i.e., D10 and D11, in Δpex cells were the same as in pex+ cells. Thus, the reporters D3, D8, and D9 had an element negatively regulated by Pex, but others did not. In addition, the difference between D9 and D10 (i.e., the 10 bp +23 to +33) is essential to regulation.
TABLE 2.
Relative bioluminescence of kaiA reporters in pex+ and Δpexa
| Gene or reporter | Relative bioluminescence forb
|
% pex+ | Position | |
|---|---|---|---|---|
| pex+ | Δpex | |||
| kaiA | 100 ± 3 | 469 ± 43 | 469 | −457-+402 |
| D3 | 40 ± 4 | 103 ± 6 | 258 | −44-+402 |
| D8 | 51 ± 9 | 183 ± 17 | 359 | −457-+49 |
| D9 | 74 ± 9 | 349 ± 17 | 471 | −457-+33 |
| D10 | 314 ± 49 | 346 ± 46 | 110 | −457-+23 |
| D11 | 297 ± 17 | 297 ± 9 | 100 | −457-+11 |
The reporters were recombined in the NSI genomic region.
Values are the means ± standard deviations of bioluminescence (n = 4).
Mapping the sequence for pex-related negative regulation.
We further scanned regions of kaiA to find the putative cis-element for Pex. We made base pair substitutions in segment D9 fused to the luxAB reporter gene (Fig. 3A) and examined the obtained reporter cells of the derivatives. Derivatives M1 and M2 exhibited the same expression activity as did the original, D9 (Fig. 3A), suggesting that the two substitutions apparently have no effect on the postulated negative regulation. In contrast, the activity in the reporter of M3 was three times that of D9 but the same as that of D10 (Table 2).
FIG. 3.
Mutation analysis of the kaiA promoter region fused to luxAB. (A) The kaiA reporter base pair substitution and relative bioluminescence. The reporter gene functioned at genomic site NSI. Boxes with −457 and luxAB represent the D9-type kaiA reporter. A series of mutations of the pex-related regulatory region in the kaiA reporter are superimposed. The transcription start site of kaiA is shown as +1, with an arrow. The mutations of segments are boxed. The peak in the mean relative bioluminescence, indicating the expression activity of kaiA, was on the second day (± standard deviation; n > 3). (B) Representative bioluminescence rhythms of the base-pair-substituted D9 reporters in pex+ and Δpex cells. (C) Means ± standard deviations (n = 3) relative bioluminescence of each reporter in pex+ and Δpex cells. Error bars indicate standard deviations. Filled bars, pex+; open bars, Δpex. LL, see the legend to Fig. 1.
The M1 to M3 reporters were assayed in Δpex cells, and the bioluminescence in these mutant cells was greater than three times that of pex cells. However, the M3 reporter bioluminescence levels in cells of pex+ and Δpex were similar (Fig. 3B). Therefore, the 5 bp between M1 and M2, i.e., AGAGA, likely constitutes the negative element.
Effects of repression or induction of the kaiA gene.
Since our molecular genetic approach suggested that the circadian period was in inverse proportion with the expression level of kaiA, we hypothesized that the period could respond to kaiA gene expression. To evaluate this hypothesis, we examined the period of the oscillator in cells in which kaiA gene expression was interrupted or constitutively expressed. To regulate the expression level of the gene, we made inducible constructs for the suppression or induction of kaiA gene expression in the genome. The construct was recombined into a specific site of the genome, NSII, in which DNA insertion per se did not affect the fundamental cell activity or circadian rhythm. And at NSII, the inducible promoter trc used here had functioned in Synechococcus (6, 9, 17). To control the down-regulation of kaiA expression, we used the transcription-translation interference technique, using antisense transcription in Synechococcus (25). First, we confirmed the complementation activity of kaiA DNA in NSII using a kaiA-inactivated arrhythmic mutant (Fig. 4B). The mutant was transformed with an NSII-targeting plasmid harboring a native kaiA gene (Fig. 4C), and the obtained transformed cell exhibited circadian rhythm similar to that in the control reporter cells (Fig. 4A). Thus, kaiA could function in NSII. Based on kaiA activity in NSII, we examined the effect of interference of the kaiA transgene expression by using an inverted trc promoter at the 3′ side of the gene (kaiA::trcp). We then introduced the plasmid pTS2kaiA::trcp into a kaiA-inactivated reporter cell. The obtained cell, with the addition of 10 mM IPTG, had a period of circadian rhythm extended to 27 h (Fig. 4D), suggesting that antisense transcription changed the period because it decreased kaiA transcription or translation.
FIG. 4.
Effect of repression of kaiA gene expression in the NSII genome region. The genomic regions of kaiABC and NSII in the kaiBC reporter strains are depicted in the left panels. The right panels show reporter bioluminescence profiles. Boxes show the functional kai gene on the genome. ΔA, the kaiA gene with nonsense mutation; NSII, a targeting site for kaiA; arrow, direction of gene transcription. “Period” is the mean circadian period ± standard deviation (n = 6). Panels A through D represent the kai genes and the rhythms of the reporter strains of kaiABC+ (A), the inactivated kaiA gene (Β), the inactivated kaiA and kaiA+ genes (C), and kaiA::trcp (D). The trcp is located downstream of the recombined kaiA in an inverted direction. Ten-millimolar IPTG inducer was used for antisense transcription from trcp. LL, see the legend to Fig. 1.
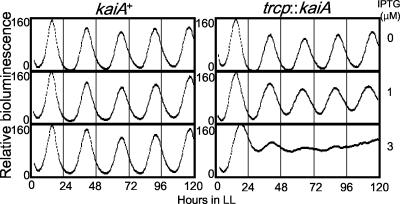
We also examined the effect of constitutive expression of kaiA by making a reporter strain harboring a trcp::kaiA fusion gene, through which kaiA mRNA was produced (Fig. 5). Although cells with trcp::kaiA exhibited a rhythm period (24.9 h) similar to that of the wild type (25.2 h) on an agar plate without IPTG, it exhibited a 1-h shorter rhythm period (24.2 h) than that of kaiA+ (25.2 h), consistent with the period in the Δpex genotype. The addition of 3 μM IPTG resulted in a lowered amplitude of the rhythm. However, the peaks of the rhythms with low amplitude clearly advanced every cycle because of their shortened period. Therefore, the constitutive expression of kaiA shortens the period of the circadian rhythm in a dose-dependent manner. Our results are consistent with the abnormal level of kaiA expression observed in pex genotype-related period mutants.
FIG. 5.
Effect of the induction of kaiA in the NSII genome region. Bioluminescence rhythm of a kaiBC expression reporter in kaiA+ and trcp::kaiA genotypes with or without the inducer IPTG. Without IPTG, the reporter cells of trcp::kaiA showed a rhythm similar to that of kaiA+. The mean period was 24.9 ± 0.1 h (± standard deviation) (n = 6). The addition of 1 μM IPTG to induce the transcription of trcp::kaiA in NSII resulted in a further short-period phenotype (24.2 ± 0.2 h; n = 6). The mean period of the rhythm in kaiA+ with 0 to 3 μM IPTG was 25.2 ± 0.1 h (n = 6). LL, see the legend to Fig. 1.
DISCUSSION
In the Synechococcus circadian oscillator, the clock protein KaiC exhibits phosphorylated or dephosphorylated forms in a circadian manner under conditions of no transcription-translation (32). This cycle requires physical interaction(s) with KaiA and KaiB (12, 14); in vitro, the three proteins plus ATP establish a reaction of KaiC with a period of about 24 h (21). In this reaction, KaiA promotes phosphorylation (11), suggesting that KaiA has a role in regulating the circadian period within a short range. Consistent with this explanation, most of the period mutants harboring a mutation in the kaiA locus exhibited longer periods of the rhythm than did the wild-type strain (23).
We found stronger kaiA expression and more significant accumulation of kaiA mRNA in the Δpex strain than in the pex+ strain (Fig. 1). This result suggests that the short-period phenotype in the Δpex strain is caused by an increase in kaiA expression and the acceleration of phosphorylation of the KaiC protein. Further analysis to quantify kaiA expression levels should be carried out to confirm this. A short period in the strain constitutively expressing the kaiA transgene (trcp::kaiA), using a trc promoter at an intermediate concentration of the inducer IPTG (Fig. 5), provided further support for this conclusion. Another circadian resetting-related mutant, ldpA (light-dependent period) (10, 13), named after one of its mutant phenotypes for the loss of the period response to light intensity in free-running conditions, showed the same circadian period as that in Δpex cells. KaiA is more abundant in the ldpA mutant than in the wild type, but ldpA is a different gene from pex and encodes a protein with Fe4S4 motifs that sense the cellular redox state (10). Thus, deficiency in the pex or ldpA gene causes similar abnormalities in kaiA expression and the period of the rhythm.
Our primer extension and in vivo bioluminescence reporter analyses indicated the transcription start site and the negative regulatory region upstream the kaiA gene. Within this region, a 5-bp sequence (AGAGA) (Fig. 3A) was essential for negative regulation by Pex. Together with our previous in vitro data, in which Pex specifically bound to the double-strand DNA of the kaiA regulatory region (1), it is plausible that Pex directly binds to this region and functions as a repressor of the kaiA gene in vivo.
Using the cyanobacterial genome database Cyanobase (Kazusa DNA Research Institute, Chiba, Japan), we estimated the 5-bp essential sequence of kaiA in other cyanobacterial species whose genomes have pex orthologs to understand the conservation and significance of the regulation. A nitrogen-fixing multicellular cyanobacterium (Anabaena sp. strain PCC 7120), a thermophilic cyanobacterium (Thermosynechococcus elongatus BP-1), and a marine cyanobacterium (Synechococcus sp. strain WH8102) have a 5-bp AGACA motif at 73, 64, and 67 bp, respectively, upstream of each kaiA translation initiation codon, whereas Synechococcus sp. strain PCC 7942 had the 5-bp motif 41 bp upstream of the gene. Therefore, the 5-bp motif and adjacent region of kaiA in the three cyanobacteria might be the binding site of Pex for negative regulation of the gene.
The clock of the Δpex strain subjected to diurnal light-dark cycles shows a phase advance of about 3 h compared to that of the pex+ strain (31). Thus, Pex affects a specific step in the oscillator. We assume that the negative regulation of kaiA by Pex is related to the delay function in the oscillator under a light-dark cycle. If this assumption is correct, kaiA expression should decrease in light-dark conditions.
X-ray diffraction analysis of the Pex crystal structure showed that it is a winged-helix dimer protein (1). A representative winged-helix repressor in Synechococcus, SmtB, derepresses the transcription of the smtA gene in response to the heavy metal cadmium, and the dimer binds to several sites in the smtA promoter region. Therefore, it will be important to demonstrate the existence of the Pex dimer in vivo.
In summary, Pex was required for the negative regulation of kaiA and the circadian period was dependent on the kaiA expression level. These in vivo results and the demonstrated in vitro binding between Pex and upstream DNA of kaiA (1) demonstrate that Pex is a direct kaiA regulator in cyanobacteria that maintains the circadian period.
Acknowledgments
We thank colleagues in our laboratories at Nagoya University and Yokohama City University; in particular, we thank Kazuhisa Okamoto (Nagoya University) for technical assistance with the bioluminescence measurements at Yokohama and Chiaki Inouye (Nagoya University) for the construction of the deletion mapping of the kaiA upstream region.
This research was supported by grants to S.K. from a research fellowship of the Japan Society for the Promotion of Science and in part by a Japan Society for the Promotion of Science Grant-in-Aid for Encouraging Young Scientists (no. 30315824).
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Arita, K., H. Hashimoto, K. Igari, M. Akaboshi, S. Kutsuna, M. Sato, and T. Shimizu. 2007. Structural and biochemical characterization of a cyanobacterium circadian clock-modifier protein. J. Biol. Chem. 282:1128-1135. [DOI] [PubMed] [Google Scholar]
- 2.Barthelmebs, L., B. Lecomte, C. Divies, and J. F. Cavin. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bustos, S. A., and S. S. Golden. 1991. Expression of the psbDII gene in Synechococcus sp. strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol. 173:7525-7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, T., T.-L. Chen, L.-M. Hung, and T. C. Hung. 1991. Circadian rhythm in amino acid uptake by Synechococcus sp. RF-1. Plant Physiol. 97:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlap, J. C., J. J. Loros, and P. J. DeCoursey (ed.). 2004. Chronobiology: biological timekeeping. Sinauer, Sunderland, MA.
- 6.Geerts, D., A. Bovy, G. D. Vrieze, M. Borrias, and P. Weisbeek. 1995. Inducible expression of heterologous genes targeted to a chromosomal platform in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology 141:831-841. [DOI] [PubMed] [Google Scholar]
- 7.Grobblelarr, N., T. C. Huang, H. Y. Lin, and T. J. Chow. 1986. Dinitrogen-fixing endogenous rhythm in Synechococcus RF-1. FEMS Microbiol. Lett. 37:173-177. [Google Scholar]
- 8.Gury, J., L. Barthelmebs, N. P. Tran, C. Divies, and J. F. Cavin. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 70:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishiura, M., S. Kutsuna, S. Aoki, H. Iwasaki, C. R. Andersson, A. Tanabe, S. S. Golden, C. H. Johnson, and T. Kondo. 1998. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281:1519-1523. [DOI] [PubMed] [Google Scholar]
- 10.Ivleva, N. B., M. R. Bramlett, P. A. Lindahl, and S. S. Golden. 2005. LdpA: a component of the circadian clock senses redox state of the cell. EMBO J. 24:1202-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki, H., T. Nishiwaki, Y. Kitayama, M. Nakajima, and T. Kondo. 2002. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA 99:15788-15793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kageyama, H., T. Kondo, and H. Iwasaki. 2003. Circadian formation of clock protein complexes by KaiA, KaiB, KaiC, and SasA in cyanobacteria. J. Biol. Chem. 278:2388-2395. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, M., T. Kondo, J. Xiong, and S. S. Golden. 2003. ldpA encodes an iron-sulfur protein involved in light-dependent modulation of the circadian period in the cyanobacterium Synechococcus elongatus PCC 7942. J. Bacteriol. 185:1415-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitayama, Y., H. Iwasaki, T. Nishiwaki, and T. Kondo. 2003. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 22:2127-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo, T., C. A. Strayer, R. D. Kulkarni, W. Taylor, M. Ishiura, S. S. Golden, and C. H. Johnson. 1993. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA 90:5672-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo, T., N. F. Tsinoremas, S. S. Golden, C. H. Johnson, S. Kutsuna, and M. Ishiura. 1994. Circadian clock mutants of cyanobacteria. Science 266:1233-1236. [DOI] [PubMed] [Google Scholar]
- 17.Kutsuna, S., T. Kondo, S. Aoki, and M. Ishiura. 1998. A period-extender gene, pex, that extends the period of the circadian clock in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 180:2167-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutsuna, S., Y. Nakahira, M. Katayama, M. Ishiura, and T. Kondo. 2005. Transcriptional regulation of the circadian clock operon kaiBC by upstream regions in cyanobacteria. Mol. Microbiol. 57:1474-1484. [DOI] [PubMed] [Google Scholar]
- 19.Mitsui, A., S. Kumazawa, A. Takahashi, H. Ikemoto, and T. Arai. 1986. Strategy by which nitrogen fixing unicellular cyanobacteria grow photoautotrophically. Nature 323:720-722. [Google Scholar]
- 20.Nakahira, Y., M. Katayama, H. Miyashita, S. Kutsuna, H. Iwasaki, T. Oyama, and T. Kondo. 2004. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc. Natl. Acad. Sci. USA 101:881-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakajima, M., K. Imai, H. Ito, T. Nishiwaki, Y. Murayama, H. Iwasaki, T. Oyama, and T. Kondo. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308:414-415. [DOI] [PubMed] [Google Scholar]
- 22.Nikaido, H. 1998. Multiple antibiotic resistance and efflux. Curr. Opin. Microbiol. 5:516-523. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura, H., Y. Nakahira, K. Imai, A. Tsuruhara, H. Kondo, H. Hayashi, M. Hirai, H. Saito, and T. Kondo. 2002. Mutations in KaiA, a clock protein, extend the period of circadian rhythm in the cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 148:2903-2909. [DOI] [PubMed] [Google Scholar]
- 24.Porter, R. D. 1988. DNA transformation. Methods Enzymol. 167:703-712. [DOI] [PubMed] [Google Scholar]
- 25.Ramasubramanian, T. S., F. Pu, and J. W. Golden. 1995. Isolation of the Anabaena sp. strain PCC 7120 sigA gene in a transcriptional-interference selection. J. Bacteriol. 177:6676-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 27.Ronnenberg, T., and E. J. Carpenter. 1993. Daily rhythm of O2-evolution in the cyanobacterium Trichodesmium thiebautii under natural and constant conditions. Mar. Biol. 117:693-697. [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Stal, L. J., and W. E. Krumbein. 1985. Nitrogenase activity in the non-heterocystous cyanobacterium Oscillatoria sp. grown under alternating light-dark cycles. Arch. Microbiol. 143:67-71. [Google Scholar]
- 30.Sweeney, B. M., and M. B. Borgese. 1989. A circadian rhythm in cell division in a prokaryote, the cyanobacterium Synechococcus WH7803. J. Phycol. 25:183-186. [Google Scholar]
- 31.Takai, N., S. Ikeuchi, K. Manabe, and S. Kutsuna. 2006. Expression of the circadian-clock-related gene pex in cyanobacteria increases in darkness and is required to delay the clock. J. Biol. Rhythms 21:1-10. [DOI] [PubMed] [Google Scholar]
- 32.Tomita, J., M. Nakajima, T. Kondo, and H. Iwasaki. 2005. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307:251-254. [DOI] [PubMed] [Google Scholar]