Abstract
Fruiting body formation of Myxococcus xanthus, like biofilm formation of many other organisms, involves the production of an extracellular matrix (ECM). While the polysaccharide component has been studied, the protein component has been largely unexplored. Proteins associated with the ECM were solubilized from purified ECM by boiling with sodium dodecyl sulfate and were identified by liquid chromatography-tandem mass spectrometry of tryptic fragments. The ECM is enriched in proteins of novel function; putative functions were assigned for only 5 of the 21 proteins. Thirteen putative ECM proteins had lipoprotein secretion signals. The genes for many ECM proteins were disrupted in the wild-type (WT), fibA, and pilA backgrounds. Disruption of the MXAN4860 gene had no effect in the WT or fibA background but in the pilA background resulted in a 24-h delay in aggregation and sporulation compared to its parent. The results of this study show that the M. xanthus ECM proteome is diverse and novel.
The transition from planktonic bacteria to biofilm-associated cells involves changes in gene expression (22, 47, 55) and is mediated at least in part by intercellular communication (21). Biofilm formation by prokaryotic organisms begins with the production of an extracellular matrix (ECM), an extracellular structure that creates shared space within a cellular community. In prokaryotes, ECM is typically composed of carbohydrate polymers, proteins, and sometimes nucleic acids (15, 50, 54). ECM proteins have not been extensively studied but could potentially play significant roles in both structural and intercellular signaling functions, similar to the capacities of ECM proteins in mammals. Mammalian cells are connected in tissues by a proteinaceous ECM. Remodeling of ECM by proteinases is regulated by signaling pathways (25, 60) and stimulates wound healing, the inflammatory response, and angiogenesis (33, 37, 38). Many pathological conditions arise from misregulated ECM proteinase activity, such as periodontitis, cancer metastasis, and cartilage degradation (10, 11, 38, 48, 51). By analogy, prokaryotic ECM proteins may play structural and signaling roles to regulate motility, cell attachment, and immune evasion.
Myxococcus xanthus is a member of the deltaproteobacteria, where it lives in soil as a microbial predator. Under starvation conditions, approximately 50,000 cells aggregate into a multicellular fruiting body, wherein some cells differentiate into dormant myxospores. M. xanthus produces an ECM like those of other biofilm-forming organisms. During swarming, cell-cell proximity induces ECM formation (4), which aids social (S) motility (35, 49). ECM biogenesis is strongly induced during starvation and may be essential for fruiting body formation, since dsp and dif mutants, which are completely deficient in ECM production, are also deficient in fruiting body formation (1, 2, 18, 36, 59). Signals controlling ECM production utilize PilA, the structural protein of type IV pili (8, 12), and the Dif chemosensory system (6, 9, 13, 59), which is composed of DifA (a methyl-accepting chemotaxis protein homolog), DifC (a CheW coupling protein homolog), and DifE (a CheA histidine kinase homolog). The Dif system is also required for development. Development in difACE mutants can be restored with exogenously supplied M. xanthus ECM (17, 59).
M. xanthus ECM is composed of 55% carbohydrate and 45% protein (3). The carbohydrate portion contains primarily glucose and glucosamine, with galactose, rhamnose, and xylose as lesser components (3). The ECM proteins are tightly associated with the exopolysaccharide, requiring detergent and boiling to remove them (5). FibA, the only known ECM protein, is a zinc metalloprotease of the elastase family (5, 29) and the most abundant protein associated with the ECM (5). FibA is required for chemotaxis to lipids and, under certain conditions, fruiting body development. During starvation, M. xanthus responds to membrane phospholipids containing the fatty acid 16:1ω5c (20, 31). A FibA active-site mutant is incapable of chemotaxis towards this molecule (12, 29), although the substrate and role of this enzyme in chemotaxis remain unknown. While disruption of fibA has no obvious effect on development, disruption of both fibA and pilA, the gene encoding the major pilus structural protein (34), abolishes development. It is unclear whether development involves lipid chemotaxis or another as yet unknown FibA-mediated process.
Deletion of pilA results in a lack of S motility and a delay in development (57); however, these cells still form fruiting bodies using A motility. The pilT mutant forms pili but is unable to retract them and also lacks S motility (58). While the fibA pilA mutant is unable to develop (12), a fibA pilT mutant develops normally (12). This result suggests that the PilA requirement for development is due not to its role in S motility but rather to its requirement for ECM biogenesis. Strains carrying a disruption in pilT (which causes hyperpiliation [58]) overproduce ECM (8). dif mutations are epistatic on pil mutations, showing that pilus signaling functions upstream of the Dif system in ECM biogenesis (8). These results argue that PilA or the structural pilus serves as a sensor for signal transduction leading to ECM biogenesis. Given that characterized signaling processes for both FibA and PilA pass through the Dif system and that dif mutations abolish development, one interpretation of these results is that the DifACE signaling pathway has two sensory inputs, one involving PilA (or the pilus) and the other involving FibA, that are functionally redundant for fruiting body development (12). The specific signals sensed, the method of signal transduction into the DifACE system, and the specific outputs that impact development remain unknown.
Proteomic analysis was used to identify matrix-associated proteins and to examine their possible roles in development. A number of novel proteins are associated with the matrix, but only one protein could be added to the DifACE pathway.
MATERIALS AND METHODS
Strains and growth conditions.
M. xanthus strains (Table 1) were grown at 32°C in CYE (1.0% Bacto Casitone, 0.5% Difco yeast extract, 10 mM 3-[N-morpholino]propanesulfonic acid [MOPS], pH 7.6, and 0.1% MgSO4) broth with vigorous shaking (16). Cultures were grown on plates containing CYE with 1.5% Difco agar. For selective growth, kanamycin was added to a final concentration of 100 μg ml−1 (CYE Km).
TABLE 1.
Strains used in this study
| Strain | Genotype or description | Source, reference, or strain backgrounda |
|---|---|---|
| DK1622 | Wild type | 27 |
| DK10410 | ΔpilA | 58 |
| LS2429 | ΔfibA | Lawrence Shimkets |
| LS2333 | MXAN4860 Kmr | DK1622 × pPDC9 → Kmr |
| LS2337 | MXAN4860 ΔpilA Kmr | DK10410 × LS2333 → Kmr |
| LS2341 | MXAN4860 ΔfibA Kmr | LS2429 × LS2333 → Kmr |
| LS2344 | MXAN2791 Kmr | DK1622 × pPDC18 → Kmr |
| LS2345 | MXAN5684 Kmr | DK1622 × pPDC19 → Kmr |
| LS2347 | MXAN2791 ΔpilA Kmr | DK10410 × LS2344 → Kmr |
| LS2348 | MXAN5684 ΔpilA Kmr | DK10410 × LS2345 → Kmr |
| LS2355 | MXAN0075 ΔpilA Kmr | DK10410 × pPDC26 → Kmr |
| LS2356 | MXAN7023 Kmr | DK1622 × pPDC40 → Kmr |
| LS2357 | MXAN5686 Kmr | DK1622 × pPDC39 → Kmr |
| LS2358 | MXAN6985 Kmr | DK1622 × pPDC30 → Kmr |
| LS2360 | MXAN5391 Kmr | DK1622 × pPDC24 → Kmr |
| LS2361 | MXAN2375 Kmr | DK1622 × pPDC36 → Kmr |
| LS2362 | MXAN2710 Kmr | DK1622 × pPDC42 → Kmr |
| LS2363 | MXAN0793 Kmr | DK1622 × pPDC38 → Kmr |
| LS2364 | MXAN1493 Kmr | DK1622 × pPDC41 → Kmr |
| LS2365 | MXAN1424 Kmr | DK1622 × pPDC27 → Kmr |
| LS2366 | MXAN4915 Kmr | DK1622 × pPDC43 → Kmr |
| LS2367 | MXAN0075 Kmr | DK1622 × pPDC26 → Kmr |
| LS2369 | MXAN6985 ΔpilA Kmr | DK10410 × LS2358 → Kmr |
| LS2370 | MXAN5686 ΔpilA Kmr | DK10410 × LS2357 → Kmr |
| LS2371 | MXAN7023 ΔpilA Kmr | DK10410 × LS2356 → Kmr |
| LS2372 | MXAN2375 ΔpilA Kmr | DK10410 × LS2361 → Kmr |
| LS2373 | MXAN1493 ΔpilA Kmr | DK10410 × LS2364 → Kmr |
| LS2374 | MXAN5391 ΔpilA Kmr | DK10410 × LS2360 → Kmr |
| LS2375 | MXAN2710 ΔpilA Kmr | DK10410 × LS2362 → Kmr |
| LS2376 | MXAN0793 ΔpilA Kmr | DK10410 × LS2363 → Kmr |
| LS2377 | MXAN1424 ΔpilA Kmr | DK10410 × LS2365 → Kmr |
| LS2378 | MXAN4915 ΔpilA Kmr | DK10410 × LS2366 → Kmr |
| LS2381 | MXAN5686 ΔfibA Kmr | LS2429 × LS2357 → Kmr |
| LS2382 | MXAN2710 ΔfibA Kmr | LS2429 × LS2362 → Kmr |
| LS2383 | MXAN2375 ΔfibA Kmr | LS2429 × LS2361 → Kmr |
| LS2384 | MXAN1424 ΔfibA Kmr | LS2429 × LS2365 → Kmr |
| LS2385 | MXAN1493 ΔfibA Kmr | LS2429 × LS2364 → Kmr |
| LS2386 | MXAN5684 ΔfibA Kmr | LS2429 × LS2345 → Kmr |
| LS2387 | MXAN0075 ΔfibA Kmr | LS2429 × LS2367 → Kmr |
| LS2388 | MXAN0793 ΔfibA Kmr | LS2429 × LS2363 → Kmr |
| LS2389 | MXAN7023 ΔfibA Kmr | LS2429 × pPDC40 → Kmr |
| LS2390 | MXAN5391 ΔfibA Kmr | LS2429 × LS2360 → Kmr |
| LS2391 | MXAN6985 ΔfibA Kmr | LS2429 × LS2358 → Kmr |
| LS2392 | MXAN2791 ΔfibA Kmr | LS2429 × pPDC18 → Kmr |
| LS2393 | MXAN4915 ΔfibA Kanr | LS2429 × pPDC43 → Kmr |
The first strain listed in the column is the recipient, and the second strain is the source of DNA used to modify the recipient.
Enrichment of ECM material.
ECM was enriched from 24-h developing cells using 0.5% sodium dodecyl sulfate (SDS) (5, 30). Solubilization of ECM-associated proteins was developed in this study. Attempts to solubilize ECM proteins with 9.5 M urea, 2% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonic acid, 2% ethylene glycol octyl phenyl ether (Triton X-100), and 350 mM NaCl, boiling for extended periods of time, and many combinations thereof failed, as did digestion of isolated ECM material with trypsin (data not shown). Boiling with a high concentration of SDS and a reducing agent solubilized at least some of the proteins. A protease inhibitor cocktail was necessary to prevent digestion of proteins by proteases in the sample.
To induce development, 7.5 ml of 5 × 109 cells ml−1 were plated on TPM agar (10 mM Tris HCl, 8 mM MgSO4, 1 mM K2HPO4-KH2PO4, 1.5% Difco agar, pH 7.6) in a 33- by 22-cm tray and incubated at 32°C for 24 h. Fruiting bodies were harvested by scraping with a razor blade. ECM material was collected using a modification of the method described by Behmlander and Dworkin (5). Five milliliters of TNE buffer (10 mM Tris [pH 7.5], 100 mM NaCl, 5 mM EDTA [4]) was added to the cell suspension and stirred for 10 min at room temperature. Five milliliters of TNE containing 1.0% SDS was then added and stirred for 30 min. The solution was centrifuged at 12,000 × g for 10 min at 4°C and the supernatant discarded. The pellet was resuspended in 5.0 ml TNE containing 0.5% SDS and stirred for 1 h at room temperature. The suspension was centrifuged at 12,000 × g at 4°C for 10 min and the supernatant discarded. The pellet was washed once with 5.0 ml TNE, twice with 5.0 ml 10 mM MOPS, and twice with 5.0 ml cohesion buffer (10 mM MOPS [pH 6.8], 1 mM CaCl2, 1 mM MgCl2). Finally, the pellet was resuspended in 1.0 ml of cohesion buffer containing 5× complete EDTA-free protease inhibitor cocktail (Roche) and stored at 4°C. A variation of this procedure using 0.1% SDS in the two initial wash steps (5) contained a wider variety of proteins, including more membrane proteins.
Extraction of matrix-associated proteins.
Proteins were solubilized from the ECM by boiling in SDS. Solubilized proteins were separated from the insoluble ECM material by SDS-polyacrylamide gel electrophoresis (PAGE) long enough for the proteins to enter the resolving gel but not separate into distinct bands. The SDS was removed from the gel by washing, allowing digestion of proteins within the gel slice with trypsin. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used in the identification of tryptic fragments of ECM proteins.
Two milliliters of purified ECM material containing 127 μg ml−1 protein was pelleted by centrifuging at 12,000 × g for 10 min at room temperature. The pellet was resuspended in 400 μl cohesion buffer containing 3% SDS. Dithiothreitol (DTT) was added to a final concentration of 100 mM and protease inhibitor to a 2.5× concentration. The suspension was boiled for 30 min and nonsolubilized material pelleted by centrifugation at 12,000 × g for 10 min. The supernatant was collected and centrifuged at 62,000 × g for 30 min at 4°C. The supernatant was collected and concentrated using a Microcon YM-3 centrifugation filter (3,000-molecular-weight cutoff) for 1.5 h, resulting in a fivefold concentration of protein (approximately 80 μl volume). To this, 40 μl of sample buffer (52.5 mM Tris-HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue, 100 mM DTT) was added, and the mixture was boiled for 15 min. The total solution was then loaded on a 12% SDS polyacrylamide gel over multiple lanes. The gel was run at 50-mA current until the proteins migrated through the stacking gel and began to enter the resolving gel. Protein detection was performed using Silver Stain Plus (Bio-Rad). It is difficult to judge the efficiency with which the protein extraction method worked due to the presence of DTT, which is detrimental to standard protein quantification assays.
The portion of the gel containing protein (approximately 1.0 by 1.5 cm) was excised, cut into smaller fragments, destained with 200 μl 15 mM potassium ferricyanide-50 mM sodium thiosulfate, and washed three times with 200 μl water. The gel pieces were dehydrated by washing them three times with 200 μl of 20 mM ammonium bicarbonate containing 50% (vol/vol) acetonitrile for 15 min each, followed by one 15-min wash with 200 μl acetonitrile. The gel slices were dried under vacuum and rehydrated with 100 μl of 10 mM DTT in 40 mM ammonium bicarbonate at 55°C for 1 h, at which point the solution was exchanged for 100 μl of 55 mM iodoacetamide-40 mM ammonium bicarbonate and incubated for 45 min at room temperature. The gel pieces were washed three times with 200 μl of 20 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile and once with 100% acetonitrile for 15 min and then dried under vacuum. The gel pieces were rehydrated with 100 μl of 10 ng μl−1 proteomics-grade trypsin (Sigma) in 40 mM ammonium bicarbonate and incubated on ice for 45 min and then with 100 μl of 40 mM ammonium bicarbonate, with incubation at 37°C overnight. Solutions from the trypsin digestion were pooled. The gel slices were washed with 150 μl of 2.5% trifluoroacetic acid in 50% acetonitrile three times for 10 min each. The washes were combined with the solutions from the previous step and taken to dryness under vacuum.
Proteomic identification of ECM proteins.
The peptide samples obtained from proteolytic digestion were analyzed on an Agilent 1100 capillary LC (Palo Alto, CA) interfaced directly to a LTQ linear ion trap mass spectrometer (Thermo Electron, San Jose, CA). Mobile phases A and B were H2O-0.1% formic acid and acetonitrile-0.1% formic acid, respectively. The peptide samples were loaded for 30 min using positive N2 pressure on a PicoFrit 8-cm by 50-μm column (New Objective, Woburn, MA) packed with 5-μm-diameter C18 beads. The peptides were then desalted for 10 min with 0.1% formic acid using positive N2 pressure. Peptides were eluted from the column into the mass spectrometer during a 90-min linear gradient from 5 to 60% of total solution composed of mobile phase B at a flow rate of 200 ηl min−1. The instrument was set to acquire MS/MS spectra on the nine most abundant precursor ions from each MS scan with a repeat count of 3 and repeat duration of 15 s. Dynamic exclusion was enabled for 20 s. Raw tandem mass spectra were converted into the mzXML format and then into peak lists using ReAdW software followed by mzMXL2Other software (40). The peak lists were then searched using Mascot 1.9 software (Matrix Science, Boston, MA).
Database searching and protein identification.
Two sequence databases were constructed. The first database (normal) consisted of annotated proteins from M. xanthus genes (as annotated by The Institute for Genome Research and provided by Roy Welch). A decoy database (random) was constructed by reversing the sequences in the normal database. Database searches were performed against the normal and random databases using the following parameters: full tryptic enzymatic cleavage with three possible missed cleavages, peptide tolerance of 500 parts-per-million, fragment ion tolerance of 0.6 Da, and a variable modification due to carboxyamidomethylation (+57 Da). The identified peptides were grouped into proteins and statistically validated using PROVALT software (53). Only proteins meeting with a protein false-discovery rate of less than 1% were considered to be statistically significant. Protein sequences were analyzed using LipoP (44), pfam (http://pfam.janelia.org/hmmsearch.shtml), and Prosite (http://us.expasy.org/tools/scanprosite/).
Mutagenesis of ECM proteins.
Genes for 14 putative ECM proteins were disrupted by insertion of a plasmid containing an internal fragment of each gene. The internal fragments were generated by PCR (see the supplemental material for primers). Primers were designed to result in as large a 3′ deletion as possible. PCR products were separated on 1.0% agarose, excised, extracted using the Gel Extraction kit (QIAGEN), and cloned into pCR2.1-TOPO (Invitrogen). The identity of each insert was verified by DNA sequencing. Each plasmid was electroporated in M. xanthus DK1622 with selection on CYE containing 100 μg ml−1 kanamycin. Genomic DNA was purified from each transformant using the Easy DNA extraction kit (Invitrogen). To verify the mutations, primers were used to amplify genomic DNA from sites outside of the internal fragment (see diagnostic primers in the supplemental material). Plasmid integration causes a mobility shift of the wild-type amplicon such that disrupted transformants have PCR products the size of the wild-type allele plus the size of the plasmid (3.9 kb).
In order to transfer the mutations to the pilA and fibA strains, approximately 3.75 μg of genomic DNA from each mutant strain was electroporated into strains DK10410 and LS2429 (28, 52), except for strains LS2355, LS2389, LS2392, and LS2393, where plasmids were used directly for disruption. Selection and screening were performed as before.
Development assay.
Cells from exponentially growing cultures were pelleted by centrifugation and resuspended to cell densities of 5 × 108, 2.5 × 109, 5 × 109, and 7.5 × 109 cells ml−1 in CYE broth. Ten microliters of each cell suspension was spotted in duplicate on TPM agar plates and incubated at 32°C for 5 days. The cell spots were viewed using a Wild Heerbrugg dissecting microscope, and images were captured every 24 h with a Spot Insight 2 camera using the Spot v4.5 software (Diagnostic Instruments, Inc.).
Spore assay.
Cells from exponentially growing cultures were resuspended in CYE broth at a density of 5 × 109 cells ml−1, and 10-μl spots were plated on TPM agar. After incubation at 32°C for 5 days, fruiting bodies were collected with a sterile razor blade and resuspended in 0.5 ml TPM buffer. The fruiting bodies were then sonicated at a 60% duty cycle for 10 s on an Ultrasonic Processor sonicator (Heat Systems- Ultrasonics Inc.) and incubated at 55°C for 2 h. Refractile myxospores were quantified using a Petroff-Hausser counting chamber. Spores were diluted and plated on CYE or CYE Km to enumerate viable spores. Spore production was examined for select strains every 24 h for 5 days.
FibA antibody and Western blot analysis.
fibA was cloned into vector pET-TOPO 102/D to create a six-His C-terminal fusion, and the construct was expressed in Escherichia coli strain BL21 Star DE3 (Invitrogen). Purification was performed using the ProBond purification system (Invitrogen). Polyclonal antibodies were generated by Antibodies, Inc. Cell lysates were prepared from 5 × 107 mid-log phase DK1622 and LS2429 cells. ECM material was prepared as described. Cell lysates and 5 μg ECM protein were separated on a 12.5% SDS-PAGE gel in triplicate. Proteins were electroblotted on nitrocellulose. Each set was probed with different primary antibodies. The polyclonal antibody was diluted 1:7,500, and the Mab2105 (4) and multiple-antigen peptide (12) antibodies were diluted 1:500. All dilutions were prepared in PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, and 0.05% Tween 20) containing 2.5% dry milk. Secondary antibodies horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G and horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G were diluted 1:5,000 in PBST. The immunoblot was developed with the ECL luminescence detection kit (Amersham Pharmacia, Piscataway, NJ).
RESULTS
Little is known about the structure and function of ECM proteins in bacterial biofilms. The purpose of this study was to identify the ECM proteins by mass spectrometry and ascertain their function in strains containing a gene disruption. The ECM was enriched following solubilization of cells with 0.5% SDS. Proteins were liberated from the insoluble ECM by boiling in 3% SDS and 100 mM dithiothreitol and were separated from the insoluble matrix by SDS-PAGE. Proteolytic fragments were released by trypsin digestion and identified using LC-MS/MS and the M. xanthus genome sequence (23).
Identification of ECM proteins.
A total of 41 proteins were identified with 99% confidence. Putative ECM proteins were selected from the list based on the presence of a Sec-dependent secretion signal using the LipoP program (26), the absence of transmembrane helices, and the lack of conserved membrane domain structures. While integral membrane proteins may serve as points of attachment of ECM components to the cell surface or channels for their secretion, there is no direct evidence that the ECM contains a lipid bilayer, so putative integral membrane proteins were removed from the list. These criteria reduced the number to 21 putative ECM proteins (Table 2). Eighteen of the twenty-one putative ECM proteins were also found in another ECM sample prepared with 0.1% SDS instead of 0.5% SDS (data not shown). The sample prepared with 0.1% SDS contained a higher proportion of non-ECM proteins, and for that reason it was less specific in extracting ECM proteins. FibA has been shown to be associated with the ECM using immunogold labeling (4), and identification of FibA in the present study (Table 2) suggests that the ECM isolation technique enriches for ECM proteins.
TABLE 2.
Putative ECM proteins in M. xanthus
| Genea | No. of peptidesb | Size of product (kDa) | Lipoprotein signalc | Identity of product |
|---|---|---|---|---|
| MXAN6106 | 16 | 79.82 | + | FibA |
| MXAN2183 | 12 | 46.45 | +/− | Conserved hypothetical |
| MXAN5391 | 11 | 47.26 | +/− | Hypothetical |
| MXAN4860 | 9 | 30.88 | + | Hypothetical |
| MXAN5684 | 7 | 22.60 | + | Conserved hypothetical, Curli protein |
| MXAN2710 | 6 | 56.89 | + | Conserved hypothetical |
| MXAN6985 | 4 | 52.78 | + | Hypothetical |
| MXAN0793 | 4 | 57.95 | + | Hypothetical |
| MXAN1493 | 4 | 28.88 | − | Hypothetical |
| MXAN3885 | 3 | 18.44 | − | Spore coat protein U |
| MXAN5685 | 3 | 30.77 | − | Hypothetical |
| MXAN6720 | 2 | 11.44 | + | Hypothetical |
| MXAN5686 | 2 | 19.81 | − | Hypothetical |
| MXAN2791 | 1 | 27.52 | + | Protease B |
| MXAN1424 | 1 | 29.91 | − | Hypothetical |
| MXAN4915 | 1 | 36.41 | − | Hypothetical |
| MXAN7023 | 1 | 78.34 | +(+2E) | Conserved hypothetical (COG4880) |
| MXAN0075 | 1 | 124.17 | − | Amidohydrolase domain protein |
| MXAN2375 | 1 | 47.97 | − | Amidohydrolase |
| MXAN0235 | 1 | 15.72 | + | Hypothetical |
| MXAN1657 | 1 | 12.22 | + | Hypothetical |
All but MXAN0793, MXAN1493, and MXAN0235 were identified in a second experiment with a sample extracted with 0.1% SDS instead of 0.5% SDS.
Number of peptides matched from each protein by LC-MS/MS.
Plus symbols (+) indicate a predicted lipoprotein secretion signal using the LipoP program (26). Minus symbols (−) indicate a predicted Sec-dependent secretion signal. The +/− symbol for MXAN2183 and MXAN5391 indicates that the program predicted both a lipoprotein secretion signal and a nonlipoprotein signal with equal probability. The +(+2E) symbol for MXAN7023 indicates that the residue following the acylated cysteine (+1) is an aspartate residue. In E. coli, aspartate at the +2 position directs proteins to the periplasmic face of the inner membrane (44), though this has not been demonstrated for M. xanthus.
Of the 21 putative ECM proteins, 13 are predicted to have lipoprotein secretion signals (Table 2), though MXAN2183 and MXAN5391 are predicted to have both lipoprotein and normal secretion signals with equal probability. Only five of the putative ECM proteins have predicted functions. The MXAN6106 protein is the zinc metalloprotease FibA, the most abundant and only previously identified ECM protein (5, 29). MXAN3885 is spore coat protein U, a protein known to be assembled on spores (24). MXAN2791 is a zinc metalloprotease known as protease B that has been identified in two studies examining milk-clotting ability by E. coli containing M. xanthus genomic DNA libraries (41, 42). A C-terminal portion of MXAN0075 and the entire length of MXAN2375 share homology to the amidohydrolase family, subgroup 1 (pfam01979). The remaining putative ECM proteins are produced by either hypothetical or conserved hypothetical genes, suggesting that the M. xanthus ECM is enriched for proteins with novel functions.
Other proteins identified in this fraction that are unlikely to be associated with the ECM are listed in Table 3. The presence of PilQ suggests that ECM enrichment also contains material from cell poles. PilQ forms the outer membrane pore for polarly localized pili. The membranes at M. xanthus cell poles appear to be difficult to separate (46), and it is possible that 0.5% SDS was unable to completely solubilize these polar complexes. Additionally, the LC-MS/MS technique is very sensitive, which may have allowed for identification of highly abundant intracellular proteins.
TABLE 3.
Non-ECM proteins identified by proteomic analysis and putative cellular location
| Genea | No. of peptidesb | Size of product (kDa) | Identity of product |
|---|---|---|---|
| Outer membrane | |||
| MXAN5572 | 13 | 96.36 | PilQ |
| MXAN5855 | 9 | 52.56 | Probable outer membrane porin |
| MXAN7203 | 7 | 26.54 | Conserved hypothetical membrane protein |
| MXAN2514 | 4 | 94.11 | XcpQ |
| MXAN3106 | 4 | 51.89 | CpaC homolog |
| MXAN7407 | 3 | 21.64 | Hypothetical outer membrane protein |
| MXAN1450 | 2 | 122.47 | OAR |
| Inner membrane | |||
| MXAN5402 | 13 | 52.84 | Cytochrome c |
| MXAN3729 | 7 | 109.47 | Pyrrolo quinoline quinine-containing dehydrogenase |
| Intracellular | |||
| MXAN6601 | 21 | 80.86 | Prolyl endopeptidase |
| MXAN4808 | 15 | 56.12 | DUF87 protein of unknown function |
| MXAN4494 | 12 | 83.96 | Conserved hypothetical: phage tail sheath protein |
| MXAN4807 | 11 | 18.00 | DUF770 protein of unknown function |
| MXAN5407 | 9 | 45.48 | Choloylglycine hydrolase |
| MXAN2815 | 4 | 36.49 | Glyceraldehyde-3-phosphate dehydrogenase |
| MXAN6090 | 4 | 34.81 | DUF481 protein of unknown function |
| MXAN5012 | 3 | 85.62 | Tyrosine decarboxylase |
| MXAN5582 | 3 | 37.72 | Hypothetical |
| MXAN4495 | 2 | 16.42 | Conserved hypothetical: phage tail region protein |
| MXAN1394 | 2 | 36.38 | Zinc-dependent hydrolase of the β-lactamase fold |
Membrane proteins were assigned based on the presence of a Sec-dependent secretion signal and the cellular location of the closest homologs. Intracellular proteins were assigned based on the absence of a Sec-dependent secretion signal as determined by LipoP analysis.
Number of peptides matched from each protein by LC-MS/MS.
Mutagenesis of putative ECM genes.
Each gene encoding a putative ECM protein was disrupted by insertion of a plasmid containing an internal fragment of the gene. A single homologous recombination integrates the entire plasmid into the chromosome and creates two defective copies of the gene. Not all genes were disrupted: the MXAN3885, MXAN6720, MXAN0235, and MXAN1657 genes are less than 600 bp in length, and the 3′ truncations may be too small to cause a gene disruption (Table 2). Repeated attempts to disrupt the MXAN2183 gene were unsuccessful, suggesting that it or the smaller hypothetical gene located downstream may be essential. MXAN5686, MXAN5685, and MXAN5684, the first three genes in a six-gene operon, were all detected in the ECM proteome. Disruption of MXAN5686 is expected to create a polar mutation on the rest of the operon. MXAN5684 was disrupted, but the protein product should also be missing in the strain containing the MXAN5686 insertion.
Genes encoding ECM proteins were disrupted in the wild type (DK1622) and screened for fruiting body defects after spotting of 5 × 106 cells for 72 h (see the supplemental material for fruiting body assays). All mutants developed at similar rates and had a fruiting body distribution similar to that of DK1622. Previous work provided genetic evidence that FibA and PilA are components of partially redundant pathways for fruiting body morphogenesis (12). While fibA and pilA mutants form fruiting bodies, the double mutant neither aggregates nor sporulates (12). Therefore, each mutation was placed in pilA (DK10410) and fibA (LS2429) backgrounds to determine whether these proteins function in a particular branch of the pathway. The fruiting bodies of the pilA mutant are more numerous and closely packed. With the exception of the MXAN4860 mutation, strains containing each mutation in combination with pilA develop comparably to the pilA parent strain. Strains containing each mutation in combination with fibA demonstrate the same timing and distribution of development as the wild type.
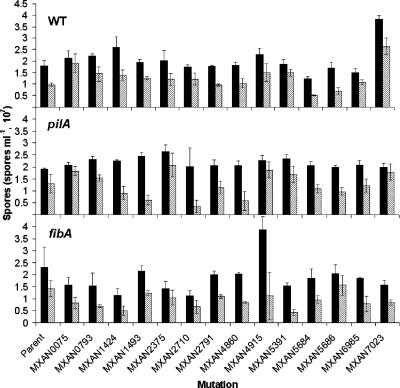
All strains were assessed for spore production and viability (Fig. 1). In order to obtain enough spores to facilitate enumeration, spore assays were performed using 10-fold more cells than the fruiting body assays (see the supplemental material). The majority of the mutants produced spores within 1.5-fold levels of each other and the parent strain, and approximately 40 to 75% of the spores were viable. There are some exceptions worth noting. Disruption of the MXAN7023 gene in DK1622 caused a nearly twofold increase in spore production and a more modest increase in viable spores. Similarly, the MXAN4915 fibA mutant produced nearly twofold more spores than the fibA parent, but viable spore production was similar to that of the parent strain. Disruption of the MXAN2710 gene in the pilA background caused no defect in total spore production but a 10-fold decrease in spore viability. Similarly, disruption of the MXAN5391 gene in fibA reduces viable spore production more substantially that total spore production.
FIG. 1.
Spore production and viability of ECM mutants. Total spores from 5 × 107 cells incubated for 5 days were enumerated microscopically (solid bars). Viable spores were determined by plating spores on CYE agar (hatched bars). Error bars indicate the standard deviation. The top-left area of each panel displays the genetic background containing each mutation: WT, wild type, DK1622; pilA, ΔpilA, DK10410; fibA, ΔfibA, LS2429.
MXAN4860.
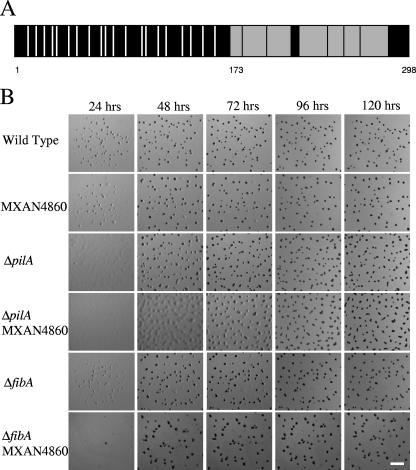
The MXAN4860 protein was identified by nine peptides in the C-terminal half, covering 27% of the total protein (Fig. 2A, shaded boxes), suggesting that this region is more trypsin sensitive than the cysteine-rich, N-terminal domain (Fig. 2A, white bars). Neither the whole protein nor the individual domains have known homologs. While the downstream gene MXAN4859 is transcribed in the same direction, it is separated by 146 bp from the MXAN4860 gene and does not appear to be operonic.
FIG. 2.
Primary structure analysis of MXAN4860 and developmental timing of MXAN4860 disruptions in wild-type and pilA strains. (A) Schematic of the MXAN4860 protein product. Vertical white bars indicate positions of 20 cysteine residues found in the N-terminal portion of the protein. Gray boxes indicate the tryptic peptides detected by mass spectrometry. The numbers indicate amino acid positions: 1, the start of the protein; 173, the start of the first peptide detected by mass spectrometry; 298, the end of the protein. (B) Developmental time course of fruiting body morphogenesis. Cells (5 × 106) were spotted in 10 μl on TPM agar and photographed every 24 h. Strains with the MXAN4860 mutation develop normally unless it is coupled with a pilA mutation, in which case there is an approximately 24-h delay in development between the 48- and 72-h time points. Bar = 1 mm.
Disruption of the MXAN4860 gene causes a lengthy delay in fruiting body formation in combination with pilA but ultimately results in normal fruiting bodies (Fig. 2B). The pilA parent undergoes aggregation between 24 and 48 h, and sporulation begins at approximately 48 h. The MXAN4860 pilA strain begins to aggregate at approximately the same time as the pilA parent, but development arrests at 48 h when the cells are in large, translucent mounds. This arrest lasts approximately 24 h until sporulation begins within the large mounds at approximately 72 h. Sporulation is completed by 96 h. Both the pilA and MXAN4860 pilA strains display more elongated fruiting bodies than the wild type. This result likely stems from the loss of S motility as has been noted elsewhere (12, 34). The same disruption in the pilT background results in no developmental defect (data not shown), indicating that the defect is due to the absence of the pilus rather than the loss of S motility, similar to defects observed with strains harboring fibA disruptions. The phenotype of the MXAN4860 fibA double mutant is comparable to that of the fibA parent (Fig. 2B). These results are consistent with MXAN4860 functioning in the FibA branch of the proposed model for development.
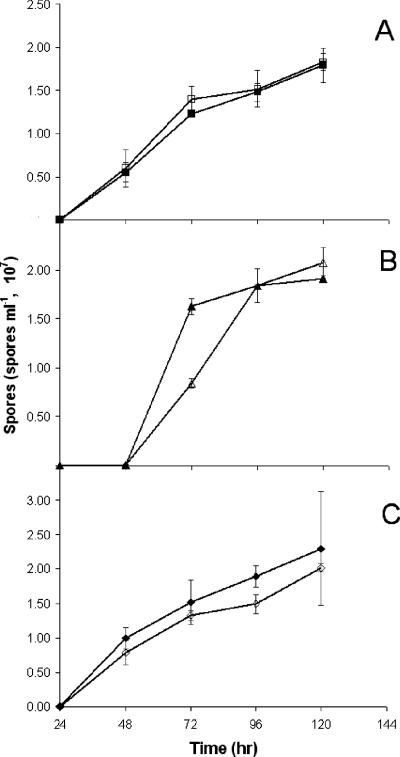
Spore yields for all MXAN4860 mutants as well as parent strains were measured every 24 h for 5 days (Fig. 3). The MXAN4860 and MXAN4860 fibA mutants had timing and spore yields similar to those of their respective parents. Sporulation in the pilA strain was delayed 24 h compared to that of the wild type; however, by 72 h, spore production had reached wild-type levels. At 72 h, the MXAN4860 pilA mutant showed 46% sporulation compared to that of its parent. By 96 h, the MXAN4860 pilA mutant produced the same number of spores as the pilA strain. The sporulation levels suggested an approximately 18-h delay in sporulation, corroborating the observed delay in fruiting body morphogenesis.
FIG. 3.
Sporulation time courses of MXAN4860 mutants compared to wild-type, pilA, and fibA strains. Cells (5 × 107) of each strain were plated on TPM and harvested at 24-h intervals. Spores were quantified using a Petroff-Hausser counting chamber. (A) Wild-type (DK1622, filled squares) and MXAN4860 (open squares) strains. (B) pilA (DK10410, filled triangle) and MXAN4860 pilA (open triangle) strains. (C) fibA (LS2429, filled diamond) and MXAN4860 fibA (open diamond) strains. Error bars indicate standard deviations.
FibA processing.
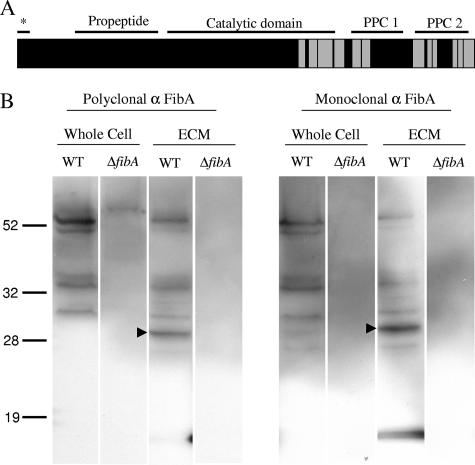
FibA isolated from ECM is found in multiple forms (4, 12, 29). Edman degradation and Western blotting suggest the C-terminal portion is found in the ECM, but the fate of the catalytic domain is unknown. The processing of FibA in the ECM was analyzed by two methods. First, peptides detected by mass spectrometry were coordinated with their location in the primary amino acid sequence and predicted domains. All peptides were found in the terminal repeats and C-terminal portion of the catalytic domain (Fig. 4A). Second, a polyclonal FibA antibody was used in Western blot analysis. The banding patterns were compared to those detected by two monoclonal FibA antibodies (3, 12). Fragments detected by the polyclonal antibody in both whole-cell extract and ECM material match those found by Mab2105 (Fig. 4B), suggesting that the monoclonal antibody is an accurate indicator of all FibA fragments found in the ECM. The size of the FibA portion containing the catalytic domain and PPC repeats is predicted to be 53.4 kDa, which matches the largest fragment detected in wild-type samples. The size of the FibA portion containing the peptides detected by mass spectrometry (amino acids 455 to 744) is predicted to be 30.8 kDa and matches the most prominent fragmentation product (Fig. 4B, arrowhead).
FIG. 4.
Analysis of FibA fragmentation patterns. (A) Primary structure analysis of FibA. Horizontal lines indicate the lipoprotein secretion signal (*; amino acids 1 to 20), propeptide domain (amino acids 93 to 228), catalytic domain (amino acids 244 to 517), PPC domain 1 (amino acids 542 to 626), and PPC domain 2 (647 to 734) (12). Gray boxes indicate peptides detected by mass spectrometry. (B) Western blot using anti-FibA (α FibA) polyclonal and monoclonal antibodies. Wild-type (WT) and ΔfibA whole-cell extracts were prepared from 5 × 107 exponentially growing cells. ECM material containing 5 μg protein was also analyzed. Arrowheads indicate the major degradation product.
DISCUSSION
Initially, matrix-assisted laser desorption ionization-time-of-flight MS was utilized for protein identification. However, poor protein coverage led to ambiguous identification. The LC-MS/MS technique negates the need for greater peptide coverage, because a single peptide can often identify a protein with 99% confidence. The lack of peptide coverage may be due to the abundance of extracellular proteases. ECM proteins are constantly exposed to proteases and may have evolved protease resistance, leading to inadequate digestion by trypsin. A specific example is the MXAN4860 protein. All the peptides detected by LC-MS/MS were derived from the C-terminal portion of MXAN4860 (Fig. 2A), suggesting that the cysteine-rich N-terminal domain is trypsin resistant. Similar results are observed with FibA (Fig. 4). The lack of peptide coverage with other proteins suggests that protease resistance may be important for protein stability and function in the extracellular milieu.
The unusual N-terminal domain may offer an insight into the function of the MXAN4860 protein. MXAN4860 shares some characteristics with Wnt proteins, a large family of eukaryotic proteins controlling development in organisms from nematodes to mammals (56). Wnt proteins are approximately 350 to 390 amino acids in length and have 22 conserved cysteines. They are secreted, bind tightly with glycosaminoglycans in the ECM, and are difficult to extract from matrix fractions (14, 43). Interaction between Wnt and a cell surface receptor (such as Fz or LRP5/6) transduces an extracellular signal into intracellular responses (19), including the planar cell polarity pathway regulating tissue polarity, cell migration, and cytoskeleton arrangement. Another eukaryotic protein involved in some of the same pathways as Wnt is Norrin. Norrin, like Wnt, is a secreted signaling protein (approximately 130 amino acids in length with 11 conserved cysteines), interacting with specific Fz and LRP receptors (7). Though Norrin is cysteine rich like Wnt, they are unrelated, suggesting that cysteine richness may have a conserved function in intercellular signaling in unrelated proteins. Interestingly, some Wnts are acylated, and it has been shown in at least one case that acylation is necessary for signaling (32). MXAN4860 is predicted to have a lipoprotein secretion signal and therefore may also be acylated.
Thirteen of twenty-one putative ECM proteins (62%) have lipoprotein secretion signals. The structural function of acylation is to anchor proteins to either the outer membrane or the outer face of the inner membrane. While membrane targeting of these proteins would seem to preclude secretion to the ECM, FibA is a lipoprotein that is associated with the inner membrane, likely in the acylated, catalytically inactive proform (46), but is also associated with the ECM (4). It is possible that temporary targeting of ECM proteins to the inner membrane may be a method of keeping ECM proteins inactive until they are secreted.
Proteomic analysis of FibA may offer new insight into its function. FibA contains a lipoprotein secretion signal, a propeptide domain to regulate catalytic function, the catalytic domain, and two C-terminal PPC repeat domains of unknown function (Fig. 4A) (12). The epitope for FibA monoclonal antibody Mab2105 is proposed to be located in the last C-terminal repeat, within the final 107 amino acids of the protein (12). Western blots using this antibody against proteins liberated from the ECM reveal bands of different sizes, indicating that FibA is proteolytically processed into several pieces (5, 12) (Fig. 4B). Most peptide fragments detected by LC-MS/MS are from the C-terminal repeats and C-terminal side of the catalytic domain (Fig. 4A). This result corroborates the Western results showing that the PPC repeats are found associated with the ECM (12). C-terminal repeats in eukaryotic matrix metalloproteinases confer substrate specificity by anchoring the enzyme next to the substrate (for a review, see reference 39), and by analogy FibA repeats may interact with the FibA substrate(s) and/or anchor FibA to the ECM. Western blot results also indicate that the catalytic domain is present in the ECM (29) (Fig. 4B). The lack of peptide detection from the majority of the catalytic domain may indicate that the catalytic domain is protease resistant or less abundant than the repeats.
While MXAN4860 and MXAN4860 fibA mutants develop, the MXAN4860 pilA mutant is delayed in development compared to the pilA mutant. This pattern of phenotypes is consistent with MXAN4860 functioning in the FibA mediated developmental process. However, if the MXAN4860-FibA process were linear, disruption of MXAN4860 in the pilA background would abolish development like a fibA pilA disruption. Instead only a partial defect was observed, suggesting that multiple processes integrate into or out of FibA including an MXAN4860-mediated process affecting developmental timing. One possibility is that FibA may be part of a protease cascade since FibA containing a mutation in a critical active site residue is still processed in vivo (12). In humans, membrane-associated matrix metalloproteinase 1 proteolytically activates extracellular matrix metalloproteinase 2 (gelatinase A) and is regulated by a prostaglandin-cAMP pathway (45). By analogy, FibA may activate other extracellular proteins in a protease cascade to effect signaling.
None of the genes encoding putative ECM proteins are essential for cell attachment, adventurous or social motility, or fruiting body formation and sporulation (outside of MXAN4860 and MXAN2710). Some of these processes have been shown to be dependent on the ECM (1, 2, 35, 36, 59), which would suggest that the polysaccharide component of the matrix performs the bulk of the ECM functions. However, so far it has been impossible to separate the functions of polysaccharide and protein in the ECM as no mutants or conditions have been described where developing cells produce polysaccharide without associated protein. The lack of striking phenotypes in ECM protein disruptions may be due in some cases to functional redundancy of protein components, which has been well established for this organism. The fibA and pilA deletions cause little loss of development unless they are combined. It is also possible that critical components of the ECM proteome were not detected in this analysis. Phenol extraction of 0.5% SDS-isolated ECM removed only 57% of the protein content (data not shown), indicating that a significant portion of ECM protein may be either covalently coupled or difficult to extract. Identification and characterization of more tightly associated ECM proteins may reveal new signaling and structural capacities of the ECM and increase the understanding of previously identified ECM-mediated functions.
Supplementary Material
Acknowledgments
We thank Jon Amster and Dennis Phillips for their advice, expertise, and generous help with proteomics, Aurelio Moraleda-Muñoz for generating the FibA His-tagged fusion protein used to produce polyclonal antibodies, Natalie Iacopelli for her assistance with making mutations, and Roy Welch for supplying annotated versions of the genome.
This material is based on work supported by the National Science Foundation under grant no. 0343874 to L. Shimkets.
Footnotes
Published ahead of print on 31 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arnold, J. W., and L. J. Shimkets. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold, J. W., and L. J. Shimkets. 1988. Inhibition of cell-cell interactions in Myxococcus xanthus by Congo red. J. Bacteriol. 170:5765-5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behmlander, R. M., and M. Dworkin. 1994. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6295-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behmlander, R. M., and M. Dworkin. 1991. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J. Bacteriol. 173:7810-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behmlander, R. M., and M. Dworkin. 1994. Integral proteins of the extracellular matrix fibrils of Myxococcus xanthus. J. Bacteriol. 176:6304-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellenger, K., X. Ma, W. Shi, and Z. Yang. 2002. A CheW homologue is required for Myxococcus xanthus fruiting body development, social gliding motility, and fibril biogenesis. J. Bacteriol. 184:5654-5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, W., D. van de Pol, M. Warburg, A. Gal, L. Bleeker-Wagemakers, H. de Silva, A. Meindl, T. Meitinger, F. Cremers, and H. H. Ropers. 1992. Mutations in the candidate gene for Norrie disease. Hum. Mol. Genet. 1:461-465. [DOI] [PubMed] [Google Scholar]
- 8.Black, W. P., Q. Xu, and Z. Yang. 2006. Type IV pili function upstream of the Dif chemotaxis pathway in Myxococcus xanthus EPS regulation. Mol. Microbiol. 61:447-456. [DOI] [PubMed] [Google Scholar]
- 9.Black, W. P., and Z. Yang. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 186:1001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blom, A. B., P. L. van Lent, S. Libregts, A. E. Holthuysen, P. M. van der Kraan, N. van Rooijen, and W. B. van den Berg. 2007. Crucial role of macrophages in matrix metalloproteinase-mediated cartilage destruction during experimental osteoarthritis: involvement of matrix metalloproteinase 3. Arthritis Rheum. 56:147-157. [DOI] [PubMed] [Google Scholar]
- 11.Bodet, C., F. Chandad, and D. Grenier. 2007. Inhibition of host extracellular matrix destructive enzyme production and activity by a high-molecular-weight cranberry fraction. J. Periodontal. Res. 42:159-168. [DOI] [PubMed] [Google Scholar]
- 12.Bonner, P. J., W. P. Black, Z. Yang, and L. J. Shimkets. 2006. FibA and PilA act cooperatively during fruiting body formation of Myxococcus xanthus. Mol. Microbiol. 61:1283-1293. [DOI] [PubMed] [Google Scholar]
- 13.Bonner, P. J., Q. Xu, W. P. Black, Z. Li, Z. Yang, and L. J. Shimkets. 2005. The Dif chemosensory pathway is directly involved in phosphatidylethanolamine sensory transduction in Myxococcus xanthus. Mol. Microbiol. 57:1499-1508. [DOI] [PubMed] [Google Scholar]
- 14.Bradley, R. S., and A. M. Brown. 1990. The proto-oncogene int-1 encodes a secreted protein associated with the extracellular matrix. EMBO J. 9:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 16.Campos, J. M., J. Geisselsoder, and D. R. Zusman. 1978. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J. Mol. Biol. 119:167-178. [DOI] [PubMed] [Google Scholar]
- 17.Chang, B. Y., and M. Dworkin. 1994. Isolated fibrils rescue cohesion and development in the Dsp mutant of Myxococcus xanthus. J. Bacteriol. 176:7190-7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang, B. Y., and M. Dworkin. 1996. Mutants of Myxococcus xanthus dsp defective in fibril binding. J. Bacteriol. 178:697-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cong, F., L. Schweizer, and H. Varmus. 2004. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development 131:5103-5115. [DOI] [PubMed] [Google Scholar]
- 20.Curtis, P. D., R. Geyer, D. C. White, and L. J. Shimkets. 2006. Novel lipids in Myxococcus xanthus and their role in chemotaxis. Environ. Microbiol. 8:1935-1949. [DOI] [PubMed] [Google Scholar]
- 21.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 22.Domka, J., J. Lee, T. Bansal, and T. K. Wood. 2007. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ. Microbiol. 9:332-346. [DOI] [PubMed] [Google Scholar]
- 23.Goldman, B. S., W. C. Nierman, D. Kaiser, S. C. Slater, A. S. Durkin, J. A. Eisen, C. M. Ronning, W. B. Barbazuk, M. Blanchard, C. Field, C. Halling, G. Hinkle, O. Iartchuk, H. S. Kim, C. Mackenzie, R. Madupu, N. Miller, A. Shvartsbeyn, S. A. Sullivan, M. Vaudin, R. Wiegand, and H. B. Kaplan. 2006. Evolution of sensory complexity recorded in a myxobacterial genome. Proc. Natl. Acad. Sci. USA 103:15200-15205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gollop, R., M. Inouye, and S. Inouye. 1991. Protein U, a late-developmental spore coat protein of Myxococcus xanthus, is a secretory protein. J. Bacteriol. 173:3597-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabellus, F., K. Worm, and K. W. Schmid. 2007. Induction of the matrix metalloproteinase-2 activation system in arteries by tensile stress. Involvement of the p38 MAP-kinase pathway. Pathol. Res. Pract. 203:135-143. [DOI] [PubMed] [Google Scholar]
- 26.Juncker, A. S., H. Willenbrock, G. Von Heijne, S. Brunak, H. Nielsen, and A. Krogh. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser, D. 1979. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 76:5952-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashefi, K., and P. L. Hartzell. 1995. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF− defect. Mol. Microbiol. 15:483-494. [DOI] [PubMed] [Google Scholar]
- 29.Kearns, D. B., P. J. Bonner, D. R. Smith, and L. J. Shimkets. 2002. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J. Bacteriol. 184:1678-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kearns, D. B., B. D. Campbell, and L. J. Shimkets. 2000. Myxococcus xanthus fibril appendages are essential for excitation by a phospholipid attractant. Proc. Natl. Acad. Sci. USA 97:11505-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearns, D. B., A. Venot, P. J. Bonner, B. Stevens, G.-J. Boons, and L. J. Shimkets. 2001. Identification of a developmental chemoattractant in Myxococcus xanthus through metabolic engineering. Proc. Natl. Acad. Sci. USA 98:13990-13994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurayoshi, M., H. Yamamoto, S. Izumi, and A. Kikuchi. 2007. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochem. J. 402:515-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, D. Q., and S. C. Pflugfelder. 2005. Matrix metalloproteinases in corneal inflammation. Ocul. Surf. 3:S198-S202. [DOI] [PubMed] [Google Scholar]
- 34.Li, Y., R. Lux, A. E. Pelling, J. K. Gimzewski, and W. Shi. 2005. Analysis of type IV pilus and its associated motility in Myxococcus xanthus using an antibody reactive with native pilin and pili. Microbiology 151:353-360. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., H. Sun, X. Ma, A. Lu, R. Lux, D. Zusman, and W. Shi. 2003. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100:5443-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, A., K. Cho, W. P. Black, X. Y. Duan, R. Lux, Z. Yang, H. B. Kaplan, D. R. Zusman, and W. Shi. 2005. Exopolysaccharide biosynthesis genes required for social motility in Myxococcus xanthus. Mol. Microbiol. 55:206-220. [DOI] [PubMed] [Google Scholar]
- 37.Lund, L. R., J. Romer, T. H. Bugge, B. S. Nielsen, T. L. Frandsen, J. L. Degen, R. W. Stephens, and K. Dano. 1999. Functional overlap between two classes of matrix-degrading proteases in wound healing. EMBO J. 18:4645-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik, M. T., and S. S. Kakar. 2006. Regulation of angiogenesis and invasion by human Pituitary tumor transforming gene (PTTG) through increased expression and secretion of matrix metalloproteinase-2 (MMP-2). Mol. Cancer 5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Overall, C. M. 2002. Molecular determinants of metalloproteinase substrate specificity: matrix metalloproteinase substrate binding domains, modules, and exosites. Mol. Biotechnol. 22:51-86. [DOI] [PubMed] [Google Scholar]
- 40.Pedrioli, P. G., J. K. Eng, R. Hubley, M. Vogelzang, E. W. Deutsch, B. Raught, B. Pratt, E. Nilsson, R. H. Angeletti, R. Apweiler, K. Cheung, C. E. Costello, H. Hermjakob, S. Huang, R. K. Julian, E. Kapp, M. E. McComb, S. G. Oliver, G. Omenn, N. W. Paton, R. Simpson, R. Smith, C. F. Taylor, W. Zhu, and R. Aebersold. 2004. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22:1459-1466. [DOI] [PubMed] [Google Scholar]
- 41.Poza, M., M. Prieto-Alcedo, C. Sieiro, and T. G. Villa. 2004. Cloning and expression of clt genes encoding milk-clotting proteases from Myxococcus xanthus 422. Appl. Environ. Microbiol. 70:6337-6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quillet, L., L. Bensmail, S. Barray, and J. Guespin-Michel. 1997. Cloning and sequencing of two genes, prtA and prtB, from Myxococcus xanthus, encoding PrtA and PrtB proteases, both of which are required for the protease activity. Gene 198:135-140. [DOI] [PubMed] [Google Scholar]
- 43.Reichsman, F., L. Smith, and S. Cumberledge. 1996. Glycosaminoglycans can modulate extracellular localization of the wingless protein and promote signal transduction. J. Cell Biol. 135:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seydel, A., P. Gounon, and A. P. Pugsley. 1999. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol. Microbiol. 34:810-821. [DOI] [PubMed] [Google Scholar]
- 45.Shankavaram, U. T., W. C. Lai, S. Netzel-Arnett, P. R. Mangan, J. A. Ardans, N. Caterina, W. G. Stetler-Stevenson, H. Birkedal-Hansen, and L. M. Wahl. 2001. Monocyte membrane type 1-matrix metalloproteinase. Prostaglandin-dependent regulation and role in metalloproteinase-2 activation. J. Biol. Chem. 276:19027-19032. [DOI] [PubMed] [Google Scholar]
- 46.Simunovic, V., F. C. Gherardini, and L. J. Shimkets. 2003. Membrane localization of motility, signaling, and polyketide synthetase proteins in Myxococcus xanthus. J. Bacteriol. 185:5066-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternlicht, M. D., and Z. Werb. 2001. How matrix metalloproteinases regulate cell behavior. Annu. Rev. Cell Dev. Biol. 17:463-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, H., D. R. Zusman, and W. Shi. 2000. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10:1143-1146. [DOI] [PubMed] [Google Scholar]
- 50.Sutherland, I. W. 2001. The biofilm matrix—an immobilized but dynamic microbial environment. Trends Microbiol. 9:222-227. [DOI] [PubMed] [Google Scholar]
- 51.Tjaderhane, L., T. Hotakainen, S. Kinnunen, M. Ahonen, and T. Salo. 2007. The effect of chemical inhibition of matrix metalloproteinases on the size of experimentally induced apical periodontitis. Int. Endod. J. 40:282-289. [DOI] [PubMed] [Google Scholar]
- 52.Vlamakis, H. C., J. R. Kirby, and D. R. Zusman. 2004. The Che4 pathway of Myxococcus xanthus regulates type IV pilus-mediated motility. Mol. Microbiol. 52:1799-1811. [DOI] [PubMed] [Google Scholar]
- 53.Weatherly, D. B., J. A. Atwood, T. A. Minning, C. Cavola, R. L. Tarleton, and R. Orlando. 2005. A heuristic method for assigning a false-discovery rate for protein identifications from mascot database search results. Mol. Cell. Proteomics 4:762-772. [DOI] [PubMed] [Google Scholar]
- 54.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 55.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 56.Wodarz, A., and R. Nusse. 1998. Mechanisms of Wnt signaling in development. Annu. Rev. Cell Dev. Biol. 14:59-88. [DOI] [PubMed] [Google Scholar]
- 57.Wu, S. S., and D. Kaiser. 1996. Markerless deletions of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacB gene. J. Bacteriol. 178:5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, S. S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 59.Yang, Z., X. Ma, L. Tong, H. B. Kaplan, L. J. Shimkets, and W. Shi. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J. Bacteriol. 182:5793-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaragoza, R., A. Gimeno, V. J. Miralles, E. R. Garcia-Trevijano, R. Carmena, C. Garcia, M. Mata, I. R. Puertes, L. Torres, and J. R. Vina. 2007. Retinoids induce MMP-9 expression through RARα during mammary gland remodeling. Am. J. Physiol. Endocrinol. Metab. 292:E1140-E1148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.