Abstract
Soluble (S-type) pyocins are Pseudomonas aeruginosa bacteriocins that kill nonimmune P. aeruginosa strains via a specific receptor. The genes coding for pyocin Sa (consisting of a killing protein and an immunity protein) were cloned and expressed in Escherichia coli. Sequence analysis revealed that Sa is identical to pyocin S2. Seventy-nine strains of P. aeruginosa were tested for their sensitivity to pyocins S1, S2, and S3, and their ferripyoverdine receptors were typed by multiplex PCR. No strain was found to be sensitive to both S2 and S3, suggesting that the receptors for these two pyocins cannot coexist in one strain. As expected, all S3-sensitive strains had the type II ferripyoverdine receptor fpvA gene, confirming our previous reports. S1 killed strains irrespective of the type of ferripyoverdine receptor they produced. All S2-sensitive strains had the type I fpvA gene, and the inactivation of type I fpvA in an S2-sensitive strain conferred resistance to the S2 pyocin. Accordingly, complementation with type I fpvA in trans restored sensitivity to S2. Some S2-resistant type I fpvA-positive strains were detected, the majority (all but five) of which had the S1-S2 immunity gene. Comparison of type I fpvA sequences from immunity gene-negative S2-sensitive and S2-resistant strains revealed only a valine-to-isoleucine substitution at position 46 of type I FpvA. However, both type I fpvA genes conferred the capacity for type I pyoverdine utilization and sensitivity to S2. When these two type I fpvA genes were introduced into strain 7NSK2 carrying mutations in type II fpvA (encoding the type II pyoverdine receptor) and fpvB (encoding the alternative type I receptor), growth in the presence of type I pyoverdine was observed and the strain became sensitive to S2. We also found that type I pyoverdine could signal type II pyoverdine production via the type I FpvA receptor in 7NSK2.
Pseudomonas aeruginosa is a ubiquitous gram-negative gammaproteobacterium able to cause severe nosocomial infections and lung infections in cystic fibrosis patients (21). P. aeruginosa is also notorious for its high level of resistance to several antibiotics (17). Like other aerobes, P. aeruginosa produces siderophores to fulfill its needs for iron (9, 29, 37). The major siderophore of P. aeruginosa is the fluorescent pyoverdine, which has a conserved chromophore and a variable peptide chain (29, 37). Both the chromophore and the peptide chain are synthesized by nonribosomal peptide synthetases (29, 37). Three different pyoverdine types in P. aeruginosa can be distinguished, each having a specific receptor that coevolves with the nonribosomal peptide synthetase genes (8, 10, 23, 34, 35). Pyocins are bacteriocins produced by P. aeruginosa that kill other strains from the same species (24, 26). P. aeruginosa pyocins can be subdivided into insoluble (particular phage-like) R and F pyocins and soluble S pyocins, and only the S pyocins are protease and heat sensitive (24). Soluble pyocins have an N-terminal receptor binding domain, a translocation domain, and a C-terminal killing domain (24). The majority of S-type pyocins (AP41, S1, S2, and S3) cause cell death by DNA breakdown due to an endonuclease C-terminal domain (24), while pyocin S4 is predicted to have tRNase activity and S5 is predicted to have pore-forming activity (24, 26). Pyocin-producing strains are protected from their own toxin by an immunity protein encoded by an adjacent gene (24). It is known that iron limitation greatly increases the killing activity of some S pyocins, including S2, S3, and Sa (11, 25, 33). Ohkawa et al. (25) first reported that mutants resistant to S2 fail to produce an iron-repressed outer membrane protein but did not establish the link with the ferripyoverdine receptor. Smith et al. (33) suggested that the pyocin Sa receptor could be the ferripyoverdine receptor, although they observed that a receptor-negative mutant, although resistant to Sa, still had some considerable residual pyoverdine utilization capacity, an observation that was not understood at that time but was later explained by the discovery of a second alternative type I ferripyoverdine receptor, FpvB (13). Later on, it was found that the receptor for type II ferripyoverdine is used by S3 to kill sensitive P. aeruginosa strains (1, 10). In this work, we show that pyocin Sa is in fact identical to pyocin S2 and that type I FpvA (the major type I ferripyoverdine receptor) is the receptor for this pyocin. The results presented here also suggest mechanisms of resistance to soluble pyocins other than the production of immunity proteins.
MATERIALS AND METHODS
Strains and plasmids.
Bacterial strains and plasmids used in this study are shown in Table 1. Strains were grown at 37°C in rich Luria broth (LB) medium (Life Technologies) or iron-poor Casamino Acids (CAA) medium (Difco Laboratories) in a New Brunswick Innova 4000 shaker at 200 rpm. The antibiotics spectinomycin, gentamicin, chloramphenicol, and ampicillin were used. The concentrations used in different media for Escherichia coli and P. aeruginosa are mentioned below.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| O:9 | Wild-type pyocin Sa-sensitive indicator strain | 33 |
| O:9fpvA− | fpvA mutant of O:9 | This work |
| W15Dec1 | Wild-type pyocin S2-sensitive strain | 27 |
| W15Dec1fpvA− | fpvA mutant of W15Dec1 | This work |
| W15Dec30 | Wild-type pyocin S2-resistant strain | This work |
| W15Apr21 | Wild-type pyocin S2-resistant strain | This work |
| W15Apr38.4 | Wild-type pyocin S2-resistant strain | This work |
| PAO29P | Wild-type pyocin S2-resistant strain | J. P. Pirnay |
| J1003 | Wild-type pyocin Sa-producing strain | 14 |
| 7NSK2fpvA−fpvB− | Pyoverdine receptor-negative mutant; Gmr Tcr | 13 |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15 recA hsdR17 recA1 endA1 gyrA96 thi-1 relA1) | 16 |
| GJ23 | JC2692(pGJ28)(R64drd11); Kmr Smr Tcr | 36 |
| S17.1 (λ pir) | Tpr Smr recA thi pro hsdRM+ RP4::2-Tc::Mu::Km Tn7 λ pir | 32 |
| Plasmids | ||
| pBR325 | ColE1 vector; Ampr Cmr Tcr | 4 |
| pBR325::fpvAI::gm | pBR325 with a 1,200-bp BamHI-EcoRI type I fpvA fragment disrupted with a Gm cassette; Cmr Ampr Gmr | This work |
| pBBR1mcs | Wide-host-range cloning vector; Cmr | 20 |
| pRG930-cat | Derivative of cosmid vector pRG930 with Cmr cassette; wide host range | 22 |
| pPYS3-3 | Pyocin S3 gene-containing plasmid | 11 |
| pYMSS11 | Pyocin S1 gene-containing plasmid | 31 |
| pYMPS1 | Pyocin S2 gene-containing plasmid | 31 |
| pPVR2 | pAK1900 derivative carrying fpvA on a 4.6-kb SphI fragment | 28 |
Gmr, Tcr, Kmr, Smr, Ampr, and Cmr indicate gentamicin, tetracycline, kanamycin, spectinomycin, ampicillin, and chloramphenicol resistance cassettes, respectively.
Sequence determination and analysis.
Sequencing was done at the VIB Genetic Service Facility (University of Antwerp, Antwerp, Belgium), and sequences were compared with entries in the P. aeruginosa database (http://v2.pseudomonas.com).
Multiplex PCR for the identification of type I, II, and III fpvA genes and fpvB.
The different fpvA genes were identified by multiplex PCR as described previously by De Chial et al. (10). Primers for the fpvB gene were included (13).
IEF and CAS overlay.
An isoelectric focusing (IEF) and Chrome Azurol S (CAS) assay of partially purified pyoverdines from different P. aeruginosa isolates was performed as described by Koedam et al. (19) in order to differentiate type I, II, and III pyoverdines (23).
Isolation of pyoverdines.
Pyoverdines for IEF and CAS analysis were partially purified from 4-ml cultures grown for 48 h (19). Type I pyoverdine for growth stimulation experiments was purified as described previously (23). The concentration of pyoverdine in a solution was estimated by measuring the absorbance at 400 nm (ɛ = 2 × 104 M−1 cm−1) (23).
Pyoverdine growth stimulation assays.
Growth stimulation by type I pyoverdine on CAA with 0.5 mg of ethylenediaminedihydroxyphenylacetic acid (EDDHA) ml−1 was done by plating 5 × 106 cells of a P. aeruginosa culture per ml onto the agar surface. A filter disk impregnated with 5 μl of a 10 mM type I pyoverdine solution was deposited on top of the bacterial cell layer. Plates were incubated at 37°C, and growth was recorded after 24 and 48 h. Plates were photographed with a Fuji digital camera (Finepix S1 Pro).
Construction of a genomic library of P. aeruginosa J1003 and screening of S-type-pyocin-producing clones.
Total DNA of P. aeruginosa J1003 was isolated using the blood and cell culture DNA midi kit (QIAGEN). Partially Sau3A-digested genome fragments were cloned into the BamHI site of pRG930-cat (22). Ligated DNA was packaged using the Gigapack III Gold-4 kit (Stratagene). About 2,200 clones were selected on LB agar plates containing 50 μg of spectinomycin ml−1. To screen for S-type-pyocin-producing clones, the genomic library was replicated on LB agar plates containing 50 μg of spectinomycin ml−1 and grown for 12 h. Cells were killed by chloroform vapors over a period of 20 min, and plates were kept open for another 20 min to eliminate all traces of chloroform. An overlay of soft LB agar (5 g of agar liter−1) containing 5 × 106 cells of the pyocin Sa-sensitive indicator strain P. aeruginosa O:9 ml−1 was used to screen for pyocin-producing clones. After incubation for 12 h, a clearance zone was seen. To distinguish S-type-pyocin-producing clones from others (those producing R- and F-type pyocins), a secondary screening of pyocin-producing clones was carried out with the addition of 100 μg of proteinase K ml−1 to the agar overlay. Wild-type strain J1003 and E. coli DH5α carrying the empty pRG930-cat vector were used as control strains.
Subcloning of S-type-pyocin-expressing fragments from cosmid 2H12.
Self-ligated PstI-digested cosmid 2H12 generated a subclone containing a ∼10-kb insert and still producing an S-type pyocin. The 10-kb fragment was gel purified with a kit from QIAGEN, cut with HindIII, and subsequently ligated into the PstI-HindIII site of pBBR1mcs, yielding an approximately 4.5-kb fragment that was still able to express the pyocin. This fragment was further analyzed by sequencing.
Generation of ΔfpvA deletion mutants by allelic exchange.
A 1,200-bp fpvA fragment was amplified using wild-type P. aeruginosa PAO1 genomic DNA and primers FpvABamHI (5′-GTG GGA TCC CAG TGC GAT CAA GGG CAA-3′) and FpvASalI (5′-GTG GTC GAC GCA GGT TCC AGT AGC TCT-3′). The PCR was performed using Taq polymerase (QIAGEN) with the following parameters: 30 s at 94°C, 30 s at 52°C, and 2 min at 72°C for 30 cycles. The PCR product purified by using a QIAGEN kit was ligated into BamHI-SalI-restricted pBluescript KS(+) (Stratagene). The constructed plasmid was digested at a unique EcoRI site and ligated with a Gm cassette. This Gm cassette was obtained by PCR with transposon miniTn5phoA3 (pGV4692) (10) as a template and primers GmFw1 (5′-GTG GAA TTC AAC GGA TGA AGG CAC GAA-3′) and GmRv1 (5′-GTG CTT AAG CGA TCT CGG CTT GAA CGA-3′). The PCR was carried out using Taq polymerase and the following parameters: 30 s at 94°C, 30 s at 51°C, and 1 min at 51°C for 30 cycles, followed by a terminal extension step for 10 min at 72°C. The construct was checked by sequencing.
The disrupted fpvA gene was then transferred into BamHI-SalI-digested pBR325 (Stratagene), competent E. coli DH5α cells were transformed with the plasmid, and chloramphenicol- and gentamicin-resistant clones were selected. Additional screening was performed by digestion. The selected clones were transferred into competent E. coli GJ23 cells for mobilization of the disrupted fpvA gene into S2-sensitive acceptor P. aeruginosa strain W15Dec1 by conjugation. Recombinants were selected on CAA agar plates containing 100 μg of gentamicin ml−1 and 50 μg of spectinomycin ml−1 to counterselect for E. coli. Double recombinants were selected as those clones growing on CAA plates with 100 μg of gentamicin ml−1 and not in the presence of 300 μg of chloramphenicol ml−1. Candidates were confirmed by PCR with primers FpvABamHI and FpvASalI, which resulted in the amplification of a 1,900-bp fragment from the mutants.
Construction of the type I fpvA(V46) and fpvA(I46) expression vectors.
For the expression of type I FpvA carrying valine at position 46 [FpvA(V46)] in trans, a 3,956-bp PstI-ApaI fragment of pPVR2 was ligated into pBBR1mcs. Type I FpvA(I46) was generated by PCR using genomic DNA of P. aeruginosa PAO29p as the template and primers FpvAPstI (5′-GTG CTG CAG CGA CCA TCA TCG AGA TCA-3′) and FpvAXbaI (5′-GTG TCT AGA GGT AGC CGA GAA GCA GAA-3′). The PCR was performed using ProofStart polymerase in combination with Taq polymerase (according to the QIAGEN protocol) and the following parameters: 30 cycles of 10 s at 94°C, 1 min at 51°C, and 3 min at 68°C, with a final extension of 10 min at 68°C. The purified PCR product was then ligated into the XbaI-PstI site of pBBR1mcs. Constructs were confirmed by sequencing.
Complementation and heterologous expression of type I fpvA.
Biparental matings with E. coli S17.1 λ pir bearing plasmid pBBR1mcs::fpvA (expressing FpvA with V46 or I46) were performed for the complementation of P. aeruginosa strain W15Dec1 carrying a mutation in fpvA (mutant strain W15Dec1fpvA−) or for the heterologous expression of the ferripyoverdine receptor in strain 7NSK2 carrying mutations in type II fpvA and fpvB (mutant strain 7NSK2fpvA−fpvB−). The complemented mutants were selected on LB supplemented with 300 μg of chloramphenicol ml−1.
SDS-PAGE analysis of outer membrane proteins.
Outer membrane proteins from bacteria grown under iron-limiting conditions (in CAA medium) were prepared by following the differential Sarkosyl solubilization method (7, 12). Cells were obtained from 250-ml cultures after 48 h of growth by centrifugation at 4°C for 10 min at 7,000 rpm in a Sorvall RC5B SLA rotor. The pellet was resuspended in 4 ml of GTE (50 mM glucose, 25 mM Tris-HCl [pH 8.0], and 10 mM EDTA containing 4 mg of lysozyme per ml). After 30 min of incubation on ice, 20 ml of PMSF-Sarkosyl solution (2 mM phenylmethylsulfonyl fluoride, 2% N-lauroylsarkosine) was added. Next, the mixture was sonicated for 15 min or until it reached homogeneity. The total outer membranes were pelleted by ultracentrifugation at 28,000 rpm for 2 h at 4°C in a Beckman TL-100 centrifuge equipped with a Ti 1003 rotor. Finally, the pellet was resuspended in 1 ml of 25 mM Tris-HCl, pH 7.5. The protein content was determined by using the QuantiPro bicinchoninic acid assay kit (Sigma), and proteins were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE).
Pyocin sensitivity assays: spot method and CFU counting.
To check the sensitivities of P. aeruginosa strains to pyocins S1, S2, and S3, 10 μl of pyocin lysate was spotted onto a bacterial cell layer containing 5 × 106 cells ml−1. Pyocins were partially purified from E. coli DH5α bearing plasmid pYMSS11, pYMPS1, or pPYS3-3 expressing pyocin S1, S2, or S3, respectively (11, 31).
Alternatively, bacterial cells (5 × 106 cells ml−1) were incubated at 37°C for 30 min with different amounts of pyocin S2. One hundred microliters of a 5 × 103 dilution was plated in triplicate onto CAA plates, and surviving colonies were counted after growth at 37°C for 24 h.
Screening for S-type pyocin production by P. aeruginosa isolates.
To determine the type of S pyocin produced by different P. aeruginosa isolates, products from PCRs with the following primers specific for the pyocin types were assessed: S1S2imm-Fw (5′-CACAAGGGAGGGAAGTGA-3′) and S1S2imm-Rv (5′-CGGCCTTAAAGCCAGGAA-3′) for pyocins S1 and S2 and pycS3RB-fw (5′-CGTATCACGAGACAGGCA-3′) and pycS3RB-rv (5′-TGCCGCTTCTTCCGCTTT-3′) for pyocin S3. Because pyocins S1 and S2 share the same immunity protein, primers corresponding to the region encoding this protein were chosen. The PCR was done with Taq polymerase and the following conditions: 30 cycles of 30 s at 94°C, 30 s at 52°C, and 30 s at 72°C.
RESULTS
Molecular cloning of pyocin Sa.
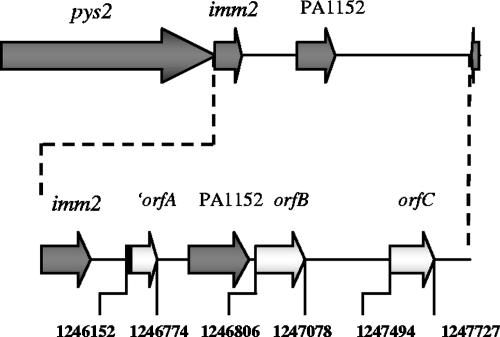
A genomic library of the Sa pyocin-producing strain J1003 was made in the wide-host-range cosmid vector pRG930-Cm (22). The recombinant E. coli clones were screened for their pyocin killing activity by using the Sa-sensitive P. aeruginosa O:9 strain as the indicator. In order to differentiate killing by R and F pyocins from killing by S pyocins, replicas were made with or without proteinase K in the top agar containing the sensitive strain. Out of 2,200 colonies, 38 displayed killing activity, 15 of which produced a pyocin that was proteinase K sensitive. Ten clones with the highest level of protease-sensitive killing activity were selected, and their cosmids were extracted and restricted with SalI or PstI. Their restriction profiles were very similar, and two clones were selected for further work. After subcloning, a 4,589-bp PstI-HindIII fragment that conferred full killing activity was obtained. Sequencing of this fragment revealed two open reading frames corresponding to the pyocin killer protein and the immunity protein. Surprisingly, these open reading frames were 100% identical to the sequences annotated as pys2 (PA1150) and imm2 (PA1151), encoding pyocin S2 and the immunity protein, respectively, of P. aeruginosa PAO1 (Fig. 1). The fragment also contained the genes PA1152 (encoding a hypothetical protein) and PA1153 (carrying the COG3617 prophage antirepressor motif), as annotated in the P. aeruginosa PAO1 genome (http://www.pseudomonas.com). Via a BLASTX analysis, we found an incomplete reading frame (′orfA) corresponding to part of the ofn44 gene (Q4W1R3_PSEAE) from the IncP-6 plasmid Rsm149 (15) in the intergenic region between the imm2 gene and the PA1152 gene, which codes for a conserved hypothetical protein corresponding to the Tn3 family of transposons (Fig. 1). Two other open reading frames were found downstream of the PA1152 gene, one (orfB) encoding a putative colicin immunity protein (Q66CV8_YERPS) and the other (orfC) encoding a hypothetical protein (Q3KAI8_PSEPF). Based on these results, we can conclude that pyocin Sa is pyocin S2, since no other cosmid with protease-sensitive pyocin activity was found. Therefore, from now on this pyocin will be designated pyocin S2.
FIG. 1.
pys2-PA1153 locus. The open reading frames in dark gray are those annotated in the PAO1 genome (http://www.pseudomonas.com), while those in white are predicted based on a BLASTX analysis. The first one (orfA) corresponds to part of a conserved hypothetical protein gene of the Tn3 family of transposons. The two other open reading frames (orfB and orfC) encode a putative colicin immunity protein and a hypothetical protein, respectively. The region between imm2 and PA1153 is shown below in more detail. The genes are not to scale, but the numbers represent the nucleotide coordinates in the P. aeruginosa PAO genome.
Determination of the specificity of pyocins S1, S2, and S3.
Seventy-nine genotypically different P. aeruginosa strains from a previously molecularly typed collection (27) were screened for their sensitivities to three different S pyocins: S1, S2, and S3 (Table 2; see also Table S1 in the supplemental material). In parallel, the same isolates were subjected to a multiplex PCR analysis to type the ferripyoverdine receptor genes (type I, IIa, IIb, and III fpvA and fpvB) (13). No strain was sensitive to both S2 and S3 pyocins, indicating that the respective receptors for these two pyocins are not expressed in the same host (Table 2; see also Table S1 in the supplemental material). All 26 S3-sensitive strains had the type II pyoverdine fpvA gene (type IIa or IIb fpvA), confirming our previous results (1, 10). Ten strains were sensitive to S1 and S2 at the same time, eight were sensitive to S1 and S3, and S1 killed strains independently of the type of FpvA they produced (Table 2). All S2-sensitive strains (17 in total) had the type I ferripyoverdine fpvA gene. Intriguingly, 14 strains that were clearly shown by multiplex PCR to have the type I ferripyoverdine receptor gene were nevertheless resistant to S2 (Table 2). Nine of these S2-resistant strains carrying type I fpvA were shown to have the gene for immunity to S1 and S2, which is the same for the two pyocins (24, 31). The remaining five S2-resistant strains carrying type I fpvA were not positive by PCR for the S1 and S2 immunity gene, meaning that their resistance had another origin. Three of these strains were positive for type I fpvA but did not produce pyoverdine, which may result in reduced expression of the receptor (37). Interestingly, the well-studied and genome-sequenced type I pyoverdine producer PA14 had no S1-S2 immunity gene but was resistant to S2 (results not shown).
TABLE 2.
Phenotypes of sensitivity or resistance to pyocins S1, S2, and S3 of 79 P. aeruginosa isolatesa
| Pyocin and phenotype | No. of isolates with:
|
No. of nontypeable isolatesc | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Type I fpvA | Type I fpvA and S1-S2 imm | Type I fpvA and V→I mutation | Type II fpvA | Type II fpvA and S1-S2 imm | Type II fpvA and S3 imm | Type III fpvA | Type III fpvA and S1-S2 imm | ||
| Pyocin S1 | |||||||||
| Sensitive | 14 | 9 | 6 | ||||||
| Resistant | 18 | 9 | 20 | 9 | 9 | 4 | 3 | ||
| Pyocin S2 | |||||||||
| Sensitive | 17 | ||||||||
| Resistant | 15 | 9b | 6b | 29 | 9 | 15 | 3 | ||
| Pyocin S3 | |||||||||
| Sensitive | 26 | ||||||||
| Resistant | 32 | 3 | 3 | 15 | 3 | ||||
| Total | 32 | 9 | 10 | 29 | 9 | 3 | 15 | 4 | 3 |
In addition to the sensitivity testing, all strains were analyzed for the presence of type I, II, and III fpvA and fpvB by multiplex PCR (13). The presence of the S1-S2 and S3 immunity (imm) genes was checked by PCR as well. V→I indicates the presence of an isoleucine in place of a valine at position 46 of type I FpvA. Boldface indicates strains having fpvAI and fpvAII and sensitive to pyocins S2 and S3, respectively.
Four strains with type I fpvA have both the S1-S2 immunity gene and the V→I mutation at residue 46.
One strain could be identified as a type II pyoverdine producer by IEF but could not be typed by multiplex PCR.
Sequence analysis of the type I fpvA receptor from S2-resistant strains.
Although our screening suggested that type I FpvA could be the receptor recognized by S2, we were intrigued by the existence of some S2-resistant strains having the gene for this receptor and most likely producing it, since pyoverdine production is dependent on the interaction of pyoverdine with the receptor (37). The full type I fpvA genes from P. aeruginosa strains O:9 (S2 sensitive) and PAO29P (S2 resistant) were amplified and sequenced. A comparison of the two sequences revealed that there were only two nonsynonymous substitutions, resulting in amino acid changes at positions 24 (I→M) and 46 (V→I). Since position 24 is in the signal peptide, it is unlikely to have an impact on the function of the receptor, but residue 46 is in the N-terminal region of FpvA, which interacts with the FpvR antisigma factor (2, 30, 38). PA14, which is also resistant to S2 pyocin and does not have the immunity gene, has the same V→I substitution at position 46.
Effect of inactivation of type I fpvA on pyocin S2 sensitivity.
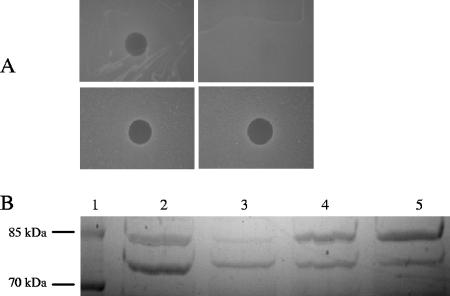
The type I fpvA gene from the S2-sensitive strain W15Dec1 was inactivated by allelic replacement, resulting in the insertion of a Gm cassette. Likewise, the type I fpvA gene in O:9 was inactivated, but work with this strain was stopped because of difficulties in maintaining plasmids in this particular host (results not shown). The W15Dec1 type I fpvA mutant was unable to grow in the presence of EDDHA but could grow when type I pyoverdine was added to the growth medium because of the presence of the fpvB gene (13). The W15Dec1 type I fpvA mutant also became fully resistant to pyocin S2 (Fig. 2A). Complementation with both type I fpvA(V46) and type I fpvA(I46) in trans restored the sensitivity to pyocin S2, suggesting that the substitution is not involved in the resistance to pyocin S2 (Fig. 2A). The production of the two forms of type I FpvA could be confirmed by SDS-PAGE analysis of outer membrane proteins extracted from cells grown in CAA medium (Fig. 2B). Growth in liquid cultures was monitored with a Bioscreen apparatus that allows the recording of the optical densities in microwell cultures. The addition of pyocin S2 alone or together with pyoverdine totally inhibited the growth of the sensitive strain W15Dec1, while the type I fpvA mutant could grow in the presence of the bacteriocin (results not shown). Again, complementation with either type I fpvA(V46) or type I fpvA(I46) restored the sensitivity to S2 (results not shown).
FIG. 2.
Effect of the inactivation of type I fpvA in the pyocin S2-sensitive strain W15Dec1 and complementation in trans with the two type I fpvA genes (encoding V46 and I46). (A) Killing activity of pyocin S2 on wild-type W15Dec1 (top left panel), W15Dec1fpvA− (top right panel), W15Dec1fpvA− complemented with type I fpvA(V46) (bottom left panel), and W15Dec1fpvA− complemented with type I fpvA(I46) (bottom right panel). (B) SDS-PAGE analysis of outer membrane proteins from CAA-grown wild-type W15Dec1 (lane 2), the W15Dec1 type I fpvA mutant (lane 3), the W15Dec1 type I fpvA mutant complemented with type I fpvA(V46) (lane 4), and the W15Dec1 type I fpvA mutant complemented with type I fpvA(I46) (lane 5). The band below the 85-kDa marker is FpvA. Lane 1, molecular mass markers.
Expression of both forms of the type I fpvA gene confers the capacity for type I pyoverdine utilization and sensitivity to pyocin S2 on P. aeruginosa 7NSK2.
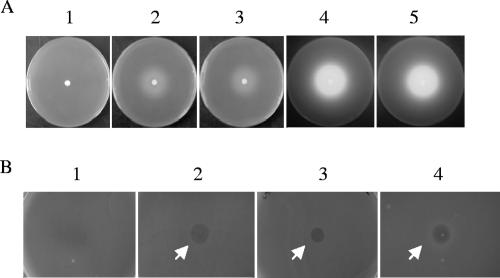
P. aeruginosa 7NSK2 is a type II pyoverdine-producing strain (10, 18). Accordingly, P. aeruginosa 7NSK2 has the type II fpvA receptor gene and is sensitive to pyocin S3 (1, 10). An 7NSK2 type II fpvA-fpvB double mutant is unable to utilize either type I ferripyoverdine (via FpvB) or type II ferripyoverdine because of the loss of the cognate receptor type II FpvA (13). Both type I fpvA genes (encoding V46 and I46) were cloned into the wide-host-range vector pBBR1mcs, and the resulting recombinant plasmids were introduced by conjugation into 7NSK2fpvA−fpvB−. In both cases, heterologous complementation with the type I fpvA gene conferred the capacity to grow in the presence of the high-affinity iron chelator EDDHA and type I pyoverdine (Fig. 3A). Surprisingly, the introduction of the heterologous type I fpvA gene not only conferred the ability to utilize externally added type I pyoverdine but also triggered the production of type II pyoverdine by 7NSK2, as judged by the intense fluorescent halo around the type I pyoverdine disk, suggesting that the type I FpvA receptor-FpvR antisigma factor interaction may signal type II pyoverdine production (Fig. 3A). Again, the expression of both variants of type I fpvA in trans rendered 7NSK2 sensitive to S2 (Fig. 3B).
FIG. 3.
Complementation of the type II pyoverdine 7NSK2fpvA−fpvB− mutant strain. (A) Growth stimulation of 7NSK2fpvA−fpvB− on CAA plates containing EDDHA around a type I pyoverdine-impregnated disk (panel 1) and the same mutant complemented with type I fpvA(V46) (panel 2) and type I fpvA(I46) (panel 3). Panels 4 and 5 are the plates from panels 2 and 3 shown under UV illumination to demonstrate the fluorescence due to type II pyoverdine production. (B) Results from an S2 pyocin killing assay of 7NSK2fpvA−fpvB− (resistant; panel 1) and the same mutant complemented with type I fpvA(V46) (sensitive; panel 2) and type I fpvA(I46) (sensitive; panel 3). Panel 4 shows the killing of the sensitive indicator strain O:9 as a positive control.
DISCUSSION
The production of pyocins by different P. aeruginosa strains suggests that these bacteria use these killer proteins to eliminate related competitors which colonize the same niche (24, 26). Normally, strains are immune to killing by their own pyocin thanks to the production of an immunity protein that binds to the nuclease C-terminal domain of the killing protein and inhibits its activity (24). The gene for the immunity protein is cotranscribed with the gene encoding the killer protein, ensuring that there is balanced production of the two proteins. However, in two recent transcriptome analyses, one of the response to H2O2 and the other of the response to the SOS-inducing antibiotic ciprofloxacin, it was observed that exposure to both agents induced the transcription of the killer pyocin S2 but repressed the transcription of the immunity gene (3, 6). It has been proposed previously that pyocins which recognize ferripyoverdine receptors are driving the evolution of these siderophore receptors and, at the same time, the diversification of pyoverdines (34). The term “Darwinian positive selection” has even been proposed (35). Our results demonstrate unambiguously that type I FpvA is needed for pyocin S2 killing. The fact that some strains (including PA14) having the FpvAI receptor but not the S1-S2 immunity gene still resist killing by S2 is puzzling. One possibility is that these strains produce an immunity protein different from that for S1 and S2 but able to recognize the killing domain of S2. A BLASTtn analysis of the PA14 genome using the S2 immunity protein sequence as a query did not reveal the existence of any immunity protein with similarity to that for S1 and S2. Another possibility is that type I FpvA is needed as the primary receptor but that a second receptor is necessary to allow the entry of the pyocin into the cell. This is the case for some E. coli colicins, which recognize the vitamin B12 receptor BtuB as the primary receptor but enter the cell, depending on the colicin, via the TolC or OmpF outer membrane porin (5). Also, more insight is needed about the way the pyocins are translocated into the cell before we can fully understand why some strains are resistant to killing. During this investigation, other interesting observations concerning pyoverdine signaling were made. First of all, we could demonstrate that the expression of type I fpvA in a type II fpvA host, in which the alternative FpvB receptor for type I ferripyoverdine is not produced, is possible and that the receptor is functional since it confers the ability to grow in the presence of EDDHA and type I pyoverdine. Even more surprising is that type I FpvA can signal the production of pyoverdine in the type II strain 7NSK2, which means that, in this strain at least, type I FpvA can interact with FpvR as efficiently as the cognate type II FpvA. In conclusion, the results presented here confirm the prediction made by Smith et al. in 1992 that the pyoverdine receptor is the receptor for Sa, and we have demonstrated that pyocin Sa is identical to the previously described pyocin S2 (25, 31, 33). The future challenge will be to understand how some strains apparently lacking an immunity gene and producing the type I S2 pyocin FpvA receptor can be resistant to killing by the S2 bacteriocin.
Supplementary Material
Acknowledgments
Sarah Denayer is the recipient of an IWT-Vlaanderen fellowship, and this research has been funded by an FWO (Fonds voor Wetenschappelijk Onderzoek Vlaanderen) grant.
We thank John Govan for giving us the strain O:9 and Jean Paul Pirnay for the strain PAO29P. We also thank Christine Baysse for the critical reading of the manuscript.
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Baysse, C., J. M. Meyer, P. Plesiat, V. Geoffroy, Y. Michel-Briand, and P. Cornelis. 1999. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J. Bacteriol. 181:3849-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. [DOI] [PubMed] [Google Scholar]
- 3.Blázquez, J., J. M. Gómez-Gómez, A. Oliver, C. Juan, V. Kapur, and S. Martins. 2006. PBP3 inhibition elicits adaptive responses in Pseudomonas aeruginosa. Mol. Microbiol. 62:84-99. [DOI] [PubMed] [Google Scholar]
- 4.Bólivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene 4:121-136. [DOI] [PubMed] [Google Scholar]
- 5.Cascales, E., S. K. Buchanan, D. Duche, C. Kleanthous, R. Lloubes, K. Postle, M. Riley, S. Slatin, and D. Cavard. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, W., D. A. Small, F. Toqhrol, and W. E. Bentley. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelis, P., A. Bouia, A. Belarbi, A. Guyonvarch, B. Kammerer, V. Hannaert, and J. C. Hubert. 1989. Cloning and analysis of the gene for the major outer membrane lipoprotein from Pseudomonas aeruginosa. Mol. Microbiol. 3:421-428. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, P., D. Hohnadel, and J. M. Meyer. 1989. Evidence for different pyoverdine-mediated iron uptake systems among Pseudomonas aeruginosa strains. Infect. Immun. 57:3491-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, P., and S. Matthijs. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4:787-798. [DOI] [PubMed] [Google Scholar]
- 10.De Chial, M., B. Ghysels, S. A. Beatson, V. Geoffroy, J. M. Meyer, T. Pattery, C. Baysse, P. Chablain, Y. N. Parsons, C. Winstanley, S. J. Cordwell, and P. Cornelis. 2003. Identification of type II and type III pyoverdine receptors from Pseudomonas aeruginosa. Microbiology 149:821-831. [DOI] [PubMed] [Google Scholar]
- 11.Duport, C., C. Baysse, and Y. Michel-Briand. 1995. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J. Biol. Chem. 270:8920-8927. [DOI] [PubMed] [Google Scholar]
- 12.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of cytoplasmic membrane of Escherichia coli by ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghysels, B., B. Thi Min Dieu, S. A. Beatson, J. P. Pirnay, U. A. Ochsner, M. L. Vasil, and P. Cornelis. 2004. FpvB, an alternative type I ferripyoverdine receptor of Pseudomonas aeruginosa. Microbiology 150:1671-1680. [DOI] [PubMed] [Google Scholar]
- 14.Govan, J. R. 1986. In vivo significance of bacteriocins and bacteriocin receptors. Scand. J. Infect. Dis. Suppl. 49:31-37. [PubMed] [Google Scholar]
- 15.Haines, A. S., K. Jones, M. Cheung, and C. M. Thomas. 2005. The IncP-6 plasmid Rsm149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187:4728-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, R. E., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist. Updat. 3:247-255. [DOI] [PubMed] [Google Scholar]
- 18.Höfte, M., S. Buysens, N. Koedam, and P. Cornelis. 1993. Zinc affects siderophore-mediated high affinity iron uptake systems in the rhizosphere Pseudomonas aeruginosa 7NSK2. Biometals 6:85-91. [DOI] [PubMed] [Google Scholar]
- 19.Koedam, N., E. Wittouck, A. Gaballa, A. Gillis, M. Höfte, and P. Cornelis. 1994. Detection and differentiation of microbial siderophores by isoelectric focusing and chrome azurol S overlay. Biometals 7:287-291. [DOI] [PubMed] [Google Scholar]
- 20.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. BioTechniques 16:800. [PubMed] [Google Scholar]
- 21.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matthijs, S., C. Baysse, N. Koedam, K. Abbaspour-Tehrani, L. Verheyden, H. Budzikiewicz, M. Schäfer, B. Hoorelbeke, J. M. Meyer, H. De Greve, and P. Cornelis. 2004. The Pseudomonas siderophore quinolobactin is synthezised from xanthurenic acid, an intermediate from the kynurenine pathway. Mol. Microbiol. 52:371-384. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, J. M., A. Stintzi, D. De Vos, P. Cornelis, R. Tappe, K. Taraz, and H. Budzikiewicz. 1997. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology 143:35-43. [DOI] [PubMed] [Google Scholar]
- 24.Michel-Briand, Y., and C. Baysse. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499-510. [DOI] [PubMed] [Google Scholar]
- 25.Ohkawa, I., S. Shiga, and M. Kageyama. 1980. Effect of iron concentration in the growth medium on the sensitivity of Pseudomonas aeruginosa to pyocin S2. J. Biochem. (Tokyo) 87:323-331. [DOI] [PubMed] [Google Scholar]
- 26.Parret, A. H., and R. De Mot. 2002. Bacteria killing their own kind: novel bacteriocins of Pseudomonas and other gamma-proteobacteria. Trends Microbiol. 10:107-112. [DOI] [PubMed] [Google Scholar]
- 27.Pirnay, J. P., S. Matthijs, H. Colak, P. Chablain, F. Bilocq, J. Van Eldere, D. De Vos, M. Zizi, L. Triest, and P. Cornelis. 2005. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbiol. 7:969-980. [DOI] [PubMed] [Google Scholar]
- 28.Poole, K., S. Neshat, K. Krebes, and D. E. Heinrichs. 1993. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175:4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravel, J., and P. Cornelis. 2003. Genomics of pyoverdine-mediated iron uptake in pseudomonads. Trends Microbiol. 11:195-200. [DOI] [PubMed] [Google Scholar]
- 30.Rédly, G. A., and K. Poole. 2005. FpvIR control of fpvA ferric pyoverdine receptor gene expression in Pseudomonas aeruginosa: demonstration of an interaction between FpvI and FpvR and identification of mutations in each compromising this interaction. J. Bacteriol. 187:5648-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sano, Y., M. Kobayashi, and M. Kageyama. 1993. Functional domains of S-type pyocins deduced from chimeric molecules. J. Bacteriol. 175:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Puhler. 1983. Broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784-791. [Google Scholar]
- 33.Smith, A. W., P. H. Hirst, K. Hughes, K. Gensberg, and J. R. Govan. 1992. The pyocin Sa receptor of Pseudomonas aeruginosa is associated with ferripyoverdin uptake. J. Bacteriol. 174:4847-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, E. E., E. H. Sims, D. H. Spencer, R. Kaul, and M. V. Olson. 2005. Evidence for diversifying selection at the pyoverdine locus of Pseudomonas aeruginosa. J. Bacteriol. 187:2138-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tümmler, B., and P. Cornelis. 2005. Pyoverdine receptor: a case of positive Darwinian selection in Pseudomonas aeruginosa. J. Bacteriol. 187:3289-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Haute, E., H. Joos, M. Maes, G. Warren, M. Van Montagu, and J. Schell. 1983. Intergeneric transfer and exchange recombination of restriction fragments cloned in pBR322: a novel strategy for the reversed genetics of the Ti plasmids of Agrobacterium tumefaciens. EMBO J. 2:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visca, P., F. Imperi, and I. L. Lamont. 2007. Pyoverdine siderophores: from biogenesis to biosignificance. Trends Microbiol. 15:22-30. [DOI] [PubMed] [Google Scholar]
- 38.Wirth, C., W. Meyer-Klaucke, F. Pattus, and D. Cobessi. 2007. From the periplasmic signaling domain to the extracellular face of an outer membrane signal transducer of Pseudomonas aeruginosa: crystal structure of the ferric pyoverdine outer membrane receptor. J. Mol. Biol. 368:398-406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.