Abstract
Streptococcus mutans, a normal inhabitant of dental plaque, is considered a primary etiological agent of dental caries. Its main virulence factors are acidogenicity and aciduricity, the abilities to produce acid and to survive and grow at low pH, respectively. Metabolic processes are finely regulated following acid exposure in S. mutans. Proteome analysis of S. mutans demonstrated that lactoylglutathione lyase (LGL) was up-regulated during acid challenge. The LGL enzyme catalyzes the conversion of toxic methylglyoxal, derived from glycolysis, to S-d-lactoylglutathione. Methylglyoxal inhibits the growth of cells in all types of organisms. The current study aimed to investigate the relationship between LGL and aciduricity and acidogenicity in S. mutans. An S. mutans isogenic mutant defective in lgl (LGLKO) was created, and its growth kinetics were characterized. Insertional inactivation of lgl resulted in an acid-sensitive phenotype. However, the glycolytic rate at pH 5.0 was greater for LGLKO than for S. mutans UA159 wild-type cells. LGL was involved in the detoxification of methylglyoxal, illustrated by the absence of enzyme activity in LGLKO and the hypersensitivity of LGLKO to methylglyoxal, compared with UA159 (MIC of 3.9 and 15.6 mM, respectively). Transcriptional analysis of lgl conducted by quantitative real-time PCR revealed that lgl was up-regulated (approximately sevenfold) during the exponential growth phase compared with that in the stationary growth phase. Gene expression studies conducted at low pH demonstrated that lgl was induced during acidic growth (∼3.5-fold) and following acid adaptation (∼2-fold).This study demonstrates that in S. mutans, LGL functions in the detoxification of methylglyoxal, resulting in increased aciduricity.
Dental caries is one of the most prevalent infectious dental diseases afflicting humans. The microorganism most strongly associated with human dental caries is Streptococcus mutans, a normal inhabitant of dental plaque. Two of the foremost cariogenic determinants of S. mutans are its acidogenicity and aciduricity (reviewed by Banas [3]). These properties are the ability to produce acid end products from the metabolism of dietary carbohydrates and the ability to survive and grow at low pH, respectively. Acidogenicity and aciduricity allow S. mutans to dramatically drop the pH within the dental plaque biofilm, which can promote the development and progression of carious lesions via its sustained dominance and acid production via glycolysis. The feast-or-famine conditions generated by the host diet can result in rapid increases in sugar that must quickly be removed from the environment. Likewise, the glycolytic intermediates must quickly be removed from the cell due to their toxicity.
Lactoylglutathione lyase (LGL) is an enzyme involved in the detoxification of methylglyoxal, a highly toxic electrophilic glycolytic by-product that reacts with and inactivates intracellular macromolecules, including both proteins and nucleic acids (9, 10). Therefore, its rapid degradation is vital for cell survival. The formation of methylglyoxal occurs via enzymatic production during glycolysis from the fragmentation of triose phosphates (4, 10). LGL is involved in methylglyoxal detoxification via the formation of S-d-lactoylglutathione from the hemimercaptal adduct that is formed nonenzymatically between glutathione and the 2-oxoaldehyde methylglyoxal (7). Glyoxalase II then converts S-d-lactoylglutathione into reduced glutathione and d-lactate (7). An examination of the S. mutans UA159 genome does not reveal the presence of a glyoxalase II homologue, suggesting an alternate pathway by which S-d-lactoylglutathione is neutralized. In Escherichia coli, methylglyoxal was accumulated under physiological conditions of uncontrolled carbohydrate metabolism (1, 6, 12) and the concentration of methylglyoxal was seen to be greater in highly metabolically active human red blood cells, where lgl expression appeared to be regulated by the rate of glycolysis (20, 23).
Salmonella enterica expressed elevated levels of lgl mRNA when engulfed in the acidic environment of the macrophage (5). Recent studies of S. mutans have suggested that the regulation of metabolic pathways is important for the survival of S. mutans in an acidic environment (15, 24, 25). Proteomic analysis by Len et al. (15) showed the increased synthesis of 70 proteins during chemostat growth at pH 5.0 relative to growth at pH 7.0. Separate two-dimensional gel electrophoresis studies also demonstrated that LGL was up-regulated in an acidic environment (15, 25). These findings suggest that LGL may be linked to the acid tolerance of S. mutans (15, 25). The goal of this study was to investigate the role that LGL may play in the aciduricity and acidogenicity of S. mutans.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. mutans wild-type strain UA159 was used in this study. The S. mutans lgl isogenic knockout mutant (LGLKO) was generated using PCR ligation mutagenesis (14). The primers used for mutagenesis are shown in Table 1. S. mutans UA159 cells were grown in Todd-Hewitt broth supplemented with 0.3% yeast extract (THYE). Erythromycin was added to a final concentration of 10 μg/ml when needed.
TABLE 1.
Primers used in this study
| Primer | Primer sequence (5′ to 3′)a | Description (reference) |
|---|---|---|
| Erm 1 | GGCGCGCCCGGGCCCAAAATTTGTTTGAT | Erythromycin cassette (14) |
| Erm 2 | GGCCGGCCAGTCGGCAGCGACTCATAGAAT | Erythromycin cassette (14) |
| LGL 1 | GGACAATCAAAATCAACCTC | lgl mutagenesis |
| LGL 2 | GGCGCGCCTAACTCATAGTCGGGTCG | lgl mutagenesis |
| LGL 3 | GGCCGGCCACTACTTCATCACAGACCCC | lgl mutagenesis |
| LGL 4 | ATGCCGACACACATAGCAAC | lgl mutagenesis |
| LGL RT 1 | CCTTGGCAATGGCTATGGTC | lgl qRT-PCR |
| LGL RT 2 | TCGGGGTCTGTGATGAAGTAG | lgl qRT-PCR |
| 16S RT 1 | CTTACCAGGTCTTGACATCCCG | 16S qRT-PCR |
| 16S RT 2 | ACCCAACATCTCACGACACGAG | 16S qRT-PCR |
Restriction sites are underlined: AscI, GGCGCGCC; FseI, GGCCGGCC.
Growth kinetics.
Overnight cultures of S. mutans UA159 and LGLKO strains were diluted (1:20) into prewarmed THYE and incubated at 37°C in an atmosphere of 5% CO2 until mid-log phase (optical density at 600 nm [OD600] of ∼0.4). The subcultures were then inoculated (1:20) in quadruplicate into microtiter plates containing THYE broth at pH 7.5 and pH 5.0. Growth was followed for 24 h using an automated growth monitor (Bioscreen C; Labsystems, Finland). From these data, growth curves were generated and doubling times were calculated (11).
ATR assay.
Overnight cultures of S. mutans UA159 and LGLKO strains were diluted (1:20) into TYE (10% tryptone, 5% yeast extract, 17.2 mM K2HPO4) supplemented with 5 mM glucose at pH 7.5 and incubated at 37°C in a 5% CO2-enhanced environment until mid-log phase (OD600 of ∼0.4). Cultures were then divided into two equal aliquots (termed “adapted” and ′“nonadapted”) and pelleted via centrifugation. Nonadapted cells were resuspended in TYE at the lethal pH value of 3.2. An aliquot was immediately removed (time zero) and serially diluted in 10 mM potassium phosphate buffer (pH 7.2), and the incubation of the culture was continued for 3 h at 37°C in an atmosphere of 5% CO2. Twenty microliters of each dilution was spotted in triplicate onto THYE agar plates and incubated at 37°C in a 5% CO2-supplemented atmosphere for 2 days. Adapted cells were first resuspended in TYE at pH 5.5 for 2 h prior to being subjected to TYE at pH 3.2. The acid tolerance response (ATR) was expressed as the percentage of cells to survive the lethal pH for 1 h, 2 h, and 3 h compared to the number of cells present at time zero.
Terminal pH determination.
Overnight cultures of S. mutans UA159 and LGLKO strains (eight independent cultures of each strain) were diluted (1:40) in fresh THYE at pH 7.5, pH 6.0, and pH 5.0 and incubated at 37°C in an atmosphere of 5% CO2 for 20 h prior to the terminal pH measurement.
Continuous-culture acidic competition assay.
Overnight cultures of UA159 and LGLKO were simultaneously inoculated (a dilution of 1:80 for each in 0.25× THYE at pH 6.0) into a chemostat biofermentor with glass rods for biofilm accumulation. Cells were continuously cultured as described previously (17) with some modification. The culture was established initially at pH 6.0 at a fresh medium flow rate of 0.1 dilution/h for 18 h. After culture establishment, the culture pH was reduced to 5.0 and the flow rate increased to 0.5 dilution/h. Aliquots of planktonic cells were removed, serially diluted, and plated onto THYE agar and THYE agar plus 10 μg/ml erythromycin. Biofilm cells were quantified by the removal of the glass rods. Biofilm cells were released by vortex, and cells were serially diluted and plated as described above.
MIC of methylglyoxal.
Commercial methylglyoxal (Sigma) was diluted in THYE and serially diluted (1:1) into 96-well microtiter plates. The microtiter plates were inoculated with 2 μl of S. mutans UA159 and LGLKO cells (at a concentration of 0.5 McFarland standard) and subsequently incubated for 24 h at 37°C in an atmosphere of 5% CO2. The MIC was defined as the lowest concentration of methylglyoxal that inhibited visible bacterial growth. To determine the minimum bactericidal concentration, bacteria from each microtiter well were streaked onto THYE agar plates that were incubated for 48 h to test for cell viability.
Measurement of glycolytic rates.
Overnight cultures of S. mutans UA159 and LGLKO strains were diluted 1:10 into THYE at pH 7.5 and incubated at 37°C in air with 5% CO2 until cultures reached mid-log phase (OD600 of ∼0.4). Cells were then harvested by centrifugation and washed twice with cold PK solution (1% peptone, 1% KCl) at either pH 7.0 or pH 5.0. Cells were then resuspended in PK solution at the appropriate pH to a final OD600 of ∼1.0. Aliquots (18 ml) of cell suspension were equilibrated in the reaction vessel at 37°C until residual glycolytic activity had diminished. Following equilibration, glucose was added to a final concentration of 200 mM, and glycolysis was monitored by the rate of addition of potassium hydroxide (10 mM KOH at pH 7.0 and 2 mM KOH at pH 5.0) required to keep the pH constant, utilizing a Radiometer ABU901 autoburette in conjunction with a PHM290 pH controller (Radiometer, Denmark). The glycolytic rate was expressed as μmol of acid neutralized per milligram (dry weight) of cells per minute. Additional experiments were conducted in the presence or absence of exogenously added 5 mM methylglyoxal.
Cell preparation for gene expression analysis and LGL enzyme activity.
S. mutans UA159 cells were grown under the following conditions prior to RNA isolation. For acid growth of cells, overnight cultures of S. mutans UA159 were diluted (1:40) in THYE at pH 7.5 and pH 5.0 and incubated at 37°C in an atmosphere of 5% CO2 until mid-log growth phase (OD600 of ∼0.4). For acid adaptation of cells, overnight cultures of S. mutans UA159 were diluted (1:20) in TYE, pH 7.5, and incubated at 37°C in a 5% CO2 atmosphere until mid-log growth phase was reached. Cultures were then divided into two aliquots, and cells were harvested via centrifugation and subsequently resuspended in TYE, pH 5.0. The first aliquot (unadapted) was processed immediately, and the second aliquot (adapted) was incubated at 37°C in a 5% CO2-supplemented atmosphere for 2 h prior to further manipulation. For growth phase assays, overnight cultures of S. mutans UA159 were diluted (1:20) into TYE at pH 7.5 and incubated at 37°C in air with 5% CO2 supplementation. An aliquot was removed when the cultures reached mid-log phase (OD600 of ∼0.4), and cells were harvested by centrifugation. The remaining cells were harvested after reaching stationary phase (20 h). For glucose response assays, cell suspensions of S. mutans UA159 were prepared at both pH 7.0 and pH 5.0 as described for the measurement of glycolytic rates. Cells were equilibrated for 20 min at 37°C to eliminate residual glycolytic activity. Subsequent to equilibration, 200 mM glucose was added to the suspensions for 15 min prior to cell harvesting. For methylglyoxal response assays, S. mutans UA159 cells were grown as described above for glucose response assays; however, cells were exposed to 10 mM methylglyoxal for 15 min before being harvested.
qRT-PCR analysis of lgl expression.
Total RNA was isolated, processed, and treated with RQ1 RNase-free DNase (Promega) as described previously (8). From this RNA, cDNA was generated via reverse transcription using a First Strand cDNA synthesis kit (MBI Fermentas) according to the manufacturer's instructions. RNA samples lacking reverse transcriptase were included as controls to ensure that results were not the product of residual DNA contamination. The single-stranded-cDNA template quantitative real-time PCRs (qRT-PCRs) were carried out using the QuantiTect SYBR green PCR kit (QIAGEN) in an Mx3005P QPCR system (Stratagene). Specific primer sequences used (Table 1) in the reactions were designed to yield 100- to 150-bp products. For each reaction, the cycle threshold (CT) was measured; this value was inversely proportional to the starting amount of DNA in each sample. All data were normalized against the expression of an internal standard, 16S rRNA. The expression fold-change was determined using the 2−ΔΔCT method (18).
Assay for glyoxalase enzyme activity.
To prepare soluble cellular proteins, S. mutans UA159 and the LGLKO strains were subjected to conditions as described above. Cells were harvested via centrifugation, after which cell pellets were resuspended in 10 mM potassium phosphate buffer (pH 7.2) and stored at −20°C. To release the intracellular soluble proteins, cells were lysed using glass beads and a FastPrep homogenizer (Savant). Glass beads and cell debris were sedimented by centrifugation at 16,000 × g for 5 min. The protein supernatant was removed and subsequently dialyzed overnight in 10 mM potassium phosphate buffer, pH 7.2, and then snap-frozen with liquid nitrogen. Protein concentrations were determined according to the manufacturer's protocol, using the Bio-Rad protein dye microassay (Bio-Rad).
Enzyme activity assays for LGL were performed in a manner similar to the protocol of Frickel et al. (7). Briefly, the reaction substrate, hemithioacetal, was prepared by incubating 4 mM each of reduced glutathione and methylglyoxal in 50 mM sodium phosphate buffer (pH 6.6) for 10 min at 37°C, and its concentration was determined spectrophotometrically (E240 = 0.44 mM−1 cm−1). The activity of LGL was analyzed by measuring the initial rate of formation of S-d-lactoylglutathione from hemithioacetal in the presence of soluble cellular protein, followed spectrophotometrically by measuring the increase in absorbance at 240 nm (S-d-lactoylglutathione E240 = 2.86 mM−1 cm−1). Rates for each reaction are expressed in μmol/min/μg of protein.
Assays for glyoxalase II activity were performed as described by Allen et al. (2) with some modifications. The initial rates of hydrolysis of 400 μM exogenously added d-lactyolglutathione by soluble cellular protein were followed spectrophotometrically by determining the decrease in absorbance at 240 nm.
RESULTS
LGL metabolizes methylglyoxal.
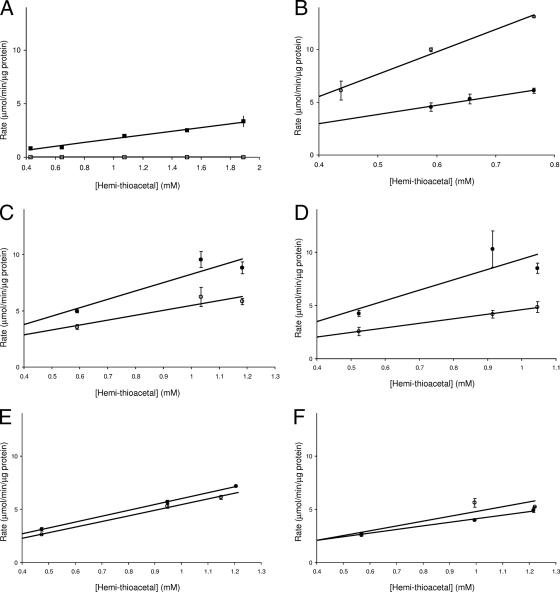
In order to determine if LGL detoxifies methyglyoxal, enzyme activity assays were carried out using soluble protein extracts isolated from both S. mutans UA159 and LGLKO strains. The UA159 protein extracts were able to efficiently convert the hemithioacetal (formed nonenzymatically between methylglyoxal and glutathione) to S-d-glutathione; in contrast, the LGLKO protein extracts had no detectable enzyme activity (Fig. 1A). These data confirmed that LGL was responsible for the detoxification of methyglyoxal in S. mutans. The involvement of LGL in methylglyoxal detoxification was further confirmed by the determination of bactericidal MICs for methylglyoxal. The LGLKO strain was unable to survive at 3.9 mM methylglyoxal, as opposed to 15.6 mM for the UA159 wild-type strain.
FIG. 1.
LGL enzyme activity in S. mutans soluble cell extracts subjected to various environmental factors. Initial rates were measured in triplicate and monitored by the increase in absorbance at 240 nm due to the conversion of hemithioacetal to S-d-lactoylglutathione. (A) UA159 (▪) and LGLKO (□); (B) S. mutans UA159 cells grown to mid-log phase at pH 7.5 (○) and pH 5.0 (•); (C) S. mutans UA159 acid-adapted cells (•) and nonadapted cells (○); (D) S. mutans UA159 cells grown to mid-logarithmic growth phase (○) and stationary growth phase (•); (E) S. mutans UA159 cells at pH 7.0 subjected to glucose starvation (○) and 200 mM glucose (•); (F) S. mutans UA159 cells at pH 5.0 subjected to glucose starvation (○) and 200 mM glucose (•).
S. mutans displays glyoxalase II activity.
A search of the S. mutans genome reveals no obvious homologue to glyoxalase II. However, S. mutans does have glyoxalase II activity, as displayed by the ability of cellular protein extracts to hydrolyze d-lactoylglutathione (data not shown).
LGL enzyme activity profile.
To corroborate qRT-PCR analysis, LGL enzyme activity was measured under the same environmental conditions used for gene expression studies. There was higher LGL-specific activity in the S. mutans protein extracts when cells were exposed to an acidic environment: when they were grown at low pH compared with neutral pH (Fig. 1B) or when they were adapted to pH 5.5 for 2 h (Fig. 1C). Growth phase also influenced LGL-specific activity, as seen by the increased activity in stationary-growth-phase cells compared with mid-logarithmic-phase cells (Fig. 1D). The presence or absence of glucose had no effect on LGL activity in a dense S. mutans cell suspension at pH 7.0 or pH 5.0 (Fig. 1E and F).
The involvement of Lgl in aciduricity.
The phenotypic effect of the lgl mutation on S. mutans acid tolerance was first quantified by measurements of growth rates at acidic pH. Growth kinetics showed that the LGLKO mutant and wild-type UA159 grew similarly in THYE at pH 7.5 (mean doubling times of 70.2 ± 1.1 min and 70.4 ± 0.2 min, respectively). However, during growth in THYE at pH 5.0, LGLKO displayed a significantly slower doubling time than UA159 (mean doubling times of 179.2 ± 5.4 min and 164.3 ± 5.2 min, respectively). The final growth yield of LGLKO after 18 h of growth in either pH 7.5 or pH 5.0 growth conditions was the same as that of the UA159 wild-type strain.
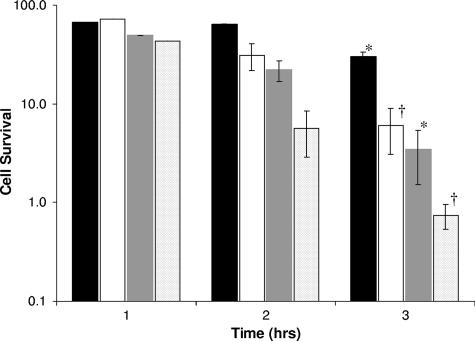
To further investigate the involvement of lgl in acid tolerance, an ATR assay was employed, again comparing S. mutans UA159 wild-type and LGLKO mutant strains (Fig. 2). The nonadapted LGLKO cells were more sensitive to acid than nonadapted UA159 wild-type cells. LGLKO cells that were adapted to acid at pH 5.5 media prior to exposure to pH 3.2 medium also had diminished survival ability compared with UA159. Interestingly, the 3-h adaptation period caused similar increases in survival for both strains: an 8.2-fold increase in survival for LGLKO and a 9.0-fold increase in survival for UA159.
FIG. 2.
ATR of S. mutans UA159 and LGLKO mutant strains. Cells were grown in TYE supplemented with glucose at pH 7.5 to mid-log phase and subjected to TYE, pH 3.2 (unadapted UA159 [dark gray bars] and LGLKO [light gray bars]) or incubated in TYE pH 5.5 for 2 h and then subjected to TYE pH 3.2 (adapted UA159 [black bars] and LGLKO [white bars]). The percentage of cell survival was calculated as the number of CFU/ml at a given time divided by the number of CFU/ml at time zero. The results are expressed as the means ± standard errors of three independent experiments. *, statistical significance comparing nonadapted cells (P < 0.05); †, statistical significance comparing adapted cells (P < 0.05).
As another indication of acid tolerance, terminal culture pHs were measured for UA159 and LGLKO, for which a small but statistically significant difference was observed. With the initial culture at pH 7.5, the LGLKO strain was unable to acidify cultures to the same extent as UA159. In addition, this trend became more pronounced as the initial culture pH was reduced to pH 5.0 [mean terminal pHs of (4.22 ± 1.7) × 10−3 and (4.26 ± 4.2) × 10−3, respectively; statistical significance determined with single-factor analysis of variance, P = 7.35 × 10−7].
Acidic continuous-culture competition.
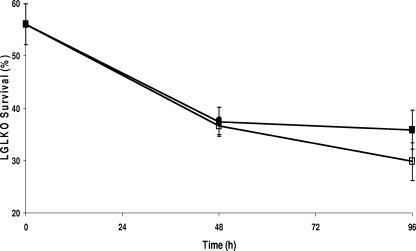
To further strengthen the argument for LGL being an important component in the aciduricity of S. mutans, the parent and mutant strains were grown simultaneously in competition with each other in an acidic environment in continuous culture (Fig. 3). The LGLKO cells were less fit in an acidic environment than the UA159 wild-type cells, as demonstrated by the significant decrease in their proportional contributions to the total culture for both planktonic and biofilm fractions. This decrease, for both fractions, increased from 2 days to 4 days. Interestingly, LGLKO cells, when grown in a biofilm, were better able to maintain themselves within the competitive culture than planktonic cells.
FIG. 3.
Simultaneous growth competition between UA159 and LGLKO strains by use of a biofermentor. Cultures were initiated at pH 6.0 prior to exposure to pH 5.0. The percentages of LGLKO survival in both biofilm (▪) and planktonic (□) culture fractions were calculated as the number of CFU/ml present on THYE agar plates plus 10 μg/ml erythromycin divided by the number of CFU/ml present on THYE agar. The results are expressed as the means ± standard errors of three independent experiments.
The effect of exogenous methylglyoxal and LGL deletion on glycolytic rates.
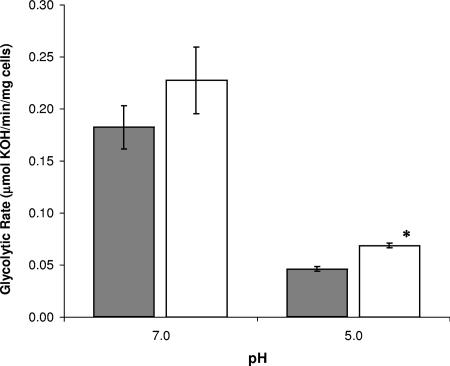
The effect of lgl deletion on the glycolytic rates of S. mutans was investigated. The absence of LGL caused no statistically significant change in glycolytic rates at pH 7.0. Interestingly, when the pH of the reaction was decreased to pH 5.0, LGLKO had increased glycolytic rates, with LGLKO able to produce acid at a rate of 68.7 ± 0.2 μmol·min−1·mg−1 compared to the ability of UA159 to produce at a rate of 46.2 ± 0.2 μmol·min−1·mg−1 (data shown in Fig. 4). To determine whether these results were caused by increased methylglyoxal concentrations, exogenous methylglyoxal was added and glycolytic rates were measured. Interestingly, the addition of methylglyoxal resulted in a modest 28% decrease in the glycolytic rate at pH 7.0 and conversely a significant 98% increase in rates at pH 5.0 (data not shown).
FIG. 4.
Glycolytic rates of S. mutans UA159 (gray bars) and LGLKO (white bars). Glycolytic rates were monitored by measuring the rate of the addition of 10 mM KOH to the cell suspension following the addition of 200 mM glucose at pH 7.0 and pH 5.0. The results are expressed as the means ± standard errors of three independent experiments. *, statistical significance compared with UA159 (pH 5.0; P < 0.05).
The expression profile of lgl.
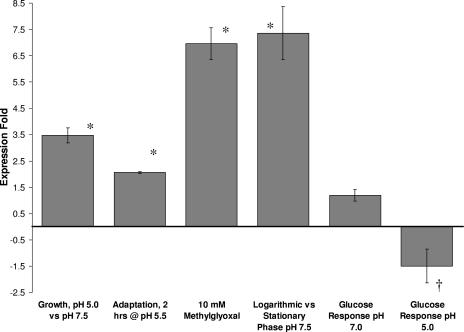
In order to compare the differential expression levels of lgl, a series of experiments with S. mutans UA159 cells grown under specific environmental conditions related to acidogenicity and aciduricity were performed. In batch cultures, the lgl mRNA levels were markedly higher, 3.5-fold, in cells grown at pH 5.0 than in cells grown at pH 7.5 (Fig. 5). The levels of lgl transcript were also increased 2.1-fold in cells exposed to pH 5.5 for 2 h. The exogenous addition of 10 mM methylglyoxal resulted in a 7.0-fold increase in lgl mRNA levels. At pH 7.0, glucose addition induced no significant change in lgl expression, whereas at pH 5.0, lgl expression levels were modestly reduced 1.5-fold upon addition of 200 mM glucose. Levels of lgl mRNA were markedly increased, 7.4-fold, during the logarithmic phase of growth compared with the stationary phase of growth (Fig. 5).
FIG. 5.
Differential gene expression levels of lgl under various environmental conditions. The increase in expression was standardized to 16S rRNA expression. The results are expressed as the means ± standard errors of at least four independent experiments. *, statistical significance, expression increase of >1 (P < 0.01); †, statistical significance, expression increase of <−1 (P < 0.01).
DISCUSSION
When living within the complex, multispecies environment of dental plaque, S. mutans rapidly metabolizes carbohydrates into organic acids. This can result in a dramatic pH drop, leading to the initiation and progression of carious lesions via the dissolution of tooth enamel. S. mutans has evolved elaborate regulatory systems affording it the abilities of rapid carbohydrate metabolism and acid survival. This investigation focused on one enzyme, LGL, its function, and its involvement in the aciduricity and acidogenicity of S. mutans.
Methylglyoxal is a strong electrophile that occurs as a natural by-product of glycolysis; therefore, cells, both eukaryotic and prokaryotic, have developed methods to neutralize its toxic effects (4, 23). One system known to detoxify methylglyoxal is the glyoxalase system in which toxic methylglyoxal is converted to d-lactate in a process involving two enzymes, the first of these enzymes being LGL. Utilizing total soluble protein obtained from S. mutans UA159 and its lgl-defective mutant, we confirmed that LGL was responsible for the conversion of methylglyoxal to S-d-lactoylglutathione in S. mutans. This conclusion was further supported by the hypersensitivity of the LGLKO strain to methylglyoxal. Isogenic inactivation of lgl proved not to be lethal, indicating that S. mutans has at least one other system capable of detoxifying methylglyoxal or that the intracellular concentrations of methylglyoxal under the conditions tested were sublethal. A possible alternative detoxification system may be comprised of aldose and aldehyde reductases, as reported for E. coli and Saccharomyces cerevisiae (13, 19, 21).
Previous investigations into the proteome-wide response of S. mutans to acid challenge have revealed a vast and diverse list of proteins with altered expression profiles (15, 16, 24, 25). Of these proteins, LGL has been shown to be up-regulated 2.6- and 51-fold under acidic conditions (15, 25). As opposed to measuring protein abundance, we investigated the “real-time” regulation of lgl mRNA expression in S. mutans, under various environmental queues relevant to acidogenicity and aciduricity. Our data clearly showed pronounced increases in lgl mRNA levels during both acidic growth and acid adaptation. The gene expression data were additionally validated by the increase in Lgl-specific enzymatic activity in cell extracts prepared from acid-grown cells and acid-adapted cells. This increased lgl expression in response to low pH at the mRNA level and the increased enzymatic activity in response to an acidic environment strongly implicate LGL as important in acid tolerance.
The involvement of LGL in the acid tolerance of S. mutans was further supported by our findings that the LGLKO strain had statistically significant longer generation times at pH 5.0 than wild-type UA159. Furthermore, LGLKO showed even greater acid sensitivity than UA159 under the conditions of the ATR assay. Although adapted LGLKO cells did not survive to the same extent as UA159 wild-type cells, interestingly, this adaptation caused nearly identical increases in survival for both strains, indicating that LGL is likely involved in intrinsic acid survival. The mutant, LGLKO, was also unable to acidify liquid culture to the same extent as UA159. These data were further supported by the inability of LGLKO to compete with UA159 when grown competitively in a biofermentor. These results definitively show the compromised competitiveness of the acid-sensitive phenotype resulting from lgl inactivation in S. mutans. We can therefore conclude that LGL is an important component of acid tolerance in S. mutans.
Generally, the concentration of methylglyoxal increases during unregulated growth and in highly glycolytically active prokaryotic and eukaryotic cells (1, 6, 12, 20, 22). Also, the formation of methylglyoxal was seen to increase following the addition of metabolites that stimulate the flux of triose phosphates, such as glucose (12). We therefore set out to determine the relationship between glucose metabolism and LGL activity in S. mutans. Remarkably, and what we believe to be a novel observation, the rate of acid production at pH 5.0 increased in response to the addition of exogenous methylglyoxal and in the absence of LGL. This suggests that excess intracellular methyglyoxal acts as a glycolytic regulator in an acidic environment. One possible mechanism by which glycolysis could be regulated is through the stimulation of pyruvate kinase. This would lead to a coordinate drop in the concentration of intracellular phosphoenolpyruvate, which is known to activate LDH activity (26). This increase in homofermentation would rob S. mutans of the additional ATP generation by heterofermentation required for robust acid tolerance, especially under conditions of limiting glucose.
Further investigation into the connection between lgl expression, glycolysis, and growth phase was undertaken by measuring lgl expression in response to glucose under neutral and acidic conditions and at different growth phases. The expression of lgl was maximal during the mid-log phase of cellular growth, whereas enzymatic activity of LGL was greater in stationary-phase cells. These somewhat contradictory results likely allude to LGL being highly stable and persisting in an active form long after being synthesized. At pH 5.0, the modestly decreased lgl expression in the presence of excess glucose was inversely proportional to the increase in glycolytic rates observed under the same conditions. Also, at pH 7.0, in the presence of excess glucose, lgl expression remained stable. Enzymatic analysis under these same conditions yielded similar results. These data suggest that lgl expression is tightly regulated and that this regulation is dependent upon more factors than acidity or exogenous glucose concentration alone.
This report examined the role of the detoxifying enzyme, LGL, in the acidogenicity and aciduricity of S. mutans. The data clearly demonstrate that LGL is an enzyme responsible for the detoxification of methylglyoxal and suggest a link between LGL and the acid tolerance afforded to S. mutans. The ability to regulate LGL may prove to be a valuable strategy to modulate S. mutans acid tolerance and hence virulence.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Ackerman, R. S., N. R. Cozzarelli, and W. Epstein. 1974. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J. Bacteriol. 119:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, R. E., T. W. Lo, and P. J. Thornalley. 1993. Purification and characterisation of glyoxalase II from human red blood cells. Eur. J. Biochem. 213:1261-1267. [DOI] [PubMed] [Google Scholar]
- 3.Banas, J. A. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267-1277. [DOI] [PubMed] [Google Scholar]
- 4.Cooper, R. A. 1984. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 38:49-68. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen., and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 6.Freedberg, W. B., W. S. Kistler, and E. C. Lin. 1971. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J. Bacteriol. 108:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frickel, E.-M., P. Jemth, M. Widersten, and B. Mannervik. 2001. Yeast glyoxalase I is a monomeric enzyme with two active sites. J. Biol. Chem. 276:1845-1849. [DOI] [PubMed] [Google Scholar]
- 8.Hanna, M. N., R. J. Ferguson, Y.-H. Li, and D. G. Cvitkovitch. 2001. uvrA is an acid-inducible gene involved in the adaptive response to low pH in Streptococcus mutans. J. Bacteriol. 183:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue, Y., and A. Kimura. 1995. Methylglyoxal and regulation of its metabolism in microorganisms. Adv. Microb. Physiol. 37:177-227. [DOI] [PubMed] [Google Scholar]
- 10.Kalapos, M. P. 1999. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 110:145-175. [DOI] [PubMed] [Google Scholar]
- 11.Khalichi, P., D. G. Cvitkovitch, and J. P. Santerre. 2004. Effect of composite resin biodegradation products on oral streptococcal growth. Biomaterials 25:5467-5472. [DOI] [PubMed] [Google Scholar]
- 12.Kim, I., E. Kim, S. Yoo, D. Shin, B. Min, J. Song, and C. Park. 2004. Ribose utilization with an excess of mutarotase causes cell death due to accumulation of methylglyoxal. J. Bacteriol. 186:7229-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ko, J., I. Kim, S. Yoo, B. Min, K. Kim, and C. Park. 2005. Conversion of methylglyoxal to acetol by Escherichia coli aldo-keto reductases. J. Bacteriol. 187:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau, P. C., C. K. Sung, J. H. Lee, D. A. Morrison, and D. G. Cvitkovitch. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193-205. [DOI] [PubMed] [Google Scholar]
- 15.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150:1353-1366. [DOI] [PubMed] [Google Scholar]
- 16.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339-1351. [DOI] [PubMed] [Google Scholar]
- 17.Li, Y.-H., M. N. Hanna, G. Svensäter, R. P. Ellen, and D. G. Cvitkovitch. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J. Bacteriol. 183:6875-6884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 19.Murata, K., Y. Fukuda, M. Simonsaka, K. Watanabe, T. Saikusa, and A. Kimura. 1985. Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent methylglyoxal-reducing enzyme from Saccharomyces cerevisiae. Eur. J. Biochem. 151:631-636. [DOI] [PubMed] [Google Scholar]
- 20.Phillips, S. A., and P. J. Thornalley. 1993. Formation of methyglyoxal and d-lactate in human red blood cells in vitro. Biochem. Soc. Trans. 21:163S. [DOI] [PubMed] [Google Scholar]
- 21.Saikusa, T., H. Rhee, K. Watanabe, K. Murata, and A. Kimura. 1987. Metabolism of 2-oxoaldehyde in bacteria: purification and characterization of methylglyoxal reductase from E. coli. Agric. Biol. Chem. 7:1893-1899. [Google Scholar]
- 22.Thornalley, P. J. 1988. Modification of the glyoxalase system in human red blood cells by glucose in vitro. Biochem. J. 254:751-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thornalley, P. J. 1996. Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification—a role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 27:565-573. [DOI] [PubMed] [Google Scholar]
- 24.Welin, J., J. C. Wilkins, D. Beighton, K. Wrzesinski, S. J. Fey, P. Mose-Larson, I. R. Hamilton, and G. Svensater. 2003. Effect of acid shock on protein expression by biofilm cells of Streptococcus mutans. FEMS Microbiol. Lett. 227:287-293. [DOI] [PubMed] [Google Scholar]
- 25.Wilkins, J. C., K. A. Homer, and D. Beighton. 2002. Analysis of Streptococcus mutans proteins modulated by culture acidic conditions. Appl. Environ. Microbiol. 68:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada, T., and J. Carlsson. 1975. Regulation of lactate dehydrogenase and change of fermentation products in streptococci. J. Bacteriol. 124:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]