Abstract
Plesiomonas shigelloides is an emerging pathogen that is widespread in the aquatic environment and is responsible for intestinal diseases and extraintestinal infections in humans and other animals. Virtually nothing is known about its genetic diversity, population structure, and evolution, which severely limits epidemiological control. We addressed these questions by developing a multilocus sequence typing (MLST) system based on five genes (fusA, leuS, pyrG, recG, and rpoB) and analyzing 77 epidemiologically unrelated strains from several countries and several ecological sources. The phylogenetic position of P. shigelloides within family Enterobacteriaceae was precisely defined by phylogenetic analysis of the same gene portions in other family members. Within P. shigelloides, high levels of nucleotide diversity (average percentage of nucleotide differences between strains, 1.49%) and genotypic diversity (64 distinct sequence types; Simpson's index, 99.7%) were found, with no salient internal phylogenetic structure. We estimated that homologous recombination in housekeeping genes affects P. shigelloides alleles and nucleotides 7 and 77 times more frequently than mutation, respectively. These ratios are similar to those observed in the naturally transformable species Streptococcus pneumoniae with a high rate of recombination. In contrast, recombination within Salmonella enterica, Escherichia coli, and Yersinia enterocolitica was much less frequent. P. shigelloides thus stands out among members of the Enterobacteriaceae. Its high rate of recombination results in a lack of association between genomic background and O and H antigenic factors, as observed for the 51 serotypes found in our sample. Given its robustness and discriminatory power, we recommend MLST as a reference method for population biology studies and epidemiological tracking of P. shigelloides strains.
Plesiomonas shigelloides is a species of rod-shaped gram-negative bacteria recently classified in the family Enterobacteriaceae and is the only oxidase-positive member of this family (32). Freshwater and estuarine water are considered to be the natural environment of P. shigelloides (36), which is often isolated from fish and seafood (30, 42). Like many Enterobacteriaceae species, P. shigelloides is also found in a wide range of hosts, including dogs, cats, sheep, cows, goats, pigs, monkeys, polecats, turtles, newts, toads, and turkey vultures (32). In humans, P. shigelloides mostly causes gastrointestinal diseases. Outbreaks are generally related to consumption of seafood or untreated water (33). Three major types of P. shigelloides gastroenteritis occur: (i) a secretory, watery type, (ii) an invasive, dysentery-like type, and (iii) a subacute or chronic form, lasting between 2 weeks and 3 months (10, 35). Extraintestinal infections, such as septicemia and meningitis, have been described in immunodeficient patients (6, 9). P. shigelloides strains can be identified using phenotypic properties (3, 34). Specific PCR amplification of the 23S rRNA and hugA genes was designed for identification purposes (26, 30).
The level of genetic diversity of P. shigelloides and its population structure are currently unknown. The existence of clones that may have distinct ecological specialization or epidemiology is an important question for the control of P. shigelloides infections. Strain diversity in this species has been demonstrated by pulsed-field gel electrophoresis (49) and random amplified polymorphic DNA analysis (28). However, these methods are difficult to standardize and provide limited information on the phylogenetic relationships among strains. To our knowledge, no other molecular method has been applied to characterization of P. shigelloides strains. A serotyping scheme based on characterization of the somatic (O) and flagellar (H) antigens has been available since 1996 and was last updated in 2000 (4, 5). This scheme describes about 100 O types and 50 H types, with a high number of combinations of these types. Correlation between some serovars and sources has been suggested (27).
Bacterial species differ widely in the rate of homologous recombination (19, 25, 50, 57). This rate is particularly relevant for understanding strain evolution. For example, high rates of recombination break down the link between the genomic background of strains and surface antigens that are important for vaccine design (43) or strain typing (8). In addition, homologous recombination renders bacterial clones unstable over time (2, 25, 31), with important consequences for the interpretation of molecular markers as applied to epidemiological follow-up of strains. For all Enterobacteriaceae species for which the impact of recombination on strain evolution was estimated previously, a low or limited impact of recombination on clonal diversification and population structure was inferred (see Discussion). However, it is not known whether a low recombination rate is a general feature of the Enterobacteriaceae family.
The population structure of bacterial species and strain evolution are best studied using standardized methods based on nucleotide sequences (1, 39, 40, 51). Multilocus sequence typing (MLST) is now widely used to study strain evolution and typing in many different species (15, 54). The resulting unambiguous and portable data allow users from different laboratories to compare data, which is necessary to get a comprehensive overview of strain diversity and distribution. In addition, these methods are suitable for studying strain phylogeny and population structure (20, 39).
In this study, we used MLST based on five genes (fusA, leuS, pyrG, recG, and rpoB) to investigate the diversity of a collection of 77 P. shigelloides strains isolated from diverse sources and different countries. We found a high level of diversity and demonstrated that P. shigelloides strains undergo high levels of homologous recombination, which eliminates congruence between trees based on single genes and disrupts correspondence between serotypes and genomic background.
MATERIALS AND METHODS
Bacterial strains.
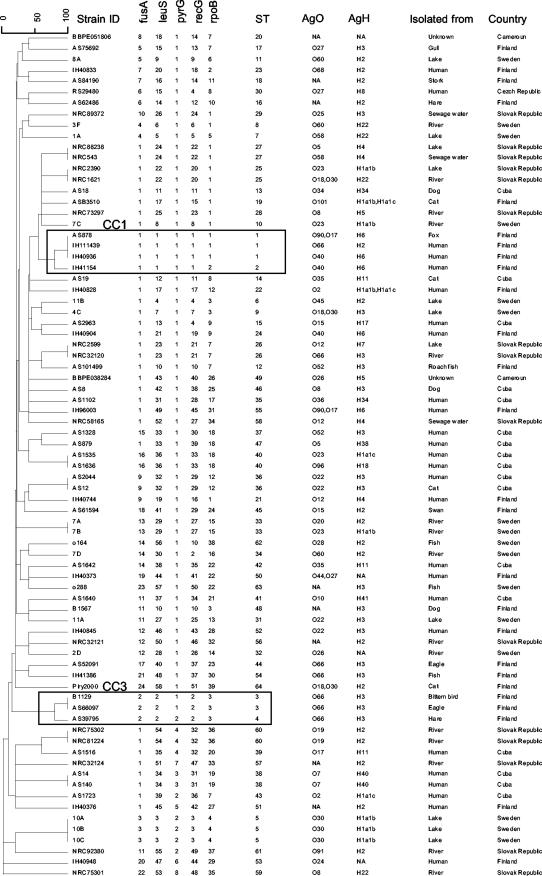
A collection of 77 strains of P. shigelloides was used. These strains were isolated from different sources in different countries and represented multiple serotypes (Fig. 1) in order to maximize the diversity of our population. Isolates that were gram negative and oxidase, indole, maltose, and glucose positive were further identified by biochemical typing using the API 20 E system (bioMerieux SA, Marcy-l'Etoile, France) and were tested by specific PCR for detection of P. shigelloides based on the 23S rRNA gene as previously described (26).
FIG. 1.
Allelic profiles, STs, O and H serotypes, sources, and countries of isolation of the 77 strains of P. shigelloides. The unweighted-pair group method using average linkage dendrogram was constructed from a distance matrix consisting of pairwise distances between allelic profiles.
Total DNA extraction.
Bacterial strains were grown on tryptone-casein-soy agar for 18 h at 37°C. One colony was resuspended in 200 μl of purified water, and the suspension was incubated at 96°C for 10 min in order to lyse the cells. After centrifugation at 13,000 rpm for 5 min, supernatants were collected and frozen until they were used.
PCR primers.
For four genes (fusA, leuS, pyrG, and recG), we designed primers suitable for P. shigelloides strain amplification by decreasing the degeneracy of universal oligonucleotides (46), using sequenced genomes of four members of the Enterobacteriaceae (Escherichia coli, Salmonella enterica, Klebsiella pneumoniae, and Yersinia pestis). For the rpoB gene, we designed primers VIC4 and VIC6 (Table 1). The five genes selected belong to a set of evolutionarily conserved genes with a single copy in 95% of sequenced genomes (46). The primers consist of two regions, a 5′ consensus region with no degeneracy and a 3′ degenerate region. Primer characteristics are shown in Table 1.
TABLE 1.
Primers and conditions used for PCR amplification of internal portions of five protein-encoding genes
| Gene | Primer | Primer sequence | Primer position in gene | PCR cycles |
|---|---|---|---|---|
| fusA | fusA3 | 5′-CAT CGG TAT CAG TGC KCA CAT CGA-3′ | 36-59 | 2 min at 94°C; 10 cycles of 1 min at 94°C, |
| fusA4 | 5′-CAG CAT CGC CTG AAC RCC TTT GTT-3′ | 814-837 | 1 min at 60°C, and 1 min 72°C; 21 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C; 5 min at 72°C | |
| leuS | leuS3 | 5′-CAG ACC GTG CTG GCC AAC GAR CAR GT-3′ | 487-512 | 2 min at 94°C; 10 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; 21 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C; 5 min at 72°C |
| leuS4 | 5′-CGG CGC GCC CCA RTA RCG CT-3′ | 1274-1293 | ||
| pyrG | pyrG3 | 5′-GGG GTC GTA TCC TCT CTG GGT AAA GG-3′ | 31-56 | 2 min at 94°C; 10 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; 21 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min 72°C; 5 min at 72°C |
| pyrG4 | 5′-GGA ACG GCA GGG ATT CGA TAT CNC CKA-3′ | 434-460 | ||
| recG | recG3 | 5′-GGG CGA CGT TGG CTC MGG KAA AAC-3′ | 918-941 | 2 min at 94°C; 10 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C; 21 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C; 5 min at 72°C |
| recG4 | 5′-GGG TGC GGG GGA TSG GSG TKG C-3′ | 1315-1336 | ||
| rpoB | VIC4 | 5′-GGC GAA ATG GC(AGT) GA(AG) AAC CA-3′ | 1369-1388 | 4 min at 94°C; 30 cycles of 30 s at 94°C, |
| VIC6 | 5′-GA(AG) TC(CT) TCG AAG TTG TAA CC-3′ | 2425-2444 | 30 s at 50°C, and 30 s at 72°C; 5 min at 72°C |
DNA sequencing.
PCR cycles used for amplification are shown in Table 1. The 50-μl PCR mixtures contained 0.2 μM of each primer, 100 μM of each deoxynucleoside triphosphate, 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 0.85 U Taq DNA polymerase (Invitrogen), and 2 μl of total DNA. Amplification products were checked by agarose (1%) gel electrophoresis and ethidium bromide staining. Two molecular size markers, 1-kb and 100-bp DNA ladders (Biolabs), were used. PCR products were purified by ultrafiltration (Millipore). Both strands of the purified PCR products were sequenced using Big Dye Terminator Ready Reaction version 3.1 (Perkin-Elmer). The 10-μl sequence reaction solutions contained 1.5 μl of Big Dye sequencing buffer, 0.3 μM of each primer, and 1 μl of Ready Reaction Premix. The reaction cycles consisted of 1 min at 96°C, followed by 25 cycles of 10 s at 96°C, 5 s at 50°C, and 4 min at 60°C. Purification was performed by ethanol precipitation. Sequence reaction products were analyzed using an ABI-3700 automated DNA sequencer.
Serotype characterization.
Serotype characterization was performed at the National Reference Laboratory for Vibrionaceae Komárno and Faculty of Public Health, Bratislava, Slovak Republic, using standard protocols (4, 5) and specific Plesiomonas sera.
Data analysis.
Editing and analysis of chromatogram traces were performed using BioNumerics v4.5 (Applied Maths, Sint-Martens-Latem, Belgium). Each base of the selected template region was confirmed by at least two chromatograms (forward and reverse); if there were ambiguities for a sequence, the sequence analysis was repeated. MLST data were analyzed by the standard MLST approach; for each gene, an allele number was attributed to each allelic variant, and the sequence type (ST) of a strain corresponded to the combination of the allele numbers of the five genes. A clonal complex (CCs) was defined using eBURST (22) as a group of strains having at least four common alleles (i.e., no more than one difference) with at least one other strain of the group. Minimum spanning tree, unweighted-pair group method using average linkage, and Pearson correlation analyses were performed using BioNumerics v4.5. Nucleotide diversity and amounts of polymorphisms were calculated using DNAsp, version 4 (44).
To test for phylogenetic congruence among the genes, the 64 distinct STs were used. Neighbor-joining trees were generated using PAUP* v4 (55) for each gene individually and for the concatenated sequence of the five genes. As described by Feil et al. (21), for each gene the differences in log likelihood between the tree for that gene and the trees constructed using the other genes were computed using PAUP*, with branch lengths optimized. These differences were compared to those obtained for 100 randomly generated trees.
The relative contributions of recombination and mutation were calculated using the MultiLocus Analyzer software, which was specifically designed for this purpose (S. Brisse, unpublished) using the simplest implementation of the clonal diversification method (23, 29). Briefly, all pairs of distinct alleles for STs differing by only one or two alleles were inspected to determine the number of nucleotide differences. Differences of only a single nucleotide polymorphism were considered to be caused by mutation; differences in more than one nucleotide were attributed to recombination. No correction was made for single nucleotide differences possibly introduced by recombination. For comparison purposes, we used MLST data sets available for Neisseria (first 200 profiles available at pubmlst.org, all except 4 corresponding to Neisseria meningitidis), Streptococcus pneumoniae (first 200 profiles available at www.mlst.net), E. coli (first 200 profiles from Mark Achtman's website, http://web.mpiib-berlin.mpg.de/mlst/), and Yersinia pseudotuberculosis (50 STs from Mark Achtman's website).
Nucleotide sequence accession numbers.
The sequences of fusA, leuS, pyrG, and rpoB obtained for 31 Enterobacteriaceae species have been deposited in the GenBank/EMBL database under accession numbers EU010012 to EU010119. The sequences of the five MLST genes and corresponding STs are available at Institut Pasteur's MLST website, www.pasteur.fr/mlst.
RESULTS
MLST development and reproducibility tests.
Eight pairs of primers proposed by Santos and Ochman (46) were fine-tuned using available Enterobacteriaceae genome sequences in order to optimize PCR amplification. Four of these pairs, which target the protein-encoding genes fusA, leuS, pyrG, and recG, were selected for further analysis based on PCR success. In addition, broad-range rpoB primers VIC4 and VIC6 were designed after available sequences were aligned. The five genes were successfully PCR amplified and sequenced for the 77 P. shigelloides strains included in this study. In order to check the reproducibility of our MLST approach and to exclude sample mix-up (which could have resulted in artifactual intergenic recombination), 16 strains were selected randomly, reisolated, and analyzed a second time. The sequences were found to be identical to those initially found.
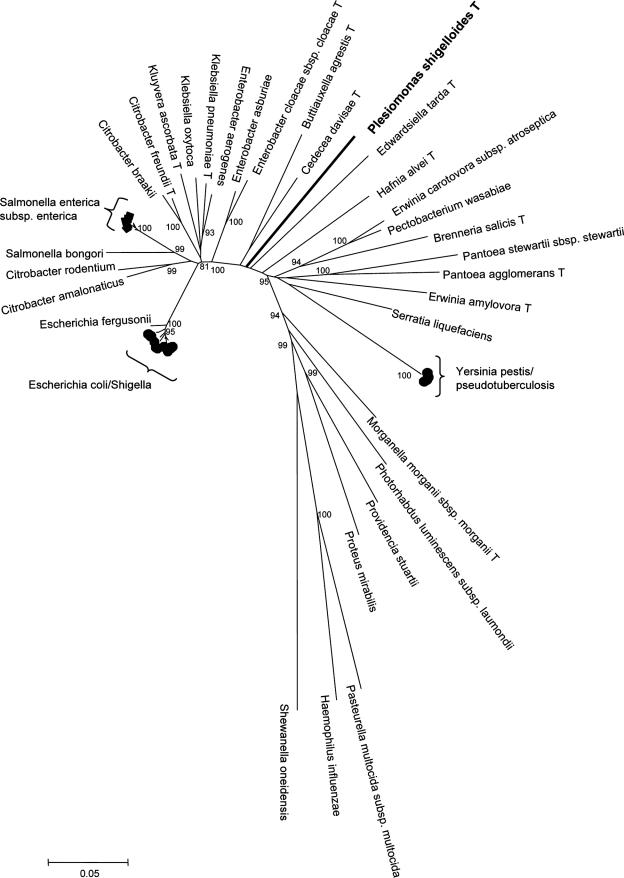
The phylogenetic relationships of P. shigelloides with other members of the family Enterobacteriaceae have not been precisely defined. In fact, based on 16S rRNA sequences, P. shigelloides was located on an external branch compared to other family members (41, 45), leading some authors to suggest that P. shigelloides could belong to a new taxonomic family (45). In order to address this question, 30 other species representing the major phylogenetic clades in the family Enterobacteriaceae (A. Deletoile, P. Grimont, and S. Brisse, unpublished) were amplified by PCR using our broad-range primers and the fusA, leuS, pyrG, and rpoB sequences were determined. Phylogenetic analysis of the concatenated sequences (Fig. 2) using three other gamma-proteobacterial species (Pasteurella multocida, Haemophilus influenzae, and Shewanella oneidensis) as outgroups clearly showed that P. shigelloides constitutes a unique branch, which is nested deep within the tree. Thus, this result firmly establishes the phylogenetic position of P. shigelloides within the family Enterobacteriaceae (11, 32).
FIG. 2.
Unrooted neighbor-joining tree of 31 Enterobacteriaceae species (type strains were sequenced) and three other Gammaproteobacteria constructed using the concatenated sequences of four loci (fusA, leuS, pyrG, and rpoB). Sequences of the following strains were retrieved from public databases: Y. pestis CO92 (accession no. NC_003143), P. multocida subsp. multocida Pm70 (NC_002663), S. oneidensis MR-1 (NC_004347), and H. influenzae Rd KW20 (NC_000907). All P. shigelloides strains were placed on the same branch as the type strain. The numbers at the nodes correspond to bootstrap values obtained with 1,000 replicates that were more than 80%.
Nucleotide diversity of P. shigelloides.
All genes showed variation, resulting in a high number of informative sites for phylogenetic analysis (Table 2). All polymorphisms corresponded to variation in nucleotide identities, as we did not observed insertion or deletion events in the selected portions of the five genes. The levels of nucleotide variation varied greatly among the genes. The most conserved gene was pyrG, with only 2.3% polymorphic sites, whereas the most variable gene was recG, with 19.6% polymorphic sites. The nucleotide diversity indices (based on the average number of nucleotide differences per site for two randomly selected strains) were 0.13 and 3.86% for pyrG and recG, respectively. The average nucleotide diversity over the five genes was 1.49%. The maximal divergence among alleles ranged from 0.53% (for pyrG) to 5.17% (for recG). Overall, these data demonstrate the high level of nucleotide diversity in the population of P. shigelloides studied. Nonsynonymous changes were not found in rpoB and pyrG and were rare for the other genes, consistent with strong selection against amino acid changes, as is typically observed for housekeeping genes (Table 2).
TABLE 2.
Polymorphism observed in five protein-encoding genes among 77 P. shigelloides strains
| Gene | Edited size (bp) | No. of polymorphic sites | No. of singletons | % of polymorphic sites | Pia | No. of synonymous changes | No. of nonsynonymous changes | Ks | Ka | Ks/Ka |
|---|---|---|---|---|---|---|---|---|---|---|
| fusA | 633 | 32 | 8 | 5.06 | 0.00635 | 26 | 6 | 0.01775 | 0.00309 | 5.74 |
| leuS | 642 | 79 | 19 | 12.31 | 0.02085 | 68 | 14 | 0.08938 | 0.0011 | 81.3 |
| pyrG | 306 | 7 | 3 | 2.29 | 0.00131 | 7 | 0 | 0.00547 | 0 | NAb |
| recG | 312 | 61 | 15 | 19.55 | 0.03856 | 56 | 7 | 0.17697 | 0.00284 | 62.3 |
| rpoB | 501 | 30 | 13 | 5.99 | 0.00719 | 33 | 0 | 0.02948 | 0 | NA |
Pi, average number of nucleotide differences per site for two randomly selected strains; Ka, number of nonsynonymous changes per nonsynonymous site; Ks, number of synonymous changes per synonymous site.
NA, not applicable.
In order to evaluate the existence of an internal phylogenetic structure in P. shigelloides, we used the concatenated sequence of the five genes to derive a tree representing the relationships among the 77 strains (see Fig. S1 in the supplemental material). It was hard to discern any clear structure in the tree obtained, as no cluster of strains was clearly separated from the remainder of the population.
Allelic and genotypic variation in P. shigelloides.
The number of distinct alleles per gene ranged from 8 (pyrG) to 57 (leuS). When the five genes were combined, 64 STs were distinguished (Fig. 1). Two STs contained three strains, and 11 STs contained two strains. The genotypic diversity index (Simpson's index) was 99.7%, which is among the highest values found so far in MLST analyses of bacterial species.
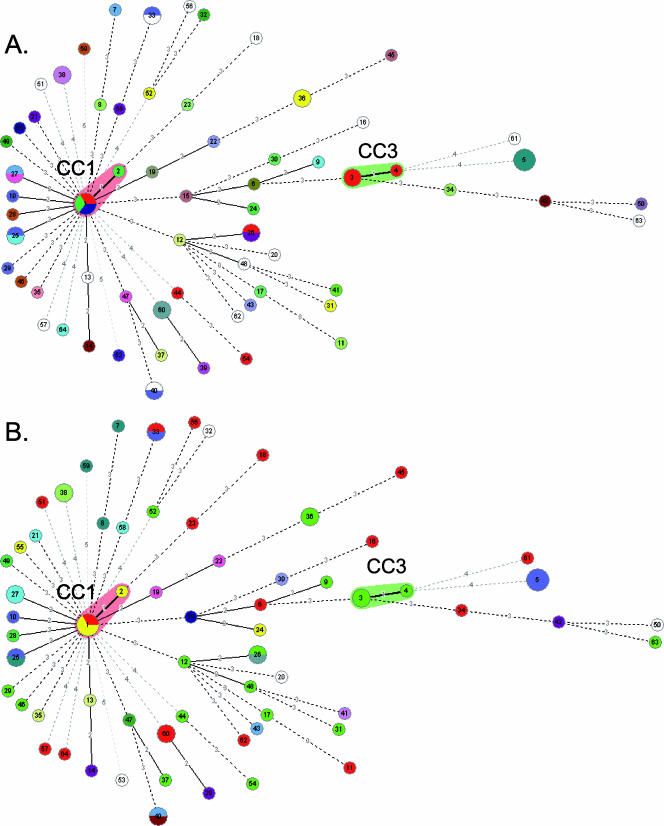
Relationships based on allelic profile codes can be more reliable than nucleotide-based phylogenies if homologous recombination occurs, as import of a single divergent allele would strongly affect the phylogenetic position of the recipient strain. A clonal complex (CC) can be defined as a group of profiles in which each profile differs by no more than one gene from at least one other profile of the group (22). STs with a single gene difference in five genes are very likely to share a common ancestry. eBURST analysis (not shown) and minimum spanning tree analysis (Fig. 3) of allelic profiles revealed only two CCs, each with only two STs. CC1 (ST1 and ST2) and CC3 (ST3 and ST4) comprised four and three strains, respectively. These two CCs each formed a single branch in the nucleotide-based phylogeny (see Fig. S1 in the supplemental material). All other STs differed by at least two of five genes from any other ST in our strain collection, and the relationships shown in Fig. 3 should not be considered reliable, as a high number of alternative branchings exist (not shown).
FIG. 3.
Minimum spanning tree analysis of the 77 strains of P. shigelloides based on the number of allelic mismatches among MLST profiles. The colors of the circles represent the serotypes of the strains. (A) O antigen. (B) H antigen. Each circle corresponds to one ST; the size of the circle indicates the number of strains in the ST. The numbers on the lines between circles are the numbers of allelic differences between the corresponding profiles.
Correspondence of STs with serotypes and sources.
The 77 strains were subjected to serotyping. For 11 of them the O and/or H antigen could not be determined; further, a few strains reacted with two sera. A total of 51 different serotypes were found (Simpson's index, 98.3%).
In some cases, there was concordance between MLST and serotyping (Fig. 3). Of the 11 pairs of strains with the same ST, 5 were of the same serotype. CC3 (which includes ST3 and ST4) included three strains with the same serotype (O66:H3).
However, in many instances, MLST and serotyping data did not coincide entirely. First, serotyping subdivided pairs or triplets of strains within a particular ST. Five of the 11 STs containing pairs of strains were heterogeneous with regard to serotype, and one ST contained strains with the same H antigen but distinct O antigens. ST1 included strains with three distinct serotypes (O40:H6, O66:H2, and O90,17:H6). Second, MLST subdivided groups of strains with the same serotype (O12:H4, O2:H1, O22:H3, O23:H1, O35:H11, O52:H3, O60:H2, O66:H3, [O90, and O17]:H6), and the distinct STs of strains of a given serotype were not closely related. Overall, the adjusted Rand coefficients for ST on the one hand and O antigen, H antigen, and both antigens on the other hand were 0.17, 0.06, and 0.26, respectively (12).
Pairs or triplets of strains sharing the same ST were always obtained in the same country. Hence, we did not identify any international ST. In addition, the two identified CCs were only found in a single country, Finland.
Strains sharing the same ST did not always come from the same host. For example, ST1 strains were isolated from a fox and two humans, ST3 strains were isolated from two distinct bird species, and ST36 strains were isolated from a cat and a human. Some pairs of environmental strains also had the same ST (Fig. 1). CC1 and CC3 were not homogeneous with respect to host (human and a fox for CC1 and three distinct host species for CC3). MLST thus deserves further evaluation, using epidemiologically well-defined samples, to determine its capacity to define the routes and modes of contamination with P. shigelloides strains.
Evidence for homologous recombination.
With time, homologous recombination causes progressive disruption of the phylogenetic signal harbored by gene sequences (18). As a consequence, phylogenies of distantly related strains derived from sequences of independent genes may not be congruent. The lack of congruence can be conveniently tested by estimating the likelihood of obtaining sequence data for a given gene by evolution with the phylogeny reconstructed using another gene (21). We calculated the congruence of the five genes with each other. Remarkably, the congruence between trees reconstructed using the five genes was no better than that between each tree and random trees, with only one exception: the likelihood of the recG data on the leuS tree was slightly higher than that on all random trees (data not shown but available upon request). These results are similar to those obtained for N. meningitidis and S. pneumoniae (21), two species with high rates of homologous recombination. Our data show that the five genes have distinct evolutionary histories in P. shigelloides and indicate that in this species homologous recombination is frequent enough to eliminate the phylogenetic signal among distantly related strains.
Short-term impact of homologous recombination.
The clonal diversification method provides a quantitative estimate of the contribution of recombination relative to mutation to the generation of genotypic diversity (23, 29). For each pair of allelic profiles that are closely related, the number of nucleotide changes in the alleles that differ is counted. In the simplest implementation, a single nucleotide difference is considered to be likely caused by mutation, whereas more than one mutation in the same portion of a gene is considered to result from recombination, as it is unlikely that two mutations would occur in the same gene while the other genes remain identical. We applied this method considering all evolutionary links between profiles sharing at least three of five alleles (i.e., number of allelic mismatches [D] = 2, corresponding to a maximal profile distance of 2/5 [40%]). The results (Table 3) indicate that P. shigelloides alleles are seven times more likely to be changed by recombination than by mutation. In addition, nucleotides are 77 times more likely to be changed by recombination than by mutation, as recombination events introduce 11 nucleotide changes at once on average. These allelic and nucleotide recombination/mutation ratios are comparable to those found at the maximal profile distance (43%) (three of seven distinct loci) in S. pneumoniae (allelic and nucleotide ratios of 7 and 66, respectively) and slightly lower than those in N. meningitidis (11 and 196, respectively) (Table 3). Remarkably, the recombination rate in P. shigelloides is much higher than that observed in the other Enterobacteriaceae species (E. coli and Yersinia enterocolitica) for which data sets are publicly available (Table 3). When only single-locus variants (D = 1) were used, the ratios were expectedly lower (7.5 and 49 for S. pneumoniae and 5.6 and 108 for N. meningitidis), consistent with previous reports (23, 24). In contrast, the nucleotide ratio for E. coli, previously estimated to be around 50 (29), was estimated to be around 5. The difference may be explained by the low number of values (only 12 strains were examined) that were used to obtain the previous estimate (29).
TABLE 3.
Recombination/mutation ratios for allelic and nucleotide replacements
| Taxon | Ratios
|
|||||
|---|---|---|---|---|---|---|
|
D = 1a
|
D = 2
|
D = 3
|
||||
| Alleles | Nucleotides | Alleles | Nucleotides | Alleles | Nucleotides | |
| P. shigelloides | NA | NA | 7 | 77 | 10.9 | 107 |
| S. pneumoniae | 7.5 | 49 | 10 | 88 | 7.3 | 66 |
| Neisseria | 5.6 | 108 | 8.29 | 153 | 11 | 196 |
| S. aureus | 0.28 | 0.9 | 0.54 | 1.88 | 1.27 | 4.89 |
| E. coli | 0.84 | 5.18 | 1.27 | 7.57 | 2.44 | 15.6 |
| Y. pseudotuberculosis | 1.46 | 6.4 | 1.4 | 5.56 | 0.85 | 3.14 |
D is the allelic profile distance criterion used for calculation of ratios.
NA, not available (too few values).
Relationship between allelic divergence and nucleotide divergence.
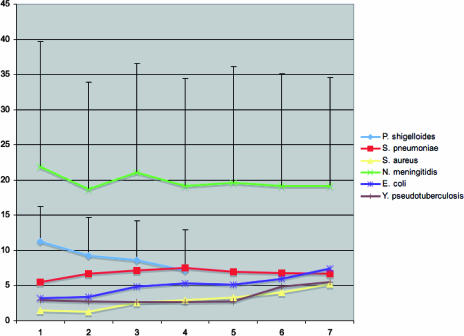
In order to extend the analysis of the impact of recombination on clonal diversification to all distinct STs rather than focusing on only the STs that are closely related, we plotted the number of nucleotide changes at nonidentical alleles (N) against the number of allelic mismatches (D) (Fig. 4) (19). Consistent with the relatively high impact of recombination, there was no positive trend in P. shigelloides between the proportion of loci with differences and the average number of nucleotide changes per locus. The average nucleotide difference values dropped slightly from 11 (for D = 2) to 7 (for D = 5), a drop that was not significant given the large standard deviations. The two pairs of P. shigelloides profiles differing by a single allele (D = 1) had one and three nucleotide differences, but the resulting average value (2), based on only two data points, is unreliable. A lack of a positive trend was also observed for N. meningitidis and S. pneumoniae data sets and contrasted with the situation observed for Staphylococcus aureus, E. coli, and Y. pseudotuberculosis. In these species, a positive trend was observed (Fig. 4), which is best explained by the progressive accumulation of mutations with time as strains diverged from their common ancestor.
FIG. 4.
Relationship between allelic profile distance (x axis) and average number of nucleotide differences (y axis) in the distinct alleles. The error bars indicate the standard deviations for N. meningitidis and P. shigelloides. A positive trend was observed for S. aureus, E. coli, and Y. pseudotuberculosis, three species that are generally considered clonal. In contrast, P. shigelloides did not show a positive trend, similar to the recombining bacteria N. meningitidis and S. pneumoniae.
DISCUSSION
The concatenated sequence of four protein-encoding genes places P. shigelloides on an internal, unique branch within the phylogenetic diversity of the Enterobacteriaceae. Therefore, P. shigelloides should not be considered an atypical, early branching member of this family, in contrast to suggestions based on the 16S rRNA gene sequence (41, 45). The fact that P. shigelloides is oxidase positive and thus different from all other Enterobacteriaceae species appears to be a derived characteristic that evolved secondarily from an oxidase-negative ancestor. Our data also rule out a previous proposal, made based on 5S rRNA sequences, to associate P. shigelloides with Proteus species (38).
We developed an MLST scheme based on five genes for evolutionary analysis and strain typing in P. shigelloides. Although most MLST schemes are based on seven genes, there is no particular reason for choosing this number of loci, especially if a lower number provides enough discrimination. Allelic diversity at five genes was enough to discriminate most P. shigelloides strains, and the proposed MLST scheme was one of the most discriminatory among those currently described (www.mlst.net; pubmlst.org; www.pasteur.fr/mlst; web.mpiib-berlin.mpg.de/mlst/). Among the advantages of the MLST method for strain typing are its accuracy and reproducibility. In addition, for P. shigelloides, the capacity to discriminate strains appears to be better than that of serotyping, and all strains could be analyzed, in contrast to serotyping analysis, where a number of strains were untypeable. We propose that MLST could become the reference method for strain tracking and global epidemiology of strains of this pathogen. To this end, a publicly available MLST website was constructed (www.pasteur.fr/mlst).
A high level of nucleotide diversity was found, even though the diversity of the sample of P. shigelloides strains is probably lower than that of the global population of the species. The allelic divergence at several loci was very high, and the nucleotide diversity values were as high as those observed for a global collection of E. coli and were much higher than those for, e.g., S. aureus (21).
The observed level of genotypic diversity, based on allelic profiles, was very high. First, genotype frequencies were widely distributed, with 49 of 77 strains representing unique STs and with no ST comprising more than three strains, resulting in a genotypic diversity index of 99.7%. Second, all but two pairs of genotypes differed by at least two alleles. Therefore, many evolutionarily intermediate genotypes are not represented, and one can expect that future studies using less limited strain samples will reveal a very high number of novel genotypes.
One possible factor contributing to the high level of genotypic diversity is rapid diversification due to high recombination rates; with the same mutation rate, genotypes diversify much more quickly in a species with a high recombination rate than in clonal species. For example, several thousand genotypes have already been described for Neisseria with a high recombination rate (pubmlst.org). We estimated that due to recombination, nucleotides in P. shigelloides evolve approximately 80 times more quickly than they would if the evolutionary process was exclusively clonal. Because the number (n = 2) of comparisons of single-locus variants was insufficient for P. shigelloides, we used comparisons of profiles differing at either one or two loci, in contrast to previous estimates (19, 23, 24) based on single-locus variants only. Therefore, the deduced rate of recombination was expected to be higher, as double-locus variants are less closely related than single-locus variants, hence increasing the probability of double or multiple mutations in the same portion of a gene. Besides, because we used only five genes instead of the seven genes used for the comparative species, the allelic distance of double-locus variants is 40%, whereas the allelic distance is only 29% for profiles that are defined by seven genes. In turn, triple-locus variants in seven-gene profiles correspond to 43% divergence. Therefore, the recombination ratio calculated using a D value of 3 may provide a more appropriate comparison. Based on this ratio, P. shigelloides shows a much higher rate of recombination than other Enterobacteriaceae species and is comparable to S. pneumoniae in both its allelic and nucleotide ratios, while N. meningitidis shows a 2.5-fold-higher nucleotide ratio. When only single-locus variants were used, we obtained values similar to those previously reported for N. meningitidis (23) and S. pneumoniae (24).
Bacterial species vary widely in their rates of homologous recombination (25, 50), and they range from very clonal species, such as Staphylococcus aureus (19), to species with high rates of recombination, such as Helicobacter pylori (53). Pathogens belonging to the Enterobacteriaceae family for which data are available (E. coli, K. pneumoniae, S. enterica, and Y. pseudotuberculosis) have not been shown to be species with high rates of recombination. S. enterica and Y. pseudotuberculosis have been considered highly clonal based on multilocus enzyme electrophoresis data (7, 16, 47), whereas E. coli shows intermediate levels of homologous recombination among housekeeping genes (48, 56, 57). Klebsiella sequence data sets that are publicly available (pubmlst.org/kpneumoniae; S. Brisse, unpublished) show very low levels of recombination. Our clonal diversification analysis shows that recombination has an impact on clonal diversification in these species (except Klebsiella), but the impact is relatively limited compared to that in P. shigelloides. Therefore, P. shigelloides stands out among Enterobacteriaceae species described so far as the member of the family that recombines most. The biological properties of P. shigelloides that could explain the higher impact of homologous recombination are currently unknown. Recombination could be favored by ecological factors, such as the free-living capacity of P. shigelloides, which can favor colocalization of donor and recipient strains, or of transducing phages and bacteria, for DNA exchange. Alternately, colonization of unidentified hosts could result in the high density of bacteria, phages, and/or DNA necessary for horizontal transfer. Other species with high rates of recombination, such as N. meningitidis and S. pneumoniae, are naturally transformable. It is not known whether P. shigelloides is naturally transformable, which would be a likely contributing factor. However, natural transformation in itself is not enough to explain high rates of recombination, as some naturally transformable species, such as H. influenzae, do not exhibit a highly recombinant population structure (21).
High rates of recombination increase the speed of genome diversification, hence affecting the interpretation of genomic differences that are observed among strains. Our results show that there is no strong correspondence between serotype and ST. This result can have several explanations, such as cross-reactivity of unrelated antigens, parallel evolution of antigenic structures, or distinct rates of evolution. However, because recombination reduces the linkage between a given genomic background and individual genes, horizontal transfer of the genetic determinants of the antigens (O-antigen gene cluster or flagellin genes) between distinct genetic backgrounds probably contributes to the weak correspondence between ST and serotypes. In addition, rapid evolution of these genetic determinants by homologous recombination can also confuse the concordance of the two kinds of markers, as classically described for S. pneumoniae (14, 17, 52). Because housekeeping genes are unlikely to be positively selected for variation, detection of recombination in these genes provides an indication that recombination is relatively frequent in the population (25). In contrast, surface-exposed antigens are likely to be under diversifying selection, and hence variants created by recombination may be more frequently retained in the population.
We found no meaningful intraspecies structure within P. shigelloides. Given the homogenizing effect of recombination, this result is consistent with our high recombination rate estimates. Horizontal transfer of genes may also result in the lack of association of genotypes or CCs with particular hosts or sources, as host adaptation factors could be transmitted frequently between genomic backgrounds. However, in N. meningitidis, some STs or CCs are known to be associated with specific epidemic or virulence properties (13, 37), with important consequences for infection control and vaccination strategies. The MLST system described here should be useful for future investigations of these important public health-related questions in P. shigelloides.
Supplementary Material
Acknowledgments
We are grateful to Virginie Passet for assistance with MLST analyses, to J. Rosinsky for serotyping of strains, and to S. Dumontet for support.
Financial support was provided by Institut Pasteur and by a generous gift from the Conny-Maeva Charitable Foundation.
Footnotes
Published ahead of print on 10 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Achtman, M. 2004. Population structure of pathogenic bacteria revisited. Int. J. Med. Microbiol. 294:67-73. [DOI] [PubMed] [Google Scholar]
- 2.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 3.Alabi, S. A., and T. Odugbemi. 1990. Biochemical characteristics and a simple scheme for the identification of Aeromonas species and Plesiomonas shigelloides. J. Trop. Med. Hyg. 93:166-169. [PubMed] [Google Scholar]
- 4.Aldovà, E., and R. H. W. Schubert. 1996. Serotyping of Plesiomonas shigelloides—a tool for understanding ecological relationship. Med. Microbiol. Lett. 5:33-39. [Google Scholar]
- 5.Aldova, E., and T. Shimada. 2000. New O and H antigens of the international antigenic scheme for Plesiomonas shigelloides. Folia Microbiol. (Prague) 45:301-304. [DOI] [PubMed] [Google Scholar]
- 6.Ampofo, K., P. Graham, A. Ratner, L. Rajagopalan, P. Della-Latta, and L. Saiman. 2001. Plesiomonas shigelloides sepsis and splenic abscess in an adolescent with sickle-cell disease. Pediatr. Infect. Dis. J. 20:1178-1179. [DOI] [PubMed] [Google Scholar]
- 7.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, A. Cravioto, et al. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bik, E. M., A. E. Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billiet, J., S. Kuypers, S. Van Lierde, and J. Verhaegen. 1989. Plesiomonas shigelloides meningitis and septicaemia in a neonate: report of a case and review of the literature. J. Infect. 19:267-271. [DOI] [PubMed] [Google Scholar]
- 10.Brenden, R. A., M. A. Miller, and J. M. Janda. 1988. Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloides infections in humans. Rev. Infect. Dis. 10:303-316. [DOI] [PubMed] [Google Scholar]
- 11.Brenner, D. J. 2006. Enterobacteriaceae, p. 587-607. In G. Garrity et al. (ed.), Bergey's manual of systematic bacteriology. Springer, New York, NY.
- 12.Carrico, J. A., F. R. Pinto, C. Simas, S. Nunes, N. G. Sousa, N. Frazao, H. de Lencastre, and J. S. Almeida. 2005. Assessment of band-based similarity coefficients for automatic type and subtype classification of microbial isolates analyzed by pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:5483-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caugant, D. A., B. E. Kristiansen, L. O. Froholm, K. Bovre, and R. K. Selander. 1988. Clonal diversity of Neisseria meningitidis from a population of asymptomatic carriers. Infect. Immun. 56:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 15.Diancourt, L., V. Passet, J. Verhoef, P. A. D. Grimont, and S. Brisse. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolina, M., and R. Peduzzi. 1993. Population genetics of human, animal, and environmental Yersinia strains. Appl. Environ Microbiol. 59:442-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Linares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dykhuizen, D. E., and L. Green. 1991. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 173:7257-7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feil, E. J., J. E. Cooper, H. Grundmann, D. A. Robinson, M. C. Enright, T. Berendt, S. J. Peacock, J. M. Smith, M. Murphy, B. G. Spratt, C. E. Moore, and N. P. Day. 2003. How clonal is Staphylococcus aureus? J. Bacteriol. 185:3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feil, E. J., and M. C. Enright. 2004. Analyses of clonality and the evolution of bacterial pathogens. Curr. Opin. Microbiol. 7:308-313. [DOI] [PubMed] [Google Scholar]
- 21.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil, E. J., M. C. Maiden, M. Achtman, and B. G. Spratt. 1999. The relative contributions of recombination and mutation to the divergence of clones of Neisseria meningitidis. Mol. Biol. Evol. 16:1496-1502. [DOI] [PubMed] [Google Scholar]
- 24.Feil, E. J., J. M. Smith, M. C. Enright, and B. G. Spratt. 2000. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics 154:1439-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil, E. J., and B. G. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Rey, C., S. B. Svenson, L. Bravo, J. Rosinsky, I. Ciznar, and K. Krovacek. 2000. Specific detection of Plesiomonas shigelloides isolated from aquatic environments, animals and human diarrhoeal cases by PCR based on 23S rRNA gene. FEMS Immunol. Med. Microbiol. 29:107-113. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Rey, C., S. B. Svenson, L. Bravo, A. Siitonen, V. Pasquale, S. Dumontet, I. Ciznar, and K. Krovacek. 2004. Serotypes and anti-microbial susceptibility of Plesiomonas shigelloides isolates from humans, animals and aquatic environments in different countries. Comp. Immunol. Microbiol. Infect. Dis. 27:129-139. [DOI] [PubMed] [Google Scholar]
- 28.Gu, W., C. Gonzalez-Rey, K. Krovacek, and R. E. Levin. 2006. Genetic variability among isolates of Plesiomonas shigelloides from fish, human clinical sources and fresh water, determined by RAPD typing. J. Food Biotechnol. 1:1-12. [Google Scholar]
- 29.Guttman, D. S., and D. E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380-1383. [DOI] [PubMed] [Google Scholar]
- 30.Herrera, F. C., J. A. Santos, A. Otero, and M. L. Garcia-Lopez. 2006. Occurrence of Plesiomonas shigelloides in displayed portions of saltwater fish determined by a PCR assay based on the hugA gene. Int. J. Food Microbiol. 108:233-238. [DOI] [PubMed] [Google Scholar]
- 31.Holmes, E. C., R. Urwin, and M. C. Maiden. 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 32.Janda, J. M. 2005. Family I. Enterobacteriaceae, genus XXVII. Plesiomonas Habs and Schubert 1962, p. 740-744. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer, New York, NY. [Google Scholar]
- 33.Kain, K. C., and M. T. Kelly. 1989. Clinical features, epidemiology, and treatment of Plesiomonas shigelloides diarrhea. J. Clin. Microbiol. 27:998-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly, M. T., and K. C. Kain. 1991. Biochemical characteristics and plasmids of clinical and environmental Plesiomonas shigelloides. Experientia 47:439-441. [PubMed] [Google Scholar]
- 35.Khardori, N., and V. Fainstein. 1988. Aeromonas and Plesiomonas as etiological agents. Annu. Rev. Microbiol. 42:395-419. [DOI] [PubMed] [Google Scholar]
- 36.Krovacek, K., L. M. Eriksson, C. Gonzalez-Rey, J. Rosinsky, and I. Ciznar. 2000. Isolation, biochemical and serological characterisation of Plesiomonas shigelloides from freshwater in northern Europe. Comp. Immunol. Microbiol. Infect. Dis. 23:45-51. [DOI] [PubMed] [Google Scholar]
- 37.Lancellotti, M., A. Guiyoule, C. Ruckly, E. Hong, J. M. Alonso, and M. K. Taha. 2006. Conserved virulence of C to B capsule switched Neisseria meningitidis clinical isolates belonging to ET-37/ST-11 clonal complex. Microbes Infect. 8:191-196. [DOI] [PubMed] [Google Scholar]
- 38.MacDonell, M. T., and R. R. Colwell. 1985. Phylogeny of the Vibrionaceae, and recommendation for two new genera, Listonella and Shewanella. Syst. Appl. Microbiol. 6:171-182. [Google Scholar]
- 39.Maiden, M. C. 2006. Multilocus sequence typing of bacteria. Annu. Rev. Microbiol. 60:561-588. [DOI] [PubMed] [Google Scholar]
- 40.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Murcia, A. J., S. Benlloch, and M. D. Collins. 1992. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA-DNA hybridizations. Int. J. Syst. Bacteriol. 42:412-421. [DOI] [PubMed] [Google Scholar]
- 42.Miller, W. A., M. A. Miller, I. A. Gardner, E. R. Atwill, B. A. Byrne, S. Jang, M. Harris, J. Ames, D. Jessup, D. Paradies, K. Worcester, A. Melli, and P. A. Conrad. 2006. Salmonella spp., Vibrio spp., Clostridium perfringens, and Plesiomonas shigelloides in marine and freshwater invertebrates from coastal California ecosystems. Microb. Ecol. 52:198-206. [DOI] [PubMed] [Google Scholar]
- 43.Porat, N., A. Arguedas, B. G. Spratt, R. Trefler, E. Brilla, C. Loaiza, D. Godoy, N. Bilek, and R. Dagan. 2004. Emergence of penicillin-nonsusceptible Streptococcus pneumoniae clones expressing serotypes not present in the antipneumococcal conjugate vaccine. J Infect. Dis. 190:2154-2161. [DOI] [PubMed] [Google Scholar]
- 44.Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer, and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496-2497. [DOI] [PubMed] [Google Scholar]
- 45.Ruimy, R., V. Breittmayer, P. Elbaze, B. Lafay, O. Boussemart, M. Gauthier, and R. Christen. 1994. Phylogenetic analysis and assessment of the genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas deduced from small-subunit rRNA sequences. Int. J. Syst. Bacteriol. 44:416-426. [DOI] [PubMed] [Google Scholar]
- 46.Santos, S. R., and H. Ochman. 2004. Identification and phylogenetic sorting of bacterial lineages with universally conserved genes and proteins. Environ. Microbiol. 6:754-759. [DOI] [PubMed] [Google Scholar]
- 47.Selander, R. K., P. Beltran, N. H. Smith, R. M. Barker, P. B. Crichton, D. C. Old, J. M. Musser, and T. S. Whittam. 1990. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect. Immun. 58:1891-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selander, R. K., and B. R. Levin. 1980. Genetic diversity and structure in Escherichia coli populations. Science 210:545-547. [DOI] [PubMed] [Google Scholar]
- 49.Shigematsu, M., M. E. Kaufmann, A. Charlett, Y. Niho, and T. L. Pitt. 2000. An epidemiological study of Plesiomonas shigelloides diarrhoea among Japanese travellers. Epidemiol. Infect. 125:523-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, J. M., N. H. Smith, M. O'Rourke, and B. G. Spratt. 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90:4384-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spratt, B. G. 2004. Exploring the concept of clonality in bacteria. Methods Mol. Biol. 266:323-352. [DOI] [PubMed] [Google Scholar]
- 52.Stanhope, M. J., S. L. Walsh, J. A. Becker, L. A. Miller, T. Lefebure, P. Lang, P. D. Bitar, and H. Amrine-Madsen. 2007. The relative frequency of intraspecific lateral gene transfer of penicillin binding proteins 1a, 2b, and 2x, in amoxicillin resistant Streptococcus pneumoniae. Infect. Genet. Evol. 7:520-534. [DOI] [PubMed] [Google Scholar]
- 53.Suerbaum, S., J. M. Smith, K. Bapumia, G. Morelli, N. H. Smith, E. Kunstmann, I. Dyrek, and M. Achtman. 1998. Free recombination within Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:12619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sullivan, C. B., M. A. Diggle, and S. C. Clarke. 2005. Multilocus sequence typing: data analysis in clinical microbiology and public health. Mol. Biotechnol. 29:245-254. [DOI] [PubMed] [Google Scholar]
- 55.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimony (and other methods), 4.0b10 ed. Sinauer, Sunderland, MA.
- 56.Whittam, T. S., H. Ochman, and R. K. Selander. 1983. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA 80:1751-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wirth, T., D. Falush, R. Lan, F. Colles, P. Mensa, L. H. Wieler, H. Karch, P. R. Reeves, M. C. Maiden, H. Ochman, and M. Achtman. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.