Abstract
We performed the first broad study aiming at the reconstruction of the evolutionary history of vibrios by means of multilocus sequence analysis of nine genes. Overall, 14 distinct clades were recognized using the SplitsTree decomposition method. Some of these clades may correspond to families, e.g., the clades Salinivibrio and Photobacteria, while other clades, e.g., Splendidus and Harveyi, correspond to genera. The common ancestor of all vibrios was estimated to have been present 600 million years ago. We can define species of vibrios as groups of strains that share >95% gene sequence similarity and >99.4% amino acid identity based on the eight protein-coding housekeeping genes. The gene sequence data were used to refine the standard online electronic taxonomic scheme for vibrios (http://www.taxvibrio.lncc.br).
Vibrios are widespread in the aquatic environment, occupying a variety of ecological niches, such as the human and animal gut, the surface of chitinous organisms, most notably copepods, and the coral mucus layer. A better understanding of the ecology and the patterns of distribution of vibrios relies on the online electronic taxonomy. Polyphasic taxonomic studies of vibrios performed in recent years have underpinned this new paradigm in studies of the biodiversity and systematics of this group (16, 17, 19). Currently, we recognize 78 species of vibrios distributed into five phylogenetic robust clades corresponding to the genera Vibrio, Photobacterium, Salinivibrio, Enterovibrio, and Grimontia based on 16S rRNA gene sequences (16, 17, 19). Both genome content and architecture indicate that these genera share a common ancestor (12). In addition, the genera within vibrios are defined on the basis of their shared sequence similarities in different loci. Species within the genus Vibrio share at least 85% gene sequence similarity in recA, rpoA, and pyrH (18).
Species of vibrios are defined as clusters of strains with high phenotypic and genotypic similarities. Clusters comprise strains with highly similar genomes as determined by multilocus sequence analysis (MLSA), amplified fragment length polymorphism analysis, and DNA-DNA hybridization (DDH) (16, 17, 19). Formal delineation of bacterial species still relies on DDH, with a cutoff level of >70% DDH similarity, but this technique is time-consuming and can be performed in relatively few laboratories and, more importantly, the DDH data are not cumulative in online databases. Clearly, a reliable and straightforward alternative is the use of MLSA. The usefulness of MLSA in the taxonomy of vibrios was described in previous papers (e.g., references 15 and 18). Overall, species form discrete clusters on the basis of recA, rpoA, and pyrH, with a species cutoff level of >94% gene sequence similarity (18). However, some groups of species, e.g., the Vibrio splendidus and Vibrio harveyi species groups, were somewhat fuzzy on the basis of recA, gyrB, and gapA (15, 18). Thus, it is very important to evaluate additional genetic markers that can distinguish closely related species of vibrios.
DNA sequences may also be useful in unraveling the nature of the speciation processes in vibrios. Some studies suggest that recombination might have occurred between different sister species, such as between V. cholerae and V. mimicus and between V. harveyi and V. campbellii, but it is not clear how prevalent and widespread this process is when all vibrio species groups are analyzed simultaneously. The rationale of this study is that by analyzing partial sequences of nine genes (i.e., ftsZ, gapA, gyrB, mreB, pyrH, recA, rpoA, topA, and 16S rRNA), we will be able to establish a more robust inference of the evolutionary history of vibrios. Clearly, we will also enhance and refine the framework of the online electronic taxonomy (5-19). This framework will allow prompt identification and classification of vibrios through the Internet.
The GenBank accession numbers for the gapA, ftsZ, mreB, topA, and gyrB gene sequences are listed in Table S1 of the supplemental material (see also references 6 and 21).
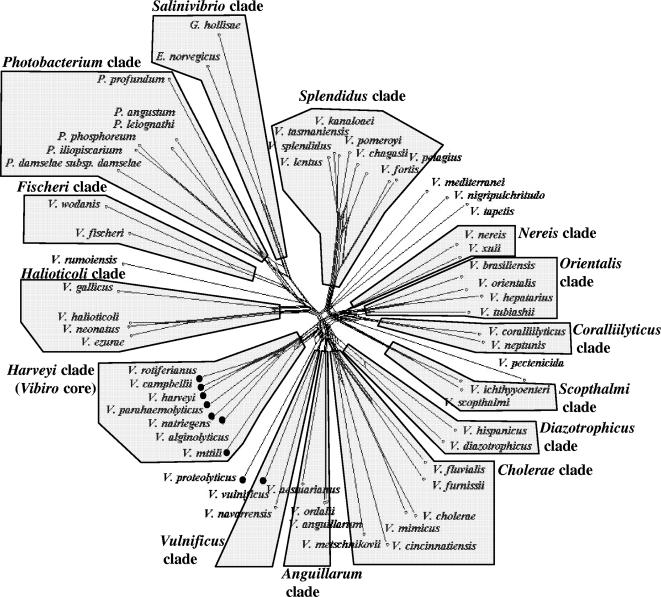
The sequences of the ftsZ, gapA, gyrB, mreB, pyrH, recA, rpoA, topA, and 16S rRNA genes of 78 type strains were concatenated and analyzed by the ClustalX program, MEGA version 3.0, and split decomposition (1, 7, 11, 20) (Fig. 1). Clearly, there were at least 14 monophyletic clades showing split signals with significant bootstrap support (Fig. 1; see also Fig. S2 in the supplemental material [a bifurcating tree]). These clades were always retained in the analysis even when the number of genes was reduced to five loci, i.e., ftsZ, gapA, gyrB, mreB, and topA (see Fig. S3 and S4 in the supplemental material). The species within each clade shared >20% DDH, <5% GC variation (mol%), >85% MLSA sequence similarity, and >89% average amino acid identity (AAI) (Table 1).
FIG. 1.
Concatenated split network tree based on nine gene loci. The ftsZ, gapA, gyrB, mreB, pyrH, recA, topA, and 16S rRNA gene sequences (6,050 bp) from 58 taxa were concatenated and reconstructed using the SplitsTree4 program. Vibrios defined as a vibrio core group (3, 9) are marked with a closed circle. Other clades, which were not included in the analysis, were determined based on five to six gene loci (see Fig. S3 and S4 in the supplemental material). Domains used to construct the phylogenetic tree were regions of ftsZ, gapA, gyrB, mreB, pyrH, recA, rpoA, topA, and 16S rRNA genes of Vibrionaceae, positions 133 to 625, 226 to 893, 441 to 1030, 387 to 892, 89 to 533, 304 to 915, 70 to 903, 155 to 1209 and 451 to 1076 (V. cholerae O1 El tor N16961 [AE003852] numbering), respectively. Sequence similarities were corrected using the Jukes-Cantor correction. All nodes were supported by 100 bootstrap replications.
TABLE 1.
Clades and subclades proposed by means of MLSA for vibrios
| Clade or subclade | Described species included | No. of species | DDH value (%)a | GC (mol%)b | MLSA concatenated similarity (%) | AAI (%)d | Phi teste (P value) | Estimated radiation time(s) for representative pair(s) (108 yrs)f | Habitat |
|---|---|---|---|---|---|---|---|---|---|
| Vibrio clades | 57 | 85.4-98.6 | 89.0-99.8 | ||||||
| Anguillarum | V. anguillarum, V. aestuarianus, and V. ordalii | 3 | >30 | 43-46 | 88.9-98.6 | 95.7-99.6 | 0.56 (V. anguillarum/V. ordalii), 3.76 (V. anguillarum/ V. aestuarianus) | Brackish water, seawater, and fish | |
| Cholerae | V. cholerae, V. cincinnatiensis, V. furnissii, V. fluvialis, V. metschnikovii, and V. mimicus | 6 | NA | 44-50 | 85.4-94.7 | 92.8-99.6 | <0.1 | 0.30 (V. cholerae/V. mimicus), 1.79 (V. fluvialis/V. furnissii), 3.59 (V. cincinnatiensis/ V. metschnikovii) | Brackish water and seawater |
| Coralliilyticus | V. coralliilyticus and V. neptunis | 2 | >64 | 45-46 | 95.6 | 99.6 | NT | Seawater, bivalves, and rotifers | |
| Diazotrophicus | V. diazotrophicus and V. hispanicus | 2 | NA | 43-47 | 91.2 | 97.0 | NT | Brackish water and seawater | |
| Gazogenes | V. aerogenes, V. gazogenes, and V. ruber | 3 | >32 | 46-47 | NT | NT | NT | Estuary and salt marsh mud | |
| Fischeri | V. fischeri, V. logei, V. salmonicida, and V. wodanis | 4 | >36 | 39-42 | 89.8-94.4b | 95.2 | <0.1b | 3.90 (V. fischeri/V. logei) | Seawater, squid, and fish |
| Halioticoli | V. halioticoli, V. ezurae, V. gallicus, V. neonatus, and V. superstes | 5 | >22 | 39-42 | 88.0-97.7 | 94.7-99.5 | <0.1 | 0.23 (V. halioticoli/V. neonatus), 1.11 (V. halioticoli/V. ezurae), 3.65 (V. halioticoli/V. gallicus) | Gut of abalone |
| Harveyi | V. harveyi, V. alginolyticus, V. campbellii, V. mytili, V. natriegens, V. parahaemolyticus, and V. rotiferianus | 8 | >25 | 42-48 | 90.1-96.2 | 97.2-99.4 | <0.1 | 0.39 (V. harveyi/V. campbellii), 1.05 (V. harveyi/ V. parahaemolyticus), 1.81 (V. harveyi/V. mytili) | Seawater, salt marsh mud, and marine animals |
| Nereis | V. nereis and V. xuii | 2 | >30 | 39-47 | 91.2 | 96.8 | NT | Seawater and shrimp | |
| Nigripulchritudo | V. nigripulchritudo and V. penaeicida | 2 | >36 | 46-47 | 89.0c | NT | NT | Seawater and shrimp | |
| Orientalis | V. orientalis, V. brasiliensis, V. hepatarius and V. tubiashii | 4 | >24 | 43-46 | 91.2-94.2 | 97-97.9 | <0.1 | 3.64 (V. orientalis/V. tubiashii) | Brackish water and seawater |
| Scopthalmi | V. scophthalmi and V. ichthyoenteri | 2 | >32 | 43-44 | 95.5 | 99.4 | NT | Gut of flat fish | |
| Splendidus | V. splendidus, V. chagasii, V. crassostrea, V. cyclitrophicus, V. fortis, V. gigantis, V. kanaloaei, V. lentus, V. pelagius, V. pomeroyi, and V. tasmaniensis | 12 | >30 | 39-47 | 90.6-96.5 | 96.5-99.8 | <0.1 | 0.17 (V. splendidus/V. tasmaniensis), 1.61 (V. splendidus/V. chagasii), 2.59 (V. splendidus/ V. pelagius) | Seawater and marine animals |
| Vulnificus | V. vulnificus and V. navarrensis | 2 | >30 | 45-48 | 88.6 | 96.4 | NT | Sewage, seawater, eel, and oyster | |
| Photobacterium subclades | 10 | 87.5-95.8 | 98.6-99.4 | ||||||
| Damselae | P. damselae | 1 | 42 | Seawater and fish | |||||
| Leiognathi | P. leiognathi and P. angustum | 2 | >44 | 40-44 | 94.0 | 98.6 | 1.07 | Seawater and luminous organs | |
| Phosphoreum | P. phosphoreum, P. frigidiphilum, and P. iliopiscarius | 3 | NA | 38-43.8 | 94.24-95.8b | 99.4 | 0.61 (P. phosphoreum/P. iliopiscarius) | Seawater and fish | |
| Profundum | P. profundum, P. indicum, and P. lipolyticum | 3 | NA | 40-42 | 87.5-92.3b | NT | Deep sea | ||
| Rosenbergii | P. rosenbergii | 1 | 47.6-47.9 | Seawater and coral |
Calculated based on six genes.
Calculated based on five genes.
Raw average amino acid identity (10).
The phi test was conducted for clades that included at least four species.
The radiation time was calculated based on average amino acid substitution results (see also Table S3 in the supplemental material). NT, not tested.
Radiation times were estimated for different sister species based on the rate of amino acid substitutions in the eight housekeeping protein genes. These rates were normalized with the known radiation time between Escherichia coli and Salmonella enterica (ca. 1 million years) (8, 13). The lowest radiation time value was calculated for the pair V. splendidus and V. tasmaniensis (Table 1; see also Table S3 in the supplemental material). These species have highly related genomes, with 61% DDH similarity, and may occupy very similar niches (19). The time span of speciation in the well-known closely related species pairs (e.g., V. anguillarum and V. ordalii, V. cholerae and V. mimicus, V. halioticoli and V. neonatus, and V. harveyi and V. campbellii) was estimated as 23 to 56 million years. A common ancestor in some Vibrio clades might have occurred at 360 to 390 million years (e.g., V. anguillarum and V. aestuarianus, V. fischeri and V. logei, or V. halioticoli and V. gallicus) (Table 1; see also Table S3 in the supplemental material), corresponding the Devonian era of vigorous diversification of marine fish. The common ancestor of Salinivibrio, Enterovibrio, and Grimontia might have occurred 580 to 620 million years ago (see Table S3 in the supplemental material), corresponding to the era of Cambrian explosion. Diversification of vibrios may have occurred during this period. Major branches showing distinct split signals represent species groups, some of which (e.g., halioticoli, splendidus, and cholerae) may share ecological niches.
All Photobacterium species formed a single clade that may well correspond to a family on its own. Some structuring was observed within this clade though, with at least five subclades. Split decomposition clearly separated Salinivibrio costicola, Enterovibrio norvegicus, Enterovibrio corallii, Grimontia hollisae, and Vibrio calviensis from the other vibrios. Salinivibrio seems to be the ancestor of the vibrios. The clades Salinivibrio and Photobacterium may correspond to families, while other clades, e.g., Splendidus and Harveyi, correspond to genera. The Fischeri clade appeared in an intermediate position between the Photobacterium and Halioticoli clades, suggesting that the V. fischeri species group may represent a genus on its own.
Overall, the species found in each clade have related genomes. The clades disclosed in this study are congruent with former polyphasic taxonomic work (Table 1). For instance, the species in the Anguillarum clade have a GC content ranging between 43 and 46 mol%. V. anguillarum and V. ordalii have at least 58% mutual DDH similarity and around 30% DDH similarity with V. aestuarianus (5). The Cholerae clade comprises six species which show a broad GC content range. Most of the species within this clade cause diarrhea, but only V. cholerae harbors epidemic and pandemic strains. High DDH values (>65%) between the pair V. cholerae and V. mimicus and between the pair V. fluvialis and V. furnisii were reported, suggesting that these species have closely related genomes. The Cholerae clade includes species with lower Na+ requirements. For instance, the Na+ requirements of V. cholerae, V. metchinikovii, and V. fluvialis range between 5 and 40 mM (2, 4, 5). The so-called Vibrio core group (3, 9) forms the Harveyi clade. Distinguishing species and strains within this clade remains a hard task in taxonomy (20). Recombination between closely related species may partially explain this fact.
Recombination was detected in the Cholerae, Fischeri, Halioticoli, Harveyi, Orientalis, and Splendidus clades (Table 1). The phi test implemented in SplitsTree4 pointed to recombination within the concatenated sequences of vibrios (P = 5.0 × 10−5). Recombination was detected in gyrB (P = 6.4 × 10−3), rrn (P = 7.3 × 10−12), gapA (P = 6.8 × 10−7), and topA (P = 4.5 × 10−2), in agreement with the conflicting phylogenetic splits (parallelograms) observed on the basis of the SplitsTree program (Fig. 1). Recombination was observed in the Cholerae, Fischeri, Halioticoli, Harveyi, Orientalis, and Splendidus clades, at least (Table 1). The recombination analysis suggests that genes responsible for different essential functions in the cell may be targets of recombination, but we cannot rule out the possibility that the recombination tests are providing false positive results. Detecting recombination is basically a statistical endeavor and ideally in vitro experimental work should be carried out in order to confirm the ability of vibrios to carry out recombination in the loci analyzed in this study.
We can define species of vibrios as groups of strains that share >95% gene sequence similarity and >99.4% AAI based on the eight protein-coding housekeeping genes. This definition is an alternative to the standard definition of Vibrio species based on DDH values. We used the gene sequences generated in this study to refine the current standard online electronic taxonomic scheme for vibrios (http://www.taxvibrio.lncc.br) (15). This work will underpin further analyses of fresh isolates of vibrios. In one test case, we analyzed several presumptive V. harveyi strains isolated from diseased animals in Tasmania and found that, in fact, these strains formed a tight new cluster that represents a new species within the Harveyi clade (J. Carson et al., unpublished data).
Supplementary Material
Acknowledgments
We are grateful to Shinichi Koizumi and Yasuhiro Tsuruya for technical assistance. We thank C. Vereecke (LMG Culture Collection, Ghent University, Ghent, Belgium) for providing bacterial strains. We also thank K. Kogure and M. Wada (Tokyo University) for helpful comments.
This work was supported by the Institute of Fermentation Osaka. This work was partly supported by an invitation program of the Goho Life Science Foundation and by the JSPS program to F. L. Thompson (S0614). F. L. Thompson acknowledges grants from IFS and CNPq.
Footnotes
Published ahead of print on 17 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bandelt, H.-J., and A. W. M. Dress. 1992. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol. Phylogenet. Evol. 1:242-252. [DOI] [PubMed] [Google Scholar]
- 2.Baumann, P., L. Baumann, S. S. Bang, and M. J. Woolkalis. 1980. Reevaluation of the taxonomy of Vibrio, Beneckea, and Photobacterium: abolition of the genus Beneckea. Curr. Microbiol. 4:127-132. [Google Scholar]
- 3.Dorsch, M., D. Lane, and E. Stackebrandt. 1992. Toward a phylogeny of the genus Vibrio based on 16S rRNA sequences. Int. J. Syst. Bacteriol. 42:58-63. [DOI] [PubMed] [Google Scholar]
- 4.Farmer, J. J., III, and J. M. Janda. 2005. Vibrionaceae Veron 1965, p. 491-494. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part B. Springer, New York, NY. [Google Scholar]
- 5.Farmer, J. J., III, J. M. Janda, F. W. Brenner, D. N. Cameron, and K. M. Birkhed. 2005. Vibrio Pacini 1854, 411AL, p. 494-546. In D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part B. Springer, New York, NY. [Google Scholar]
- 6.Gil, R., F. J. Silva, J. Pereto, and A. Moya. 2004. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68:518-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huson, D. H., and D. Bryant. 2005. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 8.Hyma, K. E., D. W. Lacher, A. M. Nelson, A. C. Bumbaugh, J. M. Janda, N. A. Strocjbine, V. B. Young, and T. S. Whittam. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 187:619-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kita-Tsukamoto, K., H. Oyaizu, K. Nanba, and U. Shimizu. 1993. Phylogenetic relationships of marine bacteria, mainly members of the family Vibrionaceae, determined on the basis of 16S rRNA sequences. Int. J. Syst. Bacteriol. 43:8-19. [DOI] [PubMed] [Google Scholar]
- 10.Konstantinidis, K. T., and J. M. Tiedje. 2005. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187:6258-6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 12.Okada, K., T. Iida, K. Kita-Tsukamoto, and T. Honda. 2005. Vibrios commonly possess two chromosomes. J. Bacteriol. 187:752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whitam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 14.Scola, B. L., Z. Zeaiter, A. Khamis, and D. Raoult. 2003. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 11:318-321. [DOI] [PubMed] [Google Scholar]
- 15.Thompson, F. L., B. Gomez-Gil, A. T. R. Vasconcelos, and T. Sawabe. 2007. Multilocus sequence analysis reveals that Vibrio harveyi and V. campbellii form distinct species. Appl. Environ. Microbiol. 73:4279- 4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson, F. L., B. Hoste, K. Vandemeulebroecke, and J. Swings. 2001. Genomic diversity amongst Vibrio isolates from different sources determined by fluorescent amplified fragment length polymorphism. Syst. Appl. Microbiol. 24:520-538. [DOI] [PubMed] [Google Scholar]
- 17.Thompson, F. L., T. Iida, and J. Swings. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, F. L., D. Gevers, C. C. Thompson, P. Dawyndt, S. Naser, B. Hoste, C. B. Munn, and J. Swings. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, F. L., and J. Swings. 2006. Taxonomy of the vibrios, p. 29-43. In F. L. Thompson, B. Austin, and J. Swings (ed.), The biology of vibrios. ASM Press, Washington, DC.
- 20.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeigler, D. R. 2003. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int. J. Syst. Evol. Microbiol. 53:1893-1900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.