Abstract
The conjugative tetracycline resistance plasmid pCW3 is the paradigm conjugative plasmid in the anaerobic gram-positive pathogen Clostridium perfringens. Two closely related FtsK/SpoIIIE homologs, TcpA and TcpB, are encoded on pCW3, which is significant since FtsK domains are found in coupling proteins of gram-negative conjugation systems. To develop an understanding of the mechanism of conjugative transfer in C. perfringens, we determined the role of these proteins in the conjugation process. Mutation and complementation analysis was used to show that the tcpA gene was essential for the conjugative transfer of pCW3 and that the tcpB gene was not required for transfer. Furthermore, complementation of a pCW3ΔtcpA mutant with divergent tcpA homologs provided experimental evidence that all of the known conjugative plasmids from C. perfringens use a similar transfer mechanism. Functional genetic analysis of the TcpA protein established the essential role in conjugative transfer of its Walker A and Walker B ATP-binding motifs and its FtsK-like RAAG motif. It is postulated that TcpA is the essential DNA translocase or coupling protein encoded by pCW3 and as such represents a key component of the unique conjugation process in C. perfringens.
Clostridium perfringens is the causative agent of several important histotoxic and enterotoxic diseases of humans and animals (48, 58, 62). Integral to the virulence of C. perfringens is its large repertoire of toxins, several of which are encoded on plasmids that appear to be conjugative (8, 11, 33, 40, 52). Extensive restriction endonuclease analysis of conjugative tetracycline resistance plasmids from C. perfringens has shown that the 47-kb plasmid pCW3 is the prototype conjugative plasmid in this bacterium (2, 3). Analysis of pCW3 previously focused on the inducible tet(P) operon, which confers tetracycline resistance (1, 27, 57). More recent studies have involved determination of the complete sequence of pCW3 and identification of its unique replication protein (8).
Analysis of the pCW3 sequence identified a locus that encodes several gene products with low-level similarity to conjugation proteins from the conjugative transposon Tn916 (8). This region was designated the transfer clostridial plasmid (tcp) locus and is required for transfer, as shown by the isolation of independent tcpF and tcpH mutants and subsequent complementation studies (8). Since the region that encompasses the tcp locus is conserved in all conjugative plasmids from C. perfringens (2, 3, 8, 11, 40), it is likely that the conjugative transfer of both antibiotic resistance and toxin plasmids from this bacterium utilizes a common mechanism.
The mechanism of conjugative transfer between gram-negative cells has been studied extensively (13, 32). In this process the movement of the transferred DNA from DNA-processing proteins, such as the relaxase, to the export proteins that make up the mating pair formation (MPF) complex is facilitated by a coupling protein. Although this precise mechanism has yet to be demonstrated with conjugative plasmids from gram-positive bacteria, bioinformatic analysis has identified considerable similarity between proteins encoded by these plasmids and conjugation systems from gram-negative bacteria (23). Significant similarity has been observed between conjugative plasmids, such as the streptococcal plasmid pIP501 (25, 31), the staphylococcal plasmid pSK41 (18), the lactococcal plasmid pMRC01 (15), and pheromone-induced plasmids from enterococci, such as pAD1 and pAM373 (19). This similarity has allowed identification of key conjugation proteins, such as putative mating channel proteins, relaxases, and coupling proteins, and therefore has led to the hypothesis that gram-positive conjugation systems utilize a mechanism similar to that of their counterparts in gram-negative bacteria (23).
Coupling proteins have two N-terminal transmembrane domains (TMDs) and a C-terminal cytoplasmic region that contains consensus Walker A and B ATP-binding sites (20). These sites are located within conserved VirD4 (COG3505), TraG (pfam02534), and TrwB (cd01127) domains. The cytoplasmic domain of TrwB (TrwBΔN70), the coupling protein from R388, has been crystallized, and its structure has been elucidated. Like F1-ATPases, it is a homohexameric protein complex with a large (20-Å) central channel, through which single-stranded DNA (ssDNA) may be pumped during conjugative transfer (22).
Coupling proteins belong to the same superfamily as DNA translocases such as FtsK and SpoIIIE. FtsK is a bifunctional protein in which the N-terminal domain is involved in cell division and the C-terminal domain is essential for correct chromosomal segregation (34, 65). To modulate chromosomal segregation, FtsK forms a ring-shaped multimeric DNA-binding complex that uses its ATPase activity to move along the double-stranded DNA (dsDNA) (9, 16). SpoIIIE is a DNA export protein that acts as a dsDNA pump to transfer DNA from the mother cell to the forespore during sporulation in Bacillus subtilis (56). FtsK-like DNA translocases are large proteins (800 to 1,200 amino acids [aa]) with five N-terminal TMDs and three conserved regions associated with ATP binding and hydrolysis, namely, the Walker A and B ATP-binding motifs (63) and an RAAG motif (gR-GxhLxxatQ) (16). FtsK-like DNA translocases differ from coupling proteins, all of which have an α-helical domain (AAD) inserted between the Walker A and Walker B motifs. Despite these differences, the structure of these proteins includes a common fold, and FtsK-like DNA translocases and coupling proteins may have similar mechanisms of action (16).
Bioinformatic analysis of pCW3 identified two potential proteins, TcpA and TcpB, that have FtsK-like domains (8). These putative DNA translocases may be involved in the movement of DNA and therefore may perform a role similar to that of the coupling proteins in other conjugation systems. TcpA was predicted to be an integral inner membrane protein with an N-terminal region containing two putative TMDs and a C-terminal cytoplasmic region containing a conserved FtsK/SpoIIIE domain (8). The FtsK/SpoIIIE domain of TcpA encompasses the Walker A and Walker B motifs, as well as the RAAG motif. TcpB, a predicted cytoplasmic protein, also carries the FtsK/SpoIIIE domain identified in TcpA, although it is less well conserved. Therefore, TcpB also has the potential to act as a DNA translocase during the conjugative transfer of pCW3.
To investigate the role of the tcpA and tcpB genes in the conjugative transfer of pCW3, we isolated a pCW3ΔtcpAB mutant that was shown to be conjugation deficient and by performing complementation studies demonstrated that tcpA was essential for conjugative transfer. A pCW3ΔtcpA mutant was also constructed and used to confirm the relationship between various conjugative plasmids from C. perfringens by demonstrating the functional interchangeability of highly divergent tcpA homologs in complementation studies. Functional genetic studies of the conserved TcpA domains revealed that the FtsK-like motifs were essential for the function of TcpA and the conjugative transfer of pCW3.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The C. perfringens strains used in this study are described in Table 1 and were cultured at 37°C in TPG broth (49), brain heart infusion broth (Oxoid), FTG medium (Difco), or nutrient agar (46) supplemented with tetracycline (10 μg/ml), rifampin (10 μg/ml), nalidixic acid (10 μg/ml), thiamphenicol (10 μg/ml), or streptomycin (200 μg/ml), when needed. When required, 1% (vol/vol) saturated potassium chlorate was included. C. perfringens agar cultures were incubated in an atmosphere containing 10% H2, 10% CO2, and 80% N2. The Escherichia coli host strain used was DH5α (Life Technologies), which was grown at 37°C in 2×YT medium (39) supplemented with erythromycin (150 μg/ml) or chloramphenicol (30 μg/ml). Plasmids are also listed in Table 1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| C. perfringens strains | ||
| JIR325 | Strain 13 derivative, Rifr Nalr | 36 |
| JIR4195 | JIR325(pCW3) Rifr Nalr Tcr | P. Johanesen, D. Lyras, and J. Rood, unpublished |
| JIR4394 | Strain 13 derivative, Smr Chlr | 8 |
| JIR4974 | JIR325(pJIR3101) Rifr Nalr Tcr Emr | This study |
| JIR4975 | JIR325(pJIR3102) Rifr Nalr Tcr Emr | This study |
| JIR12051 | JIR325(pJIR3213) Rifr Nalr Tcr Emr | This study |
| JIR12052 | JIR325(pJIR3214) Rifr Nalr Tcr Emr | This study |
| Plasmids | ||
| pT7Blue | E. coli cloning vector, bla+, f1 origin, pUC origin, lacZ α-peptide | Novagen |
| pCW3 | Confers conjugative tetracycline resistance | 8, 49 |
| pCPF4969 | CPE toxin plasmid from F4969 | 40 |
| pMRS4969 | pCPF4969cpe::catP | 50 |
| pJGS1495 | β-Toxin plasmid from JGS1495 | J. G. Songer (University of Arizona) |
| pJGS1721 | ɛ-Toxin plasmid from JGS1721 | J. G. Songer (University of Arizona) |
| pJIR26 | pJIR27ΔTn4452 | 3 |
| pJIR750 | E. coli-C. perfringens shuttle vector, catP+lacZ α-peptide | 7 |
| pJIR1909 | pCW3 10,358-bp ClaI (bp 41533 to 4628) plus pCW3 9,744-bp ClaI (bp 4628 to 14372) | 8 |
| pJIR2715 | Base plasmid for construction of C. perfringens suicide vectors, erm(Q)+catP+oriT+ | 8 |
| pJIR2774 | Conjugative lincomycin resistance plasmid | D. Lyras, J.G. Songer, and J. Rood, unpublished |
| pJIR3093 | pT7Blue (EcoRV) ΩJRP2025/JRP2026 PCR product (1,940 bp) (upstream of tcpA) | This study |
| pJIR3094 | pT7Blue (EcoRV) ΩJRP2027/JRP2028 PCR product (2,291 bp) (downstream of tcpA) | This study |
| pJIR3095 | pT7Blue (EcoRV) ΩJRP2029/JRP2030 PCR product (2,152 bp) (downstream of tcpB) | This study |
| pJIR3096 | pT7Blue(EcoRV) ΩJRP2044/JRP2074 PCR product (2,126 bp) (upstream of tcpB, tcpA+) | This study |
| pJIR3097 | pJIR2715 (XhoI/SacI) ΩpJIR3095 (XhoI/SacI; 1,940 bp) (3′ tcpAB suicide vector) | This study |
| pJIR3099 | pJIR3097 (SphI/BamHI) ΩpJIR3095 (SphI/BamHI; 2,152 bp) (tcpAB suicide vector) | This study |
| pJIR3101 | pCW3ΔtcpAB1 | Transformation with pJIR3099 |
| pJIR3102 | pCW3ΔtcpAB2 | Transformation with pJIR3099 |
| pJIR3104 | pT7Blue (EcoRV) ΩJRP1370/JRP2356 PCR product (1,659 bp) (tcpB+) | This study |
| pJIR3105 | pJIR750 (SphI/BamHI) ΩpJIR3104 (SphI/BamHI; 1,659 bp) (tcpB+ complementation vector) | This study |
| pJIR3207 | pT7Blue (EcoRV) ΩJRP2244/JRP2356 PCR product (3,145 bp) (tcpA+B+) | This study |
| pJIR3209 | pJIR750 (SphI/BamHI) ΩpJIR3207 (SphI/BamHI; 3,145 bp) (tcpA+B+ complementation vector) | This study |
| pJIR3211 | pJIR3099 (SphI/BamHI) ΩpJIR3094 (SphI/BamHI; 2,291 bp) (tcpA suicide vector) | This study |
| pJIR3212 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP2274 PCR product (SphI/BamHI; 2,126 bp) (tcpA+ complementation vector) | This study |
| pJIR3213 | pCW3ΔtcpA1 | Transformation with pJIR3211 |
| pJIR3214 | pCW3ΔtcpA2 | Transformation with pJIR3211 |
| pJIR3333 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP3433 PCR product (SphI/BamHI; 2,126 bp) (tcpApJGS1495 complementation vector) | This study |
| pJIR3334 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP3433 PCR product (SphI/BamHI; 2,126 bp) (tcpApJIR26 complementation vector) | This study |
| pJIR3335 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP3432 PCR product (SphI/BamHI; 2,126 bp) (tcpApCPF4969 complementation vector) | This study |
| pJIR3336 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP2645 PCR product (SphI/BamHI; 1,861 bp) (tcpA1-469 complementation vector) | This study |
| pJIR3337 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP2824 PCR product (SphI/BamHI; 1,546 bp) (tcpA1-365 complementation vector) | This study |
| pJIR3338 | pJIR750 (SphI/BamHI) ΩJRP2244/JRP2825 PCR product (SphI/BamHI; 1,400 bp) (tcpA1-316 complementation vector) | This study |
| pJIR3339 | pJIR3096 tcpAK242A | Site-directed mutagenesis |
| pJIR3340 | pJIR750 (SphI/BamHI) ΩpJIR3339 (SphI/BamHI; 2,126 bp) | This study |
| pJIR3341 | pJIR3096 tcpADE334/5AA | Site-directed mutagenesis |
| pJIR3342 | pJIR750 (SphI/BamHI) ΩpJIR3341 (SphI/BamHI; 2,126 bp) | This study |
| pJIR3343 | pJIR3096 tcpAQ379A | Site-directed mutagenesis |
| pJIR3344 | pJIR750 (SphI/BamHI) ΩpJIR3343 (SphI/BamHI; 2,126 bp) | This study |
| pJIR3345 | pJIR3096 tcpAΔ46aa-69 | Site-directed mutagenesis |
| pJIR3346 | pJIR750 (SphI/BamHI) ΩpJIR3345 (SphI/BamHI; 2,057 bp) | This study |
| pJIR3347 | pJIR3096 tcpAΔ79aa-104 | Site-directed mutagenesis |
| pJIR3348 | pJIR750 (SphI/BamHI) ΩpJIR3347 (SphI/BamHI; 2,051 bp) | This study |
| pJIR3349 | pJIR3096 tcpAΔ46aa-104 | Site-directed mutagenesis |
| pJIR3350 | pJIR750 (SphI/BamHI) ΩpJIR3349 (SphI/BamHI; 1,952 bp) | This study |
Rifr, Nalr, Tcr, Emr, TmR, Smr, and Chlr, resistance to rifampin, nalidixic acid, tetracycline, erythromycin, thiamphenicol, streptomycin, and potassium chlorate, respectively.
Molecular techniques.
E. coli plasmid DNA was isolated by an alkaline lysis method performed according to the manufacturer's instructions (QIAGEN). Purified C. perfringens DNA was obtained as described previously (47). For PCR amplification Taq DNA polymerase (Roche) and 0.5 μM of each primer were used. Denaturation (94°C for 30 s), annealing (50°C for 30 s), and extension (72°C for 3 to 5 min) steps were carried out for 30 cycles. Sequence analysis of constructs was performed with an Applied Biosystems 3730S capillary sequencer. Sequence data were analyzed using Vector NTI (Invitrogen) in conjunction with the Sanger Institute freeware Artemis, release 6. TcpA sequences were aligned using ClustalW (61) and T Coffee (42). Details concerning oligonucleotides are shown in Table S1 in the supplemental material.
Construction of C. perfringens mutants by allelic exchange.
C. perfringens suicide vectors contained ca. 2 kb of sequence upstream and downstream of the gene to be mutated. These regions were generated by PCR and cloned into the pT7Blue cloning vector (Novagen). Each product was then subcloned sequentially into the E. coli vector pJIR2715 (8) (Table 1). This vector, which replicates in E. coli but not in C. perfringens, encodes chloramphenicol (or thiamphenicol) and erythromycin resistance. To isolate a derivative of pCW3 from which both tcpA and tcpB were deleted, a ΔtcpAB suicide vector was constructed as follows. The bp 23973 to 25913 pCW3 region upstream of tcpA was cloned upstream of erm(Q), and the bp 28420 to 30572 region downstream of tcpB was cloned downstream of erm(Q) to generate the suicide vector pJIR3099. Similarly, a ΔtcpA suicide vector was constructed by cloning the bp 23973 to 25913 and bp 27476 to 29767 regions of pCW3 into pJIR2715 upstream and downstream, respectively, of erm(Q) to form pJIR3211. The suicide vectors were independently introduced into JIR4195 [i.e., JIR325(pCW3)] by electroporation (55). In these experiments, transformation of these suicide vectors into C. perfringens and selection for erythromycin resistance selected for strains in which the plasmid had integrated into the host genome, presumably within pCW3. Screening for thiamphenicol sensitivity identified putative mutants derived from double-crossover events. DNA was purified from potential erythromycin-resistant thiamphenicol-sensitive recombinants, and PCR and Southern blotting (59) were used to confirm the replacement of the target gene(s) with the erm(Q) cassette and loss of the suicide plasmid.
Construction of complementation vectors.
PCR products carrying the pCW3-derived wild-type tcpA, tcpB, and tcpAB gene regions were generated using the primer pairs JRP2244/JRP2274, JRP1370/JRP2356, and JRP2244/JRP2356, respectively, and were cloned into the C. perfringens-E. coli shuttle vector pJIR750 via pT7Blue, generating pJIR3212, pJIR3105, and pJIR3209, respectively. The tcpA gene was also amplified from C. perfringens plasmids pJGS1495, pJIR26, and pMRS4969, which is a derivative of pCPF4969 (50). Due to sequence differences the same 5′ primer but different 3′ primers were used. The resulting PCR products were cloned directly into pJIR750 to generate pJIR3333, pJIR3334, and pJIR3335, respectively. Various site-directed mutants or internal deletions of the tcpA gene were isolated using a QuikChange mutagenesis kit (Stratagene). The starting vector in these experiments was a pT7BluetcpA+ derivative, pJIR3096. Oligonucleotide pairs JRP3406/JRP3407, JRP3408/JRP3409, and JRP3410/JRP3411 were used to construct the tcpA mutants K242A, DE334/335AA, and Q379A, respectively. Plasmids encoding the TMD deletion derivatives TcpAΔ46-69, TcpAΔ79-104, and TcpAΔ46-104 were also generated by QuickChange mutagenesis of pJIR3096 using oligonucleotide pairs JRP2827/JRP2828, JRP2829/JRP2830, and JRP2831/JRP2832, respectively. Each pJIR3096 derivative was then subcloned into pJIR750 to generate the mutated complementation vectors pJIR3340, pJIR3342, and pJIR3344, as well as the TMD deletion complementation vectors pJIR3346, pJIR3348, and pJIR3350, respectively. tcpA derivatives encoding TcpA1-469, TcpA1-365, and TcpA1-316 were constructed using PCR products amplified using JRP2244 and reverse primers JRP2645, JRP2824, and JRP2825, respectively. Each C. perfringens strain that contained one of these complementation vectors was confirmed by restriction endonuclease analysis of PCR products and sequence analysis where appropriate.
Conjugations.
Matings on solid media were carried out as described previously (46, 47). Nutrient agar supplemented with tetracycline, streptomycin, and potassium chlorate was used to select for transconjugants when strain JIR4394 was used as the recipient. The efficiency of conjugative transfer was expressed as the number of transconjugants per donor cell.
RESULTS
TcpA protein is a member of the FtsK superclade.
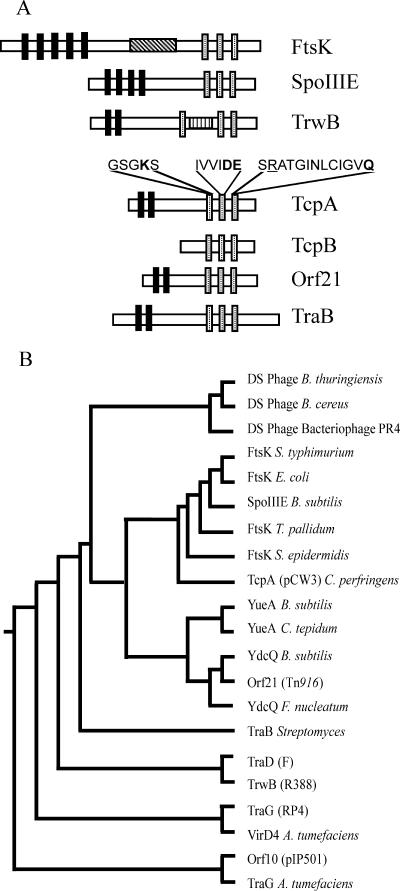
Bioinformatic analysis of the putative tcpA gene product identified ATP-binding Walker boxes A and B within an FtsK-like conserved domain (COG1674) and a similar, although less well conserved, domain within TcpB (8). FtsK domains are also present within coupling proteins, but further analysis of the 530-aa TcpA protein did not identify any domains specific to coupling proteins. To investigate the relationship between TcpA, TcpB, DNA translocases, and coupling proteins, their amino acid sequences were aligned (data not shown) and their putative domain structures were compared (Fig. 1A). The results indicated that TcpA shares similar domain features with FtsK and several coupling proteins.
FIG. 1.
Bioinformatic analysis of TcpA and TcpB. (A) Domain structure analysis of TcpA and TcpB. The analysis was performed using Sosui (24) and Conserved Domain Database (37) searches, and the results were compared with related FtsK-HerA ATPase proteins. TMDs are represented by solid bars; coiled-coil domains are represented by the box with diagonal stripes; the AAD region is represented by a box with vertical stripes; and the Walker A, Walker B, and RAAG motifs are represented by dotted bars. Residues of the TcpA motifs targeted by amino acid substitutions are in bold type, and the conserved arginine residue of the RAAG motif is underlined. (B) Dendrogram constructed using ClustalW alignments of TcpA, TcpB, and FtsK-HerA members and the PHYLIP algorithm (17).
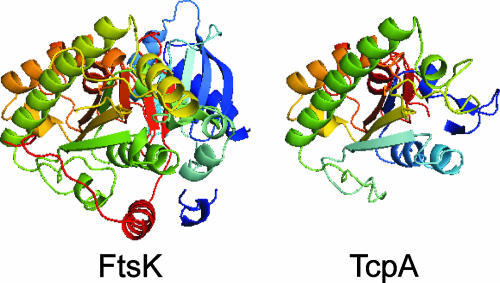
Multiple alignments of TcpA with various members of the FtsK-HerA superfamily suggested that TcpA was a member of this superfamily and was most similar to the FtsK clade. Members of this clade lack an AAD domain and typically have a coiled-coil region and a glycine before the core ATPase domain (26). The FtsK clade has been divided into three families, the classical FtsK family, the YueA family, and the YdcQ family. The distinction between the classical FtsK and YdcQ families is based on conserved residues, while YueA members contain three tandem ATPase domains. Searches of TcpA using these conserved features did not clearly identify TcpA as a member of any one of these families. To further define the relationships between the FtsK clade proteins and TcpA, ClustalW multiple alignments containing a diverse range of FtsK-HerA members were used. The resultant alignments indicated that TcpA branches with members of the FtsK clade (Fig. 1B). By using the crystal structure of an ATPase mutant of FtsKc, a derivative of FtsK that contains the C-terminal region (38), a putative model (Fig. 2) of the central section of TcpA (aa 212 to 420) could be constructed using Swiss Model (54). This analysis supported the conclusion that TcpA is a member of the FtsK family. Note that TcpA could not be modeled on the known TrwB structure.
FIG. 2.
Model of TcpA structure. Swiss Model (54) was used to construct a putative model of the central region of TcpA (aa 212 to 420). The template structure was a C-terminal region of FtsK (aa 818 to 1329; PDB: 2iusA) (38).
The tcpB gene, located directly downstream of tcpA, encodes a predicted 327-aa protein. Although conserved domain searches and BlastP analysis identified an FtsK domain within TcpB, the level of similarity was lower than that of TcpA. In addition, unlike other members of the FtsK family and the coupling proteins, TcpB lacks potential TMDs. Multiple alignments of TcpB with members of the FtsK-HerA superfamily indicated that the three conserved motifs were not well conserved in TcpB. The phylogenetic tree constructed using this alignment did not place TcpB within any of the described FtsK-HerA clades or families (data not shown), and TcpB could not be modeled on the FtsK structure.
TcpA is essential for the conjugative transfer of pCW3.
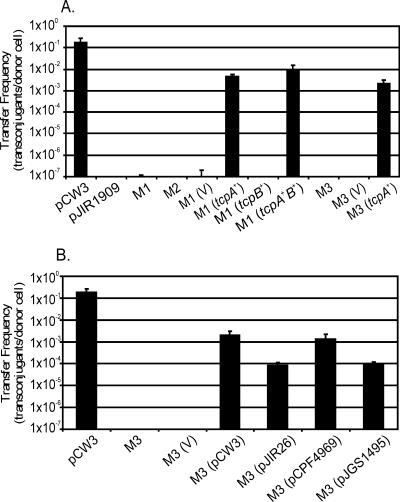
Since tcpA and tcpB both encoded putative proteins with FtsK-like domains, pCW3ΔtcpAB deletion mutants were constructed by allelic exchange to investigate the role of these genes in the conjugative transfer of pCW3. After transformation of JIR325(pCW3) with the suicide vector pJIR3099, two independently derived deletion derivatives were obtained, and their pCW3ΔtcpAB genotypes were confirmed by PCR analysis and Southern blotting (data not shown). In mixed plate matings these pCW3 derivatives (pJIR3101 and pJIR3102) were unable to confer conjugative transfer (Fig. 3). In these experiments the same host strain carrying pCW3 or pJIR1909 were used as positive and negative controls, respectively. Since both plasmids behaved in the same way, all further studies were carried out with pJIR3101.
FIG. 3.
Complementation of pCW3ΔtcpAB and pCW3ΔtcpA mutants. (A) Conjugation-deficient pCW3ΔtcpAB and pCW3ΔtcpA mutants were complemented with functional copies of tcpAB, tcpA, and tcpB and tested for the ability to transfer in mixed plate matings. (B) tcpA orthologs from pJIR26, pCPF4969, and pJGS1495 were tested for conjugative proficiency using mixed plate mating and compared to the wild-type pCW3 derivative. pCW3, wild-type positive control; pJIR1909, nontransferable negative control; M1, pCW3ΔtcpAB1; M2, pCW3ΔtcpAB2; M3, pCW3 ΔtcpA; V, pJIR750 vector control. The transfer efficiencies were expressed as the number of transconjugants per donor cell.
To confirm that the loss of function was due to a specific mutation in pCW3, complementation analysis was performed. The wild-type tcpA and tcpB genes were cloned independently into the C. perfringens-E. coli shuttle vector pJIR750, as was a wild-type region containing both genes. The plasmids subsequently were introduced into JIR325(pJIR3101), which carried the mutated ΔtcpAB::erm(Q) region. Note that the replication region of pJIR750 is derived from the nonconjugative bacteriocin plasmid pIP404 and its derivatives are compatible with pCW3. Complementation with the wild-type tcpAB genes restored conjugative transfer, although not to wild-type levels (Fig. 3A). When the mutation was complemented with the wild-type tcpA gene, the conjugation frequency was not significantly different from that observed after complementation with the tcpA+B+ vector. By contrast, complementation with the wild-type tcpB gene alone did not restore conjugative transfer (Fig. 3A).
To confirm the essential role of the tcpA gene in conjugation, a pCW3ΔtcpA mutant was constructed by allelic exchange, shown to be nonconjugative, and then complemented with the tcpA+ complementation vector. The transfer frequencies of pCW3ΔtcpAB and pCW3ΔtcpA, when complemented with the wild-type tcpA gene, were indistinguishable (Fig. 3A). Based on these results it was concluded that tcpA was essential for the conjugative transfer of pCW3 but that tcpB was not an essential conjugation gene.
Various tcpA homologs can complement the pCW3ΔtcpA mutation.
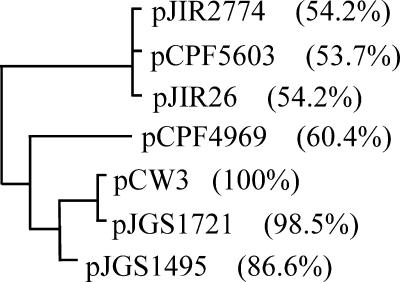
The TcpA protein is the most variable gene product encoded in the C. perfringens tcp locus, and the levels of TcpA amino acid sequence identity vary from 54 to 98% (Fig. 4). In addition, several deletion and insertion events have been observed in the region between the tcpA and tcpC genes (8). The specificity of conjugation systems in gram-negative bacteria is determined by the interaction between the coupling proteins and the cognate relaxase (12). On this basis the abilities of several tcpA homologs to complement the pCW3ΔtcpA mutant were examined to investigate the functional relationship between the tcp loci of the most divergent conjugative plasmids from C. perfringens. A comparative analysis was carried out with TcpA homologs from the conjugative plasmids pCW3, pJIR2774 (which encodes lincomycin resistance), pJIR26 (which encodes tetracycline resistance), and pCPF4969 (which encodes the enterotoxin CPE), as well as plasmids pCPF5603 (CPE), pJGS1495 (β-toxin), and pJGS1721 (ɛ-toxin), which carry the tcp locus (8, 40) but whose conjugative abilities have not been tested. The resultant ClustalW alignments identified significant sequence identity in the N-terminal region of these proteins, as well as in the central region that encompasses the Walker boxes, although the last 70 aa were more variable (data not shown). The phylogenetic tree constructed from these alignments separated these TcpA homologs into four distinct groups (Fig. 4), which correlated with the genetic organization of the tcp region (8).
FIG. 4.
Phylogenetic tree of TcpA homologs. The amino acid sequences of seven TcpA orthologs from a range of C. perfringens conjugative plasmids were aligned using ClustalW. The level of identity of each protein to TcpA from pCW3 is indicated in parentheses. The accession numbers are as follows: pCW3, DQ366035; pCPF4969, NC007772; pJIR2774, DQ338473; pCPF5603, NC007773; and pJIR26, DQ338471. The sequences of pJGS1495 and pJGS1721 were obtained from G. Myers, I. Paulsen, J. Songer, B. McClane, R. Titball, J. Rood, and S. Melville (personal communication).
The genes encoding divergent TcpA homologs from pJIR26, pCPF4969, and pJGS1495 were cloned into pJIR750, and the resultant derivatives were introduced into C. perfringens and used to complement the pCW3ΔtcpA mutant in mixed plate matings. The tcpA gene from the CPE plasmid pCPF4969 yielded a transfer frequency that was indistinguishable from that obtained with the pCW3-derived tcpA gene. The genes from the tetracycline resistance plasmid pJIR26 and the β-toxin plasmid pJGS1495 also complemented the mutation, but at a frequency 1 order of magnitude lower (Fig. 3B). Comparative analysis could not attribute this reduction in transfer proficiency to any specific amino acid sequence variations.
Identification of the regions of TcpA that are essential for conjugation.
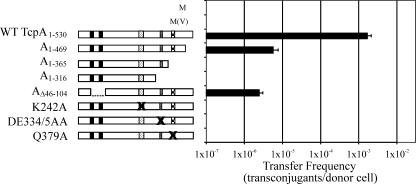
To identify regions of TcpA that were required for the transfer of pCW3, a series of C-terminal deletion derivatives were constructed and tested for the ability to complement the pCW3ΔtcpA mutant (Fig. 5). We sequentially deleted the last 61 aa of TcpA (TcpA1-469) and then the RAAG motif (TcpA1-365) and the Walker B box (TcpA1-316) specifically to investigate the role of these regions in the transfer of pCW3. Only one C-terminal deletion derivative, tcpA1-469, restored transfer in the mutant, although not to levels observed with the full-length wild-type gene (Fig. 5). The other deletion derivatives, tcpA1-365 and tcpA1-316, were unable to complement the tcpA mutant. To investigate the role of the TMDs, derivatives of TcpA were constructed in which the putative TMDs were deleted individually and together. Both tcpAΔ46-69 and tcpAΔ76-104 were unable to complement the mutation (data not shown). However, a derivative with both TMDs deleted, tcpAΔ46-104, was able to restore transfer, although not to wild-type levels (Fig. 5).
FIG. 5.
Effect of deletion and site-directed mutations on TcpA activity. Isogenic shuttle vector derivatives were used to complement the pCW3ΔtcpA mutant (M), and the conjugation frequencies of the resultant derivatives were determined. The positive control carried the wild-type tcpA gene (WT TcpA1-530), and the negative control carried the shuttle vector [M(V)]. The mutated tcpA derivatives encoded TcpA1-469, TcpA1-365, TcpA1-316, TcpAΔ46-69, TcpAΔ79-104, TcpAΔ46-104, TcpA K242A, TcpA DE334/5AA, and TcpA Q379A. The solid bars represent TMDs, and the Walker A box, the Walker B box, and the RAAG motif are represented by dotted bars, bars with horizontal stripes, and checked bars, respectively. The crosses indicate the motifs targeted by site-directed mutagenesis.
Since the TcpA Walker boxes were conserved, it was predicted that, like DNA translocases and coupling proteins, TcpA may have a requirement for ATPase activity. To investigate the role of the Walker boxes and the RAAG motif, individual site-directed mutants with substitutions in each motif were constructed and tested for the ability to complement the pCW3ΔtcpA mutant. No detectable transfer was observed for the derivatives containing alanine substitutions in the Walker A motif (K242A), the Walker B motif (DE334/335AA), or the RAAG motif (Q379A) (Fig. 5). Based on these results it was concluded that all three motifs are essential for the functional integrity of the TcpA protein.
DISCUSSION
Several C. perfringens toxin genes are associated with putative conjugative plasmids (8, 33, 40, 52), which has significant implications not only for the toxin-typing scheme used to type C. perfringens strains but also for pathogenesis. Despite this finding, very little is known about the effect of transfer on pathogenesis or the mechanism by which transfer occurs. To contribute to our understanding of the mechanism of C. perfringens conjugative transfer, we investigated the role of two genes in the conjugative transfer of the paradigm conjugative plasmid, pCW3. These genes, tcpA and tcpB, encode putative proteins with domains associated with DNA translocases, which suggests that either protein may be involved in the movement of DNA during conjugative transfer. In this study mutation and complementation analysis showed that pCW3ΔtcpAB mutants were unable to transfer by conjugation but could be complemented in trans with just the wild-type tcpA gene, providing evidence that loss of the ability to transfer was not the result of polarity effects. Furthermore, a pCW3ΔtcpA mutant was also transfer deficient and could be complemented with the tcpA gene. Based on these experiments we concluded that the tcpA gene has an essential role in the conjugative transfer of pCW3. tcpA is the third gene in the tcp locus shown to be essential for conjugation since previous studies have shown that both tcpF and tcpH are also essential conjugation genes (8).
Complementation of the pCW3ΔtcpAB mutants with the wild-type tcpAB genes had the same effect on conjugation as complementation with the tcpA gene alone. In addition, complementation with the tcpB gene alone did not restore conjugative transfer. These results provide good evidence that tcpB is not essential for transfer. The results of a comparative analysis of the tcp region from various conjugative plasmids from C. perfringens were in agreement with this conclusion since not all of these conjugative plasmids carry tcpB. Previous studies showed that tcpB is absent from the conjugative tetracycline resistance plasmid pJIR26, as well as from the β-toxin plasmid pJGS1495 (8). Furthermore, our analysis and that of other workers identified several potential recombination events within the tcpAB region, including the presence of a third FtsK-like gene that is located between tcpA and tcpB in the conjugative CPE plasmid pCPF4969 and encodes a putative 203-aa protein (8, 40). It is postulated that tcpB is a truncated version of tcpA that arose from a gene duplication event and subsequent deletion events, although attempts to identify potential recombination sites by bioinformatic analysis have not been successful.
To investigate the relationship between TcpA, coupling proteins, and DNA translocases, multiple alignment was performed using members of the FtsK-HerA superfamily of ATPases. This family encompasses several families of proteins involved in the movement of DNA during bacterial replication, sporulation, conjugation, or phage packaging (26). Extensive analysis of this superfamily has identified conserved sequence blocks that encompass the ATPase domain and can be used to separate members of the superfamily into two superclades based on the presence of the AAD domain between the Walker A and Walker B boxes. Further classification of these superclades into clades and then families was carried out using residues located in and around the conserved sequence blocks. As determined by this approach, coupling proteins, which have an AAD domain, belong to a different superclade than the FtsK-SpoIIIE family of DNA translocases, which do not have an AAD domain (26). TcpA does not have the AAD region that is associated with coupling proteins and is most similar to members of the FtsK clade, to which part of TcpA can also be modeled. Members of this clade include proteins from gram-positive plasmids, bacteriophages, and conjugative transposons. These proteins are thought to function in cis as DNA pumps for the transfer of plasmid DNA during cell division or for packaging of DNA into the bacteriophage (26). ORF21 from the enterococcal conjugative transposon Tn916 and the Streptomyces TraB protein are also FtsK-like proteins that lack the AAD region and are structurally similar to TcpA (Fig. 1). Although Tn5 mutagenesis of ORF21 has implicated this protein in conjugative transfer (64), no data are available to shed light on its precise functional role.
In Streptomyces, TraB is a central factor in a conjugative transfer mechanism that is distinct from that of gram-negative bacteria. TraB is unique in that it is the only protein required for transfer (28). In this process, TraB binds specifically to clt, an essential 50-bp noncoding region adjacent to the traB gene (45). Analysis of denatured TraB-DNA complexes indicated that TraB does not process the DNA during binding (45), and furthermore, it is thought that dsDNA, not ssDNA, is transferred to the recipient cell (44). Since TcpA, like TraB, is also an FtsK-like protein and no pCW3-encoded relaxase has been identified, we cannot rule out the possibility that dsDNA rather than ssDNA is transferred during pCW3-mediated conjugation. However, pCW3 is unlikely to use a Streptomyces-like mechanism of transfer since the transfer of pCW3 also requires TcpH, a putative membrane-spanning protein, and TcpF, a putative cytoplasmic ATPase (8). It is postulated that although TcpA is more closely related to members of the FtsK family of dsDNA translocases, it is still a functional coupling protein that links the DNA to be transferred to the MPF apparatus.
Sequence analysis of plasmids that have the tcp locus has revealed that TcpA is the most variable tcp gene product (8). However, complementation with even the most divergent TcpA homologs was able to restore transfer proficiency to a pCW3ΔtcpA mutant (Fig. 3B). This result provides experimental evidence that supports the sequence data predictions that all of the conjugative plasmids that have been identified from C. perfringens use a similar transfer mechanism. Comparative analysis of these TcpA homologs identified significant C-terminal sequence variation, which may be attributable to recombination events that clearly occur in the tcpAB region. In the F plasmid system, changes to the C-terminal domain of TraD are associated with a decrease in the specificity of TraD, such that interactions occur with noncognate relaxases, although these changes also lead to a reduction in transfer efficiency (51). The tcpA genes from pJIR26 and pJGS1495 were less efficient at complementing a pCW3ΔtcpA mutant than the equivalent genes from pCW3 and pCPE4969. However, since the C-terminal region of TcpA from pJGS1495 is very similar to that from pCPF4969, sequence differences outside this region may also be responsible for the observed functional differences.
In other conjugation systems protein-protein interactions have been demonstrated between the relaxosome components TraM (F), TraI (RP4), and both TrwC and TrwA (R388) and their cognate coupling proteins (10, 35, 53). Interaction between TraM and the coupling protein TraD is dependent on the last 38 aa of TraD (10). To further investigate the role of the C terminus of TcpA in the transfer of pCW3, the deletion derivative TcpA1-469 was constructed and analyzed. The results showed that removal of the last 61 aa led to a reduction in transfer frequency of at least 2 orders of magnitude (Fig. 4). Although this region of TcpA is not absolutely required for transfer, it clearly plays a major role in the conjugation process. Evidence for any involvement of this region with interactions between TcpA and the putative relaxase must await identification of the latter protein.
The Walker A box of the cytoplasmic domains of coupling proteins is essential for ATP binding and conjugative transfer (6, 30, 41). For example, in the Agrobacterium tumefaciens type IV secretion system the active transfer of the ssDNA-relaxase intermediate from VirB11 to the core subunits VirB6 and VirB8 is dependent on the Walker A box of VirD4 (4). Functional analysis of the Walker B motif in TraG (RP4) also has indicated that this motif is essential for conjugative transfer (6). The aspartate residue targeted for substitution in TraG (and in TcpA in this study) is predicted to be required for coordination of the magnesium cation (Mg2+) involved in nucleoside triphosphate hydrolysis, whereas the neighboring glutamate primes a water molecule for a nucleophilic attack on the γ-phosphate of the bound ATP (60). To date no functional analysis of the RAAG motif of coupling proteins has been reported; however, comparative analysis of the structural similarity between TrwB and F1-ATPase has indicated that the conserved arginine of this motif may function as an arginine finger (21). In AAA+ ATPases such as F1-ATPase the arginine finger is thought to interact with the C-terminal phosphate of an ATP molecule that is bound to the preceding protomer in the hexameric ring and to affect oligomerization, ATP recognition, and ATP hydrolysis (14, 43). The conserved glutamine appears to be required for sensing the triphosphate moiety of the bound nucleotide, triggering its hydrolysis (29, 60). In this study, substitutions in each of these three motifs in TcpA eliminated conjugative transfer, providing evidence that TcpA-mediated ATPase activity is essential for the conjugation process.
The TMDs of TraG are essential for transfer of RP4; they are required for oligomerization in vitro, as well as protein-protein interactions with relaxosome components (53). By contrast, the TMDs of FtsK are not essential for its ATPase activity, multimer formation, or DNA translocase activity (5). Similarly, we have found that the TMDs of TcpA are not essential for transfer, although deletion of these TMDs led to a very significant reduction in transfer frequency (Fig. 5). Protein-protein interactions between the TMD deletion derivative TcpAΔ46-104 and other conjugation proteins may still allow correct localization of TcpA at the MPF apparatus, although at a greatly reduced efficiency.
In this study we demonstrated that TcpA is essential for the conjugative transfer of pCW3. Functional analysis of TcpA confirmed the role of the Walker A and B boxes, the RAAG motif, and the TMDs and suggested that efficient transfer depends upon the ATPase activity of TcpA and its membrane localization. Given the relationship between TcpA and the FtsK-HerA family of pumping ATPases, we propose that TcpA acts as a functional coupling protein and therefore is involved in the movement of DNA during conjugative transfer. By carrying out similar studies with the other Tcp proteins and analyzing their abilities to interact with TcpA, we aim to develop a model that describes the conjugation process in C. perfringens. Since all of the known conjugative plasmids from C. perfringens have the tcp locus and therefore appear to transfer by the same Tcp-mediated mechanism and since the tcp locus is also present on several other toxin plasmids not yet demonstrated to transfer (8, 33, 40, 52), the development of this model is critical for our understanding of how toxin genes and antibiotic resistance determinants can move between strains of this important human and animal pathogen.
Supplementary Material
Acknowledgments
We thank Wee Lin Teng for helpful discussions.
This research was supported by a grant from the Australian Research Council to the ARC Centre of Excellence in Structural and Functional Microbial Genomics and by grant AI056177-03 from the U.S. National Institute of Allergy and Infectious Diseases. J. A. Parsons was the recipient of a postgraduate scholarship awarded by the ARC Centre of Excellence and the Department of Microbiology.
Footnotes
Published ahead of print on 24 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abraham, L. J., and J. I. Rood. 1985. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 13:155-162. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, L. J., and J. I. Rood. 1985. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J. Bacteriol. 161:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham, L. J., A. J. Wales, and J. I. Rood. 1985. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 14:37-46. [DOI] [PubMed] [Google Scholar]
- 4.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aussel, L., F.-X. Barre, M. Aroyo, A. Stasiak, A. Z. Stasiak, and D. Sherratt. 2002. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108:195-205. [DOI] [PubMed] [Google Scholar]
- 6.Balzer, D., W. Pansegrau, and E. Lanka. 1994. Essential motifs of relaxase (TraI) and TraG proteins involved in conjugative transfer of plasmid RP4. J. Bacteriol. 176:4285-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannam, T. L., and J. I. Rood. 1993. Clostridium perfringens-Escherichia coli shuttle vectors that carry single antibiotic resistance determinants. Plasmid 29:233-235. [DOI] [PubMed] [Google Scholar]
- 8.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 188:4942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beranek, A., M. Zettl, K. Lorenzoni, A. Schauer, M. Manhart, and G. Koraimann. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J. Bacteriol. 186:6999-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabezon, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 13.Christie, P. J. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey, M. J., D. Jeruzalmi, J. Kuriyan, and M. O'Donnell. 2002. Motors and switches: AAA+ machines within the replisome. Nat. Rev. Mol. Cell Biol. 3:826-835. [DOI] [PubMed] [Google Scholar]
- 15.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 16.Errington, J., J. Bath, and L. J. Wu. 2001. DNA transport in bacteria. Nat. Rev. Mol. Cell Biol. 2:538-545. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 2005. PHYLIP—phylogeny interference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 18.Firth, N., K. P. Ridgway, M. E. Byrne, P. D. Fink, L. Johnson, I. T. Paulsen, and R. A. Skurray. 1993. Analysis of a transfer region from the staphylococcal conjugative plasmid pSK41. Gene 136:13-25. [DOI] [PubMed] [Google Scholar]
- 19.Francia, M. V., and D. B. Clewell. 2002. Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45:375-395. [DOI] [PubMed] [Google Scholar]
- 20.Gomis-Ruth, F. X., F. de la Cruz, and M. Coll. 2002. Structure and role of coupling proteins in conjugal DNA transfer. Res. Microbiol. 153:199-204. [DOI] [PubMed] [Google Scholar]
- 21.Gomis-Ruth, F. X., G. Moncalian, F. de la Cruz, and M. Coll. 2002. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277:7556-7566. [DOI] [PubMed] [Google Scholar]
- 22.Gomis-Ruth, F. X., G. Moncalian, R. Perez-Luque, A. Gonzalez, E. Cabezon, F. de la Cruz, and M. Coll. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637-641. [DOI] [PubMed] [Google Scholar]
- 23.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in Gram positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 25.Horodniceanu, T., D. H. Bouanchaud, G. Bieth, and Y. A. Chabbert. 1976. R plasmids in Streptococcus agalactiae (group B). Antimicrob. Agents Chemother. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iyer, L. M., K. S. Makarova, E. V. Koonin, and L. Aravind. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosomal segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32:5260-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johanesen, P. A., D. Lyras, T. L. Bannam, and J. I. Rood. 2001. Transcriptional analysis of the tet(P) operon from Clostridium perfringens. J. Bacteriol. 183:7110-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataoka, M., T. Seki, and T. Yoshida. 1991. Five genes involved in self-transmission of pSN22, a Streptomyces plasmid. J. Bacteriol. 173:4220-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelley, J. A., and K. L. Knight. 1997. Allosteric regulation of RecA protein function is mediated by Gln194. J. Biol. Chem. 272:25778-25782. [DOI] [PubMed] [Google Scholar]
- 30.Kumar, R. B., and A. Das. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523-1532. [DOI] [PubMed] [Google Scholar]
- 31.Kurenbach, B., C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann. 2003. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 50:86-93. [DOI] [PubMed] [Google Scholar]
- 32.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 33.Li, J., K. Miyamoto, and B. A. McClane. 2007. Comparison of virulence plasmids among Clostridium perfringens type E isolates. Infect. Immun. 75:1811-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, G., G. C. Draper, and W. D. Donachie. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol. 29:893-903. [DOI] [PubMed] [Google Scholar]
- 35.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 37.Marchler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massey, T. H., C. P. Mercogliano, J. Yates, D. J. Sherratt, and J. Lowe. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23:457-469. [DOI] [PubMed] [Google Scholar]
- 39.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Miyamoto, K., D. J. Fisher, J. Li, S. Sayeed, S. Akimoto, and B. A. McClane. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 188:1585-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moncalian, G., E. Cabezon, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goni, and F. de la Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 42.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 43.Ogura, T., S. W. Whiteheart, and A. J. Wilkinson. 2004. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 146:106-112. [DOI] [PubMed] [Google Scholar]
- 44.Possoz, C., C. Ribard, J. Gagnat, J. Pernodet, and M. Guerineau. 2001. The integrative element pSAM2 from Streptomyces: kinetics and mode of conjugal transfer. Mol. Microbiol. 42:159-166. [DOI] [PubMed] [Google Scholar]
- 45.Reuther, J., C. Gekeler, Y. Tiffert, W. Wohlleben, and G. Muth. 2006. Unique conjugation mechanism in mycelial streptomycetes: a DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol. Microbiol. 61:436-446. [DOI] [PubMed] [Google Scholar]
- 46.Rood, J. I. 1983. Transferable tetracycline resistance in Clostridium perfringens strains of porcine origin. Can. J. Microbiol. 29:1241-1246. [DOI] [PubMed] [Google Scholar]
- 47.Rood, J. I., E. A. Maher, E. B. Somers, E. Campos, and C. L. Duncan. 1978. Isolation and characterization of multiply antibiotic-resistant Clostridium perfringens strains of porcine origin. Antimicrob. Agents Chemother. 13:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rood, J. I., and B. A. McClane. 2002. Clostridium perfringens: enterotoxaemic diseases, p. 1117-1139. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, London, United Kingdom.
- 49.Rood, J. I., V. N. Scott, and C. L. Duncan. 1978. Identification of a transferable tetracycline resistance plasmid (pCW3) from Clostridium perfringens. Plasmid 1:563-570. [DOI] [PubMed] [Google Scholar]
- 50.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 51.Sastre, J. I., E. Cabezon, and F. de la Cruz. 1998. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J. Bacteriol. 180:6039-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sayeed, S., J. Li, and B. A. McClane. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect. Immun. 75:2391-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schroder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185:4371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott, P. T., and J. I. Rood. 1989. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene 82:327-333. [DOI] [PubMed] [Google Scholar]
- 56.Sharp, M. D., and K. Pogliano. 2002. Role of cell-specific SpoIIIE assembly in polarity of DNA transfer. Science 295:137-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sloan, J., L. M. McMurry, D. Lyras, S. B. Levy, and J. I. Rood. 1994. The Clostridium perfringens TetP determinant comprises two overlapping genes: tetA(P), which mediates active tetracycling efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol. Microbiol. 11:403-415. [DOI] [PubMed] [Google Scholar]
- 58.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Southern, E. 1975. Detection of specific sequences among DNA fragments separated by gel-electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 60.Story, R. M., I. T. Weber, and T. A. Steitz. 1992. The structure of the E. coli RecA protein monomer and polymer. Nature 355:318-325. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Titball, R. W., and J. I. Rood. 2002. Clostridium perfringens: wound infections, p. 1875-1903. In M. Sussman (ed.), Molecular medical microbiology. Academic Press, London, United Kingdom.
- 63.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto, M., J. M. Jones, E. Senghas, C. Gawron-Burke, and D. B. Clewell. 1987. Generation of Tn5 insertions in streptococcal conjugative transposon Tn916. Appl. Environ. Microbiol. 53:1069-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, X.-C., E. K. Weihe, and W. Margolin. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 180:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.