Abstract
The facultative aerobe Escherichia coli K-12 can use respiratory nitrate ammonification to generate energy during anaerobic growth. The toxic compound nitric oxide is a by-product of this metabolism. Previous transcript microarray studies identified the yeaR-yoaG operon, encoding proteins of unknown function, among genes whose transcription is induced in response to nitrate, nitrite, or nitric oxide. Nitrate and nitrite regulate anaerobic respiratory gene expression through the NarX-NarL and NarQ-NarP two-component systems. All known Nar-activated genes also require the oxygen-responsive Fnr transcription activator. However, previous studies indicated that yeaR-yoaG operon transcription does not require Fnr activation. Here, we report results from mutational analyses demonstrating that yeaR-yoaG operon transcription is activated by phospho-NarL protein independent of the Fnr protein. The phospho-NarL protein binding site is centered at position −43.5 with respect to the transcription initiation site. Expression from the Shewanella oneidensis MR-1 nnrS gene promoter, cloned into E. coli, similarly was activated by phospho-NarL protein independent of the Fnr protein. Recently, yeaR-yoaG operon transcription was shown to be regulated by the nitric oxide-responsive NsrR repressor (N. Filenko et al., J. Bacteriol. 189:4410-4417, 2007). Our mutational analyses reveal the individual contributions of the Nar and NsrR regulators to overall yeaR-yoaG operon expression and document the NsrR operator centered at position −32. Thus, control of yeaR-yoaG operon transcription provides an example of overlapping regulation by nitrate and nitrite, acting through the Nar regulatory system, and nitric oxide, acting through the NsrR repressor.
Escherichia coli K-12, a facultative aerobe, is able to respire with a variety of electron acceptors, including oxygen (O2), nitrate (NO3−), and nitrite (NO2−). Synthesis of the corresponding respiratory enzymes is subject to hierarchical control to ensure use of the preferred electron acceptor. The top level of this control is mediated by the Fnr transcription activator, which senses the absence of oxygen through its iron-sulfur cluster (27). The second level of hierarchical control is mediated by the NarL and NarP response regulators, which, when phosphorylated, bind DNA to activate or repress transcription. The NarX and NarQ sensors control NarL and NarP phosphorylation in response to nitrate and nitrite (53).
Several operons require both Fnr and phospho-NarL or -NarP proteins for maximal transcription. For the narGHJI, narK, and fdnGHI operons, Fnr protein, bound near position −41.5 with respect to the transcription initiation site, acts synergistically with phospho-NarL protein bound to sites further upstream (53). For the napFDAGHBC operon, Fnr protein, bound at position −64.5, acts synergistically with phospho-NarP protein bound at position −44.5 (15, 17). For the nirBDC and nrfABCDEFG operons, Fnr protein, bound near position −41.5, activates transcription maximally only when phospho-NarL or -NarP protein is bound further upstream to block inhibition by other proteins (2, 8, 59). Although transcription of many other operons is known to be activated by the Fnr protein acting alone (12, 26), to date there are no examples of Fnr-independent transcription activation by the phospho-NarL or -NarP protein.
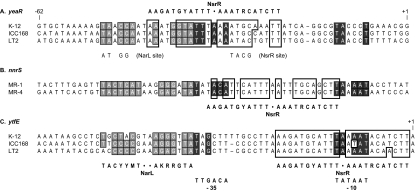
In preliminary transcript microarray experiments, we observed that the levels of yeaR-yoaG operon transcripts (encoding proteins of unknown function) are increased during growth with nitrate only in a narL+ strain (23). Sequence inspection revealed a likely binding site for phospho-NarL protein but no obvious site for binding of Fnr protein (Fig. 1A). Therefore, we were interested in characterizing the control of yeaR-yoaG operon transcription in more detail. Our results, reported here, suggest that this is an example of Fnr-independent transcription activation by phospho-NarL protein. Furthermore, the transcriptional control region for the nnrS gene from Shewanella oneidensis MR-1 has architecture similar to that of the E. coli yeaR-yoaG operon (Fig. 1B). Our results suggest that S. oneidensis nnrS transcription from a construct introduced into E. coli likewise is activated by phospho-NarL protein independent of the Fnr protein.
FIG. 1.
Transcription control regions for the yeaR-yoaG operon (A), nnrS gene (B), and ytfE gene (C). The nucleotide sequences are the sequences of E. coli K-12, C. rodentium ICC168, S. enterica LT2, S. oneidensis MR-1, and Shewanella sp. MR-4. The experimentally determined transcription initiation sites, designated +1, are shown for the E. coli yeaR-yoaG operon (this study) and for the E. coli ytfE gene (6). Consensus sequences are shown for the promoter −10 and −35 regions and for the NarL and NsrR protein binding sites. Nucleotides that match the promoter and NarL binding site consensus sequences are indicated by black and gray backgrounds, respectively. Nucleotides that match the NsrR binding site consensus sequence are enclosed in boxes. Site-specific alterations in the yeaR-yoaG operon control region sites for NarL and NsrR proteins are shown. Dashes indicate gaps introduced to align the sequences with respect to their −35 and −10 elements.
Meanwhile, other transcript microarray experiments identified yeaR-yoaG operon induction in response to nitric oxide (NO) (25) and in response to nitrate or nitrite (12). The latter study found that nitrate elicits a large increase in yeaR-yoaG transcripts in a narL+ narP+ strain, a small increase in a ΔnarL null narP+ strain, and no increase in a ΔnarL ΔnarP double null strain. Furthermore, nitrate- and nitrite-stimulated levels of yeaR-yoaG transcripts are increased in a Δfnr null strain (12).
Recently, the NsrR repressor has been identified as a factor mediating a transcriptional response to nitric oxide (6, 40, 45). Nitric oxide is formed from nitrite by cytochrome c nitrite reductase (NrfABCD enzyme) and by NADH-nitrite reductase (NirBD enzyme) in E. coli (13, 58). Very recently, nitrite induction of yeaR-yoaG operon transcription has been shown to result at least in part from control by the NsrR repressor (20). Our results, reported here, indicate that overall nitrate- and nitrite-responsive control of yeaR-yoaG operon transcription results from a combination of activation by the phospho-NarL protein and repression by the NsrR protein.
MATERIALS AND METHODS
Strains and plasmids.
Strains and plasmids are listed in Table 1. Standard methods were used for restriction endonuclease digestion, ligation, transformation, and PCR amplification of DNA (29).
TABLE 1.
E. coli K-12 strains and plasmids
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| VJS632 | F− λ− prototroph | 52 |
| VJS676 | As VJS632 but Δ(argF-lacIZYA)U169 | 52 |
| VJS2197 | As VJS676 but λΦ(narG-lacZ) | 39 |
| VJS8364 | As VJS632 but ΔlacZ | This study |
| BW25113 | lacIqrrnB hsdR ΔlacZ ΔaraBAD ΔrhaBAD | 18 |
| Derivatives of strain VJS8364 | ||
| VJS9563 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} | This study |
| VJS10505 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} Δfnr-275 | This study |
| VJS10506 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} ΔnarL261 | This study |
| VJS10507 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} ΔnarP262 | This study |
| VJS10508 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} ΔnsrR | This study |
| VJS10513 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} Δfnr-275 ΔnsrR | This study |
| VJS10516 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} ΔnarL261 ΔnarP262 | This study |
| VJS10519 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} ΔnarL261 ΔnarP262 ΔnsrR | This study |
| VJS10520 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} Δfnr-275 ΔnarL261 ΔnarP262 | This study |
| VJS10522 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} Δfnr-275 ΔnarL261 ΔnarP262 ΔnsrR | This study |
| VJS9556 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ62]} | This study |
| VJS9571 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ62]} ΔnarL261 | This study |
| VJS9572 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ62]} ΔnarP262 | This study |
| VJS9573 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ62]} ΔnsrR | This study |
| VJS9581 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ62]} ΔnarL261 ΔnarP262 | This study |
| VJS9584 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ62]} ΔnarL261 ΔnarP262 ΔnsrR | This study |
| VJS9557 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} (NarL site mutant) | This study |
| VJS9565 | λ− Δ(attλ-lom)::bla {Φ(yeaR-lacZ) [Δ175]} (NsrR site mutant) | This study |
| VJS9438 | λ− Δ(attλ-lom)::bla {Φ(nnrS-lacZ) [Δ268]} | This study |
| VJS9545 | λ− Δ(attλ-lom)::bla {Φ(nnrS-lacZ) [Δ268]} Δfnr-275 | This study |
| VJS9546 | λ− Δ(attλ-lom)::bla {Φ(nnrS-lacZ) [Δ268]} ΔnarL261 | This study |
| VJS9547 | λ− Δ(attλ-lom)::bla {Φ(nnrS-lacZ) [Δ268]} ΔnarP262 | This study |
| VJS9548 | λ− Δ(attλ-lom)::bla {Φ(nnrS-lacZ) [Δ268]} ΔnsrR | This study |
| Plasmids | ||
| pKD13 | Apr Kmr; source of FRT-kan-FRT cassette | 18 |
| pKD46 | Apr Tcs; Red recombinase expression plasmid | 18 |
| pRS414 | Apr; lacZ gene fusion vector | 47 |
| pRS415 | Apr; lacZ operon fusion vector | 44 |
| pVJS3253 | Apr; Δ(lacY lacA cynX tet) derivative of pRS414 | 50 |
| pVJS3266 | Apr; Δ(lacY lacA cynX tet) derivative of pRS415 | 23 |
| pVJS4533 | As pVJS3253 but Φ(nnrS-lacZ) [Δ268] | This study |
| pVJS4701 | As pVJS3266 but Φ(yeaR-lacZ) [Δ175] | This study |
| pVJS4702 | As pVJS3253 but Φ(yeaR-lacZ) [Δ62] | This study |
| pVJS4705 | As pVJS3253 but Φ(yeaR-lacZ) [Δ175] | This study |
We used a bacteriophage λ Red recombination procedure (18) to construct an in-frame ΔnsrR deletion. Briefly, a DNA fragment was PCR amplified with oligonucleotide primers 5′-TGCAGTTAACGAGTTTCACTGATTACGGATTACGTATTCCGGGGATCCGTCGACC and 5′-CACCAGCAATAATTTATAAAGCGGTTGATTCTCTTGTGTAGGCTGGAGCTGCTTC, each including a 35-nucleotide (nt) nsrR homology extension and a 20-nt priming sequence (underlined) for the kanamycin resistance gene in plasmid pKD13. The ∼1.4-kb PCR product was electrotransformed into the Red+ strain BW21153 carrying pKD46, and the resulting nsrR::kan allele was confirmed by PCR analysis. The nsrR::kan allele was introduced into other strains by bacteriophage P1-mediated generalized transduction, whereupon the kan gene was removed by FLP recombinase-mediated excision. The deletion was designed so that the residual “scar” sequence remaining after FLP recombination was in frame with the nsrR coding sequence. Codons 13 to 128 were removed from the nsrR coding region (141 codons) and replaced with an in-frame scar sequence consisting of 27 codons.
Similarly, we constructed an in-frame ΔlacZ deletion with oligonucleotide primers 5′-GATTCACTGGCCGTCGTTTTACAACGTCGTGACTGGATTCCGGGGATCCGTCGACC and 5′-ATGGTAGCGACCGGCGCTCAGCTGGAATTCCGCCGATAGTGTAGGCTGGAGCTGCTTC including 36- to 38-nt lacZ homology extensions and a 20-nt priming sequence (underlined) for the kanamycin resistance gene in plasmid pKD13. Codons 18 to 994 were removed from the lacZ coding region (1,016 codons) and replaced with an in-frame scar sequence consisting of 27 codons.
Work in our laboratory has resulted in analogous deletion alleles of the fnr, narL, and narP genes, details of which will be reported elsewhere.
Culture media and conditions.
Defined, complex, and indicator media for genetic manipulations were used as described previously (29). Defined medium to grow cultures for enzyme assays was buffered with 3-(N-morpholino)propanesulfonic acid (MOPS) as previously described (52). The initial pH of this medium was adjusted to 8.0 to ameliorate nitrite toxicity. Because the pKa of MOPS is 7.2, the buffering capacity of this medium continually increased as acidic fermentation products accumulated. At the time of harvest, cultures typically had a pH of about 7.5. Glucose (40 mM for aerated cultures and 80 mM for anaerobic cultures) was provided as a carbon source. To prepare enriched medium, MOPS-glucose medium was mixed at a 1:1 ratio with TY medium (0.8% tryptone, 0.5% yeast extract, 0.5% NaCl). The respiratory oxidants NaNO3 (40 mM) and NaNO2 (5 mM) and the nitric oxide-generating compound sodium nitroprusside (SNP) (100 μM) were added as indicated below. Aerated cultures were harvested with chloramphenicol to prevent adaptation to anaerobiosis (37).
Cultures were grown at 37°C to the early exponential phase, about 25 to 35 Klett units (see Fig. 2A). Culture densities were monitored with a Klett-Summerson photoelectric colorimeter (Klett Manufacturing Co., New York, NY) equipped with a number 66 (red) filter. Anaerobic cultures for enzyme assays and for RNA extraction were grown in screw-cap tubes as described previously (52).
FIG. 2.
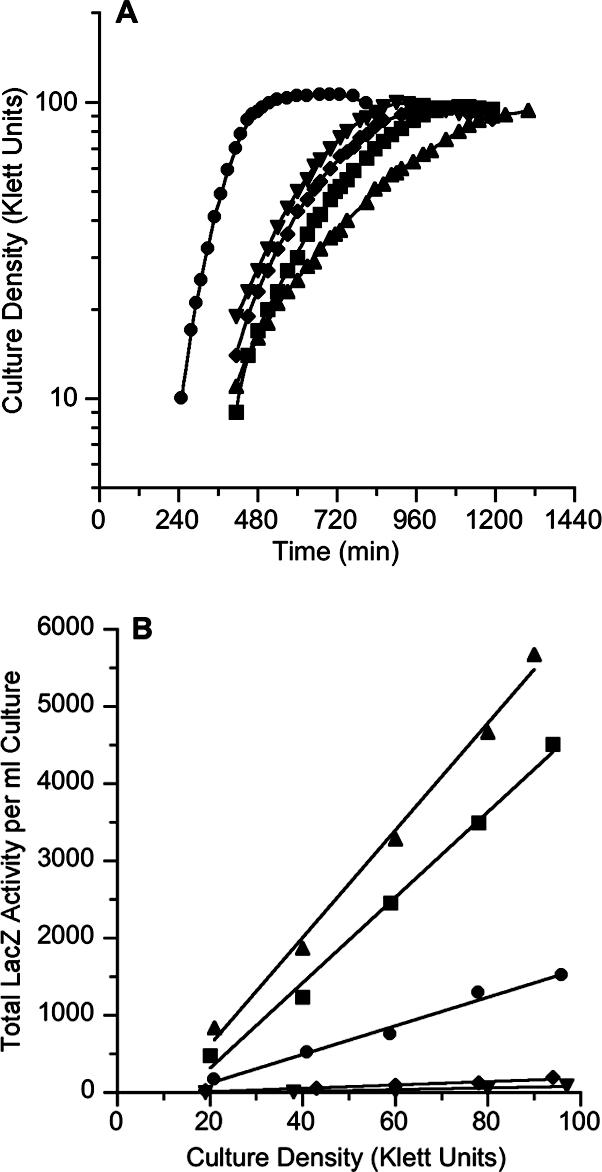
(A) Growth curves and (B) rates of β-galactosidase synthesis for Φ(yeaR-lacZ) strains cultured anaerobically in defined medium with nitrate (40 mM). •, VJS9563 (wild type); ▪, VJS10505 (Δfnr); ▴, VJS10513 (Δfnr ΔnsrR); ▾, VJS10520 (Δfnr ΔnarL ΔnarP); ⧫, VJS10522 (Δfnr ΔnarL ΔnarP ΔnsrR). Total LacZ enzyme activities per volume were determined as described in Materials and Methods. Similar results were obtained in independent experiments. Time refers to minutes after inoculation.
Gene fusions.
Plasmid pVJS4705 contains the yeaR-yoaG operon control region on a 308-bp DNA fragment from an engineered EcoRI site at position −175 to an engineered BamHI site downstream of yeaR codon 17, whereas pVJS4702 contains the yeaR-yoaG operon control region from an engineered EcoRI site at position −62. The control region cassettes were recloned into the vector pVJS3253, a Δ(lacYA) derivative of plasmid pRS414. The resulting Φ(yeaR-lacZ) gene fusions were transferred to bacteriophage λ and integrated into the chromosome of strain VJS8364 as described previously (7, 50). A similar strategy was used to construct the Φ(nnrS-lacZ) gene fusion. Plasmid pVJS4533 contains the nnrS operon control region on a 368-bp DNA fragment from an engineered EcoRI site at position −268 (including the termination codon from the upstream gene, locus tag SO2804) with respect to the hypothetical transcription initiation site to an engineered BamHI site downstream of nnrS codon 10. The veracity of each cloned insert was confirmed by DNA sequencing.
Site-specific mutagenesis.
Oligonucleotide-directed site-specific mutagenesis was used to introduce substitutions into the yeaR-yoaG operon control region. Mutagenesis was performed using the QuickChange protocol (Stratagene Cloning Systems, La Jolla, CA), as described previously (1). The oligonucleotide primers used for the phospho-NarL and NsrR protein binding sites were 5′-GCTGATATGGTGCTAAAAAGATAGGAATAAATGGTATTTAAAATG and 5′-ACCAATAAATGGTATTTAAATACGAAATTATCAGGCGTACCCTG, respectively.
Transcription initiation analysis.
A strain carrying the Φ(yeaR-lacZ) operon fusion plasmid pVJS4701 was used. Analysis by rapid amplification of cDNA ends (5′-RACE) (41), also termed anchored PCR, was performed by using commercial reagents (5′-RACE system, version 2.0; Invitrogen Life Technologies, Carlsbad, CA), essentially as described in the manufacturer's instructions. The oligonucleotide primers used were as follows: 5′-AAGCTTAGTGAATCCGTAATCATGGTCATAG (gene-specific primer 1 for lacZ), 5′-CGGAACTGGCGGCTGTGGGATTA (gene-specific primer 2 for lacZ), and 5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTT (abridged anchor primer).
β-Galactosidase assay.
β-Galactosidase activities were determined at room temperature (approximately 21°C) by following the hydrolysis of o-nitrophenyl-β-d-galactopyranoside in CHCl3-sodium dodecyl sulfate-permeabilized cells. Specific activities are expressed in arbitrary (Miller) units (32). All cultures were assayed in duplicate, and the reported values are averages from at least two independent experiments. Differential rates of β-galactosidase synthesis (33) were determined for anaerobic cultures essentially as described previously (54). Cultures (8 ml) were grown in screw-cap tubes. Samples (200 μl) were withdrawn and mixed with 50 μl of a solution containing 250 μg chloramphenicol per ml (to inhibit further protein synthesis). Samples were stored on ice before the assay was performed. The reported activities are the total activities per ml of culture and are not normalized for culture density.
Genome database searches.
For analyses the BLAST programs (31) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) were employed. Draft genome sequence data for Citrobacter rodentium ICC168 and Klebsiella pneumoniae MGH78578 were produced by the Bacterial Genomes Sequencing Group at the Sanger Institute (http://www.sanger.ac.uk) and by the Genome Sequencing Center at Washington University, St. Louis, MO (http://www.genome.wustl.edu), respectively. Completed genome sequence data for the following organisms were accessed through GenBank: E. coli K-12 (GenBank accession number NC_000913), Salmonella enterica LT2 (NC_009137), Erwinia carotovora SCRI1043 (NC_004547), Yersinia pseudotuberculosis IP32953 (NC_006155), S. oneidensis MR-1 (NC_004347), Shewanella sp. strain MR-4 (NC_008321), and Streptococcus pneumoniae R6 (NC_003098).
RESULTS
yeaR-yoaG operon transcription control region.
We used the 5′-RACE method (41) as described in Materials and Methods to determine the 5′ end of yeaR mRNA isolated from a strain carrying a multicopy Φ(yeaR-lacZ) operon fusion. This method uses terminal deoxynucleotidyl transferase to add an A homopolymeric tail to the 5′ end of the cDNA. This analysis (data not shown) identified the 5′ end as corresponding to the G residue designated position +1 in Fig. 1A. The initiation site is preceded by σ70-dependent promoter −10 and −35 elements (42) (Fig. 1A).
Most phospho-NarL and -NarP binding sites consist of inverted heptamer sequences (consensus sequence, TACYYMT, where Y is C or T and M is A or C) separated by 2 nt (16, 30). A potential phospho-NarL and -NarP binding site is centered at position −43.5 relative to the transcription initiation site (Fig. 1A), immediately adjacent to the promoter −35 element.
While examining the yeaR control region, we noted a sequence with similarity to the NsrR protein consensus binding site (Fig. 1A), which consists of inverted hendacamer sequences (consensus sequence, AAGATGYATTT) separated by 1 nt (6, 40). This potential NsrR protein binding site is centered at position −32 relative to the transcription initiation site (Fig. 1A). The phospho-NarL and NsrR protein binding sites overlap, and the −35 motif of the yeaR promoter is close to the center of the NsrR inverted repeat (Fig. 1A). Finally, no Fnr protein binding site is evident in the yeaR control region (see below).
Comparisons of homologous regulatory regions from related species reveal conserved and nonconserved sequences, thereby implying that the conserved sequences are more likely to be functionally important for regulated gene expression (5, 10). This comparative approach has been termed phylogenetic footprinting (56). Figure 1A shows comparisons between yeaR-yoaG operon control region sequences from three close relatives: E. coli K-12, C. rodentium ICC168, and S. enterica LT2. In these sequences, the promoter elements and binding sites for phospho-NarL and NsrR proteins are well conserved.
Phylogenetic distribution and expression of the yeaR and yoaG genes.
The yeaR gene (119 codons) is separated by only 3 nt from the downstream yoaG gene (60 codons), and mRNA corresponding to the two genes is coordinately expressed (12, 23). Thus, these genes likely form the yeaR-yoaG operon. Genome database searches (as described in Materials and Methods) revealed that the yeaR-yoaG operon, along with its transcription control region, is conserved in the very closely related Escherichia-Shigella, Salmonella, and Citrobacter enterobacterial species (Fig. 1A). The operon is somewhat different in the Klebsiella and Erwinia enterobacterial species; the yeaR gene (110 codons) is separated by 8 nt from the downstream yoaG gene (112 codons), which encodes an amino-terminal extension of 52 residues. In the Klebsiella and Erwinia examples, the transcription control region is also different and contains an NsrR protein binding site (40) but no apparent phospho-NarL or -NarP protein binding site.
In other species, the YeaR domain is present as an amino-terminal extension in homologs of the E. coli TehB protein, an S-adenosylmethionine-dependent non-nucleic acid methyltransferase involved in resistance to tellurite (28). This YeaR-TehB fusion protein is annotated as “TehB” in several genome sequences (e.g., locus tag YPTB1947 in Y. pseudotuberculosis IP32953 and locus tag spr0880 in S. pneumoniae R6). Consequently, the YeaR protein itself has been designated “TehB” or “TehB^” in some annotations (40).
A yeaR homolog is immediately downstream of the norVW operon, encoding anaerobically expressed nitric oxide reductase, in some species belonging to the family Vibrionaceae (40). Presumably, here yeaR transcription is induced by nitric oxide along with norVW transcription. In Yersinia and Erwinia species, transcription of the gene encoding the YeaR-TehB fusion protein is predicted to be controlled by the NsrR protein, as is transcription of the yeaR gene in Salmonella and Klebsiella species (40). Thus, synthesis of the YeaR protein (alone or fused to the TehB-like domain) likely is induced by nitric oxide in a variety of species belonging to the class Gammaproteobacteria. However, in none of these cases was transcription predicted to be controlled also by the phospho-NarL or -NarP protein (40).
In contrast to the yeaR gene, which is broadly distributed, the yoaG gene is confined to members of the family Enterobacteriaceae in the yeaR-yoaG operon, as described above. The structure of the YoaG protein reveals that it is a soluble dimer (PDB accession code 1NEI).
Effects of nitrate and nitrite on anaerobic Φ(yeaR-lacZ) expression during growth in defined medium.
We constructed two different monocopy Φ(yeaR-lacZ) gene fusions at the chromosomal λatt site as described in Materials and Methods. One construct carries a sequence extending 175 nt upstream of the transcription initiation site, including the last 16 codons from the upstream yeaS gene, whereas the second construct carries only 62 nt (Fig. 1A). We used these constructs to monitor LacZ specific activity from strains cultured under different conditions. There was no difference in expression from the two constructs, demonstrating that all essential regulatory sequences are within 62 nt of the initiation site (Tables 2 and 3).
TABLE 2.
Effects of ΔnarL, ΔnarP, and ΔnsrR null alleles on expression from Φ(yeaR-lacZ) fusions during anaerobic growth in defined medium
| Strain | Endpointb | Genotype
|
LacZ sp act (Miller units)a
|
Activation by:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| narL | narP | nsrR | No addition | With NO3− | With NO2− | NO3− | NO2− | ||
| VJS9563 | [Δ175] | + | + | + | 6 | 3,000 | 680 | 500 | 113 |
| VJS9556 | [Δ62] | + | + | + | 6 | 3,100 | 500 | 517 | 83 |
| VJS10506 | [Δ175] | − | + | + | 3 | 180 | 420 | 60 | 140 |
| VJS9571 | [Δ62] | − | + | + | 5 | 190 | 360 | 38 | 72 |
| VJS10507 | [Δ175] | + | − | + | 5 | 3,100 | 450 | 620 | 90 |
| VJS9572 | [Δ62] | + | − | + | 6 | 3,000 | 390 | 500 | 65 |
| VJS10516 | [Δ175] | − | − | + | 3 | 22 | 61 | 7.3 | 20 |
| VJS9581 | [Δ62] | − | − | + | 6 | 25 | 85 | 4.2 | 14 |
| VJS9557 | [Δ175] (NarL site mutant) | + | + | + | 6 | 39 | 110 | 6.5 | 18 |
| VJS10508 | [Δ175] | + | + | − | 210 | 8,600 | 990 | 41 | 4.7 |
| VJS9573 | [Δ62] | + | + | − | 210 | 10,300 | 900 | 49 | 4.3 |
| VJS9565 | [Δ175] (NsrR site mutant) | + | + | + | 190 | 7,800 | 940 | 41 | 4.9 |
| VJS10519 | [Δ175] | − | − | − | 150 | 150 | 190 | 1.0 | 1.3 |
| VJS9584 | [Δ62] | − | − | − | 200 | 200 | 250 | 1.0 | 1.3 |
Strains were cultured to the early exponential phase in glucose defined medium.
The location of the upstream endpoint in each construct is in brackets.
TABLE 3.
Effects of ΔnarL, ΔnarP, and ΔnsrR null alleles on expression from Φ(yeaR-lacZ) fusions during anaerobic growth in complex medium
| Strain | Endpointb | Genotype
|
LacZ sp act (Miller units)a
|
Activation by:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| narL | narP | nsrR | No addition | With NO3− | With NO2− | With SNP | NO3− | NO2− | SNP | ||
| VJS9563 | [Δ175] | + | + | + | 5 | 460 | 97 | 210 | 92 | 19 | 42 |
| VJS9556 | [Δ62] | + | + | + | 5 | 520 | 130 | 220 | 104 | 26 | 44 |
| VJS10506 | [Δ175] | − | + | + | 3 | 34 | 85 | 220 | 11 | 28 | 73 |
| VJS9571 | [Δ62] | − | + | + | 3 | 55 | 110 | 150 | 18 | 37 | 50 |
| VJS10507 | [Δ175] | + | − | + | 4 | 290 | 110 | 160 | 73 | 28 | 40 |
| VJS9572 | [Δ62] | + | − | + | 5 | 430 | 130 | 220 | 86 | 26 | 44 |
| VJS10516 | [Δ175] | − | − | + | 2 | 3 | 15 | 24 | 1.5 | 7.5 | 12 |
| VJS9581 | [Δ62] | − | − | + | 2 | 4 | 20 | 35 | 2.0 | 10 | 18 |
| VJS9557 | [Δ175] (NarL site mutant) | + | + | + | 3 | 11 | 41 | 70 | 3.7 | 14 | 23 |
| VJS10508 | [Δ175] | + | + | − | 310 | 1,400 | 330 | 360 | 4.5 | 1.1 | 1.2 |
| VJS9573 | [Δ62] | + | + | − | 230 | 1,800 | 350 | 540 | 7.8 | 1.5 | 2.3 |
| VJS9565 | [Δ175] (NsrR site mutant) | + | + | + | 130 | 1,500 | 210 | 290 | 12 | 1.6 | 2.2 |
| VJS10519 | [Δ175] | − | − | − | 68 | 63 | 90 | 58 | 0.9 | 1.3 | 0.9 |
| VJS9584 | [Δ62] | − | − | − | 110 | 110 | 110 | 110 | 1.0 | 1.0 | 1.0 |
Strains were cultured to the early exponential phase in enriched medium with glucose.
The location of the upstream endpoint in each construct is in brackets.
We measured Φ(yeaR-lacZ) expression from strains cultured in defined medium with glucose as the carbon source. In wild-type strains, nitrate and nitrite induced expression about 500- and 100-fold, respectively (Table 2, lines 1 and 2). A ΔnarL null allele decreased nitrate induction to 60-fold but had little effect on nitrite induction (Table 2, lines 3 and 4). By contrast, a ΔnarP null allele had little effect on either nitrate or nitrite induction (Table 2, lines 5 and 6). Nevertheless, the ΔnarL and ΔnarP null alleles together reduced nitrate and nitrite induction to only about 5- and 20-fold, respectively (Table 2, lines 7 and 8). This indicates that phospho-NarL protein is sufficient for normal Φ(yeaR-lacZ) induction but that phospho-NarP protein also can contribute, at least in the absence of phospho-NarL protein.
To further examine regulation by the phospho-NarL and -NarP proteins, we introduced multiple substitutions into the upstream half-site sequence (Fig. 1A). Substitutions were designed to eliminate binding to the half-site sequence, to leave the NsrR protein binding site intact, and to maintain the overall G+C composition. The phenotype conferred by this alteration (Table 2, line 9) was indistinguishable from that conferred by the ΔnarL ΔnarP double null alleles. This confirms that the phospho-NarL and -NarP protein binding site, identified by sequence inspection, is critical for regulation by these proteins. This also shows that the influence of the ΔnarL and ΔnarP null alleles on yeaR-yoaG operon expression reflects a direct effect of phospho-NarL and -NarP proteins on transcription activation rather than an indirect effect of altered nitrate and nitrite metabolism in ΔnarL and ΔnarP null strains.
We constructed a ΔnsrR null allele as described in Materials and Methods. We designed the deletion to leave the remaining ΔnsrR sequence in frame, in order to avoid polarity effects on expression of the downstream rnr gene. In an otherwise wild-type strain background, the ΔnsrR null allele caused an approximately 40-fold increase in basal-level anaerobic expression compared to that of the nsrR+ strain during growth with no added nitrate or nitrite (Table 2, compare lines 10 and 11 to lines 1 and 2). This indicates that the NsrR protein is a negative regulator of yeaR-yoaG operon transcription. Induction by nitrate in the ΔnsrR null strain was reduced roughly 10-fold (from about 500-fold to about 50-fold), whereas induction by nitrite was reduced roughly 20-fold (from about 100-fold to about 5-fold).
To further examine regulation by the NsrR protein, we introduced multiple substitutions into the downstream NsrR half-site sequence (Fig. 1A). Substitutions were designed to eliminate binding to the half-site sequence, to leave the promoter −35 element and the phospho-NarL and -NarP protein binding site intact, and to maintain the overall G+C composition. The phenotype conferred by this alteration (Table 2, line 12) was indistinguishable from that conferred by the ΔnsrR null allele. This confirms that the NsrR protein binding site, identified by sequence inspection, is critical for regulation by this protein. This also shows that the influence of the ΔnsrR null allele on yeaR-yoaG operon expression reflects a direct effect of NsrR protein on transcription repression.
Finally, we examined Φ(yeaR-lacZ) expression in a ΔnarL ΔnarP ΔnsrR triple null strain. The basal-level expression was similar to that in the ΔnsrR single null strain (Table 2, compare lines 13 and 14 to lines 10 and 11), and expression was not affected during growth with nitrate or nitrite. This demonstrates that no other regulatory protein is essential for nitrate or nitrite control of yeaR-yoaG operon expression. It also shows that the 5- to 20-fold residual induction by nitrate and nitrite in the ΔnarL ΔnarP double null strain was due to control by the NsrR repressor, presumably responding to the resultant nitric oxide (see below).
Effects of nitrate, nitrite, and SNP on anaerobic Φ(yeaR-lacZ) expression during growth in complex medium.
We amended the glucose defined medium with tryptone and yeast extract in order to study Φ(yeaR-lacZ) expression in response to SNP during anaerobic growth (Table 3). SNP nitrosates thiols in enriched medium, which then release nitric oxide (38), and so SNP provides a convenient means for examining the response to nitric oxide. Evidence suggests that transcriptional responses to SNP and nitric oxide are similar (34).
The overall patterns of Φ(yeaR-lacZ) expression in complex medium (Table 3) were similar to those in defined medium (Table 2), except that the induced levels of Φ(yeaR-lacZ) expression were roughly fivefold lower in the complex medium and so the level of induction by nitrate was correspondingly lower (Tables 2 and 3). We do not know why nitrate induction was less efficient during growth in complex medium.
The responses to nitrite were very similar to those to SNP (Table 3). The results are congruent with the hypothesis that the NsrR protein mediates nitric oxide regulation of yeaR-yoaG operon transcription and that the NsrR-dependent transcriptional response to nitrate and nitrite is a consequence of the conversion of these compounds to nitric oxide (13, 45, 58).
Fnr protein is not needed for Nar-dependent Φ(yeaR-lacZ) expression.
All previously characterized phospho-NarL- or phospho-NarP-activated promoters also require the Fnr activator for expression (see Introduction). Fnr protein binding sites consist of inverted pentamer sequences (consensus sequence, TTGAT) separated by 4 nt (46). However, the yeaR-yoaG operon control region contains no sequence with any recognizable similarity to the Fnr protein DNA binding consensus sequence within 62 nt upstream (Fig. 1A) or 50 nt downstream (not shown) of the transcription initiation site. Other workers have also found no Fnr protein binding site in the yeaR-yoaG operon control region (12, 40).
We examined Φ(yeaR-lacZ) expression in Δfnr null strains by monitoring differential rates of LacZ enzyme synthesis during anaerobic exponential growth in defined medium supplemented with nitrate. The results are shown in Fig. 2. The rate of LacZ enzyme synthesis in the fnr+ Φ(yeaR-lacZ) strain was approximately 20 U per Klett unit (Fig. 2), whereas the rate in the Δfnr null strain was about 53 U per Klett unit. Thus, expression was increased more than twofold in the Δfnr null strain. By contrast, the rate of LacZ enzyme synthesis in the Δfnr ΔnarL ΔnarP triple null strain was less than 1 U per Klett unit (Fig. 2). Together, these data demonstrate that Nar-dependent Φ(yeaR-lacZ) expression does not require the Fnr activator.
The expression rates in ΔnsrR null strains were slightly higher than those in the corresponding nsrR+ strains, about 65 U per Klett unit in the ΔnsrR Δfnr double null strain (compared to 53 U per Klett unit in the nsrR+ Δfnr null strain) and about 2 U per Klett unit in the ΔnsrR Δfnr ΔnarL ΔnarP quadruple null strain (compared to less than 1 U per Klett unit in the corresponding nsrR+ Δfnr ΔnarL ΔnarP triple null strain). Thus, the phenotypes of the ΔnsrR and the Δfnr null mutants were similar.
Nitrate induction of yeaR-yoaG operon expression in aerated cultures.
We next studied the response to culture aeration. As a control, we also measured expression from a Φ(narG-lacZ) gene fusion known to be activated by both the Fnr and phospho-NarL proteins (53). As expected, Φ(narG-lacZ) expression was induced more than 20-fold by anaerobiosis and, during anaerobic growth, an additional 100-fold by nitrate (Table 4). Nitrate did not activate expression in aerated cultures. This regulatory pattern reflects the requirement of Fnr protein for phospho-NarL protein activation in this and other known phospho-NarL-activated regulatory regions.
TABLE 4.
Effects of culture aeration and nitrate on expression from Φ(narG-lacZ) and φ(yeaR-lacZ) fusionsa
| Strain | Fusion | LacZ sp act (Miller units)b
|
Activation by:
|
|||||
|---|---|---|---|---|---|---|---|---|
| With O2
|
Without O2
|
Anaerobiosis | NO3−
|
|||||
| No addition | With NO3− | No addition | With NO3− | With O2 | Without O2 | |||
| VJS2197 | Φ(narG-lacZ) | <1 | 4 | 26 | 2,300 | >26 | <4.0 | 88 |
| VJS9563 | Φ(yeaR-lacZ) | 2 | 160 | 5 | 2,400 | 2.5 | 80 | 480 |
| VJS9557 | Φ(yeaR-lacZ) (NarL site mutant) | 6 | 8 | 7 | 30 | 1.2 | 1.3 | 4.3 |
| VJS9565 | Φ(yeaR-lacZ) (NsrR site mutant) | 80 | 3,200 | 160 | 8,000 | 2.0 | 40 | 50 |
All φ(yeaR-lacZ) fusions were [Δ175].
Strains were cultured to the early exponential phase in MOPS medium (defined medium with 40 mM glucose).
Expression from the Φ(yeaR-lacZ) fusion differed in two respects. First, expression was induced only slightly (two- to threefold) by anaerobiosis (Table 4). Second, nitrate induced expression about 80-fold in aerated cultures. During anaerobic growth, expression was induced about 500-fold by nitrate (Table 4), as noted above (Table 2, lines 1 and 2).
We next measured expression from the Φ(yeaR-lacZ) fusion, in which the phospho-NarL protein binding site was destroyed. Nitrate failed to induce expression in aerated cultures (Table 4) and only weakly induced expression during anaerobic growth (compare Table 4 to Table 2, line 9). Thus, phospho-NarL protein is responsible for nitrate-activated expression in aerated cultures. It has been established that the NarX-NarL system responds to nitrate in aerated cultures (50, 51).
Finally, we measured expression from the Φ(yeaR-lacZ) construct in which the NsrR protein binding site was destroyed. Overall expression was derepressed, but expression was still responsive to nitrate in both aerated and anaerobic cultures (compare Table 4 to Table 2, line 12). The anaerobic expression was about twice the aerobic expression both in the absence and in the presence of nitrate.
Expression from the S. oneidensis nnrS control region in E. coli.
We constructed a monocopy Φ(nnrS-lacZ) gene fusion at the chromosomal λatt site as described in Materials and Methods. We used this fusion to monitor LacZ specific activity from strains cultured under different conditions.
In contrast to Φ(yeaR-lacZ) expression, nitrite and SNP were very weak inducers of Φ(nnrS-lacZ) expression during growth in defined and complex medium (Tables 5 and 6). Furthermore, the ΔnsrR null allele had very little influence on Φ(nnrS-lacZ) expression (Tables 5 and 6, lines 1 and 4). We concluded that transcription from the S. oneidensis MR-1 nnrS regulatory region in E. coli is not subject to repression by the NsrR protein.
TABLE 5.
Effects of ΔnarL, ΔnarP, ΔnsrR and Δfnr null alleles on expression from a Φ(nnrS-lacZ) fusion during anaerobic growth in defined medium
| Strain | Genotype
|
LacZ sp act (Miller units)a
|
Activation by:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| narL | narP | nsrR | fnr | No addition | With NO3− | With NO2− | NO3− | NO2− | |
| VJS9438 | + | + | + | + | 43 | 1,000 | 94 | 23 | 2.2 |
| VJS9546 | − | + | + | + | 46 | 95 | 82 | 2.1 | 1.8 |
| VJS9547 | + | − | + | + | 52 | 990 | 91 | 19 | 1.8 |
| VJS9548 | + | + | − | + | 63 | 900 | 86 | 14 | 1.4 |
| VJS9545 | + | + | + | − | 60 | 1,100 | 300 | 18 | 5.0 |
Strains were cultured to the early exponential phase in glucose defined medium.
TABLE 6.
Effects of ΔnarL, ΔnarP, ΔnsrR, and Δfnr null alleles on expression from a Φ(nnrS-lacZ) fusion during anaerobic growth in complex medium
| Strain | Genotype
|
LacZ sp act (Miller units)a
|
Activation by:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| narL | narP | nsrR | fnr | No addition | With NO3− | With SNP | NO3− | SNP | |
| VJS9438 | + | + | + | + | 54 | 870 | 54 | 16 | 2.0 |
| VJS9546 | − | + | + | + | 54 | 85 | 54 | 1.6 | 1.3 |
| VJS9547 | + | − | + | + | 64 | 610 | 64 | 10 | 1.9 |
| VJS9548 | + | + | − | + | 71 | 630 | 71 | 9 | 1.4 |
| VJS9545 | + | + | + | − | 130 | 1,000 | 130 | 8 | 3.9 |
Strains were cultured to the early exponential phase in enriched medium with glucose.
On the other hand, the nitrate regulation of Φ(nnrS-lacZ) expression in nsrR+ strains (Tables 5 and 6, lines 1 to 3) was similar to that of Φ(yeaR-lacZ) expression in ΔnsrR null strains (Tables 2 and 3, lines 10, 11, 13 and 14). During anaerobic growth, nitrate induced Φ(nnrS-lacZ) expression by about 20-fold in the wild-type strain, by about 2-fold in the ΔnarL null strain, and by about 10- to 20-fold in the ΔnarP null strain (Tables 5 and 6, lines 1 to 3). Thus, phospho-NarL protein was the predominant activator of nnrS gene expression in E. coli. Induction by nitrate was influenced little by the Δfnr null allele (Tables 5 and 6, line 5).
DISCUSSION
Enterobacteria can use respiratory nitrate ammonification, in which energy is conserved by sequential reduction of nitrate through nitrite to ammonium (11, 43, 55). Nitric oxide, a highly reactive and toxic compound (19, 36), is generated in measurable amounts as a by-product of respiratory nitrite ammonification (13, 58). Enzymes involved in nitrate and nitrite respiration are synthesized in response to nitrate and nitrite (53), whereas enzymes involved in nitric oxide metabolism are synthesized in response to nitric oxide (45). It is now apparent that these two stimulons (35) overlap (12, 20). Regulation of yeaR-yoaG operon expression provides one example of overlap between the nitrate- and nitrite-responsive Nar regulon and the nitric oxide-responsive NsrR regulon.
Regulated yeaR-yoaG operon expression required no more than 62 nt upstream of the transcription initiation site, which included the phylogenetically conserved regulatory elements: a site for binding phospho-NarL and -NarP activators and an overlapping site for binding the NsrR repressor (Fig. 1A). Since nitric oxide is formed as a by-product of nitrite respiration, transcriptional response to added nitrate and nitrite can be either direct (via the Nar regulatory systems) or indirect (through the NsrR repressor). We employed mutant analysis to separate the relative contributions of the individual regulators.
Fnr-independent transcription activation by the phospho-NarL protein.
The contribution of the Nar regulatory systems to Φ(yeaR-lacZ) expression is revealed in strains where the NsrR repressor does not function, due either to a ΔnsrR null allele or to multiple substitutions in the NsrR operator sequence (Fig. 1A). Similar results were observed in both cases. Induction by nitrate and nitrite in these NsrR− strains was eliminated upon introduction of both ΔnarL and ΔnarP null alleles. Further results, obtained with NsrR+ strains, indicate that phospho-NarL protein is responsible for most of this induction. These results extend those of Constantinidou et al. (12). A minor contribution by phospho-NarP protein, revealed only in ΔnarL null strains, is of uncertain physiological significance. The phenotype conferred by multiple alterations in the phospho-NarL binding site (Fig. 1A) was similar to that of the ΔnarL ΔnarP double null mutant, demonstrating that this site is essential for phospho-NarL-activated yeaR-yoaG operon expression.
As noted in the Introduction, previously studied examples of Nar-dependent transcription activation require the oxygen-responsive Fnr activator. Consequently, maximal expression of the operons is observed only during anaerobic growth with nitrate. However, sequence inspection failed to reveal an apparent Fnr protein binding site in the yeaR-yoaG operon control region (Fig. 1A) (12, 40), and microarray analysis revealed Fnr-independent induction of yeaR transcription in response to nitrite (12).
Expression of the Φ(yeaR-lacZ) NsrR operator mutant was induced efficiently by nitrate during either aerobic or anaerobic growth, and the nitrate-stimulated Φ(yeaR-lacZ) expression in the Δfnr null strain was even higher than that in the fnr+ strain. Together, these results indicate that the phospho-NarL protein can activate transcription independent of the Fnr protein.
In the yeaR-yoaG operon control region, the phospho-NarL binding site is immediately adjacent to the promoter −35 element (Fig. 1A), so activation likely operates through direct contacts with RNA polymerase (a class II mechanism) as defined initially for activation by the cyclic AMP receptor protein (9).
Transcription repression by NsrR protein.
The contribution of the NsrR repressor to Φ(yeaR-lacZ) expression is revealed in strains where the Nar regulatory systems do not function, due either to ΔnarL and ΔnarP null alleles or to multiple substitutions in the phospho-NarL and -NarP binding sequence (Fig. 1A). Similar results were observed in both cases. Induction by nitrate, nitrite, and SNP in these Nar− strains was eliminated upon introduction of a ΔnsrR null allele. The phenotype conferred by multiple alterations in the NsrR operator (Fig. 1A) was similar to that of the ΔnsrR null mutant, demonstrating that this site is essential for NsrR-mediated yeaR-yoaG operon repression.
Is the Fnr protein a direct repressor of yeaR-yoaG operon transcription? No Fnr protein binding sequence is evident either upstream or downstream of the yeaR-yoaG operon transcription initiation site, suggesting that the Fnr protein is an indirect negative regulator in this case (20, 45). Nitrate-stimulated Φ(yeaR-lacZ) expression was similar in the Δfnr null and Δfnr ΔnsrR double null strains, consistent with the idea that the NsrR repressor is partially inactive in Δfnr null strains (45). Alternatively, the Δfnr null strain may be less able to metabolize small amounts of the NsrR inducer, nitric oxide. Further studies are necessary to determine the interactions between the Fnr and NsrR regulators.
Expression from the S. oneidensis nnrS control region in E. coli.
We wished to find a second example of Fnr-independent phospho-NarL transcription activation. We chose the S. oneidensis nnrS gene for four reasons. First, the upstream control region resembles the E. coli yeaR-yoaG control region, with an apparent phospho-NarL binding site immediately upstream of an apparent promoter −35 element (Fig. 1B). No Fnr site is evident. Second, transcript microarray analysis revealed that nnrS transcription is strongly induced during anaerobic growth with nitrate (4). Third, Shewanella spp. are close relatives of the enterobacteria, making E. coli a potential surrogate host for studying expression from Shewanella promoters (49). Finally, S. oneidensis is a well-studied model for understanding anaerobic respiration and its control (14, 21, 57). NnrS is a heme- and copper-containing membrane protein of unknown function (3) that is present in many species of nitrate-respiring bacteria (40).
Results indicate that Nar-dependent induction of Φ(nnrS-lacZ) expression requires phospho-NarL protein but not Fnr protein, as found also for Φ(yeaR-lacZ) expression. Again, this indicates that phospho-NarL protein can activate transcription independent of other regulators, such as the Fnr protein. In E. coli, phospho-NarP protein apparently played a relatively minor role in Φ(nnrS-lacZ) expression, similar to the role in Φ(yeaR-lacZ) expression. This is noteworthy because S. oneidensis encodes the NarQ-NarP but not the NarX-NarL regulatory system (48).
In striking contrast to Φ(yeaR-lacZ) expression, neither NsrR protein nor SNP greatly affected Φ(nnrS-lacZ) expression (Tables 5 and 6) despite the presence of an apparent NsrR operator site spanning the nnrS promoter −35 and −10 elements (Fig. 1B) (40). Assuming that this operator site is authentic, this may indicate that E. coli and S. oneidensis NsrR proteins differ in their specificity determinants for operator recognition.
Control of E. coli ytfE (dnrN) gene expression.
Recent studies have documented NsrR and Fnr regulation of ytfE (dnrN) gene expression patterns similar to those reported here for the yeaR-yoaG operon: NsrR-dependent induction by nitrite and nitric oxide (or compounds that generate nitric oxide) and enhanced expression in Δfnr null strains (6, 20, 24). Here, the NsrR operator overlaps the promoter −10 element (6), and a predicted site for binding the phospho-NarL activator (40) is centered at position −45.5 with respect to the ytfE gene transcription initiation site (6). Thus, it is likely that regulation of ytfE gene transcription is also subject to dual control by the NsrR repressor and the NarL activator.
We noted that the NsrR operator sequence occupies three different positions with respect to the promoter elements (Fig. 1): overlapping the −35 element (yeaR-yoaG operon), between the −35 and −10 elements (nnrS gene), and overlapping the −10 element (ytfE gene). Analogous observations for the position of the TrpR operators for the aroH, trpEDCBA, and trpR operons led to the suggestion that these operators evolved independently (60).
Overlapping Nar and NsrR regulons.
The discovery of the NsrR regulon (6, 20, 22, 40) has added new challenges to our understanding of transcriptional responses to nitrogen oxides. Overlapping regulation by the Nar regulatory systems and the NsrR repressor likely controls transcription of at least five operons: hcp-hcr, nrfABCDEFG (20), yeaR-yoaG, ytfE and, in Shewanella spp., nnrS. The first two operons are also activated by the Fnr protein, whereas the last three are not. Some of the resulting proteins may be involved in protecting the aerobic respiratory chain from inhibition by nitric oxide (22).
Induction by the Nar regulatory systems, which control anaerobic respiratory gene expression in response to nitrate and nitrite (53), may reflect the generation of substantial nitric oxide as a by-product of respiratory nitrate ammonification (13, 58). Thus, synthesis of proteins to protect against nitric oxide would be induced concomitantly with that of proteins that generate nitric oxide (12). As nitric oxide accumulated, release from NsrR repression would provide further synthesis of protective proteins.
Acknowledgments
We are grateful to Li-Ling Chen for invaluable help with and advice concerning construction of the ΔnsrR null allele and to Li-Ling Chen and Alice V. Lin for providing the ΔlacZ and Δfnr null alleles, respectively. We thank Stephen Spiro and J. Alex Appleman for teaching us how and why to use SNP. We appreciate helpful comments and suggestions from Stephen Busby and Stephen Spiro.
This study was supported by Public Health Service grant GM036877 from the National Institute of General Medical Sciences. H.-Y.L. thanks the Food Science Graduate Group for generous financial support.
Footnotes
Published ahead of print on 24 August 2007.
REFERENCES
- 1.Appleman, J. A., and V. Stewart. 2003. Mutational analysis of a conserved signal-transducing element: the HAMP linker of the Escherichia coli nitrate sensor NarX. J. Bacteriol. 185:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, A., A. Wolfe, and S. Busby. 2004. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 7:102-108. [DOI] [PubMed] [Google Scholar]
- 3.Bartnikas, T. B., Y. Wang, T. Bobo, A. Veselov, C. P. Scholes, and J. P. Shapleigh. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology 148:825-833. [DOI] [PubMed] [Google Scholar]
- 4.Beliaev, A. S., D. M. Klingeman, J. A. Klappenbach, L. Wu, M. F. Romine, J. M. Tiedje, K. H. Nealson, J. K. Fredrickson, and J. Zhou. 2005. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187:7138-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloch, M. A., and O. Raibaud. 1986. Comparison of the malA regions of Escherichia coli and Klebsiella pneumoniae. J. Bacteriol. 168:1220-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188:874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browning, D. F., D. J. Lee, A. J. Wolfe, J. A. Cole, and S. J. Busby. 2006. The Escherichia coli K-12 NarL and NarP proteins insulate the nrf promoter from the effects of integration host factor. J. Bacteriol. 188:7449-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busby, S., and R. H. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 10.Buvinger, W. E., and M. Riley. 1985. Regulatory region of the divergent Klebsiella pneumoniae lac operon. J. Bacteriol. 163:858-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. A., and C. Brown. 1980. Nitrite reduction to ammonia by fermentative bacteria: a short circuit in the biological nitrogen cycle. FEMS Microbiol. Lett. 7:65-72. [Google Scholar]
- 12.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4815. [DOI] [PubMed] [Google Scholar]
- 13.Corker, H., and R. K. Poole. 2003. Nitric oxide formation by Escherichia coli. Dependence on nitrite reductase, the NO-sensing regulator Fnr, and flavohemoglobin Hmp. J. Biol. Chem. 278:31584-31592. [DOI] [PubMed] [Google Scholar]
- 14.Cruz-García, C., A. E. Murray, J. A. Klappenbach, V. Stewart, and J. M. Tiedje. 2007. Respiratory nitrate ammonification by Shewanella oneidensis MR-1. J. Bacteriol. 189:656-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darwin, A. J., and V. Stewart. 1995. Nitrate and nitrite regulation of the Fnr-dependent aeg-46.5 promoter of Escherichia coli K-12 is mediated by competition between homologous response regulators (NarL and NarP) for a common DNA-binding site. J. Mol. Biol. 251:15-29. [DOI] [PubMed] [Google Scholar]
- 16.Darwin, A. J., K. L. Tyson, S. J. Busby, and V. Stewart. 1997. Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement. Mol. Microbiol. 25:583-595. [DOI] [PubMed] [Google Scholar]
- 17.Darwin, A. J., E. C. Ziegelhoffer, P. J. Kiley, and V. Stewart. 1998. Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180:4192-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang, F. C. 2004. Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat. Rev. Microbiol. 2:820-832. [DOI] [PubMed] [Google Scholar]
- 20.Filenko, N., S. Spiro, D. F. Browning, D. Squire, T. W. Overton, J. Cole, and C. Constantinidou. 2007. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J. Bacteriol. 189:4410-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fredrickson, J. K., and M. F. Romine. 2005. Genome-assisted analysis of dissimilatory metal-reducing bacteria. Curr. Opin. Biotechnol. 16:269-274. [DOI] [PubMed] [Google Scholar]
- 22.Gilberthorpe, N. J., M. E. Lee, T. M. Stevanin, R. C. Read, and R. K. Poole. 2007. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-γ-stimulated J774.2 macrophages. Microbiology 153:1756-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goh, E. B., P. J. Bledsoe, L. L. Chen, P. Gyaneshwar, V. Stewart, and M. M. Igo. 2005. Hierarchical control of anaerobic gene expression in Escherichia coli K-12: the nitrate-responsive NarX-NarL regulatory system represses synthesis of the fumarate-responsive DcuS-DcuR regulatory system. J. Bacteriol. 187:4890-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Justino, M. C., C. C. Almeida, V. L. Goncalves, M. Teixeira, and L. M. Saraiva. 2006. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 257:278-284. [DOI] [PubMed] [Google Scholar]
- 25.Justino, M. C., J. B. Vicente, M. Teixeira, and L. M. Saraiva. 2005. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J. Biol. Chem. 280:2636-2643. [DOI] [PubMed] [Google Scholar]
- 26.Kang, Y., K. D. Weber, Y. Qiu, P. J. Kiley, and F. R. Blattner. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiley, P. J., and H. Beinert. 1998. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22:341-352. [DOI] [PubMed] [Google Scholar]
- 28.Liu, M., R. J. Turner, T. L. Winstone, A. Saetre, M. Dyllick-Brenzinger, G. Jickling, L. W. Tari, J. H. Weiner, and D. E. Taylor. 2000. Escherichia coli TehB requires S-adenosylmethionine as a cofactor to mediate tellurite resistance. J. Bacteriol. 182:6509-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maloy, S. R., V. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Maris, A. E., M. Kaczor-Grzeskowiak, Z. Ma, M. L. Kopka, R. P. Gunsalus, and R. E. Dickerson. 2005. Primary and secondary modes of DNA recognition by the NarL two-component response regulator. Biochemistry 44:14538-14552. [DOI] [PubMed] [Google Scholar]
- 31.McGinnis, S., and T. L. Madden. 2004. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32:W20-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Monod, J., A. M. Pappenheimer, and G. Cohen-Bazire. 1952. La cinétique de la biosynthèse de la β-galactosidase chez Escherichia coli considérée comme fonction de la croissance. Biochim. Biophys. Acta 9:648-660. [DOI] [PubMed] [Google Scholar]
- 34.Moore, C. M., M. M. Nakano, T. Wang, R. W. Ye, and J. D. Helmann. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 186:4655-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidhardt, F. C. 1987. Multigene systems and regulons, p. 1313-1317. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 36.Poole, R. K. 2005. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33:176-180. [DOI] [PubMed] [Google Scholar]
- 37.Poole, R. K., M. F. Anjum, J. Membrillo-Hernandez, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36:775-783. [DOI] [PubMed] [Google Scholar]
- 39.Rabin, R. S., and V. Stewart. 1993. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaefer, B. C. 1995. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 227:255-273. [DOI] [PubMed] [Google Scholar]
- 42.Shultzaberger, R. K., Z. Chen, K. A. Lewis, and T. D. Schneider. 2007. Anatomy of Escherichia coli sigma 70 promoters. Nucleic Acids Res. 35:771-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, J. 2002. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26:285-309. [DOI] [PubMed] [Google Scholar]
- 44.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 45.Spiro, S. 2007. Regulators of bacterial responses to nitric oxide. FEMS Microbiol. Rev. 31:193-211. [DOI] [PubMed] [Google Scholar]
- 46.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6:399-428. [DOI] [PubMed] [Google Scholar]
- 47.Stewart, G. S., S. Lubinsky-Mink, C. G. Jackson, A. Cassel, and J. Kuhn. 1986. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid 15:172-181. [DOI] [PubMed] [Google Scholar]
- 48.Stewart, V. 2003. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria. Biochem. Soc. Trans. 31:1-10. [DOI] [PubMed] [Google Scholar]
- 49.Stewart, V., and P. J. Bledsoe. 2005. Fnr-, NarP-, and NarL-dependent regulation of transcription initiation from the Haemophilus influenzae Rd napF (periplasmic nitrate reductase) promoter in Escherichia coli K-12. J. Bacteriol. 187:6928-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart, V., and P. J. Bledsoe. 2003. Synthetic lac operator substitutions for studying the nitrate- and nitrite-responsive NarX-NarL and NarQ-NarP two-component regulatory systems of Escherichia coli K-12. J. Bacteriol. 185:2104-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart, V., L. L. Chen, and H. C. Wu. 2003. Response to culture aeration mediated by the nitrate and nitrite sensor NarQ of Escherichia coli K-12. Mol. Microbiol. 50:1391-1399. [DOI] [PubMed] [Google Scholar]
- 52.Stewart, V., and J. Parales, Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart, V., and R. S. Rabin. 1995. Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli, p. 233-252. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, DC.
- 54.Stewart, V., and C. Yanofsky. 1986. Role of leader peptide synthesis in tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 167:383-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strohm, T. O., B. Griffin, W. G. Zumft, and B. Schink. 2007. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 73:1420-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tagle, D. A., B. F. Koop, M. Goodman, J. L. Slightom, D. L. Hess, and R. T. Jones. 1988. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 203:439-455. [DOI] [PubMed] [Google Scholar]
- 57.Tiedje, J. M. 2002. Shewanella: the environmentally versatile genome. Nat. Biotechnol. 20:1093-1094. [DOI] [PubMed] [Google Scholar]
- 58.Weiss, B. 2006. Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J. Bacteriol. 188:829-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu, H., K. L. Tyson, J. A. Cole, and S. J. Busby. 1998. Regulation of transcription initiation at the Escherichia coli nir operon promoter: a new mechanism to account for co-dependence on two transcription factors. Mol. Microbiol. 27:493-505. [DOI] [PubMed] [Google Scholar]
- 60.Yanofsky, C. 1984. Comparison of regulatory and structural regions of genes of tryptophan metabolism. Mol. Biol. Evol. 1:143-161. [DOI] [PubMed] [Google Scholar]