Abstract
The sequences of the capsular biosynthetic (cps) loci of 90 serotypes of Streptococcus pneumoniae have recently been determined. Bioinformatic procedures were used to predict the general functions of 1,973 of the 1,999 gene products and to identify proteins within the same homology group, Pfam family, and CAZy glycosyltransferase family. Correlating cps gene content with the 54 known capsular polysaccharide (CPS) structures provided tentative assignments of the specific functions of the different homology groups of each functional class (regulatory proteins, enzymes for synthesis of CPS constituents, polymerases, flippases, initial sugar transferases, glycosyltransferases [GTs], phosphotransferases, acetyltransferases, and pyruvyltransferases). Assignment of the glycosidic linkages catalyzed by the 342 GTs (92 homology groups) is problematic, but tentative assignments could be made by using this large set of cps loci and CPS structures to correlate the presence of particular GTs with specific glycosidic linkages, by correlating inverting or retaining linkages in CPS repeat units with the inverting or retaining mechanisms of the GTs predicted from their CAZy family membership, and by comparing the CPS structures of serotypes that have very similar cps gene contents. These large-scale comparisons between structure and gene content assigned the linkages catalyzed by 72% of the GTs, and all linkages were assigned in 32 of the serotypes with known repeat unit structures. Clear examples where very similar initial sugar transferases or glycosyltransferases catalyze different linkages in different serotypes were also identified. These assignments should provide a stimulus for biochemical studies to evaluate the reactions that are proposed.
Streptococcus pneumoniae capsular polysaccharide (CPS) is produced by almost all isolates recovered from cases of invasive disease and is a major virulence factor (84). It is also a major immunogen, and antibodies directed against the CPS provide protective immunity. Consequently, CPS is the basis of all current pneumococcal vaccines. The serotyping scheme for S. pneumoniae is based on the immunochemical differences between CPS expressed by different strains, which are recognized by their reactivity with a set of typing sera. Currently, 91 different S. pneumoniae serotypes are recognized (31, 64), some of which show sufficient immunological similarities to be considered related and are placed within a serogroup.
The synthesis and export of bacterial polysaccharides are mediated by three pathways known as the Wzy-dependent pathway, the synthase-dependent pathway, and the ABC transporter-dependent pathway, which have been reviewed recently (77, 86). The Wzy-dependent pathway is most common in S. pneumoniae capsular biosynthesis, but the synthase-dependent pathway is also found (12, 34, 49, 90). The ABC transporter-dependent pathway has not been described for S. pneumoniae capsules.
The two known synthase-dependent pathways are present in serotypes 3 and 37 (7, 12, 48). The synthase-dependent pathway is the simplest, with a single integral inner membrane protein generally thought to be responsible for both the sequential addition of the sugars and the concurrent extrusion of the nascent polymer across the cell membrane. The repeat units with synthase-dependent pathways are generally single sugars, or two sugars may alternate, or one may form the main chain and the other a side branch on each main chain residue.
The Wzy-dependent pathway has been identified in 88 S. pneumoniae serotypes, and the cps loci are located at the same site on the chromosome, known as the CPS synthesis (cps) locus, between the dexB and aliA genes (49). The Wzy-dependent pathway involves the synthesis of the polysaccharide repeat unit on an undecaprenyl lipid carrier (UndPP) and then translocation of this oligosaccharide across the membrane by the Wzx flippase so that the oligosaccharide component, still attached to UndPP, is on the outer face of the membrane, where it is polymerized by Wzy to produce a UndPP-attached polymer (Fig. 1) (90).
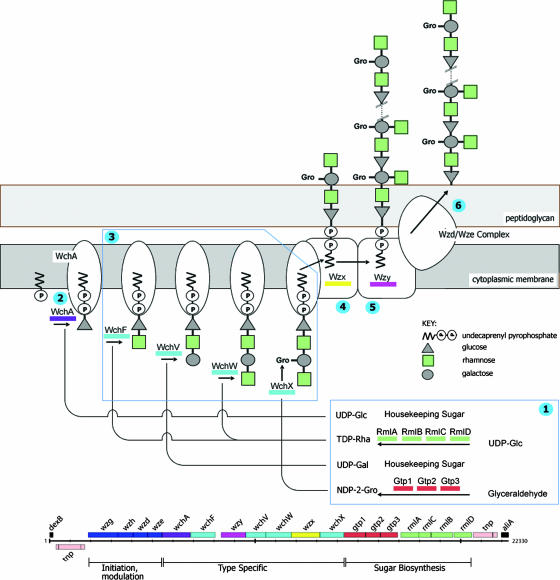
FIG. 1.
Schematic representation of the biosynthesis of CPS by the Wzy-dependent pathway. The biosynthesis of the CPS of serotype 23F is represented. UDP-linked components of the repeat CPS unit are synthesized by genes encoded within the cps locus or are available from central metabolism (1). Repeat unit biosynthesis is initiated by the transfer of glucose phosphate to the lipid carrier by the IT WchA (2), followed by sequential addition of the other components of the repeat unit, catalyzed by the GTs (3), and the lipid-linked repeat unit is transferred across the membrane by the Wzx flippase (4) and polymerized by Wzy to result in lipid-linked CPS (5). Finally, the lipid-linked CPS is linked to the cell wall by a poorly understood process involving the Wzd/Wze complex, with release of the undecaprenyl phosphate carrier (6). The genes in the representation of the cps locus and their gene products are color coded; a key is given for the repeat unit constituents (modified from reference 12).
The sequences of the cps loci from 90 S. pneumoniae serotypes have been presented recently, and the products of the genes present in these cps loci have been classified into 249 groups (homology groups [HGs]) (12). The great diversity of these products reflects the chemical diversity of the S. pneumoniae capsules, and we have presented some examples of predictions of functions by correlating gene content with known CPS structure and serology (12). Recently, we have used similarities in the sequence-weighted gene contents of the S. pneumoniae cps loci to identify closely related cps loci and have related these to their similar CPS repeat units (51). Here an analysis of the predicted functions of almost all of the 249 HGs encoded by the cps genes of the 90 serotypes is presented, along with the predicted specificity of the transferases that synthesize the CPS repeat units.
MATERIALS AND METHODS
Functional assignment of cps gene products.
The sequences of the S. pneumoniae cps loci from all 90 serotypes (GenBank accession numbers CR931632 to CR931722) and the schematic representations of the available CPS structures were those of Bentley et al. (12), incorporating the revised CPS structures of serotypes 15B, 17F, and 33F (36, 37, 45). Recently, an immunological variant within serogroup 6 that has a different CPS structure (serotype 6C) has been reported, but as the cps sequence is not available, we do not consider it further (64). The predicted cps gene products have been placed into HGs, i.e., nonoverlapping groups of proteins showing highly significant amino acid sequence similarity, by using TRIBE-MCL with a TBLASTX cutoff level of 1e−50 (12). Genes encoding members of different HGs were assigned different gene names, excepting the polymerase (wzy) and flippase (wzx) genes. The polymerases and flippases fell into multiple HGs but, to maintain prior nomenclature, were each given the same gene name and were sequentially numbered to denote the members of the different HGs (12). New names for cps genes were assigned in accordance with the Bacterial Polysaccharide Gene Database (68). Amino acid sequence alignments were performed by using CLUSTAL_X (76). BlastN and TBLASTX comparisons of the cps loci were visualized by using ACT version 6 and WebACT, an online version of the Artemis comparison tool (1, 17).
To predict a functional assignment, the cps gene products were classified into families based on hidden Markov model profiles by using the Pfam database (25; http://www.sanger.ac.uk/Software/Pfam/). If there was no Pfam hit for an HG, the GenBank database (3; http://www.ncbi.nlm.nih.gov/) and the InterPro database (5; http://www.ebi.ac.uk/interpro/) were used for searches of homologs with known function in other organisms. The MetaCyc database (18; http://metacyc.org/) was used for the identification of genes involved in biosynthetic pathways. The genomic sequences of S. pneumoniae R6 (32; GenBank accession number AE007317) and S. pneumoniae TIGR4 (75; GenBank accession number AE005672) were used for the identification of genes involved in the biosynthesis of housekeeping components of the repeat units. The families of glycosyltransferases (GTs) encoded by the S. pneumoniae cps loci were retrieved from the carbohydrate-active enzymes (CAZy) database (20; http://afmb.cnrs-mrs.fr/CAZY/). There were 26 cps gene products that did not possess any known Pfam domain and were annotated as “conserved hypothetical proteins” if they showed weak similarity to products involved in bacterial polysaccharide biosynthesis or else as “hypothetical proteins” (12). Of these 26 cps gene products, 12 were present in serotypes with known CPS structures and their function could be predicted; 8 were present in serotypes with unknown CPS structures, and the rest were pseudogenes.
Assignment of reactions catalyzed by transferases.
The approaches used to identify and assign functions to the sugar/polyalcohol phosphate transferases and pyruvyltransferases and for identification of polymerase linkages and initial sugar transferase specificities are described in Results. A number of approaches were used to tentatively assign the linkages catalyzed by the GTs, as follows.
(i) GT-linkage correlation across serotypes.
Initially, all known repeat unit structures were broken down into the triplet linkages (donor sugar-linkage-acceptor sugar) catalyzed by the GTs, and correlations between the presence of a particular GT gene with the triplet linkages present in the repeat units were made. A perfect correlation between the presence of a GT gene in the cps loci and a single triplet linkage in the CPS of the corresponding structures, that was not found in structures where the gene was absent, corresponded to a strong assignment. This procedure was started with the GT gene (and thus the HG) that was most often present within the cps loci of the serotypes whose CPS structures are known. Having assigned the linkage catalyzed by the most common GT, we repeated the process with the GT gene that is next most common in the cps loci of serotypes with known CPS structures, ignoring all incidences of GT linkages that have already been assigned by linkage correlation. This iterative process was continued to assign the linkages catalyzed by further GTs, eliminating at each cycle the linkages already assigned, until assignments became ambiguous.
(ii) Gene content-linkage comparison between similar serotypes.
Comparisons between gene content in very similar cps loci and their CPS structures can also be used to assign linkages. Should two cps loci differ only in a single GT gene and the CPS structures of those serotypes differ only in a single linkage, this can be used to assign enzyme specificity.
(iii) CAZy GT family membership and linkage status.
GTs were assigned membership, where possible, to either inverting or retaining GT families based on the CAZy database (20). Each linkage in the repeat unit is catalyzed by a GT that either retains or inverts the stereochemistry of the donor sugar. Correlating the number of inverting or retaining GTs encoded in the cps loci with the number of inverting or retaining linkages in the CPS structures can in some cases be used to assign specificity. For example, the cps locus of a serotype may encode a single inverting GT, as predicted by CAZy family membership, and the CPS structure contains a single linkage that requires an inverting GT. We can therefore assign the specificity of the inverting GT to the synthesis of the inverting linkage.
(iv) Combination of the methods described above.
In practice, the methods described above were often used in combination to assign GT specificities. For example, the cps locus of a serotype may possess three GT genes. One of these may have already been assigned by GT-linkage correlation and another by CAZy membership and linkage status. The third GT can then be assigned to the remaining linkage.
RESULTS AND DISCUSSION
S. pneumoniae capsules synthesized by the synthase-dependent pathway.
The type 37 synthase (Tts) directs the synthesis of serotype 37 CPS with a glucose main chain and an additional glucose side branch in each repeat unit (47, 48). The synthase gene of serotype 37 is not located within the cps locus, but a defective type 33F cps locus is present between dexB and aliA (12, 48). In the serotype 37 strain sequenced by Bentley et al. (12), the defective type 33F cps locus has frameshift mutations in the wciC and wciG genes and a stop codon in glf, compared to the cps locus of a serotype 33F isolate (GenBank accession no. CR931702).
The type 3 synthase (WchE; also known as Cap3B) directs the synthesis of serotype 3 CPS with glucose and glucuronic acid alternating (7, 82). WchE transfers glucose (Glc) from UDP-Glc to phosphatidylglycerol to form glucosephoshatidylglycerol, which serves as a primer for the addition of Glc and glucuronic acid (GlcA) (15, 26), and UDP-sugar substrate concentrations modulate the polysaccharide chain length (80). The type 3 synthase gene is within a defective cps locus along with three genes (ugd, galU, and pgm, also known as cap3A, cap3C, and cap3D, respectively) involved in the synthesis of sugar precursors (6, 12).
S. pneumoniae capsules synthesized by the Wzy-dependent pathway.
Excluding putative transposase genes, there are 1,502 cps gene products encoded by the cps loci of the 88 S. pneumoniae serotypes whose capsules are synthesized by the Wzy-dependent pathway (Table 1). As mentioned previously, the cps loci of the 88 pneumococcal serotypes that use the Wzy pathway include the polymerase (wzy) and flippase (wzx) genes, and although their gene products fall into a number of different HGs, they can be readily identified by homology searches and by their characteristic hydrophobicity patterns, providing a useful predictor of the presence of the Wzy-dependent pathway (12).
TABLE 1.
Gene products, classified into functional groups, and their Pfam families, encoded by the 88 cps loci of S. pneumoniae that use the Wzy-dependent pathway of CPS biosynthesisa
| Functional group | No. of genes (pseudogenes) | Pfam family (or families)b |
|---|---|---|
| Processing, regulation, export | ||
| Wzg | 88 (0) | (PF02916-PF03816) |
| Wzh | 88 (0) | PF02811 |
| Wzd | 88 (0) | PF02706 |
| Wze | 88 (0) | |
| Polymerization and export | ||
| Wzy | 90 (2) | PF04932 |
| Wzx | 88 (0) | PF01943, PF03023 |
| Biosynthetic pathways | ||
| RmlA | 40 (0) | PF00483 |
| RmlC | 40 (1) | PF00908 (PF01073-PF01370-PF02719-PF04321) |
| RmlB | 41 (0) | (PF01073-PF01370-PF04321) |
| RmlD | 41 (1) | (PF01073-PF01370-PF02719) |
| Glf | 58 (23) | PF03275 (PF00984-PF01210-PF03720-PF03721) |
| Ugd | 17 (0) | (PF00984-PF03720) |
| Gla | 4 (0) | (PF01073-PF01370-PF02719-PF04321) |
| MnaA | 15 (0) | PF02350 |
| MnaB | 5 (0) | (PF00984-PF03720-PF03721) |
| FnlA | 8 (1) | (PF01073-PF01370-PF02719-PF04321-PF08485) |
| FnlB | 8 (0) | PF01370 |
| FnlC | 8 (0) | PF02350 |
| Gct | 12 (2) | PF01467 |
| Abp1 | 6 (1) | PF01128 (PF01073-PF01370-PF02719-PF04321) |
| Abp2 | 6 (0) | (PF01073-PF01370) |
| Mnp1 | 3 (0) | (PF00483-PF01128) |
| Mnp2 | 3 (0) | (PF01073-PF01370) |
| Gtp1 | 9 (0) | PF01761 |
| Gtp2 | 9 (0) | PF00483 |
| Gtp3 | 9 (0) | PF00702 |
| RbsF | 7 (0) | PF00702 |
| Transferases | ||
| ITs | 88 (1) | PF02397 |
| GTs | 342 (8) | PF00534, PF00535, PF04488, PF03808, PF02485, PF05704, PF01501 (PF00535-PF05704) |
| Acetyltransferases | 78 (12) | PF00132 |
| PF01757 | ||
| Sugar phosphate transferases | 69 (3) | PF04991, PF04464, PF01066 |
| Pyruvyltransferases | 2 (0) | PF04230 |
| Others | ||
| Group II intron proteins | 4 (0) | (PF00078-PF08388) |
| Long repetitive protein | 1 (0) | (PF00746-PF05738) |
| GT enhancers | 13 (0) | |
| Hypothetical proteins | 26 (7) | |
| Total | 1,502 (62) |
Predicted S. pneumoniae cps gene products for 88 serotypes whose CPS is synthesized by the Wzy-dependent pathway. Products of transposase genes (n = 224) are not included.
Pfam domains found in the cps gene products are shown. Different Pfam domains are found in different HGs of a functional group, and some contain more than one Pfam domain, as indicated by parentheses.
There are also four conserved genes (wzg, wzh, wzd, and wze) at the 5′ end of all S. pneumoniae cps loci that use the Wzy pathway (see the next section) (12, 34, 49, 90). The cps loci also include genes whose products are involved in the biosynthesis of nonhousekeeping components (cps-specific biosynthetic pathway genes; see below), initiation of capsule biosynthesis (initial sugar transferase genes), and transfer of sugars or other moieties and their assembly in the repeat unit (GT, acetyltransferase, sugar phosphate transferase, and pyruvyltransferase genes) (Fig. 1) (12).
Processing, regulation, and export of S. pneumoniae capsules.
The highly conserved wzg, wzh, wzd, and wze genes (also known as cpsA, cpsB, cpsC, and cpsD, respectively) at the 5′ end of the cps locus are known to be involved in the processing, regulation, and export of CPS (56). These four genes constitute a regulatory system for CPS production via tyrosine phosphorylation of Wze (CpsD) (10, 56, 59), and recent findings suggest that this mechanism is important for attachment of the CPS to the cell wall (58).
Biosynthetic pathways for precursors of S. pneumoniae capsules.
The precursors for the known components of the repeat units are all thought to be transferred from nucleotide diphospho (NDP) derivatives, with the exception of O-acetyl and pyruvate moieties, which are transferred from acetyl coenzyme A and phosphoenolpyruvate, respectively (Fig. 2 and 3). We consider first those CPS components for which NDP derivatives are generally present in S. pneumoniae because they are used for other pathways (housekeeping components); as expected, genes for these pathways are not present in the cps loci. Thirty-six of the pneumococcal CPS structures are unknown, and there could be additional housekeeping components besides those mentioned below. Genes should be present within cps loci for the biosynthesis, from available intermediates, of activated forms of the sugars or other CPS components that are not normally present in S. pneumoniae (nonhousekeeping or CPS-specific components). If some of the eight cps gene products without assigned functions (see Materials and Methods) are involved in biosynthetic pathways, there could also be additional nonhousekeeping components.
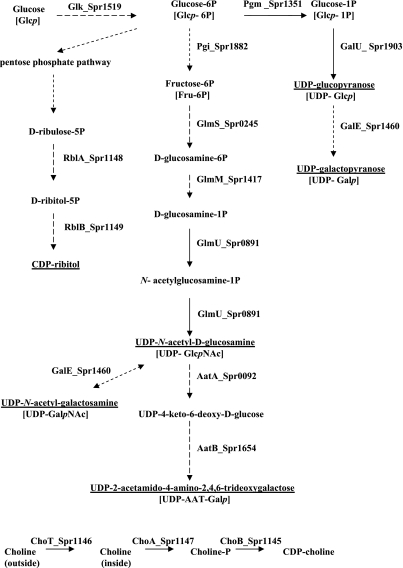
FIG. 2.
Biosynthetic pathways of housekeeping components of S. pneumoniae CPS. Putative pathways are denoted by a dotted line, and the constituents of the repeat units are underlined. Glk, glucokinase; Pgi, glucose-6-phosphate isomerase; Pgm, phosphoglucomutase; GlmS, l-glutamine-d-fructose-6-phoshate amidotransferase; GlmM, phosphoglucosamine mutase; GlmU, UDP-N-acetyl-glucosamine pyrophosphorylase; GalU, UTP-glucose-1P uridylyltransferase; GalE, UDP-N-acetylglucosamine-4-epimerase/UDP-glucose-4-epimerase; RblA, ribitol-5-phosphate dehydrogenase; RblB, d-ribitol-5-phosphate cytidylyltransferase; AatA, UDP-GlcNAc 4,6-dehydratase; AatB, UDP-4-keto-6-deoxy-d-glucose 4-transaminase; ChoT (also known as LicB), choline transporter; ChoA (also known as LicA), choline kinase; ChoB (also known as LicC), cholinephosphate cytidylyltransferase. Spr numbers refer to the annotation of the S. pneumoniae R6 genome sequence.
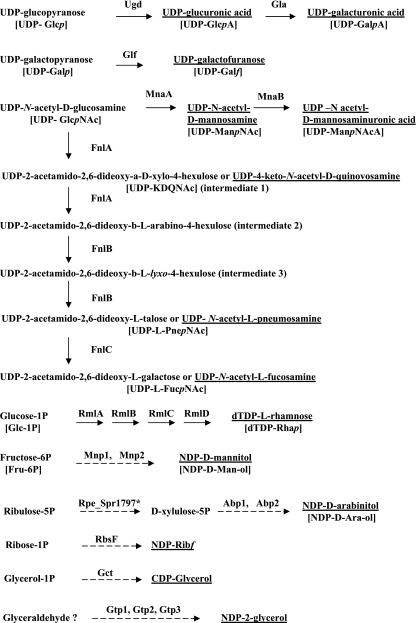
FIG. 3.
Biosynthetic pathways of nonhousekeeping (CPS-specific) components of S. pneumoniae CPS. Putative pathways are denoted by a dotted line, and the constituents of the repeat units are underlined. All gene products are encoded within the cps loci, excepting ribulose phosphate 3-epimerase (Rpe), which is chromosomally encoded and marked by an asterisk. Ugd, UDP-glucose 6-dehydrogenase; Gla, UDP-galacturonate 4-epimerase; Glf, UDP-galactopyranose mutase; MnaA, UDP-N-acetylglucosamine-2-epimerase; MnaB, UDP-N-acetylmannosamine dehydrogenase; FnlA, steps 1 and 2 of UDP-FucNAc synthesis; FnlB, steps 3 and 4 of UDP-FucNAc synthesis; FnlC, step 5 of UDP-FucNAc synthesis; RmlA, glucose-1-phosphate thymidylyltransferase; RmlB, dTDP-d-glucose 4,6-dehydratase; RmlC, dTDP-4-keto-6-deoxy-d-glucose 3,5-epimerase; RmlD, dTDP-4-keto-l-rhamnose reductase; Mnp1, putative nucleotidyltransferase (NDP-mannitol pathway); Mnp2, putative reductase (NDP-mannitol pathway); RbsF, putative epimerase/dehydratase (NDP-ribose biosynthesis); Gct, CDP-glycerol biosynthetic protein; Gtp1-3, NDP-2-glycerol pathway; Abp1, putative nucleotidyltransferase (NDP-arabinitol pathway); Abp2, putative reductase (NDP-arabinitol pathway).
(i) Housekeeping components.
There are no gene products present in the S. pneumoniae cps loci for the biosynthesis of seven components in the known CPS structures (glucopyranose, N-acetylglucosamine, galactopyranose, N-acetylgalactosamine, 2-acetamido-4-amino-2,4,6-trideoxy-d-galactopyranose, ribitol-phosphate and phosphorylcholine), and their precursors are presumably available for CPS synthesis from S. pneumoniae general metabolism. The first four are known to have UDP derivatives as precursors, whereas CDP-d-ribitol-5P and CDP-choline are the precursors for ribitol-phosphate and phosphorylcholine, respectively.
The genes expected for the synthesis of six of these components are present in the S. pneumoniae genome, and the biosynthetic pathways are well documented (68, 70), as shown in Fig. 2. There are also genes for a putative pathway for the other component, UDP-2-acetamido-4-amino-2,4,6-trideoxy-d-galactose (UDP-AAT-Galp), as discussed below. Glucopyranose, N-acetylgalactosamine, AAT-Galp, ribitol-phosphate, and phosphorylcholine are components of S. pneumoniae teichoic and lipoteichoic acids, often called C-polysaccharide and F-polysaccharide, respectively (9, 38, 40).
Glucopyranose (Glcp) and galactopyranose (Galp).
UDP-Glcp is the precursor of Glcp, synthesized from Glcp-6P (Fig. 2) in two steps catalyzed by the cellular phosphoglucomutase Pgm and UTP-Glc-1P uridylyltransferase GalU (14, 54). Although these two genes are present on the pneumococcal chromosome (14, 30), they also are present within the type 3 cps locus but can be deleted, presumably due to this redundancy, and type 3 CPS synthesis depends mostly on the chromosomally encoded Pgm activity (29).
Exogenous galactose can be used for the synthesis of UDP-Galp via the enzymes galactokinase (GalK) and Gal-1P uridylyltransferase (GalT) (27, 88). UDP-Glcp can also be converted to UDP-Galp by UDP-Glc-4-epimerase (GalE) (28). Putative carbohydrate-specific ABC transporter genes and the galE, galT, and galK genes are present in the S. pneumoniae R6 genome (GenBank accession no. AE007317). Therefore, S. pneumoniae should be able to use exogenous galactose, although this has not been shown experimentally.
N-Acetylglucosamine (GlcpNAc).
Fructose-6P (Fru-6P) is housekeeping in S. pneumoniae as part of the glycolytic pathway (Fig. 2) and is a substrate for the synthesis of N-acetylglucosamine-1-phosphate (GlcNAc-1P); the first two steps of the pathway are catalyzed by l-glutamine-d-Fru-6P amidotransferase (GlmS) and phosphoglucosamine mutase (GlmM). The two genes appear to be present in the S. pneumoniae genome (spr0245 and spr1417, respectively; 32). The resultant GlcpN-1P is converted to UDP-GlcpNAc in a two-step reaction by the bifunctional enzyme GlmU, which possesses a pyrophosphorylase and an acetyltransferase domain (74). The latter sugar is also the substrate for the synthesis of some CPS nonhousekeeping components (Fig. 3).
N-Acetylgalactosamine (GalpNAc).
A specific UDP-GlcpNAc- 4-epimerase (Gne) is required for the conversion of UDP-GlcpNAc to UDP-GalpNAc in Escherichia coli and Yersinia enterocolitica (11, 83). For a long time, it was assumed that GalE, a UDP-Glc-4-epimerase, was also a UDP-GlcpNAc-4-epimerase. It is now recognized that there are three classes of GalE-like proteins (33), i.e., epimerases that preferentially catalyze the conversion between UDP-Glcp and UDP-Galp (group 1), epimerases that do not show a preference for either UDP-Glcp/UDP-Galp or UDP-GlcpNAc/UDP-GalpNAc (group 2), and epimerases that preferentially convert between UDP-GlcpNAc and UDP-GalpNAc (group 3).
Several S. pneumoniae capsules contain GalpNAc, but none of their cps loci have a galE/gne homolog. Nevertheless, Spr1460/NP_346051 (GenBank accession numbers AE007317 and AE005672), encoded within the S. pneumoniae genome, is closely related to the Y. enterocolitica AAC60777 and Bacillus subtilis NP_391765 enzymes, which are known to epimerize both UDP-Glc/UDP-Gal and UDP-GlcNAc/UDP-GalNAc. We conclude that Spr1460 is the product of a group 2 galE gene responsible for the synthesis of GalNAc in teichoic acid and CPS and Gal in CPS in the absence of exogenous galactose.
2-Acetamido-4-amino-2,4,6-trideoxy-d-galactose (AAT-Galp).
The uncommon sugar AAT-Galp is a component of the CPS of S. pneumoniae serotype 1, of S. pneumoniae lipoteichoic and teichoic acids (38), and of Streptococcus mitis biovar I teichoic acid (13). It is also found in the Bacteroides fragilis capsules, the Shigella sonnei form I antigen (where it is called 4n-d-FucNAc) and Plesiomonas shigelloides serotype O17 antigen (21, 72, 87). To the best of our knowledge, these are the only occurrences in bacteria. As the latter three species lack teichoic acids, one might expect the AAT-Galp pathway genes to be present in the polysaccharide biosynthetic loci and there are indeed appropriate genes that correlate with the presence of AAT-Galp, and this made it possible to propose a biosynthetic pathway.
In S. sonnei, the proposed pathway for UDP-AAT-Galp (87) involves a dehydratase (WbgZ) that converts UDP-GlcpNAc to 4-keto-6-deoxy-d-GlcpNAc, and then an amino group is added at carbon 4 by an aminotransferase (WbgX) to give UDP-AAT-Galp. Finally, as AAT-Galp is assumed to be the initial sugar of the repeat unit, UndPP-AAT-Galp is produced by WbgY. In the case of S. sonnei, all three genes are present in the O-antigen gene cluster, whereas in the case of B. fragilis, the gene encoding the WbgZ homolog is located elsewhere on the chromosome (21, 87). Presumably, it is involved in other biosynthetic pathways and this would not be surprising as NDP-4-keto-6-deoxy sugars are often branch points in NDP-sugar biosynthesis (70).
In pneumococci, the AAT-Galp biosynthetic genes should be chromosomally encoded due to the presence of AAT-Galp in teichoic acid, and in the serotype 1 cps locus there are no gene products for UDP-AAT-Galp synthesis. The products of spr0092 and spr1654 in the S. pneumoniae R6 genome (32) are equivalent to WbgZ and WbgX of S. sonnei, respectively, which we are renaming aatA and aatB to reflect their probable role in the synthesis of UDP-AAT-Galp for both teichoic acids and the serotype 1 capsule (Fig. 2).
Phosphatidylcholine (Cho-P).
The CPS structures of serogroup 15 (excepting 15A), serogroup 32, and serotype 27 have been reported to include Cho-P (38), although this is now in some doubt for serogroup 15 (36; see below). Cho-P is also a component of teichoic and lipoteichoic acids in S. pneumoniae (38, 40). Choline is required for growth, but S. pneumoniae is unable to synthesize choline, obtaining it from its human host. This requirement for choline means that it is always available for CPS synthesis if needed. Genes for the uptake of choline and synthesis of the precursor of Cho-P (lic genes) have been identified in several bacteria (73) and studied in Haemophilus influenzae (85). They were first recognized to be in S. pneumoniae by Zhang et al. (91) and are in the genome as licB, licA, and licC, which we have renamed choT (choline transporter), choA (choline phosphorylation), and choB (conversion to CDP-choline), respectively, to reflect their role in choline transport and CDP-choline synthesis (Fig. 2).
Ribitol-phosphate (Rib-ol-P).
CDP-d-ribitol-5P is the precursor for the addition of d-ribitol-5P, which is also a constituent of teichoic acids and hence available for pneumococcal CPS synthesis (40). In H. influenzae, CDP-ribitol synthesis is catalyzed by the bifunctional enzyme Bcs1 in a two-step reaction (93). In B. subtilis, two separate genes, tarJ and tarI, encode a reductase that converts ribose-5P to ribitol-5P and a cytidylyltransferase that converts ribitol-5P to the precursor CDP-ribitol, respectively (44). The products of spr1148 and spr1149 in the S. pneumoniae R6 genome show a high level of sequence similarity to TarJ and TarI of B. subtilis, and as these pneumococcal homologs should have the same function, we name them rblA and rblB, respectively, to reflect their role in the synthesis of the ribitol-5P precursor (Fig. 2). Note that the single report of ribitol-1P in types 11B and 11F (38) appears to be a case of a different name for ribitol-5P.
(ii) Nonhousekeeping (CPS-specific) components.
There are 14 components in the known S. pneumoniae CPS biochemical structures that are not available from general metabolism and for which the biosynthetic genes are found in the pneumococcal cps loci (Fig. 3). Many of the genes discussed below are also found in polysaccharide biosynthetic loci of other species. There are well-documented pathways from a number of bacterial species for the synthesis of the NDP-linked precursors of rhamnose, galactofuranose, glucuronic acid, galacturonic acid, N-acetylmannosamine, N-acetylmannosaminuronic acid, N-acetylfucosamine, N-acetyl-l-pneumosamine, and glycerol-1-phosphate (70), and in pneumococci the relevant genes are always present intact in the cps locus when the sugar is present in the structure (12). The pathways for the remaining components are less well documented, but the allocation of cps genes to these pathways seems secure from the correlation between the presence of a particular gene(s) and these components in the CPS.
Rhamnose (l-Rhap)—rmlA, rmlB, rmlC, and rmlD.
Rhamnose is present in 23 CPS structures, and all of their cps loci contain the four rml genes for synthesis of the precursor dTDP-rhamnose (34) (Fig. 3). The gene order (rmlA, -C, -B, and -D) is always the same in S. pneumoniae, but there are incomplete sets of rml genes in a few other serotypes, and where the structure is known they lack rhamnose (12). For example, the cps loci of types 1 (63) and 24B have frameshifts within the rmlD and rmlC genes, respectively, whereas the cps locus of type 15F has only a partial rmlB and an rmlD gene present. A truncated UDP-galactofuranose biosynthetic gene (glf) and/or remnants of transposase genes are usually present downstream of the rml genes.
Galactofuranose (d-Galf)—glf.
The precursor of galactofuranose is UDP-galactofuranose, which is synthesized by Glf from UDP-galactopyranose (34) (Fig. 3). The presence of an intact glf gene correlates with Galf in CPS; however, type 15F is an exception as it has an intact glf gene but there is no Galf in the CPS. There are cases where two copies of the glf gene are present in the cps locus, the second copy being truncated in some cases.
Glucuronic acid (d-GlcpA) and galacturonic acid (d-GalpA)—ugd and gla.
UDP-GlcpA is the precursor of GlcpA and is synthesized by Ugd from UDP-Glcp (Fig. 3) and then is converted by Gla to UDP-GalpA, the precursor of GalpA (12, 34, 61, 63). There are 18 cps loci that have ugd, including the type 3 cps locus. Where known, the CPS structures (or CPS constituents) of these serotypes contain GlcpA, with the exception of serotype 1; the latter cps locus has both ugd and gla, and thus serotype 1 contains GalpA rather than GlcpA.
UDP-N-acetyl-d-mannosamine (d-ManpNAc) and UDP-N-acetyl-d-mannosaminuronic acid (d-ManpNAcA)—mnaA and mnaB.
The mnaA gene for the synthesis of d-ManpNAc (34, 70) (Fig. 3) is present in 15 cps loci, whereas mnaB, for the conversion of d-ManpNAc to d-ManpNAcA, is present in only 5 of these (serogroup 12 and types 44 and 46). The available CPS structures are consistent with that expected from the presence of mnaA alone or mnaA and mnaB; thus, type 4 and serogroups 9 and 19 contain d-ManpNAc, whereas types 12A and 12F contain d-ManpNAcA.
N-Acetyl-l-fucosamine (l-FucpNAc)—fnlA, fnlB, and fnlC.
The precursor of l-FucpNAc is UDP-l-FucpNAc, which is synthesized in a five-step pathway (60) (Fig. 3) by the products of three genes (FnlA, -B, and -C), the first two (FnlA and FnlB) being bifunctional. The three genes are present in eight cps loci, and l-FucpNAc is present in the CPS where the structures are known (serotypes 4, 5, 12F, 12A, and 45).
N-Acetyl-l-pneumosamine (l-PnepNAc) and 2-acetamido-2,6-dideoxy-d-xylo-hexos-4-ulose (4-keto-N-acetyl-d-quinovosamine, KDQNAc).
The type 5 CPS contains l-PnepNAc and KDQNAc, in addition to l-FucpNAc, and the fnlA, fnlB, and fnlC genes are present in the cps locus. The precursor of l-PnepNAc is UDP-PnepNAc, a UDP-FucpNAc intermediate synthesized by FnlB, and UDP-KDQNAc, the likely precursor of KDQNAc, is the first intermediate before undergoing further modification by FnlA (60) (Fig. 3). Both l-PnepNAc and KDQNAc are uniquely present in type 5, and the need to divert some of the intermediate of the first FnlA reaction to a KDQNAc transferase reaction may explain the much greater sequence divergence between the fnlA gene of type 5 and those of the other serotypes than observed for fnlB and fnlC (data not shown).
Glycerol-1P (Gro-1P)—gct.
The precursor of glycerol-1P is CDP-glycerol, which is synthesized by Gct (34) (Fig. 3), and the gct gene is present in 13 cps loci. An intact gct gene correlates with the presence of Gro-1P in the CPS of serogroup 18 and types 11A, 11C, and 45, whereas types 11B and 11F have a truncated gct gene and Gro-1P is not found in their CPS, being replaced by Rib-ol-1P.
Glycerol-2P (Gro-2P)—gtp1, gtp2, and gtp3.
The gtp1, gtp2, and gtp3 genes are present in the cps loci of serogroups 15, 23, and 28, and their products are thought to be responsible for the synthesis of NDP-2-glycerol (Fig. 3). Gro-2P is rarely present in bacteria and was originally only reported to be present in types 15A and 23F, with Cho-P replacing it in types 15F, 15B, and 15C (38). However, recent nuclear magnetic resonance (NMR) analysis indicates that type 15B contains glycerol and not choline (36). As the earlier NMR spectra of types 15B and 15C were superimposable, this most likely also applies to types 15C and 15F, the reported presence of Cho-P being due to contamination of the CPS with teichoic acid.
The sugar phosphate transferase for Gro-2P is proposed to be encoded by wchX (see below), and the presence of Gro-2P in the CPS of types 15A and 23F correlates with the presence of wchX and gtp1 to -3 in their cps loci (12, 55, 67). We consider the balance of the evidence to favor the presence of Gro-2P in the CPS of all four serotypes within serogroup 15, as all possess wchX and gtp1 to -3 and there are no differences in their cps loci that would account for the different side groups in the structures reported earlier.
Mannitol-6P (Man-ol-6P)—mnp1 and mnp2.
The presence of mannitol-6P in the CPS of type 35A correlates with the presence of two genes (mnp1 and -2), which are also present in the cps loci of types 35C and 42, for which there are no available CPS structures. Mnp1 possesses the PF00483 family domain of NTP-transferases, i.e., enzymes that transfer nucleotides onto phosphosugars. Mnp2 possesses the PF01370 family domain of NAD-dependent epimerases/dehydratases. Mnp1 and −2 could be responsible for the synthesis of NDP- Man-ol-6P from d-fructose-6P by a two-step pathway parallel to that for CDP-ribitol-5P formation from ribulose-5P by TarJ and TarI (see above). The two steps are addition of the nucleoside monophosphate to give NDP-fructose-6P and reduction of the keto group on carbon 2. However, the order of the two reactions is not known and there is no biochemical support for this pathway as yet. The genes will be named mnpA and mnpB in order of function if this proposal is confirmed.
Arabinitol-1P (Ara-ol-1P)—abp1 and abp2.
Arabinitol-1P is only reported in the CPS of type 17F. The abp1 and −2 genes present in the type 17F cps locus encode proteins containing the PF00483 and PF01370 family domains of NTP-transferases and NAD-dependent epimerase/dehydratase family domains, respectively; thus, they are proposed to be involved in the biosynthesis of NDP-Ara-ol-1P. d-Xylulose-5P is produced from ribulose-5P (present as an intermediate of the pentose phosphate pathway) by a ribulose phosphate 3-epimerase (Rpe) (2), and the rpe gene (spr1797) is present in the S. pneumoniae R6 genome (GenBank accession number AE007317). NDP-Ara-ol-1P is proposed to be derived from d-xylulose-5P in a two-step pathway catalyzed by the products of the abp1 and abp2 genes. As there is no biochemical support for the pathway and the order of the two reactions of the abp1 and −2 gene products is not known, the genes will be renamed abpA and abpB in order of function if this proposal is confirmed.
The abp1 and abp2 genes are also present in the cps loci of serogroup 24 and types 13 and 48. Of these, only the structure of type 13 CPS is available and contains Rib-ol-5P but not Ara-ol-1P. However, the structure is old and there are no recent NMR data to confirm the presence of Rib-ol-5P rather than Ara-ol-1P. Alternatively, the absence of Ara-ol-1P could be attributed to the sequence divergence of the type 13 abp1 and abp2 genes compared to those present in the other cps loci, but new NMR data are required to address this issue.
Ribofuranose (Ribf)—rbsF.
ADP-ribose (ADP-ribofuranose) is believed to be available from the recycling of NAD. It is a component of the E. coli O114 antigen (23), but no gene for ADP-ribose biosynthesis is present in this O-antigen gene cluster (24). However, in S. pneumoniae types 19B and 19C, the rbsF (also known as cps19R) gene was proposed to be responsible for the synthesis of NDP-ribose (57). RbsF possesses the PF01370 family domain of NAD-dependent epimerases/dehydratases. The rbsF gene is also present in the cps loci of types 7B, 7C, 24B, 24F, and 40. The CPS of type 7B and the constituents of type 24F also include Ribf (constituents of the other serotypes are not known), which adds support to the proposed function of RbsF.
Assignment of initial sugar transferases.
Repeat unit synthesis mediated by the Wzy pathway is initiated by transfer of a sugar phosphate to a lipid carrier (Fig. 1) (77, 86). This reaction is catalyzed by an initial transferase (IT) and is reversible (66), whereas the reactions of the GTs that add the other sugars are, in practical terms, irreversible. This can be important in the allocation of gene function if an IT is able to transfer more than one sugar phosphate. In such cases, the specificity of the second sugar transferase determines which sugar is incorporated into the repeat unit, as this reaction is irreversible. Determination of the CPS repeat unit structure does not identify which glycosidic link is made by the Wzy polymerase, and repeat units are generally depicted chemically as circular molecules. Identification of the initial sugar (IT specificity) allows us to identify the linkage made by Wzy and to represent the repeat units as they are synthesized on the lipid carrier.
In most S. pneumoniae cps loci, the fifth gene appears to encode the proposed IT (WchA, WciI, WcjG, and WcjH; see below), as each possesses the same Pfam domain (PF02397) (12) (Table 1; see Fig. S1 in the supplemental material). In a few cps loci, the IT gene is not immediately downstream of the fourth regulatory gene (wze) because of the presence of an intervening gene or gene fragment and/or a rearrangement in the order of the first four cps genes. Thus, the gene order differs in the closely related cps loci of types 25A, 25F, and 38 and a defective transposase gene is between wze and wzg. Similarly, there is a defective transposase gene upstream of the IT gene (wciI) in the cps loci of types 12A and 46.
In S. pneumoniae type 1, there is no cps gene product harboring the Pfam domain PF02397 found in other ITs. Furthermore, none of the predicted gene products, other than Wzx and Wzy, have putative transmembrane segments, making it unlikely that the IT is encoded within the serotype 1 cps locus. The constituents of the type 1 CPS repeat unit are an AAT-Galp and two α-d-GalpA residues (38). The cps locus possesses the first four cps genes (wzg, wzh, wzd, and wze), the wzy and wzx genes, the ugd and gla genes for the biosynthesis of GalpA, and a set of rml genes (as mentioned earlier, rmlD is frameshifted and therefore rhamnose is absent from the CPS) (12, 63). Type 1 CPS is acetylated, and accordingly, there is also a putative acetyltransferase gene (wchC). The products of the remaining genes (wchB and wchD) both possess the PF00534 domain found in GTs and are presumably involved in the assembly of the trisaccharide repeat unit. Thus, there is no candidate gene that could encode an IT.
As mentioned previously, WbgY in S. sonnei and WcfS in B. fragilis are thought to be the ITs for AAT-Galp (21, 87), and in the S. pneumoniae genome, there is a WbgY/WcfS homolog (Spr1655; GenBank accession number AE007317) which possesses the PF02397 family domain found in ITs encoded within the other cps loci. Therefore, it is assumed that this chromosomally encoded IT provides the lipid-linked-AAT-Galp required for repeat unit synthesis in serotype 1 pneumococci. Coyne et al. (21) suggested that the pneumococcal WbgY homolog makes lipid-linked-AAT-Galp for initiating teichoic acid synthesis, but AAT-Galp does not appear to be the initial sugar in teichoic acid repeat units (9). We assume that WbgY is present on the chromosome as there is some other noncapsular pneumococcal polysaccharide that has AAT-Galp as the initial sugar.
WchA.
The IT WchA has been shown to transfer Glcp-1P from UDP-Glcp to a lipid carrier in the S. pneumoniae capsules of types 8, 9V, and 14 (42, 65, 78). The data obtained for serotype 2 (15) made it nearly certain that UndPP is the lipid carrier for initiation of capsule synthesis in pneumococci, as shown for several other bacterial polysaccharides. WchA has been shown to be essential for CPS biosynthesis in S. pneumoniae serotype 8 as unencapsulated variants possess frameshift mutations within wchA (also known as cap8E) (81). Glucose is assumed to be the donor sugar used by WchA in all of the S. pneumoniae serotypes where this IT is present. This is supported by the observation that glucose is present in the repeat units of all serotypes that possess wchA (see Fig. S1 in the supplemental material). Where there are two glucose residues in the main chain of the repeat unit (serogroup 18, serogroup 32, and serotypes 2, 7B, 8, 20, 9A, 9L, and 9V), the initial sugar can be determined by GT-linkage correlation of the second sugar transferase (see GT section below.)
WciI.
Serogroup 12 and serotypes 4, 5, 25A, 25F, 38, 44, 45, and 46 possess the putative IT gene wciI in their cps loci (see Fig. S1 in the supplemental material). Structures are known for types 4, 5, 12F, 12A, and 45, and for type 46 the sugar composition is known. l-FucpNAc is the only sugar common to all of these serotypes, but serotypes 25A, 25F, and 38 lack the l-FucpNAc pathway genes, suggesting that their CPS lacks this sugar. However, all but serotype 5 contain GalpNAc and/or GlcpNAc (serotypes 4, 12F, and 45 contain only GalpNAc, and only GlcpNAc is present in serotype 12A), and a possible scenario is that both GalpNAc-P and GlcpNAc-P can be transferred by WciI. There is a precedent in the IT WecA of E. coli and Y. enterolitica (4, 50, 92) that transfers GlcpNAc-P to initiate synthesis of the repeat unit of the enterobacterial common antigen but also acts as the IT for O antigens in E. coli that contain GlcpNAc or GalpNAc. In the case of WecA, the gene is outside of the O-antigen gene cluster and it is unlikely that WecA itself determines if GlcpNAc or GalpNAc is the initial sugar. However, as already mentioned, the transfer of a sugar phosphate to UndP to give a pyrophosphate linkage is reversible, and it is envisaged that both UndPP-GlcpNAc and UndPP-GalpNAc are formed, the next transferase determining which is permanently incorporated into the repeat unit. The reversibility of the first reaction ensures that no UndP is locked up as UndPP-GlcpNAc or UndPP-GalpNAc if it is not used in further reactions.
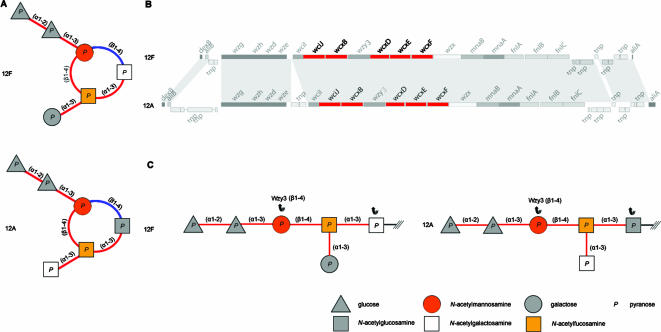
Besides correlating the presence of a particular sugar in the CPS repeat unit with the presence of an IT gene (as above), it is in some cases possible to predict the initial sugar (and thus the polymerase linkage) by correlating the nature of the linkages (either retaining or inverting) in the circularized repeat unit with the proposed nature of the GTs encoded by the cps locus from their CAZy family membership (see Materials and Methods). For example, the repeat units of serotypes 12F and 12A can be depicted as in Fig. 4A. The circular chemical structures each contain six linkages, three main chain and three side branch linkages. Of these six linkages, five are retaining and one is inverting. One of the main chain linkages is the Wzy linkage and if identified would also specify the initial sugar.
FIG. 4.
Assignment of Wzy polymerase linkage and IT specificity. (A) Circularized representation of CPS repeat unit structures for serotypes 12F and 12A, where the polymerase linkage and the initial sugar are not indicated. Each sugar is represented symbolically as in the report by Bentley et al. (12), and the nature of the linkage is represented by red if it is retaining and blue if it is inverting. Each circular structure contains five retaining linkages and a single inverting linkage. (B) Excepting remnant transposases, the cps clusters of serotypes 12F and 12A are syntenic, and both encode five GTs, all of which are retaining based on CAZy GT family membership (colored red). Shading between the two loci represents TBLASTX amino acid sequence similarity viewed by using ACT. (C) We can infer that the single inverting linkage in the circular structure (A) corresponds to the linkage made by the polymerase, which also allows us to define the initial sugar and depict the structures biologically, with the lipid-linked initial sugar depicted to the right of the structure. ///, lipid carrier.
Excepting remnant transposases, the cps loci of serotypes 12F and 12A are syntenic and are predicted to encode five GTs, all of which are members of retaining GT families (Fig. 4B). We can therefore suggest that the single inverting linkage in the circular chemical representation of the repeat unit represents the Wzy polymerization linkage, allowing us to define the initial sugar (and hence WciI specificity) as GalpNAc in 12F and GlcpNAc in 12A and also to define the polymerase linkages (Fig. 4C). This approach also suggests that GalpNAc is the initial sugar for WciI in serotypes 4 and 45, which is further supported by the presence of FucpNAc as the second sugar (see below).
If GlcpNAc-P and GalpNAc-P are the normal substrates for WciI, we have to account for serotype 5 CPS, which contains only Glcp, FucpNAc, PnepNAc, and KDQNAc, one of which must be the substrate for WciI. As mentioned above, the specificity of the second sugar transferase can determine which sugar is incorporated into the repeat unit.
In all cps loci where wciI is the putative IT gene and FucpNAc is also presumed to be present in the CPS (due to the presence of the fnlA, fnlB, and fnlC genes in the cps loci), the sixth gene encodes the GT, wciJ. In serotypes 25F, 25A, and 38, where wciI is also the putative IT gene but the sixth gene is not wciJ, FucpNAc is not predicted to be present in the repeat unit. Thus, WciJ should be the FucpNAc transferase that transfers the second sugar in the repeat unit. This allows us to infer that the initial sugar in serotype 5 is KDQNAc. Clearly, biochemical studies of WciI are required to evaluate our proposals for the initial sugars in these serotypes.
WcjH.
The cps loci of serotypes 29, 35F, 39, 43, and 47F possess wcjH, but only the CPS structure of type 29 is available and is similar to that of type 35B. There are differences in the acetylation patterns, due to the presence of an extra acetyltransferase gene (wciG) in type 35B, but apart from these differences, the CPS of types 29 and 35B differ in only one sugar (Galp and Glcp, respectively) and their cps loci have different IT genes (wcjH and wchA, respectively); since WchA transfers Glcp, it seems clear from this comparison that in serotype 29 WcjH transfers Galp.
WcjG.
The putative IT gene wcjG is found in the cps loci of serogroup 10 and serotypes 31, 33C, and 47A (see Fig. S1 in the supplemental material), but there are biochemical structures only for 10A, 10F, and 31. Galp, Galf, GalpNAc, and Rib-ol are the constituents of the pentasaccharide backbone of type 10A and 10F CPS, differing only in the anomeric status of Galp (α in 10F and β in 10A). Galf, Rhap, and GlcpA are the constituents of type 31 CPS (38), which might suggest that WcjG transfers Galf, but we are not confident in the latter CPS structure in the absence of more recent NMR data (8). Assignment of sugar specificity to the IT WcjH (see above) is based on a single monosaccharide difference between the two almost identical CPS structures of serotypes 29 and 35B. The second sugar in both of these structures is Galf linked to the initial sugar by a β1-3 linkage. Within the cps loci where WchA is the IT, there are six serotypes where the second sugar is Galf β1-3 linked to the initial sugar Glcp (17A, 20, 33F, 35A, 35B, and 34). Within the cps of these six serotypes, as well as serotype 29 (IT-WcjG) and serotypes 10F and 10A, the only putative GT gene in common is wciB, which in all cases is the sixth gene in the cps loci. Furthermore, wciB is not found in any other serotype where we have a different second sugar. We can therefore suggest that WciB transfers Galf as the second sugar in the repeat units. Serotype 10F contains a single Galf(β1-3) linkage to Galp. Serotype 10A contains two Galf(β1-3) linkages; however, one is a side branch (d-Galf-(β1-3)-d- GalpNAc) and not part of the circular main chain; therefore, we can suggest that Galf is the second sugar and also propose with confidence that WcjG transfers Galp.
Repeat unit polymerases (Wzy) and flippases (Wzx).
The repeat unit polymerase (Wzy) and flippase (Wzx) define a polymerization/export process (Fig. 1), and in polysaccharide biosynthetic loci these genes almost invariably occur together (86). There is experimental evidence in Y. enterocolitica serotype O:8 that Wzy is strictly specific for the O unit to be polymerized, whereas Wzx has a relaxed specificity for the translocated polysaccharide (11). However, recently it has been reported that the Wzx flippases involved in biosynthesis of O antigen correlate with the first sugar of the O-specific lipopolysaccharide subunit (50) but otherwise seem to lack specificity at least in the situation where only one potential substrate is available.
There is enormous sequence divergence among the pneumococcal repeat unit polymerases (Wzy), but most have a best match to other Wzy proteins in a BLAST search and have about 10 putative transmembrane segments (12). The pneumococcal Wzy repeat unit polymerases fall into 40 HGs, which we refer to as Wzy-1 to Wzy-40. For their association with particular ITs, the putative linkages that they catalyze, and the serotypes in which they are present, see Table S1 in the supplemental material. The pneumococcal flippases (Wzx) are similarly divergent but are identified by their similarity to other Wzx proteins in BLAST searches and by having about 12 transmembrane segments (12). The pneumococcal Wzx flippases fall into 13 HGs (Wzx-1 to Wzx-13), although 40 cps loci encode a Wzx-1 (HG7) flippase, but in pneumococci there is no apparent association between the pneumococcal Wzx HGs and the IT HGs or the putative initial sugars (data not shown).
Following prediction of the polymerase linkage in the 52 known repeat unit structures that are synthesized by the Wzy-dependent pathway (see above), it can be seen that, in some cases, the same polymerase linkage is formed by members of different HGs (see Table S1 in the supplemental material). For example, the d-Glcp-(α1-3)-α-l-Rhap linkage is formed by Wzy-12 in types 6A and 6B and by Wzy-28 in type 19A, the d-Glcp-(β1-3)-α-d-Galp linkage is formed by Wzy-14 in types 15A/15F and by Wzy-16 in types 17A/17F, and the d-Glcp-(β1-4)-β-d-Galf linkage is formed by Wzy-6 in serogroup 18 and by Wzy-19 in type 23F. The IT is WchA, and Glcp is present in the CPS structures in all of these cases; therefore, we are very confident in the assignment of the above polymerization linkages.
Assignment of GTs.
GTs catalyze the formation of glycosidic bonds between the lipid-linked glycan precursor as an acceptor and a nucleotide-activated sugar as a donor. Thus, GTs determine the sequence of components of the repeat units of polysaccharides and the characterization of the specificity of GTs would enable the prediction of the constituents of repeat units where these are unknown (Fig. 1) (35, 39).
Functional characterization of pneumococcal GTs is extremely limited, however, and has only been reported for types 8 and 14 (42, 69). The reaction specificities of the GTs may vary for the acceptor or donor monosaccharide residues and the linkage between them, and there are instances reported where a single nucleotide replacement can alter these specificities (41). For example, in pneumococci, a single amino acid difference in a GT has been reported to be responsible for the α-1,3 linkage of rhamnose to ribitol in serotype 6A but an α-1,4 linkage in serotype 6B (52). Prediction of the linkage catalyzed by a GT is therefore problematic and must be considered tentative until biochemical data are available. However, the sequences of the cps loci of all 88 S. pneumoniae serotypes that use the Wzy-dependent pathway, along with the 52 published CPS structures from these serotypes, present an opportunity to utilize comparative methods to tentatively assign ab initio the linkages within these structures catalyzed by the GTs (see Materials and Methods) and in many cases allow us to assign slightly different linkages to the same HG with some confidence.
Given the great diversity of sugars and sugar linkages among the pneumococcal CPS repeat units, it is not surprising that the GTs form a divergent group of 92 HGs (including three hypothetical proteins, WcwD, WcrT, and WcwX, which we predict to act as GTs), the majority of which fall into three Pfam families (PF00534, PF00535, and PF05704) (Table 2). GTs have been classified into CAZy families based on high sequence similarity with one or more founding members of the family with experimentally demonstrated GT activity (20). Members of CAZy families appear to be either retaining GTs, forming glycosidic bonds with stereochemistry identical to that of the donor sugar (for example, UDP-glucose→α-glucoside) or inverting GTs which in forming the glycosidic linkage alter the stereochemistry of the donor sugar (for example, UDP-glucose→β-glucoside) (20, 46). Pfam families were highly correlated with CAZy families (Table 2), and CAZy family assignment proved a useful contributor to our putative functional assignments, as linkages that are inverting or retaining should be catalyzed by GTs that, from their CAZy family membership, are expected to be inverting or retaining.
TABLE 2.
GT genes and the linkages their gene products are predicted to catalyzea
| GT gene | Pfam | CAZy | Mechanism | No. of occurrences of GT gene | Structure (donor, linkage, acceptor) | No. of occurrences where CPS structure known | Serotype(s) possessing GT gene | No. of occurrences, where structure known, that have the linkage | No. of occurrences of linkage where GT gene is absent | Note(s) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wchF | PF00534 | 4 | RET | 27 | l-Rhap | β1-4 | β-d-Glcp | 14 | 2, 7F, 7A, 7B, 17F, 18F, 18A, 18B, 18C, 22F, 23F, 27, 32F, 32A | 14 | 0 | A1 | ||
| wciB | 24 | d-Galf | β1-3 | α-d-Galp | 10 | 10F | 1 | 0 | A2, A3 | |||||
| d-Galf | β1-3 | β-d-Galp | 10A, 29 | 2 | 1 | |||||||||
| d-Galf | β1-3 | β-d-Glcp | 17A, 20, 33F, 35A, 35B | 5 | 0 | |||||||||
| d-Galf | β1-3 | α-d-Glcp | 34 | 1 | 0 | |||||||||
| Structure uncertain | 31 | |||||||||||||
| wcrC | PF00534 | 4 | RET | 13 | d-Galp | α1-2 | d-Rib-ol | 4 | 10F, 10A, 33B, 34 | 4 | 0 | A4 | ||
| wciF | PF00535 | 2 | INV | 13 | d-GalpNAc | β1-3 | α-d-Galp | 5 | 10F, 10A | 2 | 0 | A6 | ||
| d-GlcpNAc | β1-4 | d-Rib-ol | 13 | 1 | 0 | |||||||||
| d-GalpNAc | β1-4 | α-d-Galpor | 33B | 1 | 1 | |||||||||
| d-Galf | β1-3 | β-d-GalpNAc | ||||||||||||
| d-Galp | β1-3 | α-d-Galpor | 33F | 1 | 1 | |||||||||
| d-Galf | β1-3 | β-d-Galp | ||||||||||||
| wchJ | PF08660 | 12 | GT enhancer | A7 | ||||||||||
| wchK | PF04101 | 1 | INV | 12 | d-Galp | β1-4 | β-d-Glcp | 10 | 11F, 11A, 11B, 11C, 13 14, 15F, 15A, 15B, 15C | 10 | 0 | A7 | ||
| wcrD | PF00535 | 2 | INV | 10 | d-Galf | β1-3 | β-d-GalpNAc | 3 | 10A | 1 | 1 | A5 | ||
| d-Galf | β1-4 | β-d-GlcpNAc | 13 | 1 | 0 | |||||||||
| d-Galf | β1-3 | α-d-Galp | 34 | 1 | 1 | |||||||||
| wchO | PF03808 | 26 | INV | 9 | d-ManpNAc | β1-4 | β-d-Glcp | 8 | 9A, 9L, 9N, 9V, 19B, 19C | 6 | 0 | A8 | ||
| d-ManpNAc | β1-4 | α-d-Glcp | 19F, 19A | 2 | 0 | |||||||||
| d-ManpNAc | β1-4 | α-l-Rhap | 19B, 19C | 2 | 0 | |||||||||
| wcrH | INV | 9 | d-Galf | β1-1 | d-Rib-ol | 4 | 29, 35B | 2 | 0 | A9 | ||||
| d-Galf | β1-1 | Man-ol | 35A | 1 | 0 | |||||||||
| d-Galf | β1-6 | α-d-Galp | 10F | 1 | 0 | |||||||||
| wciJ | PF00534 | 4 | RET | 8 | l-FucpNAc | α1-3 | β-d-GlcpNAc | 5 | 12A | 1 | 0 | A10 | ||
| l-FucpNAc | α1-3 | α-d-GalpNAc | 4 | 1 | 0 | |||||||||
| l-FucpNAc | α1-3 | β-d-GalpNAc | 12F, 45 | 2 | 0 | |||||||||
| l-FucpNAc | α1-3 | β-d-KDQNAc | 5 | 1 | 0 | |||||||||
| wciE | PF05704 | 32 | RET | 7 | d-Galp | α1-2 | α-d-Galp | 2 | 33F, 33B | 2 | 0 | A6 | ||
| wciU | PF00534 | 4 | RET | 7 | d-GlcpNAc | α1-3 | β-l-Rhap | 4 | 18A | 1 | 0 | A11 | ||
| d-Glcp | α1-3 | β-l-Rhap | 18F, 18B, 18C | 3 | 1 | |||||||||
| wciD | PF00535 | 2 | INV | 6 | Assignment uncertain | 3 | 20 | A25 | ||||||
| d-GalpNAc | β1-4 | α-d-Galpor | 33B | 1 | 1 | A6 | ||||||||
| d-Galf | β1-3 | β-d-GalpNAc | ||||||||||||
| d-Galp | β1-3 | α-d-Galpor | 33F | 1 | 1 | |||||||||
| d-Galf | β1-3 | β-d-Galp | ||||||||||||
| wchQ | PF00535 | 2 | INV | 6 | l-Rhap | α1-P-4 | β-d-ManpNAc | 6 | 19F, 19A, 19B, 19C | 4 | 0 | A8 | ||
| l-Rhap | α1-P-2 | β-d-ManpNAc | 32A, 32F | 2 | 0 | |||||||||
| wcxB | PF00534 | 4 | RET | 6 | d-Galp | α1-3 | α-l-FucpNAc | 3 | 12F, 45 | 2 | 0 | A12 | ||
| d-GalpNAc | α1-3 | α-l-FucpNAc | 12A | 1 | 0 | |||||||||
| wcyK | PF00534 | 4 | RET | 6 | d-Galp | α1-3 | β-d-Galp | 4 | 11F, 11A, 11B, 11C | 4 | 0 | A15 | ||
| wcwA | PF00534 | 4 | RET | 5 | d-Galp | α1-3 | β-l-Rhap | 3 | 7F, 7A | 2 | 1 | A13 | ||
| d-Glcp | α1-3 | β-l-Rhap | 22F | 1 | 1 | A14 | ||||||||
| wcxD | PF00534 | 4 | RET | 5 | Assignment uncertain | 2 | 12F, 12A | A12 | ||||||
| wcxE | PF00534 | 4 | RET | 5 | Assignment uncertain | 2 | 12F, 12A | A12 | ||||||
| wcxF | PF00534 | 4 | RET | 5 | Assignment uncertain | 2 | 12F, 12A | A12 | ||||||
| wcrL | PF04488 | 32 | RET | 5 | d-GlcpNAc | α1-4 | α-d-Galp | 4 | 11F, 11B, 11C | 3 | 0 | A15 | ||
| d-Glcp | α1-4 | α-d-Galp | 11A | 1 | 1 | |||||||||
| wchL | PF00535 | 2 | INV | 5 | d-GlcpNAc | β1-3 | β-d-Galp | 5 | 14, 15F, 15A, 15B, 15C | 5 | 0 | A16 | ||
| wchM | PF00535 | 2 | INV | 5 | d-Galp | β1-4 | β-d-GlcpNac | 5 | 14, 15F, 15A, 15B, 15C | 5 | 0 | A16 | ||
| wchN | PF05704 | 32 | RET | 5 | d-Galp | α1-2 | β-d-Galp | 4 | 15F, 15A, 15B, 15C | 4 | 0 | A17 | ||
| wciN | PF01501 | 8 | RET | 5 | d-Galp | α1-3 | α-d-Glcp | 3 | 6A, 6B | 2 | 0 | A18 | ||
| Nonfunctional | 33B | A6 | ||||||||||||
| wcjA | PF00534 | 4 | RET | 5 | Assignment uncertain | 4 | 9A, 9L, 9N, 9V | A19 | ||||||
| wcjB | 4 | Assignment uncertain | 4 | 9A, 9L, 9N, 9V | A19 | |||||||||
| wcjC | PF00534 | 4 | RET | 4 | Assignment uncertain | 4 | 9A, 9L, 9N, 9V | A19 | ||||||
| wciV | PF00535 | 2 | INV | 4 | d-Galp | β1-4 | α-d-GlcpNac | 4 | 18A | 1 | 1 | A11 | ||
| d-Galp | β1-4 | α-d-Glcp | 18F, 18B, 18C | 3 | 0 | |||||||||
| wciW | PF05704 | 32 | RET | 4 | d-Glcp | α1-2 | β-d-Galp | 4 | 18F, 18A, 18B, 18C | 4 | 0 | A11 | ||
| wcrP | PF00535 | 2 | INV | 4 | d-GlcpA | β1-3 | β-d-Galfor | 2 | 17A, 31 | 1 | 0 | A20 | ||
| l-Rhap | α1-4 | β-d-GlcpA | ||||||||||||
| wcrR | PF00535 | 2 | INV | 4 | d-GlcpA | β1-3 | β-d-Galfor | 2 | 17A, 31 | 1 | 1 | A20 | ||
| l-Rhap | α1-4 | β-d-GlcpA | ||||||||||||
| wcyS | PF05704 | 32 | RET | 4 | d-Galp | α1-4 | β-l-Rhap | 3 | 27 | 1 | 0 | A21 | ||
| d-Glcp | α1-4 | β-l-Rhap | 32F, 32A | 2 | 0 | |||||||||
| wcrW | 4 | RET | 3 | Structure uncertain | 1 | 31 | ||||||||
| wcxU | PF00535 | 2 | INV | 3 | Assignment uncertain | 1 | 7B | A22 | ||||||
| wcyA | PF00534 | 4 | RET | 3 | No structure | 0 | ||||||||
| wcyB | PF00535 | 2 | INV | 3 | No structure | 0 | ||||||||
| wcyC | PF00534 | 4 | RET | 3 | No structure | 0 | ||||||||
| wcyD | 3 | No structure | 0 | |||||||||||
| wcrQ | PF05704 | 32 | RET | 3 | d-Glcp | α1-2 | β-d-Galf | 1 | 17A | 1 | 0 | A23 | ||
| wcrI | PF00535 | 2 | INV | 3 | d-Galp | β1-3 | β-d-Galf | 1 | 35A | 1 | 0 | A24 | ||
| wcrK | PF05704 | 32 | RET | 3 | No structure | 0 | ||||||||
| wcxI | PF00535 | 2 | INV | 3 | No structure | 0 | ||||||||
| wcxJ | 3 | No structure | 0 | |||||||||||
| wcxN | 1 and 2 | INV/INV | 3 | No structure | 0 | |||||||||
| wciL | PF00534 | 4 | RET | 3 | Assignment uncertain | 3 | 4, 20, 45 | A25 | ||||||
| wcrG | PF02485 | 14 | INV | 3 | d-Galp | β1-6 | β-d-GalpNAc | 1 | 10A | 1 | 0 | A5 | ||
| wciP | PF00535 | 2 | INV | 3 | l-Rhap | α1-2 | d-Ara-ol | 3 | 17F | 1 | 0 | A26 | ||
| l-Rhap | α1-3 | d-Rib-ol | 6A | 1 | 0 | |||||||||
| l-Rhap | α1-4 | d-Rib-ol | 6B | 1 | 0 | |||||||||
| wciC | PF00534 | 4 | RET | 3 | d-Galp | α1-3 | β-d-Galf | 1 | 33F | 1 | 0 | A6 | ||
| wchV | PF00535 | 2 | INV | 3 | d-Galp | β1-4 | β-l-Rhap | 1 | 23F | 1 | 2 | A14, A27 | ||
| wchW | 3 | l-Rhap | α1-2 | β-d-Galp | 1 | 23F | 1 | 0 | A28 | |||||
| wcwI | PF00535 | 2 | INV | 3 | Assignment uncertain | 1 | 7B | A22 | ||||||
| wcwL | PF00534 | 4 | RET | 3 | Assignment uncertain | 1 | 7B | A22 | ||||||
| wcwX | 2 | Assignment uncertain | 1 | 22F | A29 | |||||||||
| wcwV | PF00534 | 4 | RET | 2 | Assignment uncertain | 1 | 22F | A29 | ||||||
| whaB | PF00535 | 2 | INV | 2 | Assignment uncertain | 1 | 22F | 1 | 1 | A29 | ||||
| wchR | PF00534 | 4 | RET | 2 | d-Ribf | β1-4 | α-l-Rhap | 2 | 19B, 19C | 2 | 1 | A8 | ||
| wchS | PF00535 | 2 | INV | 2 | l-Rhap | α1-3 | β-d-ManpNAc | 2 | 19B, 19C | 2 | 0 | A8 | ||
| wcxS | PF00535 | 2 | INV | 2 | l-Rhap | α1-3 | α-d-Galp | 1 | 45 | 1 | 0 | A30 | ||
| wcrM | PF00535 | 2 | INV | 2 | d-GalpNAc | β1-6 | β-d-Galf | 2 | 29, 35B | 2 | 0 | A31, A9 | ||
| wcrV | PF00535 | 2 | INV | 2 | d-Galp | β1-4 | β-l-Rhap | 2 | 17F, 17A | 2 | 1 | A14 | ||
| PF05704 | 32 | RET | d-Galp | α1-3 | β-l-Rhap | 17F, 17A | 2 | 2 | ||||||
| wcyE | 2 | No structure | 0 | |||||||||||
| wcwD | 2 | d-Galp | β1-2 | α-d-Galp | 2 | 7F, 7A | 1 | 0 | A32 | |||||
| wcwF | PF00535 | 2 | INV | 2 | d-GalpNAc | β1-6 | α-d-Galpor | 2 | 7F, 7A | 2 | 0 | A33 | ||
| l-Rhap | α1-4 | β-d-GalpNAc | ||||||||||||
| wcwG | 2 | d-GalpNAc | β1-6 | α-d-Galpor | 2 | 7F, 7A | 2 | 0 | A33 | |||||
| l-Rhap | α1-4 | β-d-GalpNAc | ||||||||||||
| wcwH | PF00534 | 4 | RET | 2 | d-GlcpNAc | α1-2 | α-l-Rhap | 2 | 7F, 7A | 2 | 1 | A34 | ||
| wcyM | PF00534 | 4 | RET | 2 | No structure | 0 | ||||||||
| wcyN | PF00535 | 2 | INV | 2 | No structure | 0 | ||||||||
| wcrT | 2 | l-Rhap | β1-4 | α-l-Rhap | 2 | 17F, 17A | 2 | 0 | A35 | |||||
| wcyQ | PF00534 | 4 | RET | 1 | Assignment uncertain | 1 | 45 | |||||||
| wcyT | PF00535 | 2 | INV | 1 | No structure | 0 | ||||||||
| wcyU | PF00535 | 2 | INV | 1 | No structure | 0 | ||||||||
| wcyV | 1 | No structure | 0 | |||||||||||
| wchU | PF00534 | 4 | RET | 1 | d-Glcp | β1-6 | β-d-ManpNAc | 1 | 19C | 1 | 0 | A8 | ||
| wchG | PF00535 | 2 | INV | 1 | l-Rhap | α1-3 | β-l-Rhap | 1 | 2 | 1 | 0 | A36 | ||
| wchH | PF00534 | 4 | RET | 1 | d-Glcp | α1-2 | α-l-Rhapor | 1 | 2 | 1 | 0 | A36 | ||
| d-GlcpA | α1-6 | α-d-Glcp | ||||||||||||
| wchI | PF00534 | 4 | RET | 1 | d-Glcp | α1-6 | α-l-Rhapor | 1 | 2 | 1 | 0 | A36 | ||
| d-GlcpA | α1-6 | α-d-Glcp | ||||||||||||
| wciS | PF00534 | 4 | RET | 1 | d-Galp | α1-4 | β-d-GlcpA | 1 | 8 | 1 | 0 | A37 | ||
| wciT | PF04488 | 32 | RET | 1 | d-Glcp | α1-4 | α-d-Galp | 1 | 8 | 1 | 1 | A37 | ||
| wchB | PF00534 | 4 | RET | 1 | d-GalpA d-GalpA | α1-3 α1-3 | AAT-α-d-Galpor d-GalpA | 1 | 1 | 1 | 0 | A38 | ||
| wchD | PF00534 | 4 | RET | 1 | d-GalpA d-GalpA | α1-3 α1-3 | AAT-α-d-Galpor d-GalpA | 1 | 1 | 1 | 0 | A38 | ||
| wciK | 1 | Assignment uncertain | 1 | 4 | A25 | |||||||||
| whaC | PF00535 | 2 | INV | 1 | Assignment uncertain | 1 | 5 | 1 | 0 | A39 | ||||
| whaD | PF00534 | 4 | RET | 1 | Assignment uncertain | 1 | 5 | A39 | ||||||
| whaE | 1 | Assignment uncertain | 1 | 5 | A39 | |||||||||
| whaF | PF05704 | 32 | RET | 1 | Assignment uncertain | 1 | 20 | A25 | ||||||
| whaJ | PF00535 | 2 | INV | 1 | Assignment uncertain | 1 | 20 | A25 | ||||||
| whaK | PF00535 | 2 | INV | 1 | d-GlcpNAc | β1-3 | α-d-Galp | 1 | 27 | 1 | 0 | A40 | ||
| whaM | PF00535 | 2 | INV | 1 | No structure | 0 | ||||||||
| wciQ | PF08660 | 1 | INV | 1 | GT enhancer | 1 | A7, A37 | |||||||
| wciR | PF04101 | 1 | INV | 1 | d-GlcpA | β1-4 | β-d-Glcp | 1 | 8 | 1 | 0 | A7, A37 | ||
| wcyP | 1 | No structure | 0 | |||||||||||
| wcwY | 1 | No structure | 0 | |||||||||||
| wcxT | PF00535 | 1 | No structure | 0 | ||||||||||
Shown are the Pfam domains and CAZy families of the GT genes and the linkages the gene products are predicted to catalyze. GT genes are ordered according to their frequency. The Pfam and CAZy families assigned to their products are shown, along with the linkage(s) in the repeat unit structures that they are predicted to catalyze (where an assignment can be made). In some cases, different linkages are assigned to the same GT in different serotypes. The number of times the predicted linkage is found in the CPS of those serotypes where the GT gene is present is shown, as is the number of times the linkage is found in serotypes that do not have the GT gene. The notes in the rightmost column refer to Table S2 in the supplemental material, where the reasons for the GT assignments are discussed. INV, inverting; RET, retaining.
The four main methods of assigning GTs to specific linkages are described in Materials and Methods. The linkages tentatively assigned to GTs are shown in Table 2; for the basis of these assignments, see the notes to Table S2 in the supplemental material. Using the GT-linkage correlation approach, the most prevalent GT gene in cps loci of serotypes whose CPS structures are known was wchF (14 occurrences). Among these 14 structures, there are 29 different triplet linkages in their repeat units but the only one in common is l-Rhap(β1-4)d-Glcp; as all 14 of the known structures contain this linkage, no other linkage in these structures correlates with the presence of wchF, and the above linkage is not found in any serotype that lacks wchF, we assign the synthesis of l-Rhap(β1-4)d-Glcp to WchF. There was also a perfect correlation between the presence of wchK (10 occurrences) and that of d-Galp(β1-4)-β-d-Glcp and between the presence of wchO (8 occurrences) and that of d-ManpNAc(β1-4)-β-d-Glcp.
In other cases, GTs can be assigned on the basis of the predicted retaining or inverting status of the GTs encoded within the cps loci and the inverting and retaining linkages in the CPS. For example, the repeat units of serotypes 6A and 6B each contain two GT linkages, of which one is retaining and one is inverting. The cps loci each contain a single gene, wciN, encoding a retaining GT, which is therefore assigned to the retaining linkage, d-Galp(α1-3)-α-d-Glcp, in both serotypes 6A and 6B. The inverting GT, WciP, can therefore be assigned to the inverting linkage, l-Rhap(α1-3)-d-Rib-ol in serotype 6A and l-Rhap(α1-4)-d-Rib-ol in serotype 6B, as also suggested by a completely different approach (52). Similarly, the repeat units of serogroup 15 each contain a single retaining linkage (d-Galp-(α1-2)-β-d-Galp) and their cps loci each contain a single gene predicted to encode a retaining GT (WchN).
Gene content-linkage comparisons between similar serotypes can also be used to assign GT specificity. For example, the repeat unit structures of serotypes 19B and 19C differ only by the presence of a single d-Glcp(β1-6)β-d-ManpNAc side branch in 19C, which can be attributed to the single additional GT gene in the cps locus of this serotype (wchU; see the notes to Table S2 in the supplemental material). Similarly, the cps loci of serotypes 7F and 7A are syntenic and only differ in the presence of a frameshift mutation in the GT gene wcwD in serotype 7A. The CPS structures of both are known and differ only in the presence of an additional d-Galp(β1-2)α-d-Galp side branch in 7F, which allows us to attribute the side branch linkage to the action of WcwD.
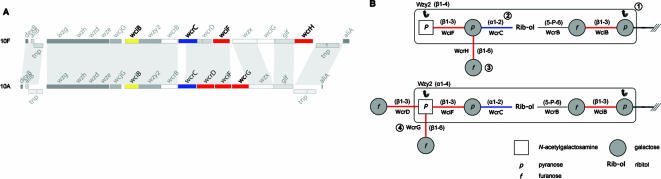
Figure 5 depicts an example where a combination of methods can assign GT specificity and therefore propose a biosynthetic pathway for the repeat unit structures of serotypes 10F and 10A. Following the identification of the polymerase linkage and therefore the initial sugar (see above; Fig. 5B-1), the three GT genes wciB, wcrC, and wciF (and the ribitol phosphate transferase gene wcrB) are common to both cps loci (Fig. 5A) and presumably account for the three GT linkages (and the ribitol phosphate linkage, which is 5-P-5 linked in 10A but 5-P-6 linked in 10F; see below) in common in the main chain of the repeat units. The single retaining linkage in the structure, Galp(α1-2)Rib-ol, can be assigned to the action of the single retaining GT, WcrC (Fig. 5B-2). WciB has been assigned as the Galf(β1-3) transferase that adds the second sugar in the repeat unit through GT-linkage correlation, and so we can assign the action of WciF to the remaining d-GalpNAc(β1-3)α-d-Galp linkage in the main chain. The side branch linkage Galf(β1-6)Galp present in 10F but not in 10A can be accounted for by the presence in 10F, but not in 10A, of the GT gene wcrH (Fig. 5B-3). The two remaining side branch linkages in 10A can be attributed to the two additional GTs, encoded by wcrD and wcrG (Fig. 5B-4), which are not found or not intact (wcrD is frameshifted in type 10F) in the cps locus of 10F. The presence of wcrD in other serotypes correlates with a Galf(β1-3) linkage, and therefore WcrD is assigned as catalyzing this linkage; WcrG, by a process of elimination, catalyzes the linkage of the other Galp(β1-6) side branch.
FIG. 5.
Assignment of GT specificity. (A) cps loci of serotypes 10A and 10F are represented linearly with genes encoding GTs colored according to their CAZy membership—red for a GT with a predicted retaining mechanism, blue for an inverting mechanism, and yellow if the GT has not been assigned to a CAZy family. Shading between the two loci represents TBLASTX amino acid sequence similarity viewed by using ACT. (B) Structural representation of CPS repeat units. Glycosidic linkages are colored according to whether they require a retaining mechanism (red) or an inverting mechanism (blue). ///, lipid carrier. See the text for methods of GT assignment.
Sometimes the evidence for GT specificity can be twofold. For example, the CPS structures of serogroup 11 contain three GT linkages, one inverting and two retaining. The cps loci each contain three GT genes which encode one inverting GT (WchK) and two retaining GTs (WcyK and WcrL). We can therefore assign WchK to the inverting linkage in the structure, and as we have already assigned this link to WchK based on GT-linkage correlation, our confidence in the assignment is further strengthened.
In some cases, the donor sugar appears to differ for GTs in the same HG. For example, WcxB uses two different sugars as donors, GalpNAc in type 12A but Galp in type 12F (12). Similarly, the donor sugar for WcrL is GlcpNAc in types 11F, 11B, and 11C but Glcp in type 11A, and WciU adds Glcp in types 18B, 18C, and 18F but GlcpNAc in type 18A (Table 2). There are also instances where the same linkage is present in the repeat units of CPS of different serogroups, but there is no common GT gene in the cps loci, suggesting that GTs of different HGs can catalyze the same linkage. For example, WciT of serotype 8 has been proposed to catalyze the d-Glcp-(α1-4)-α-d-Galp linkage (69) and the same linkage is present in serotype 11A CPS, where we have suggested it is catalyzed by WcrL (Table 2; see the notes to Table S2 in the supplemental material). The donor sugar for WcrL is Glcp in type 11A but GlcpNAc in types 11F, 11B, and 11C.
Using the methods outlined above, we have been able to assign the GTs that catalyze 145 of the 200 glycosidic linkages in the available repeat unit structures, and in 32 of the 52 structures that use the Wzy-dependent pathway we have been able to assign all of the linkages (110 in total), with the remaining structures containing some linkages that could not be assigned to a specific GT (see Table S2 in the supplemental material) by any of the methods. In some cases, this was due to the presence of a greater number of GT genes than there are GT linkages in the repeat unit; for example, serotype 33B contains four GT linkages but the cps locus encodes five GTs. Conversely, there are instances where there are fewer GT genes in the cps locus than GT linkages in the repeat unit, for example, serotype 7B. In other cases, lack of assignment is due to ambiguous linkage correlation. For example, within the type 12F and 12A repeat units there are three unassigned retaining linkages and within the cps loci three remaining genes that encode retaining GTs, WcxD, WcxE, and WcxF. No CPS structures are available for the other serotypes that contain these genes and therefore there is no method of assigning their specificity.
For a few GTs, the assignments include linkages that are different and in some cases surprisingly so. These merit further study to establish if they are correct and, if so, the basis for the variation in linkage. For example, WcrV is credited with both a β(1-4) inverting linkage and an α(1-3) retaining linkage.
It has been suggested that the order in which GTs act to synthesize the repeat unit correlates with their order in the cps locus. This study allowed a more systematic examination of this suggestion and showed that the relationship held in 25/32 (78%) cps loci where we could assign all of the GTs (see Table S2 in the supplemental material), possibly suggesting that GTs might interact as part of a protein complex (22) that synthesizes the repeat unit. However, in this paper we chose not to use this correlation to assign GTs to linkages in those serotypes where this could not be done by any of the methods we have applied, and these unassigned GTs are in parentheses (see Fig. S1 in the supplemental material) to denote that we could not assign them to the linkages.
Assignment of sugar/polyalcohol phosphate transferases.
Most GTs involved in CPS biosynthesis transfer a sugar from the NDP-linked precursor to the lipid-linked acceptor sugar, but there are several cases where there is a phosphodiester linkage and presumably a sugar phosphate is transferred. The polyalcohols Rib-ol, Ara-ol, Man-ol, Gro, and Cho are also incorporated into the repeat units via phosphodiester linkages. The cps loci in each of the above cases have a gene product that falls into one of three Pfam families (PF04464, PF04991, and PF01066), at least some of whose members are known to make phosphodiester linkages (Table 3). As most known CPS structures have a single phosphodiester linkage in the repeat unit and a single cps gene encoding a protein in one of the above Pfam families, we can often assign the sugar/polyalcohol transferase.
TABLE 3.
Distribution of sugar/polyalcohol phosphate transferases into Pfam families and HGs and occurrence in serotypes and assignment of the putative linkages that they catalyze
| Pfam family and gene | Gene frequency | Donor | Linkage | Acceptor | Serotype(s) | ||
|---|---|---|---|---|---|---|---|
| PF04991 | |||||||
| wcxG | 5 | d-Ara-ol | 1-P-3 | β-l-Rhap | 17F | ||
| No structure | 24A, 24B, 24F, 48 | ||||||
| wcrO | 4 | d-Rib-ol | 5-P-3 | β-d-Galf | 34 | ||
| No structure | 33C, 35F, 36 | ||||||
| whaI | 4 | No structure | 39, 43, 47A, 47F | ||||
| wcrB | 4 | d-Rib-ol | 5-P-5 | β-d-Galf | 10A | ||
| d-Rib-ol | 5-P-6 | β-d-Galf | 10F | ||||
| No structure | 10B, 10C | ||||||
| wchP | 4 | l-Rhap | 1-P-4 | β-d-ManpNAc | 19A, 19B, 19C, 19F | ||
| wcxK | 3 | No structure | 24A, 24B, 24F | ||||
| wcrN | 3 | Cho | P-2 | β-l-Rhap | 27 | ||
| Cho | P-3 | β-l-Rhap | 32A, 32F | ||||
| wcxQ | 3 | No structure | 16F, 28A, 28F | ||||
| wcxR | 1 | No structure | 16A | ||||
| wcyR | 1 | Gro | 1-P-6 | β-d-GlcpNAc | 45 | ||
| PF04464 | |||||||
| wchX | 7 | Gro | 2-P-3 | β-d-Galp | 15A, 15B, 15C, 15F, 23F | ||
| No structure | 23A, 23B | ||||||
| wcwU | 5 | Gro | 1-P-4 | α-d-Glcp | 11A | ||
| Gro | 1-P-4 | α-d-GlcpNAc | 11C | ||||
| Rib-ol | 1-P-4 | α-d-GlcpNAc | 11B, 11F | ||||
| No structure | 11D | ||||||
| wciY | 5 | No linkage | 14 | ||||
| Gro | 1-P-3 | β-d-Galp | 18A, 18B, 18C, 18F | ||||
| wcrJ | 5 | Rib-ol | 5-P-4 | β-d-GalpNAc | 29, 35B | ||
| Man-ol | 6-P-3 | β-d-Galp | 35A | ||||
| No structure | 35C, 42 | ||||||
| wcxP | 3 | No structure | 16F, 28A, 28F | ||||
| whaG | 1 | Rib-ol | 5-P-4 | β-d-Galp | 13 | ||
| PF01066 | |||||||
| wciO | 4 | Rib-ol | 5-P-2 | α-d-Galp | 6A, 6B | ||
| Rib-ol | 5-P-6 | β-d-Glcp | 33B | ||||
| No structure | 33D | ||||||
| No Pfam hit | |||||||
| wcwK | 5 | d-Glcp | 1-P-6 | α-d-Glcp | 7B | ||
| d-GlcpNAc | 1-P-6 | α-d-Glcp | 20 | ||||
| No structure | 7C, 21, 40 | ||||||
| wcyI | 1 | l-Rhap | 1-P-2 | a-d-Galp | 32A, 32F | ||
In some instances, phosphodiester linkages are present in the CPS structures but there were no cps gene products possessing the Pfam domains of sugar phosphate transferases. For example, the CPS structures of types 7B and 20 possess an α-Glcp-1P linkage and an α-GlcpNAc-1P linkage, respectively, and the wcwK gene is present in both cps loci, but neither WcwK nor any other cps gene product contains one of the above sugar phosphate transferase Pfam domains. The conserved hypothetical protein WcwK is also present in types 7C, 40, and 21, but their CPS structures are unknown. WcwK of type 21 is 95% similar to WefC encoded by the streptococcal receptor polysaccharide gene cluster of S. gordonii, which has been proposed to be the α-Gal-1P transferase that forms the phosphodiester linkage in the middle of the repeat unit (89). Therefore, we suggest that WcwK may function as a α-Glcp-1P or α-GlcpNAc-1P transferase.
There are two phosphodiester linkages in serogroup 32 CPS, the Cho-P-3-β-l-Rhap linkage and the α-l-Rhap-(1-P-2)-α-d-Galp linkage. The incorporation of Cho is proposed to be catalyzed by WcrN, as in serotype 27, with amino acid differences in this protein accounting for the P-3 linkage in serogroup 32 compared to the P-2 linkage in serotype 27. There are no other gene products in serogroup 32 with the Pfam domains PF04464, PF04991, and PF01066, but there is a conserved hypothetical protein (WcyI) which could function as the second sugar phosphate transferase for the formation of the α-l-Rhap-(1-P-2)-α-d-Galp linkage. The assignments of the sugar/polyalcohol phosphate transferases to the phosphodiester linkages in the CPS structures are summarized in Table 3.
Assignment of acetyltransferases.
In this section, we consider only O-acetyl groups as, in general, N-acetyl sugars are transferred from NDP-N-acetyl precursors, each of which has a defined pathway (Fig. 2), whereas O-acetyl groups are commonly added to sugars by transferases independent of the NDP-sugar pathway. Several studies indicate that the O-acetyl groups in pneumococcal repeat units can be immunologically important, for example, in serotypes 1, 11A, 15F, and 34, whereas they appear not to influence the antibody response in other cases, for example, serotypes 9V and 20 (38, 53).
There are 78 putative acetyltransferases in the pneumococcal cps loci distributed into two Pfam families, PF01757 and PF00132 (Table 1). O-acetyl groups are present in many pneumococcal repeat units, but the patterns of acetylation differ considerably, and also the site or the percentage of acetylation for several structures has been revised in recent NMR studies (36, 45). There is no obvious correlation between the acetyltransferase HG and the pattern of acetylation (data not shown). Frameshifted, inserted, or deleted acetyltransferase genes are also often present in the pneumococcal cps loci, and frameshifting arising from variation in the number of TA repeats in the acetyltransferase gene wciZ has been shown to lead to frequent serotype switching between types 15B and 15C (79).
Assignment of pyruvyltransferases.
Serotypes 4 and 27 have pyruvate moieties in their CPS (2,3-pyruvate and 4,6-pyruvate, respectively, in the S stereo configuration), and marked changes in immunological specificity have been observed by removing these moieties from the repeat units (38). Only two genes in the S. pneumoniae cps loci (Table 1) encode products, of different HGs, that possess the Pfam domain PF04230 (WciM and WhaL of types 4 and 27, respectively), which is found in other pyruvyltransferases involved in bacterial polysaccharide biosynthesis, e.g., CsaB in Bacillus anthracis, WcaK in E. coli, and AmsJ in Erwinia amylovora. It seems clear that WciM and WhaL are the expected pyruvyltransferases for type 4 and 27 CPS biosynthesis, respectively.
Transposases and similar elements within the cps loci.
IS elements or IS-like sequences and several other transposable elements in the form of conjugative transposons have been previously characterized in pneumococci, but their role in population dynamics is still unclear (62, 71). They constitute a relatively large proportion (>3.5%) of the S. pneumoniae TIGR4 genome, where two full-length group II introns and a fragment of the streptococcal conjugative plasmid Tn5252 are also present (75). In the cps loci studied here, IS630, IS1167, IS1202, IS1381, IS1670, and IS1671 elements were identified (data not shown). Group II intron genes (Table 1) are also present in the 5′ region of serotype 19F and in the 3′ region of type 25A, 25F, and 38 cps loci. The presence of these elements at the 5′ end and often at the 3′ end of the cps locus suggests that they may (or did) have a role in the horizontal transfer of cps loci. However, in the few cases where recent changes of serotype have been examined this has occurred by homologous recombination, usually with crossover points in genes at the ends of, or flanking, the cps locus (19).
Concluding remarks.
Biochemical studies of CPS biosynthesis in S. pneumoniae have been limited to serotypes 2, 3, 8, 14, and 37 (15, 16, 29, 42, 43, 47, 65, 69); thus, the vast majority of cps gene products have had to be assigned putative functions in the absence of experimental data. Assignment of function has been achieved by using bioinformatic procedures that group similar cps gene products into HGs, by seeking homologs with established functions in the databases, and by identifying domains shared by other proteins (Pfam families). The availability of the full set of cps sequences also allows the presence of particular HGs, members of Pfam families and CAZy families, to be correlated with the structures of the CPS repeat units, which can be particularly effective when comparing cps loci that are very closely related (51).
The assignment of a general function for cps gene products works well, and there were only 26 of the 1,999 gene products that had no significant database matches or which only matched other proteins of unknown function. The process of assigning specific reactions to the cps gene products also works well in many cases, and the products of biosynthetic pathway genes and most of the ITs, polymerases, sugar phosphate transferases, and pyruvyltransferases could be tentatively assigned specific functions by correlating gene distribution with repeat unit structures. The assignment of the linkages catalyzed by the large number of GTs is more problematic. Rather than simply making predictions based on database matches, we have attempted to develop a rational ab initio approach (see the notes to Table S2 in the supplemental material for the basis for the predicted assignments shown in Table 2) as a basis for stimulating further biochemical work that establishes the specific reactions catalyzed by these enzymes.
Assignments of GT specificity by correlation have a number of limitations. For example, they assume that the published CPS structures are correct, which in some instances may not be the case (for example, type 31), particularly with some of the older structures where contamination with other cell wall polysaccharides could have been a problem. Also, correlation of GTs and linkages relies on the assumption that members of a single CAZy family of GTs are invariably all retaining or inverting. See the supplemental material for outlines of cases where there are inconsistencies in functional assignments of GTs, which may reflect incorrect structures, incorrect prediction of mechanism from CAZy family membership, or unsuspected variation in the linkage catalyzed by different members of the same HG.
Our analysis highlights some areas of pneumococcal CPS biosynthesis where functional analysis is required or where efforts could be focused. Besides analysis of the specificity of the large numbers of GTs, these include studies of the biochemical reactions catalyzed by the putative GalE-like epimerase, Spr1460, and verification of the proposed AAT-Galp biosynthetic pathway. It would also be valuable to examine whether deletion of the chromosomal gene encoding the pneumococcal WbgY homolog (Spr1655) results in loss of the ability to produce the serotype 1 capsule, to determine whether the substrate of the IT, WciI, is GlcpNAc-P/GalpNAc-P and whether or not the substrate of the IT, WcjG, is Galp/Galf. Additionally, it would be valuable to determine the CPS structures for some of the 36 serotypes where these are unavailable, to reanalyze some of the older structures where the predicted functions of cps gene products do not correlate well with the existing structure (for example, serotype 31), and to reassess the presence of Cho-P in the CPS of serotypes 15F and 15C.
Supplementary Material
Acknowledgments
This work was funded by the Wellcome Trust. B.G.S. is a Wellcome Trust Principal Research Fellow.
Footnotes
Published ahead of print on 31 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abbott, J. C., D. M. Aanensen, K. Rutherford, S. Butcher, and B. G. Spratt. 2005. WebACT—an online companion for the Artemis comparison tool. Bioinformatics 21:3665-3666. [DOI] [PubMed] [Google Scholar]
- 2.Akana, J., A. A. Fedorov, E. Fedorov, W. R. Novak, P. C. Babbitt, S. C. Almo, and J. A. Gerlt. 2006. d-Ribulose 5-phosphate 3-epimerase: functional and structural relationships to members of the ribulose-phosphate binding (beta/alpha)8-barrel superfamily. Biochemistry 45:2493-2503. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amer, A. O., and M. A. Valvano. 2002. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148:571-582. [DOI] [PubMed] [Google Scholar]
- 5.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. López, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. Sigrist, and E. M. Zdobnov. 2000. InterPro—an integrated documentation resource for protein families, domains and functional sites. Bioinformatics 16:1145-1150. [DOI] [PubMed] [Google Scholar]
- 6.Arrecubieta, C., E. García, and R. López. 1995. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene 167:1-7. [DOI] [PubMed] [Google Scholar]
- 7.Arrecubieta, C., R. López, and E. García. 1996. Type 3-specific synthase of Streptococcus pneumoniae (Cap3B) directs type 3 polysaccharide biosynthesis in Escherichia coli and in pneumococcal strains of different serotypes. J. Exp. Med. 184:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batavyal, L., and N. Roy. 1983. Structure of the capsular polysaccharide of Diplococcus pneumoniae type 31. Carbohydr. Res. 119:300-302. [Google Scholar]
- 9.Behr, T., W. Fischer, J. Peter-Katalinic, and H. Egge. 1992. The structure of pneumococcal lipoteichoic acid. Improved preparation, chemical and mass spectrometric studies. Eur. J. Biochem. 207:1063-1075. [DOI] [PubMed] [Google Scholar]
- 10.Bender, M. H., R. T. Cartee, and J. Yother. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 185:6057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bengoechea, J. A., E. Pinta, T. Salminen, C. Oertelt, O. Holst, J. Radziejewska-Lebrecht, Z. Piotrowska-Seget, R. Venho, and M. Skurnik. 2002. Functional characterization of Gne (UDP-N-acetylglucosamine-4-epimerase), Wzz (chain length determinant), and Wzy (O-antigen polymerase) of Yersinia enterocolitica serotype O:8. J. Bacteriol. 184:4277-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley, S. D., D. M. Aanensen, A. Mavroidi, D. Saunders, E. Rabbinowitsch, M. Collins, K. Donohoe, D. Harris, L. Murphy, M. A. Quail, G. Samuel, I. C. Skovsted, M. S. Kaltoft, B. Barrell, P. R. Reeves, J. Parkhill, and B. G. Spratt. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergstrom, N., P. E. Jansson, M. Kilian, and U. B. Skov Sorensen. 2000. Structures of two cell wall-associated polysaccharides of a Streptococcus mitis biovar 1 strain. A unique teichoic acid-like polysaccharide and the group O antigen which is a C-polysaccharide in common with pneumococci. Eur. J. Biochem. 267:7147-7157. [DOI] [PubMed] [Google Scholar]
- 14.Bonofiglio, L., E. García, and M. Mollerach. 2005. Biochemical characterization of the pneumococcal glucose 1-phosphate uridylyltransferase (GalU) essential for capsule biosynthesis. Curr. Microbiol. 51:217-221. [DOI] [PubMed] [Google Scholar]
- 15.Cartee, R. T., W. T. Forsee, M. H. Bender, K. D. Ambrose, and J. Yother. 2005. CpsE from type 2 Streptococcus pneumoniae catalyzes the reversible addition of glucose-1-phosphate to a polyprenyl phosphate acceptor, initiating type 2 capsule repeat unit formation. J. Bacteriol. 187:7425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cartee, R. T., W. T. Forsee, and J. Yother. 2005. Initiation and synthesis of the Streptococcus pneumoniae type 3 capsule on a phosphatidylglycerol membrane anchor. J. Bacteriol. 187:4470-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422-3423. [DOI] [PubMed] [Google Scholar]
- 18.Caspi, R., H. Foerster, C. A. Fulcher, R. Hopkinson, J. Ingraham, P. Kaipa, M. Krummenacker, S. Paley, J. Pick, S. Y. Rhee, C. Tissier, P. Zhang, and P. D. Karp. 2006. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 34:D511-D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffey, T. J., M. C. Enright, M. Daniels, J. K. Morona, R. Morona, W. Hryniewicz, J. C. Paton, and B. G. Spratt. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol. Microbiol. 27:73-83. [DOI] [PubMed] [Google Scholar]
- 20.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 21.Coyne, M. J., A. O. Tzianabos, B. C. Mallory, V. J. Carey, D. L. Kasper, and L. E. Comstock. 2001. Polysaccharide biosynthesis locus required for virulence of Bacteroides fragilis. Infect. Immun. 69:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dandekar, T., B. Snel, M. Huynen, and P. Bork. 1998. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 23:324-328. [DOI] [PubMed] [Google Scholar]
- 23.Dmitriev, B. A., V. Lvov, N. V. Tochtamysheva, A. S. Shashkov, N. K. Kochetkov, B. Jann, and K. Jann. 1983. Cell-wall lipopolysaccharide of Escherichia coli 0114:H2. Structure of the polysaccharide chain. Eur. J. Biochem. 134:517-521. [DOI] [PubMed] [Google Scholar]
- 24.Feng, L., W. Wang, J. Tao, H. Guo, G. Krause, L. Beutin, and L. Wang. 2004. Identification of Escherichia coli O114 O-antigen gene cluster and development of an O114 serogroup-specific PCR assay. J. Clin. Microbiol. 42:3799-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forsee, W. T., R. T. Cartee, and J. Yother. 2006. Role of the carbohydrate binding site of the Streptococcus pneumoniae capsular polysaccharide type 3 synthase in the transition from oligosaccharide to polysaccharide synthesis. J. Biol. Chem. 281:6283-6289. [DOI] [PubMed] [Google Scholar]
- 27.Frey, P. A. 1996. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10:461-470. [PubMed] [Google Scholar]
- 28.Guo, H., L. Li, and P. G. Wang. 2006. Biochemical characterization of UDP-GlcNAc/Glc 4-Epimerase from Escherichia coli O86:B7. Biochemistry 45:13760-13768. [DOI] [PubMed] [Google Scholar]
- 29.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy, G. G., A. D. Magee, C. L. Ventura, M. J. Caimano, and J. Yother. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 33:2759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishiyama, N., C. Creuzenet, J. S. Lam, and A. M. Berghuis. 2004. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: substrate specificity in UDP-hexose 4-epimerases. J. Biol. Chem. 279:22635-22642. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, S. M., L. Wang, and P. R. Reeves. 2001. Molecular characterization of Streptococcus pneumoniae type 4, 6B, 8, and 18C capsular polysaccharide gene clusters. Infect. Immun. 69:1244-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolly, L., J. Newell, I. Porcelli, S. J. Vincent, and F. Stingele. 2002. Lactobacillus helveticus glycosyltransferases: from genes to carbohydrate synthesis. Glycobiology 12:319-327. [DOI] [PubMed] [Google Scholar]
- 36.Jones, C., and X. Lemercinier. 2005. Full NMR assignment and revised structure for the capsular polysaccharide from Streptococcus pneumoniae type 15B. Carbohydr. Res. 340:403-409. [DOI] [PubMed] [Google Scholar]
- 37.Jones, C., C. Whitley, and X. Lemercinier. 2000. Full assignment of the proton and carbon NMR spectra and revised structure for the capsular polysaccharide from Streptococcus pneumoniae type 17F. Carbohydr. Res. 325:192-201. [DOI] [PubMed] [Google Scholar]
- 38.Kamerling, J. P. 2000. Pneumococcal polysaccharides: a chemical view, p. 81-114. In A. Tomasz (ed.), Streptococcus pneumoniae: molecular biology and mechanisms of disease. Mary Ann Liebert Inc., Larchmont, NY.
- 39.Kanehisa, M., S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki, and M. Hirakawa. 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34:D354-D357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karlsson, C., P. E. Jansson, and U. B. Skov Sorensen. 1999. The pneumococcal common antigen C-polysaccharide occurs in different forms: mono-substituted or di-substituted with phosphocholine. Eur. J. Biochem. 265:1091-1097. [DOI] [PubMed] [Google Scholar]
- 41.Kawano, S., K. Hashimoto, T. Miyama, S. Goto, and M. Kanehisa. 2005. Prediction of glycan structures from gene expression data based on glycosyltransferase reactions. Bioinformatics 21:3976-3982. [DOI] [PubMed] [Google Scholar]
- 42.Kolkman, M. A., B. A. van der Zeijst, and P. J. Nuijten. 1997. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J. Biol. Chem. 272:19502-19508. [DOI] [PubMed] [Google Scholar]
- 43.Kolkman, M. A., W. Wakarchuk, P. J. Nuijten, and B. A. van der Zeijst. 1997. Capsular polysaccharide synthesis in Streptococcus pneumoniae serotype 14: molecular analysis of the complete cps locus and identification of genes encoding glycosyltransferases required for the biosynthesis of the tetrasaccharide subunit. Mol. Microbiol. 26:197-208. [DOI] [PubMed] [Google Scholar]
- 44.Lazarevic, V., F. X. Abellan, S. B. Moller, D. Karamata, and C. Mauel. 2002. Comparison of ribitol and glycerol teichoic acid genes in Bacillus subtilis W23 and 168: identical function, similar divergent organization, but different regulation. Microbiology 148:815-824. [DOI] [PubMed] [Google Scholar]
- 45.Lemercinier, X., and C. Jones. 2006. Full assignment of the 1H and 13C spectra and revision of the O-acetylation site of the capsular polysaccharide of Streptococcus pneumoniae type 33F, a component of the current pneumococcal polysaccharide vaccine. Carbohydr. Res. 341:68-74. [DOI] [PubMed] [Google Scholar]
- 46.Liu, J., and A. Mushegian. 2003. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 12:1418-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Llull, D., E. García, and R. López. 2001. Tts, a processive beta-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in Pneumococcus and other gram-positive species. J. Biol. Chem. 276:21053-21061. [DOI] [PubMed] [Google Scholar]
- 48.Llull, D., R. Muñoz, R. López, and E. García. 1999. A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide. Type 37 pneumococci are natural, genetically binary strains. J. Exp. Med. 190:241-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.López, R., and E. García. 2004. Recent trends on the molecular biology of pneumococcal capsules, lytic enzymes, and bacteriophage. FEMS Microbiol. Rev. 28:553-580. [DOI] [PubMed] [Google Scholar]
- 50.Marolda, C. L., J. Vicarioli, and M. A. Valvano. 2004. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology 150:4095-4105. [DOI] [PubMed] [Google Scholar]
- 51.Mavroidi, A., D. M. Aanensen, D. Godoy, I. C. Skovsted, M. S. Kaltoft, P. R. Reeves, S. D. Bentley, and B. G. Spratt. 2007. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 189:7841-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mavroidi, A., D. Godoy, D. M. Aanensen, D. A. Robinson, S. K. Hollingshead, and B. G. Spratt. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J. Bacteriol. 186:8181-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNeely, T. B., J. M. Staub, C. M. Rusk, M. J. Blum, and J. J. Donnelly. 1998. Antibody responses to capsular polysaccharide backbone and O-acetate side groups of Streptococcus pneumoniae type 9V in humans and rhesus macaques. Infect. Immun. 66:3705-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mollerach, M., R. López, and E. García. 1998. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J. Exp. Med. 188:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morona, J. K., D. C. Miller, T. J. Coffey, C. J. Vindurampulle, B. G. Spratt, R. Morona, and J. C. Paton. 1999. Molecular and genetic characterization of the capsule biosynthesis locus of Streptococcus pneumoniae type 23F. Microbiology 145:781-789. [DOI] [PubMed] [Google Scholar]
- 56.Morona, J. K., D. C. Miller, R. Morona, and J. C. Paton. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 189:1905-1913. [DOI] [PubMed] [Google Scholar]
- 57.Morona, J. K., R. Morona, and J. C. Paton. 1999. Comparative genetics of capsular polysaccharide biosynthesis in Streptococcus pneumoniae types belonging to serogroup 19. J. Bacteriol. 181:5355-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morona, J. K., R. Morona, and J. C. Paton. 2006. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc. Natl. Acad. Sci. USA 103:8505-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 60.Mulrooney, E. F., K. K. Poon, D. J. McNally, J. R. Brisson, and J. S. Lam. 2005. Biosynthesis of UDP-N-acetyl-l-fucosamine, a precursor to the biosynthesis of lipopolysaccharide in Pseudomonas aeruginosa serotype O11. J. Biol. Chem. 280:19535-19542. [DOI] [PubMed] [Google Scholar]
- 61.Muñoz, R., R. López, M. de Frutos, and E. García. 1999. First molecular characterization of a uridine diphosphate galacturonate 4-epimerase: an enzyme required for capsular biosynthesis in Streptococcus pneumoniae type 1. Mol. Microbiol. 31:703-713. [DOI] [PubMed] [Google Scholar]
- 62.Muñoz, R., R. López, and E. García. 1998. Characterization of IS1515, a functional insertion sequence in Streptococcus pneumoniae. J. Bacteriol. 180:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muñoz, R., M. Mollerach, R. López, and E. García. 1997. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol. Microbiol. 25:79-92. [DOI] [PubMed] [Google Scholar]
- 64.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 207:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelosi, L., M. Boumedienne, N. Saksouk, J. Geiselmann, and R. A. Geremia. 2005. The glucosyl-1-phosphate transferase WchA (Cap8E) primes the capsular polysaccharide repeat unit biosynthesis of Streptococcus pneumoniae serotype 8. Biochem. Biophys. Res. Commun. 327:857-865. [DOI] [PubMed] [Google Scholar]
- 66.Price, N. P., and F. A. Momany. 2005. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology 15:29R-42R. [DOI] [PubMed] [Google Scholar]
- 67.Ramirez, M., and A. Tomasz. 1998. Molecular characterization of the complete 23F capsular polysaccharide locus of Streptococcus pneumoniae. J. Bacteriol. 180:5273-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reeves, P. R., M. Hobbs, M. A. Valvano, M. Skurnik, C. Whitfield, D. Coplin, N. Kido, J. Klena, D. Maskell, C. R. Raetz, and P. D. Rick. 1996. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4:495-503. [DOI] [PubMed] [Google Scholar]
- 69.Saksouk, N., L. Pelosi, P. Colin-Morel, M. Boumedienne, P. L. Abdian, and R. A. Geremia. 2005. The capsular polysaccharide biosynthesis of Streptococcus pneumoniae serotype 8: functional identification of the glycosyltransferase WciS (Cap8H). Biochem. J. 389:63-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Samuel, G., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503-2519. [DOI] [PubMed] [Google Scholar]
- 71.Sánchez-Beato, A. R., E. García, R. López, and J. L. García. 1997. Identification and characterization of IS1381, a new insertion sequence in Streptococcus pneumoniae. J. Bacteriol. 179:2459-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shepherd, J. G., L. Wang, and P. R. Reeves. 2000. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) sonnei and Plesiomonas shigelloides O17: sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect. Immun. 68:6056-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sohlenkamp, C., I. M. López-Lara, and O. Geiger. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42:115-162. [DOI] [PubMed] [Google Scholar]
- 74.Sulzenbacher, G., L. Gal, C. Peneff, F. Fassy, and Y. Bourne. 2001. Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture. J. Biol. Chem. 276:11844-11851. [DOI] [PubMed] [Google Scholar]
- 75.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 76.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Valvano, M. A. 2003. Export of O-specific lipopolysaccharide. Front. Biosci. 8:s452-s471. [DOI] [PubMed] [Google Scholar]
- 78.van Selm, S., M. A. Kolkman, B. A. van der Zeijst, K. A. Zwaagstra, W. Gaastra, and J. P. van Putten. 2002. Organization and characterization of the capsule biosynthesis locus of Streptococcus pneumoniae serotype 9V. Microbiology 148:1747-1755. [DOI] [PubMed] [Google Scholar]
- 79.van Selm, S., L. M. van Cann, M. A. Kolkman, B. A. van der Zeijst, and J. P. van Putten. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect. Immun. 71:6192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ventura, C. L., R. T. Cartee, W. T. Forsee, and J. Yother. 2006. Control of capsular polysaccharide chain length by UDP-sugar substrate concentrations in Streptococcus pneumoniae. Mol. Microbiol. 61:723-733. [DOI] [PubMed] [Google Scholar]
- 81.Waite, R. D., D. W. Penfold, J. K. Struthers, and C. G. Dowson. 2003. Spontaneous sequence duplications within capsule genes cap8E and tts control phase variation in Streptococcus pneumoniae serotypes 8 and 37. Microbiology 149:497-504. [DOI] [PubMed] [Google Scholar]
- 82.Waite, R. D., J. K. Struthers, and C. G. Dowson. 2001. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variation. Mol. Microbiol. 42:1223-1232. [DOI] [PubMed] [Google Scholar]
- 83.Wang, L., S. Huskic, A. Cisterne, D. Rothemund, and P. R. Reeves. 2002. The O-antigen gene cluster of Escherichia coli O55:H7 and identification of a new UDP-GlcNAc C4 epimerase gene. J. Bacteriol. 184:2620-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Watson, D. A., D. M. Musher, and J. Verhoef. 1995. Pneumococcal virulence factors and host immune responses to them. Eur. J. Clin. Microbiol. Infect. Dis. 14:479-490. [DOI] [PubMed] [Google Scholar]
- 85.Weiser, J. N., A. A. Lindberg, E. J. Manning, E. J. Hansen, and E. R. Moxon. 1989. Identification of a chromosomal locus for expression of lipopolysaccharide epitopes in Haemophilus influenzae. Infect. Immun. 57:3045-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Whitfield, C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39-68. [DOI] [PubMed] [Google Scholar]
- 87.Xu, D. Q., J. O. Cisar, N. Ambulos, Jr., D. H. Burr, and D. J. Kopecko. 2002. Molecular cloning and characterization of genes for Shigella sonnei form I O polysaccharide: proposed biosynthetic pathway and stable expression in a live Salmonella vaccine vector. Infect. Immun. 70:4414-4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu, D. Q., J. Thompson, and J. O. Cisar. 2003. Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38. J. Bacteriol. 185:5419-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshida, Y., S. Ganguly, C. A. Bush, and J. O. Cisar. 2005. Carbohydrate engineering of the recognition motifs in streptococcal co-aggregation receptor polysaccharides. Mol. Microbiol. 58:244-256. [DOI] [PubMed] [Google Scholar]
- 90.Yother, J. 2004. Capsules, p. 30-48. In E. I. Tuomanen (ed.), The pneumococcus. ASM Press, Washington, D.C.
- 91.Zhang, J. R., I. Idanpaan-Heikkila, W. Fischer, and E. I. Tuomanen. 1999. Pneumococcal licD2 gene is involved in phosphorylcholine metabolism. Mol. Microbiol. 31:1477-1488. [DOI] [PubMed] [Google Scholar]
- 92.Zhang, L., J. Radziejewska-Lebrecht, D. Krajewska-Pietrasik, P. Toivanen, and M. Skurnik. 1997. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol. Microbiol. 23:63-76. [DOI] [PubMed] [Google Scholar]
- 93.Zolli, M., D. J. Kobric, and E. D. Brown. 2001. Reduction precedes cytidylyl transfer without substrate channeling in distinct active sites of the bifunctional CDP-ribitol synthase from Haemophilus influenzae. Biochemistry 40:5041-5048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.