Abstract
Gram-negative bacteria of the genus Serratia are opportunistic human, plant, and insect pathogens. Serratia sp. strain ATCC 39006 secretes pectinases and cellulases and produces the secondary metabolites carbapenem and prodigiosin. Mutation of a gene (pigX) resulted in an extremely pleiotropic phenotype: prodigiosin antibiotic biosynthesis, plant virulence, and pectinase production were all elevated. PigX controlled secondary metabolism by repressing the transcription of the target prodigiosin biosynthetic operon (pigA-pigO). The transcriptional start site of pigX was determined, and pigX expression occurred in parallel with Pig production. Detailed quantitative intracellular proteome analyses enabled the identification of numerous downstream targets of PigX, including OpgG, mutation of which reduced the production of the plant cell wall-degrading enzymes and virulence. The highly pleiotropic PigX regulator contains GGDEF and EAL domains with noncanonical motifs and is predicted to be membrane associated. Genetic evidence suggests that PigX might function as a cyclic dimeric GMP phosphodiesterase. This is the first characterization of a GGDEF and EAL domain protein in Serratia and the first example of the regulation of antibiotic production by a GGDEF/EAL domain protein.
Gram-negative bacteria of the genus Serratia are members of the family Enterobacteriaceae (22). Some species, such as Serratia marcescens, are becoming a major cause of nosocomial infections (25) and are often difficult to treat due to the prevalence of resistance to multiple antibiotics (48). Serratia sp. strain ATCC 39006 was originally isolated from a salt marsh in Cheesequake, NJ, by E. R. Squibb and Sons, Inc., in an effort to mine producers of novel antibiotics (39). In common with certain S. marcescens strains, Serratia strain ATCC 39006 produces the red, linear tripyrrole pigment prodigiosin (Pig; 2-methyl-3-pentyl-6-methoxyprodigiosin), a secondary metabolite with antimicrobial, immunosuppressant, and anticancer properties (56). Indeed, Pig is in preclinical anticancer trials, and a derivative is in phase I/II clinical trials as an anticancer agent (56). A simple β-lactam antibiotic, carbapenem (Car; 1-carbapen-2-em-3-carboxylic acid) (14), and plant cell wall-degrading exoenzymes (47) are also produced by Serratia strain ATCC 39006. Serratia strain ATCC 39006 is also virulent in Caenorhabditis elegans (15) and potato tuber-rotting (17) infection models, demonstrating a broad-host-range capacity for pathogenesis in insect and plant hosts.
The Pig biosynthetic pathway was recently elucidated and requires gene products from the pigA-pigO operon (24, 57), which is transcribed as a polycistronic mRNA (47). Considerable progress has been made in unraveling the complex regulatory network that governs the production of Pig in Serratia strain ATCC 39006. For example, an N-acyl homoserine lactone quorum-sensing system (55), encoded by the SmaIR locus, derepresses the transcription of the Pig biosynthetic genes in response to cell density by modulating the expression of at least three pigment regulators (20, 47, 51). In addition, molecular mechanisms involved in altering pigA-pigO expression in response to changes in inorganic phosphate and gluconate levels have been characterized (19, 47). The regulatory hierarchy of Pig biosynthesis in Serratia strain ATCC 39006 employs at least 15 genetic loci and has been reviewed recently (56).
Recently, we identified a novel master regulator, PigP, that controls the transcription of the pigA-pigO biosynthetic operon by modulating the expression of six other regulators (20). Random transposon insertion mutations within one of these members of the PigP regulon resulted in hyperpigmented phenotypes (20). Partial sequence analysis suggested that the gene disrupted in these, designated pigX, might encode a GGDEF/EAL domain protein (20). GGDEF/EAL domain proteins are involved in the synthesis and degradation of the bacterial intracellular secondary messenger bis-(3′-5′)-cyclic dimeric GMP (c-di-GMP) (32). The aim of the current study was to investigate the phenotypic consequences of the pigX mutation with respect to secondary metabolism and virulence in Serratia strain ATCC 39006. A combination of genetics and proteomic analyses enabled the dissection of pathways mediated by PigX in the control of virulence and secondary metabolism. Finally, we show that PigX contains GGDEF and EAL domains and provide genetic evidence that PigX might exert phenotypic effects by functioning as a c-di-GMP phosphodiesterase (PDE).
MATERIALS AND METHODS
Bacterial strains, plasmids, phage, and culture conditions.
Bacterial strains and plasmids are listed in Table 1. Serratia sp. strain ATCC 39006 strains were grown at 30°C and Escherichia coli strains were grown at 37°C in Luria broth (LB; 5 g/liter yeast extract, 10 g/liter Bacto tryptone, and 5 g/liter NaCl) at 300 rpm or on LB agar supplemented with 1.5% (wt/vol) agar (LBA). Bacterial growth (optical density at 600 nm [OD600]) was measured in a Unicam Heλios spectrophotometer at 600 nm. When required, LB was supplemented with antibiotics at the following concentrations: kanamycin, 50 μg/ml; spectinomycin, 50 μg/ml; ampicillin, 50 μg/ml; and chloramphenicol, 25 μg/ml. The generalized transducing phage φOT8 was used for the transduction of chromosomal mutations as previously described (51).
TABLE 1.
Bacterial strains, plasmids, and phages used in this study
| Strain/plasmid | Genotype/phenotype | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ−gyrA96 relA1 | Gibco/BRL |
| ER2566 | F− λ−fhuA2 (lon) ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::mini-Tn10-Tets)2 R(zgb-210::Tn10)(Tets) endA1 (dcm) | NEB |
| Serratia | ||
| LacA (WT) | Lac− derivative of ATCC 39006 | 51 |
| 4SOPG | pigX::mini-Tn5Sm/Sp opgG::Tn-DS1028 Spr Cmr | This study |
| 4SPAL | pigX::mini-Tn5Sm/Sp pigA::mini-Tn5lacZ1 Spr Kmr | This study |
| HSPIG66 | pigXpro::mini-Tn5lacZ1 Kmr (insertion + 16 in pigX transcript) | 20 |
| MCP2L | pigA::mini-Tn5lacZ1 Kmr | 47 |
| NW34 | opgG::Tn-DS1028; Cmr | This study |
| ROP4 | pigX::mini-Tn5lacZ1 Kmr (insertion + 207 in pigX transcript) | 20 |
| ROP4S | pigX::mini-Tn5Sm/Sp Spr (insertion + 259 in pigX transcript) | 20 |
| Phage | ||
| φOT8 | Serratia generalized transducing phage | Crow et al., unpublished |
| Plasmids | ||
| pBluescript II SK+ | Cloning vector, ColE1 replicon, Apr | Stratagene |
| pNRW94 | EAL domain (residues 77-362) of the E. coli protein YahA expression vector, pQE-80L based, Apr | This study |
| pNRW98 | opgG::Tn-DS1028 plasposon clone (digested with NsiI and self-ligated), Cmr | This study |
| pNRW104 | N-terminal and GGDEF domain of PigX expression vector, pQE-80L based, Apr | This study |
| pNRW105 | GGDEF domain of PigX expression vector, pQE-80L based, Apr | This study |
| pNRW106 | Extended EAL domain of PigX expression vector, pQE-80L based, Apr | This study |
| pNRW107 | EAL domain of PigX expression vector, pQE-80L based, Apr | This study |
| pNRW108 | GGDEF and EAL domain of PigX expression vector, pQE-80L based, Apr | This study |
| pQE-80L | Expression vector for native or N-terminal hexahistidine proteins, Apr | QIAGEN |
| pTA40 | Native PigX expression vector, pQE-80L based, Apr | This study |
| pTA41 | pTA40 derivative with 424ELI to 424ALI point mutation, pQE-80L based, Apr | This study |
| pTA48 | pTA40 derivative with 424ELI to 424AAA point mutations, pQE-80L based, Apr | This study |
DNA manipulations and sequencing.
Molecular biology techniques, unless stated otherwise, were performed by standard methods (45). Oligonucleotide primers were obtained from Sigma Genosys and are listed in Table 2. DNA sequencing was performed at the DNA Sequencing Facility, Department of Biochemistry, University of Cambridge, Cambridge, United Kingdom. The nucleotide sequence data were analyzed using GCG (Genetics Computer Group, University of Wisconsin, WI) and compared with the GenBank DNA or nonredundant protein sequence databases using BLAST (2).
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| HS34 | GCTGACTCATAAATATCTGACTG |
| NW128 | GATAATAAGCGGATGAATGGCAG |
| NW130 | CTGCATGCCGCGATATTTTCTTTCAGTAC |
| NW132 | GAAGGATCCACGATTGCTGCTCAACTGGTG |
| NW133 | GGGGGATCCTGGTGTGTGTATGACCGCCAG |
| NW139 | GTTGCATGCAACCACCTGCTTTCATTAC |
| NW147 | GGGGAATTCAGGAGGACAGGGATGATACATTGATTCGCGCTTTC |
| NW148 | CATACTGCAGTCAGCCATTCCCTCCTTGTAATAC |
| PF92 | GTATCTAGACCACTCGACAACATTAAG |
| PF93 | GCACGAGTAAAGCCTGG |
| PF94 | GCTTATGCCCGTCACG |
| PF106 | GACCACACGTCGACTAGTGCNNNNNNNNNNAGAG |
| PF107 | GACCACACGTCGACTAGTGCNNNNNNNNNNACGCC |
| PF108 | GACCACACGTCGACTAGTGCNNNNNNNNNNGATAC |
| PF109 | GACCACACGTCGACTAGTGC |
| PF111 | CAGGTTAATGATAGTGGCGGAC |
| PF112 | CGATCCACTGATTGGTATACCTC |
| PF117 | CRTTGGAAAACATGCCACG |
| PF120 | GATGAATTCAGGAGGACAGGGATGGGATTTACTG |
| PF121 | GTCTAAGCTTCTACGCCAATTCGCAC |
| PF128 | AGGCGAAGTGCATCATCGAGCATTAATCAGCCGTATTTATG |
| PF129 | CATAAATACGGCTGATTAATGCTCGATGATGCACTTCGCCT |
| PF135 | GCGAAGTGCATCATCGAGCAGCAGCCAGCCGTATTTATGATGGTT |
| PF136 | AACCATCATAAATACGGCTGGCTGCTGCTCGATGATGCACTTCGC |
| TBOL58 | CTAGAGTCGACCTGCAGGCATGCAAGC |
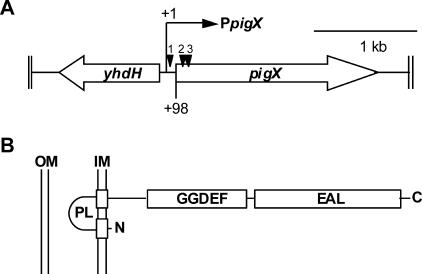
A partial sequence of pigX was determined previously (20) and was completed as described below. Random-primed PCR was performed as described previously (19), using the specific primers PF111 (first-round PCR) and PF112 (second-round PCR), which generated ∼300 bp more of the pigX sequence. To complete the pigX sequence, a degenerate primer (PF117) was designed to the 5′ end of mreB, which was predicted based on sequence alignments to be located 3′ of pigX in Serratia strain ATCC 39006. Primers PF112 and PF117 were used to amplify two independent products that were sequenced on both strands. The completed pigX sequence was assembled and is depicted schematically in Fig. 1A.
FIG. 1.
The pigX gene encodes a GGDEF/EAL domain protein. (A) Genomic location of pigX, indicating the transcriptional start site (+1) and the insertion sites of transposons (black triangles) in strains HSPIG66 (1), ROP4S (2), and ROP4 (3). (B) Predicted GGDEF/EAL protein domain structure and inner membrane (IM) localization of PigX. OM, outer membrane; PL, periplasmic loop.
The insertion site of transposon Tn-DS1028 in the opgG mutant was determined using random-primed PCR and the plasposon cloning approach. For random-primed PCR, primers facing out the right-hand side (NW128 and TBOL58) were used by following the method described previously (19). Plasposon cloning was performed as described previously (34). Plasposon pNRW98, containing the sequence flanking opgG, was generated by digesting the genomic DNA of the opgG::Tn-DS1028 mutant with NsiI followed by self-ligation. The plasposon was sequenced in a primer-walking strategy initially using primers NW127 and NW128.
Plasmid constructions.
The different domains of pigX were amplified by PCR and cloned into the expression vector pQE-80L. The primers used, and their restriction sites, were PF120 (EcoRI) and PF121 (HindIII) for the full-length PigX (pTA40), PF120 (EcoRI) and NW148 (PstI) for the N-terminal GGDEF domain (pNRW104), NW147 (EcoRI) and NW148 (PstI) for the GGDEF domain (pNRW105), NW132 (BamHI) and PF121 (HindIII) for the extended EAL domain (pNRW106), NW133 (BamHI) and PF121 (HindIII) for the EAL domain (pNRW107), and finally, NW147 (EcoRI) and PF121 (HindIII) for the GGDEF and EAL domain construct (pNRW108). Note that primers PF120 and NW147 introduced consensus ribosome-binding sites (AGGAGGA).
To generate two plasmids with amino acid substitution mutations in the EAL domain of PigX (pTA41, ELI→ALI, and pTA48, ELI→AAA), an overlap PCR strategy was used. The pigX gene was amplified in two fragments. Primer pairs PF120 and PF129 (pTA41) or PF120 and PF136 (pTA48) were used to generate the N-terminal pigX fragments. Primer pairs PF121 and PF128 (pTA41) or PF121 and PF135 (pTA48) were used to generate the C-terminal pigX fragments. Next, the N- and C-terminal fragments were mixed and used as a template in a second PCR, using primers PF120 and PF121. The resulting PCR fragments of the pigX mutants were digested with EcoRI and HindIII and cloned into pQE-80L, previously digested with the same enzymes.
A construct that enabled the expression of an EAL domain known to function as a PDE was created as outlined below. The EAL domain (residues 77 to 362) of the E. coli protein YahA was PCR amplified from chromosomal DNA of E. coli ER2566 with primers NW130 and NW139 and then digested with SphI and cloned into the SphI site of pQE-80L, generating construct pNRW94. The veracity of all plasmids was confirmed by DNA sequencing.
5′ RACE to determine transcriptional start site of pigX and primer extension studies.
Total RNA was extracted from wild-type (WT) Serratia strain ATCC 39006 using a QIAGEN RNeasy mini kit according to the manufacturer's instructions. 5′ RACE (rapid amplification of cDNA ends) was performed using a Roche 5′/3′ RACE second-generation kit, following the manufacturer's specifications. The following specific primers were used for mapping the transcriptional start of pigX: PF94, PF93, and PF92. The 5′ RACE PCR products were digested with XbaI (on PF92) and SalI (on the anchor primer) and cloned into pBluescript II SK+. To determine the transcriptional start site of pigX, a number of clones were sequenced, using primer M13-20, and analyzed. RNA extraction and primer extension analysis for the pigA transcript were performed as described previously, using primer HS34 (47). All primer extension reactions were performed with 25 μg of total RNA and 0.2 pmol of the appropriate 32P-labeled primer.
Prodigiosin, Car, exoenzyme, swimming motility, β-galactosidase, and potato virulence assays.
The assays for Pig and Car were performed as described previously (47). Pig production was plotted as (A534/ml/OD600) × 50. The activities of pectinase (Pel) and cellulase (Cel) were analyzed on agar plates containing the substrates as described previously (4) and plotted as the halo area (cm2). Swimming and β-galactosidase assays were performed as previously described (20). Potato-rotting assays were performed as described previously (8), except that potatoes were surface sterilized by submersion in 1% Virkon for 10 min and 103 CFU of each strain was used as the inocula. Experiments were performed at least seven times, and harvesting was done after 96 h. The mass of rot generated by each strain was compared, and the differences between the strains assessed for statistical significance using a two-tailed paired t test.
2D-DiGE proteomic analysis.
Cell-associated (intracellular) two-dimensional difference in gel electrophoresis (2D-DiGE) proteomics comparing the pigX mutant (ROP4S) with the WT strain was performed (biological replicates and reciprocally labeled), initially as described previously but with the detergent ASB-14 instead of CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) (16) and then in a more detailed manner as described below. LB (25 ml) was inoculated to a starting OD600 of 0.02, and four biological replicates of WT and ROP4S cultures were grown for 16 h at 30°C. Protein was harvested from the cultures as previously described (16) and quantified using a Bio-Rad protein quantification assay according to the manufacturer's instructions. The experimental approach was as described previously to allow biological variance analysis of the experimental data set using DeCyder version 5.0 (1, 36). In every gel (n = 4), 50 μg of WT and ROP4S protein samples were minimally labeled with either Cy3 or Cy5 and 50 μg of a pooled sample labeled with a third dye, Cy2, was loaded. Proteins were separated on 24-cm, pI 4 to 7, isoelectric-focusing strips and then in the second dimension on 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels (36), and the images were collected on a Typhoon 9410 (GE Healthcare) (16). To identify any spots that were significantly altered (Student's t test; P < 0.01), image analysis was performed using the DeCyder biological variance analysis software package (GE Healthcare), and spots of interest were excised and identified by tandem mass spectrometry (MS-MS) as previously described (36).
RESULTS
PigX is a GGDEF/EAL domain protein that represses prodigiosin.
Previously, the prodigiosin master regulator, PigP, was shown to negatively affect the expression of the pigX gene. Hyperpigmented random transposon mutant strains (ROP4 and ROP4S) and a decreased pigment production mutant strain (HSPIG66) were identified in a screen for regulators of Pig biosynthesis and were mapped either to within pigX (ROP4 and ROP4S) or 5′ of pigX (HSPIG66) (20). The sequence and genomic context of pigX were completed and are depicted schematically in Fig. 1A. The 649-amino-acid (aa) predicted PigX protein is similar to YhdA from E. coli (64% similarity/54% identity) and is most closely related to the predicted product of ECA0266 from Erwinia carotovora subsp. atroseptica SCRI1043 (80% similarity/73% identity). Divergently transcribed from and 5′ of pigX is a 984-bp open reading frame, which is predicted to encode a homolog of YhdH from E. coli (76% similarity/69% identity), a putative quinone oxidoreductase. PigX is predicted to have two N-terminal transmembrane helices and a central GGDEF domain (aa 222 to 384), as well as a C-terminal EAL domain (aa 398 to 633) (Fig. 1B). Therefore, it was predicted that PigX might be involved in intracellular c-di-GMP metabolism.
PigX acts as a transcriptional repressor of prodigiosin biosynthesis.
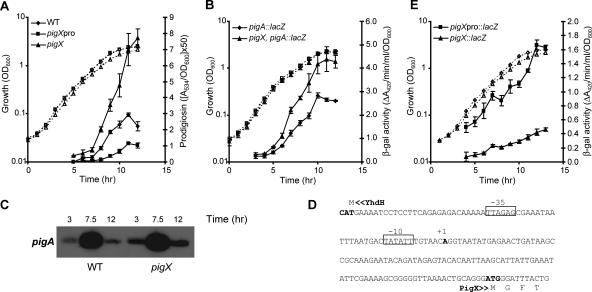
Pig production by the WT, the pigX mutant (ROP4), and the intergenic pigX mutant (HSPIG66) strains was measured throughout growth in LB. Pig production by the pigX mutant was increased by up to approximately 350% compared with the level in the WT, whereas the intergenic mutant produced between 30 and 40% of the WT Pig levels (Fig. 2A). In the intergenic mutant, the transcription of pigX is probably being driven from an internal mini-Tn5lacZ1 promoter (18), resulting in deregulated levels of the PigX repressor, causing decreased Pig production. In the pigX mutants, the pigment was confirmed as prodigiosin by liquid chromatography-MS analysis and the production of another antibiotic, carbapenem, was shown to be unaffected (data not shown). Next, we assessed the effect of PigX on the transcription of pigA-pigO. The expression of a chromosomal pigA::lacZ fusion was increased by more than 150% in the pigX mutant strain compared with its expression in the WT background (Fig. 2B). This result was confirmed by primer extension analysis of the pigA-pigO transcript, which demonstrated increased pigA-pigO mRNA levels in the pigX mutant (Fig. 2C). Finally, the introduction of a plasmid containing the entire pigX gene (pTA40) complemented Pig production in a pigX mutant strain to the levels observed in a WT strain carrying the vector control (see Fig. 5). Therefore, the GGDEF/EAL domain protein, PigX, represses the biosynthesis of Pig by decreasing the transcription of the pigA-pigO operon.
FIG. 2.
PigX represses Pig production by controlling transcription of the Pig biosynthetic operon, pigA-pigO. (A) Pig levels in WT, HSPIG66 (pigXpro), and ROP4 (pigX) strains throughout growth in LB. (B) β-Galactosidase activity was measured from a chromosomal pigA::lacZ fusion in an otherwise WT background (MCP2L) or in a strain containing a mini-Tn5Sm/Sp chromosomal insertion in pigX (4SPAL). (C) Primer extension analysis of the pigA-pigO transcript from the WT and ROP4S (pigX) strains throughout the time of growth in LB. (D) yhdH-pigX intergenic region showing the transcription start site of pigX (+1) and the predicted −10 and −35 elements. (E) β-Galactosidase activity was measured from chromosomal pigX::lacZ and pigXpro::lacZ fusion strains throughout growth in LB. Solid symbols and lines represent the results of either Pig or β-galactosidase assays, whereas the open symbols and dashed lines represent the growth curves of the corresponding strains. Data shown are the means ± standard deviations of the results of at least three independent experiments. β-gal, β-galactosidase.
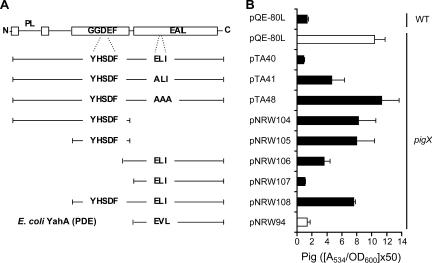
FIG. 5.
PigX is predicted to function as a c-di-GMP PDE. (A) Schematic of domain organization of PigX and regions cloned into pQE-80L. The sequences of the divergent motifs in the GGDEF and EAL domains are shown (YHSDF and ELI, respectively). Also shown (bottom) is the domain of the E. coli PDE YahA that was cloned into pQE-80L. PL, periplasmic loop. (B) Pig assays of WT and pigX mutant (ROP4) strains in the presence of the PigX plasmids and YahA plasmid shown in panel A. Data shown are the means ± standard deviations of the results of at least three independent experiments.
pigX is transcribed throughout growth, in parallel with Pig production.
The transcriptional start of pigX was determined, with only one transcriptional start (+1) predicted to be located 98 bp 5′ of the predicted translational start (ATG). Promoter −35 and −10 regions were proposed based on the E. coli σ70 consensus sequence (23) (Fig. 2D). To examine the transcriptional profile of pigX, β-galactosidase activities from both the chromosomal pigX::lacZ fusion and the fusion 5′ of the pigX ATG were measured throughout growth in LB (Fig. 2E). The expression of pigX increased in parallel with growth and continued to increase in stationary phase (Fig. 2E). Interestingly, β-galactosidase activity from the intergenic fusion was approximately fourfold higher than that from the fusion in pigX. Therefore, the transcription of pigX increases throughout growth under the culture conditions used in the current study and mirrors the expression of the target pigA-pigO operon and Pig production (Fig. 2A and B).
Numerous proteins are altered in abundance in a pigX mutant.
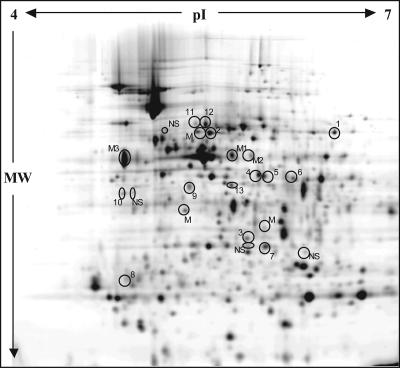
It was clear that the PigP regulon member PigX was an important secondary metabolite regulator in Serratia strain ATCC 39006 that controlled the transcription of the biosynthetic operon, pigA-pigO. To define cellular PigX targets, global proteomic analysis was used to determine the proteins that were altered in abundance in the pigX mutant in comparison to their abundance in the WT strain. In an initial 2D-DiGE proteomic analysis (“difference in gel analysis”), we detected that the PigA and OpgG (an osmoregulated periplasmic glucan [OPG] biosynthesis protein) proteins, among others, were altered between the pigX mutant (ROP4S) and the WT (data not shown). This prompted a more thorough proteome analysis. Total cell-associated proteins were prepared from the pigX mutant (ROP4S) and the WT strains and compared using 2D-DiGE. Thirty-three protein spots were identified as being significantly altered (26 were increased and 7 were decreased) in the pigX mutant compared with their abundance in the WT. Of the variously abundant proteins, 23 were visible on a Coomassie blue-stained gel and were excised for MS analysis. A Cy2 image of the pooled sample is shown in Fig. 3, and the identities of the protein spots which gave single hits are given in Table 3. Several spots gave either no significant hit or produced a mixed hit. The mixed hits are not included in Table 3, as it is not possible to determine which protein was actually altered in the mutant. These proteomic analyses demonstrated that PigX modulates the levels of multiple proteins, the significance of which will be covered in Discussion.
FIG. 3.
The 2D-DiGE intracellular protein profile in the pigX mutant strain is altered in comparison to that in WT Serratia strain ATCC 39006. Cy2 image of the pooled sample of the 2D gel (24 cm, pI 4 to 7, 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel), with proteins which were picked for identification by MS circled. NS means that no significant hits were obtained by MS analysis, and M identifies the protein spot as a mixed hit. M1, M2, and M3 contained FabB (M1 and M2) and FliC (M3) mixed with elongation factor Tu. A list of proteins that were altered in abundance is provided in Table 3. MW, molecular weight.
TABLE 3.
Intracellular proteins significantly altered in abundance in the pigX mutant compared with their levels in Serratia strain ATCC 39006
| Relative level of pigX mutant | Spot no.a | Protein identity | Nameb | Change in abundance (fold) | Bacteriumb | Accession no.b |
|---|---|---|---|---|---|---|
| Increased | 1 | Glycerol kinase | GlpK | 1.10 | Escherichia coli | gi 1134966 |
| 2 | Aldehyde dehydrogenase | PutA | 1.31 | Shewanella baltica OS155 | gi 68541744 | |
| 3 | GTP cyclohydrolase I | Sden_1513 | 1.28 | Shewanella denitrificans OS217 | gi 91792870 | |
| 4 | l-Prolyl-PCP dehydrogenase | PigA | 2.22 | Serratia strain ATCC 39006 | gi 55581716 | |
| 5 | Phosphoserine aminotransferase | SerC | 1.17 | Yersinia enterocolitica | gi 134441 | |
| 6 | ABC-type branched-chain amino acid transport systems, periplasmic component | LivK | 1.27 | Yersinia mollaretii ATCC 43969 | gi 77961773 | |
| 7 | Acyl-carrier protein, enoyl reductase (NADH) | FabI | 1.41 | Erwinia carotovora subsp. atroseptica | gi 49611426 | |
| 8 | Aminopeptidase N | PepN | 1.69 | Rubrivivax gelatinosus PM1 | gi 47573617 | |
| 9 | Predicted periplasmic lipoprotein involved in iron transport | YP_1538 | 1.13 | Yersinia pestis biovar microtus strain 91001 | gi 45436217 | |
| 10 | HBC O-methyl transferase | PigF | 2.07 | Serratia strain ATCC 39006 | gi 55581721 | |
| Periplasmic glucan biosynthesis proteinc | OpgG | 1.78 | Pseudomonas syringae pv. tomato strain DC3000 | gi 28855528 | ||
| Decreased | 11 and 12 | Aspartate ammonia-lyase 2 | AspA | −1.34 −1.25 | Erwinia carotovora subsp. atroseptica | gi 49610097 |
| 13 | Glycerol dehydrogenase | GldA | −1.26 | Erwinia carotovora subsp. atroseptica | gi 49610315 |
The spot number is the spot number in the 2D-DiGE image in Fig. 3.
Protein name, bacterium name, and accession number refer to the top hits for the Serratia strain ATCC 39006 peptides identified by MS using MASCOT.
The spot described here was picked from the original “difference in gel analysis” 2D-DiGE analysis (not shown) and identified by MS.
PigX represses exoenzyme production and virulence in planta via OpgG.
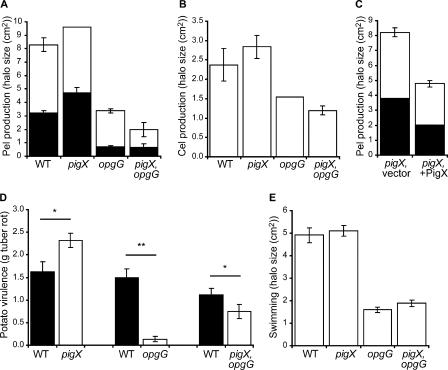
Examination of the intracellular proteome of the pigX mutant strain indicated elevated levels of a protein from Pseudomonas syringae pv. tomato strain DC3000 with similarity to OpgG from Erwinia chrysanthemi (Table 3). In Erwinia chrysanthemi, the opgGH operon is involved in the synthesis of OPGs (6), and mutation of opgG resulted in dramatic reductions in the plant cell wall-degrading enzymes and virulence (38). We predicted that the upregulated levels of OpgG in the pigX mutant might result in an increase in exoenzyme production and a hypervirulent phenotype. Therefore, phenotypes associated with plant virulence, including Cel and Pel production and potato tuber rotting, were assessed. Pel activity was increased in the pigX mutant compared with its level in the WT strain (Fig. 4A), whereas Cel production was not detectably altered (Fig. 4B). Interestingly, overexpression of PigX in trans in the pigX mutant repressed the production of Pel (Fig. 4C) but had no detectable effect on Cel (data not shown). Finally, the pigX mutant displayed a statistically significant increase in virulence in planta based on potato tuber-rotting assays (Fig. 4D). Serendipitously, in an independent study we isolated a Serratia strain ATCC 39006 opgG transposon mutant with decreased swimming motility (Fig. 4E) and no effect on growth in LB or prodigiosin production (data not shown). Sequence analysis of the opgG region in Serratia strain ATCC 39006 (Materials and Methods) demonstrated that the predicted product, OpgG, was 88% similar and 85% identical (of 450 aa aligned) to OpgG from Erwinia chrysanthemi. The Serratia opgG gene is also predicted to be in an operon with opgH, as in Erwinia and E. coli (data not shown). Pel and Cel production levels in the opgG mutant were reduced compared with their levels in the WT (Fig. 4A and B), and the opgG mutant's virulence was dramatically reduced in planta (Fig. 4D). A pigX opgG double mutant displayed Pel and Cel production phenotypes slightly lower than those of the opgG single mutant (Fig. 4A and B). The pigX opgG double mutant strain had reduced virulence compared to that of the WT strain in potato tuber-rotting assays but was more virulent than the opgG single mutant strain (Fig. 4D). Therefore, the proteomic, exoenzyme, and virulence data suggest that PigX represses the level of OpgG, which is required for full exoenzyme production and plant pathogenicity in Serratia strain ATCC 39006.
FIG. 4.
PigX represses Pel activity and virulence in a potato tuber-rotting virulence assay. (A) Pel activities of the WT (LacA), a pigX mutant (ROP4S), an opgG mutant (NW34), and a pigX opgG double mutant (4SOPG) strain. Two halos of enzyme activity were observed and are shown as black (inner halo) and white (outer halo). (B) Cel activity of the WT (LacA), a pigX mutant (ROP4S), an opgG mutant (NW34), and a pigX opgG double mutant (4SOPG) strain. (C) Pel activity of the pigX mutant strain plus vector (ROP4 and pQE-80L) and the pigX mutant strain plus PigX (ROP4 and pTA40). (D) Results of potato tuber-rotting assays comparing the amount of rot caused by the WT with the amount caused by the pigX mutant (ROP4S), the opgG mutant (NW34), and the pigX opgG double mutant (4SOPG) strain, using 103 CFU as inocula. The difference between the WT and each mutant was statistically significant using a two-tailed paired t test with a P value of either <0.05 (*) or <0.001 (**), and the data shown are the means ± standard errors of the means of the results of at least seven independent experiments. (E) Swimming motility of the WT (LacA), a pigX mutant (ROP4S), an opgG mutant (NW34), and a pigX opgG double mutant (4SOPG) strain. Data shown are the means ± standard deviations of the results of at least three independent experiments (unless otherwise stated).
PigX is predicted to function as a c-di-GMP PDE.
It was of interest to examine whether PigX could be affecting virulence and secondary metabolism by modulating the levels of the intracellular secondary messenger c-di-GMP. Recently, biochemical data have provided unequivocal evidence that GGDEF domains possess diguanylate cyclase activity responsible for the synthesis of c-di-GMP from two molecules of GTP (40, 44). Conversely, EAL domains are c-di-GMP-specific PDEs required for the turnover of this secondary messenger (12, 46, 50). PigX contains a GGDEF domain with the noncanonical active-site sequence YHSDF instead of the conserved GGDEF motif, which is necessary for the synthesis of c-di-GMP (40, 44) (Fig. 5A). Therefore, PigX is unlikely to catalyze the production of c-di-GMP. In addition, PigX does not contain the conserved sequence RXXD (X is any amino acid) of the inhibitory c-di-GMP-binding site situated adjacent to the GGDEF motif (10, 32). Analysis of the EAL domain of PigX revealed that PigX contains the amino acids ELI (Fig. 5A). Multiple functional studies have demonstrated the importance of the EAL motif in the c-di-GMP PDE activity (29, 50, 53). Our bioinformatic analyses led to the hypothesis that PigX might function (via its EAL domain) as a c-di-GMP-specific PDE.
Cultures of the WT, the pigX mutant (ROP4), and the pigXpro mutant (HSPIG66) strains were grown in the presence of 32Pi, and nucleotides were extracted and analyzed by 2D thin-layer chromatography to examine total intracellular c-di-GMP levels as previously described (5, 26, 52). Unfortunately, despite good extractions and separation of nucleotides, c-di-GMP was not detected in any of the cultures (data not shown). Therefore, a genetic strategy was used to determine the possible function of PigX.
Alanine substitution mutants were constructed in the ELI motif, resulting in two plasmids with altered forms of PigX. A plasmid with an E424A (ELI→ALI) substitution mutation could partially repress Pig production in a pigX mutant strain, demonstrating that the function of PigX was impaired (Fig. 5A and B). An E424A L425A I426A (ELI→AAA) triple amino acid substitution mutant plasmid could not complement the pigX mutant for the biosynthesis of Pig (Fig. 5A and B). These complementation experiments suggested that the ELI motif was essential for the activity of PigX. To further examine the roles of the different PigX domains, a series of plasmids was constructed encoding different domains of PigX (Fig. 5A). Strikingly, the EAL domain alone fully complemented Pig production in the pigX mutant strain back to WT levels (Fig. 5A and B). Furthermore, plasmids expressing the GGDEF domain alone or in combination with the potential membrane-spanning region had no significant effect on Pig production in the pigX mutant. In addition, the expression of a construct possessing both GGDEF and EAL domains, but not the membrane spanning regions, failed to complement the pigX mutant.
To test if the PDE activity of a biochemically characterized EAL protein could function analogously to PigX, the pigX mutant strain was transformed with a plasmid expressing the EAL domain of YahA from E. coli (46) and examined for Pig production. The EAL domain of YahA complemented Pig production in the pigX mutant to WT levels (Fig. 5A and B). These heterologous expression experiments suggest that artificial modulation of c-di-GMP levels can affect secondary metabolism in Serratia strain ATCC 39006 and support the proposed assignment of PigX as a PDE. A definitive designation awaits biochemical characterization.
DISCUSSION
The current study has investigated the role of the pleiotropic regulator, PigX, in Serratia strain ATCC 39006. PigX controlled diverse phenotypes, including virulence and the biosynthesis of prodigiosin, presumably by functioning as a PDE modulating levels of the intracellular secondary messenger, c-di-GMP. This is the first experimental report examining the physiological role of a GGDEF/EAL domain protein in a Serratia species. Furthermore, we have recently demonstrated that PigX has a key role in the regulation of a conditional biosurfactant production and swarming phenotype in Serratia strain ATCC 39006 (N. R. Williamson, P. C. Fineran, W. Ogawa, L. R. Woodley, and G. P. C. Salmond, submitted for publication).
The master secondary metabolite regulator, PigP, was shown to repress the expression of pigX (20), the predicted product of which contained GGDEF and EAL domains (Fig. 1). PigX repressed the transcription of the biosynthetic operon pigA-pigO and the production of the red-pigmented secondary metabolite Pig (Fig. 2). Furthermore, levels of the PigA and PigF pigment biosynthetic proteins were elevated in the pigX mutant strain in a proteomic analysis (Fig. 3 and Table 3). Therefore, bioassay, gene fusion, primer extension, and proteomic experiments showed that PigX repressed (presumably indirectly) the transcription of the Pig biosynthetic gene cluster to control secondary metabolism.
A proteomic strategy was devised to identify cellular changes in the absence of a functional PigX (Fig. 3 and Table 3). As mentioned above, the levels of proteins implicated in the biosynthesis of Pig were elevated in the hyperpigmented mutant strain. Furthermore, proteins involved in amino acid biosynthesis and uptake were altered in the pigX mutant strain. Increased levels of SerC, involved in serine biosynthesis; PutA, a proline utilization membrane protein; and LivK, a periplasmic component of an ABC-type branched-chain amino acid transporter, were detected in the pigX mutant. In addition, the pigX mutant displayed decreased levels of AspA, an enzyme required for l-aspartate metabolism (l-aspartate is linked to serine and glycine biosynthesis) (37). Serine and proline are precursors in the biosynthetic pathway of prodigiosin (54). It is possible that the elevated level of SerC, which catalyzes the second step of serine biosynthesis (37), provides increased serine for the overproduction of pigment in the pigX background. The multifunctional PutA flavoenzyme converts proline into glutamate in a two-step reaction and is also a DNA binding protein that represses its own expression and that of putP, a proline transporter (37). Therefore, the increased levels of PutA in the pigX mutant strain might suggest a drop in intracellular proline. However, it has been observed that proline transport mutants of Streptomyces coelicolor A3(2) can also lose the ability to degrade proline while still retaining proline biosynthesis. The result was an increase in the production of undecylprodigiosin, which was suggested to be acting as a “metabolic sink” for excess proline (30). Finally, PepN, an intracellular aminopeptidase, was increased in the pigX mutant background. In E. coli, PepN has been implicated as the major aminopeptidase involved in the degradation of cytosolic proteins and may have a role in some cellular stress responses (9). Therefore, although we have speculated about links between specific altered proteins involved in amino acid metabolism and enhanced Pig production in the pigX mutant, it is possible that PigX plays a more general role in the regulation of amino acid metabolism.
Glycerol can be phosphorylated in a GlpK-catalyzed, ATP-dependent mechanism, yielding glycerol-3-phosphate (37). Alternatively, GldA (glycerol dehydrogenase) can convert glycerol to glycerone in an NAD+-dependent reaction (37). The decreased GldA and increased GlpK levels in the pigX mutant suggest that the level of glycerol-3-phosphate, which may be funneled into either glycolysis or phospholipid biosynthesis, will be elevated.
The abundance of FabI, an enoyl-ACP reductase involved in the elongation cycle of fatty acid biosynthesis, was increased in the pigX background, and FabB (β-ketoacyl-ACP synthase I protein) was present in an elevated mixed hit in the pigX mutant strain. FabI can provide the acyl-ACP precursors of the N-AHL quorum-sensing signals (28). However, mutation of pigX had no effect on N-AHL production or SmaI expression throughout the time of growth (data not shown). It is clear that proteins involved in fatty acid metabolism are altered in the pigX mutant, and it is possible that these pathways might be linked to the biosurfactant production observed in the pigX mutant (Williamson et al., submitted).
Vitamin B6 and vitamin B9 metabolism may also be increased in the pigX strain due to elevated levels of the SerC and GTP cyclohydrolase I enzymes, respectively (37). Furthermore, a predicted periplasmic lipoprotein involved in iron transport was also increased in the pigX mutant. It is interesting to note that a transcriptomic study of a diguanylate cyclase-overexpressing strain of E. coli displayed repression of iron uptake genes (35).
Finally, increased levels of OpgG were detected in the pigX mutant. In Erwinia chrysanthemi, a number of genes, including OpgG, are involved in the synthesis of OPGs, branched glucans consisting of a β-1,2-linked glucose backbone with branched β-1,6 linkages (6, 13, 38). The exact role of OPGs is unknown, but they are important for osmoprotection, membrane integrity, and virulence in proteobacteria (6). The opgGH genes in Serratia strain ATCC 39006 were organized in a predicted operon, similar to the arrangement in Erwinia chrysanthemi (38). OpgG is a periplasmic protein, but its role in OPG synthesis is still unclear (6). However, mutation of opgG abolished the production of OPGs (38). We predicted that PigX (via opgG) may regulate OPG production, membrane integrity, exoenzyme production, and virulence. Indeed, Pel production was shown to be repressed by PigX and the pigX mutant was hypervirulent in a potato tuber-rotting model (Fig. 4). Mutation of opgG resulted in reduced exoenzyme production and decreased virulence in planta, even in the hyper-producing pigX mutant background (Fig. 4). Therefore, PigX represses levels of OpgG, which positively influences exoenzyme production and virulence in planta. A recent proteomic study was performed on an opgG mutant of Erwinia chrysanthemi (7). However, there is little overlap between those results and the ones reported here for the pigX mutant. To our knowledge, this is the first example of a proteomic study on a GGDEF/EAL mutant. These experiments revealed that, in addition to the alteration in the secondary metabolite production phenotype of a pigX mutant strain, proteins involved in primary metabolism and virulence are also affected, indicating a far more pleiotropic role of PigX than initially thought.
GGDEF and EAL domain proteins can modulate levels of the intracellular secondary messenger c-di-GMP and control multiple phenotypes, including biofilm formation, motility, and virulence (21, 32, 41). A number of lines of genetic evidence suggest that PigX functions as a PDE (via its EAL domain) and that levels of c-di-GMP might influence secondary metabolism, exoenzyme production, and virulence in Serratia strain ATCC 39006. First, overexpression of a biochemically characterized PDE protein from E. coli affected the PigX-controlled pigment and swarming (data not shown) phenotypes analogously to PigX, implicating altered levels of c-di-GMP in the modulation of Pig biosynthesis and swarming. Second, the EAL domain of PigX alone could fully complement Pig production and swarming (data not shown) in the pigX mutant strain. Finally, site-directed mutagenesis of the EAL motif of PigX abolished the function of PigX. Interestingly, during the preparation of the manuscript it was reported that the PigX homologue YhdA from E. coli (now designated CsrD) was not involved in c-di-GMP signaling and instead may be involved in binding RNA (49). In contrast to our results, the authors were unable to complement the yhdA mutant strain with the EAL domain alone, suggesting that YhdA and PigX might not have identical functions (49). It is worth noting that sequence deviation in EAL motifs can be tolerated and still allow PDE activity (42). For example, FimX from P. aeruginosa and CdgR from Salmonella enterica serovar Typhimurium possess EVL and EII motifs, respectively, yet retain PDE activity (27, 33). There is still no three-dimensional structural data of an EAL domain protein, which is likely to function as a monomer. Therefore, questions remain in our understanding of the details of c-di-GMP PDE activities. Finally, it is interesting that PDE mutations (and increased c-di-GMP) commonly result in decreased virulence, whereas in this study, mutation of pigX caused an increase in virulence.
Despite the recent advances in the biochemistry of c-di-GMP signaling, it is still unclear how this secondary messenger regulates cellular phenotypes at the molecular level. However, recent bioinformatic and biochemical analyses have led to the identification of the PilZ domain that can bind c-di-GMP specifically and therefore may be the “missing link” in these signal transduction systems (3, 11, 43). It has also been proposed that certain nonconsensus GGDEF and EAL domain proteins may bind c-di-GMP and fulfill the role of downstream c-di-GMP receptor proteins (32). No PilZ domain proteins were identified in the known Pig regulators (data not shown). Therefore, any c-di-GMP binding proteins in Serratia strain ATCC 39006 await identification.
The pleiotropic regulator PigX may be regulated at the transcriptional and/or posttranscriptional level, which would influence its effects on secondary metabolism and virulence. The transcriptional start site of pigX was determined, and its expression followed a pattern similar to that of Pig production, increasing throughout the time of growth in LB, with maximal levels present in stationary phase (Fig. 2). The pleiotropic master regulator PigP was shown to strongly repress the transcription of pigX (20). In addition to transcriptional control by PigP, the activity of the PigX protein may be modulated. PigX is predicted to be anchored in the inner membrane via two transmembrane-spanning helices (Fig. 1), which might enable signal perception leading to an alteration in the activity of PigX. A number of proteins involved in c-di-GMP signaling, such as PleD from Caulobacter crescentus and FimX from Pseudomonas aeruginosa, can have a spatially constrained membrane localization, potentially resulting in fluctuations in the local subcellular c-di-GMP pool (31, 33, 40).
In conclusion, we have shown that PigX, a GGDEF/EAL domain protein, has phenotypic impacts on virulence and secondary metabolism in Serratia strain ATCC 39006. Proteomic analyses enabled the identification of a protein (OpgG), the levels of which were modulated by PigX. Mutation of opgG had impacts on exoenzyme production and virulence in planta. Despite sequence divergence in the EAL domain, genetic evidence implicated PigX as a PDE, involved in the breakdown of the intracellular secondary messenger c-di-GMP. From this study, it is clear that GGDEF/EAL domain proteins can also regulate the transcription of genes involved in the production of secondary metabolites, such as antibiotics, in addition to the regulation of motility, biofilm formation, and virulence. Our study also demonstrates the power of proteomics in the generation of hypotheses, which can then be validated by more-traditional genetic approaches.
Acknowledgments
We thank all members of the Salmond and Welch groups for helpful discussions and I. Foulds for technical assistance. Martin Welch is acknowledged for his assistance with the 2D thin-layer chromatography analyses of c-di-GMP levels. We also thank S. Hester for performing the protein mass spectrometric analyses and H. Simonsen for the liquid chromatography-MS analysis. L. Everson is gratefully acknowledged for performing the primer extension analysis.
This work was supported by the BBSRC, United Kingdom. P.C.F. was also supported by a Bright Futures Top Achiever doctoral scholarship from the Tertiary Education Commission of New Zealand.
Footnotes
Published ahead of print on 31 August 2007.
REFERENCES
- 1.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3:36-44. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amikam, D., and M. Y. Galperin. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3-6. [DOI] [PubMed] [Google Scholar]
- 4.Andro, T., J. P. Chambost, A. Kotoujansky, J. Cattaneo, Y. Bertheau, F. Barras, F. Van Gijsegem, and A. Coleno. 1984. Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J. Bacteriol. 160:1199-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759-9769. [PubMed] [Google Scholar]
- 6.Bohin, J. P. 2000. Osmoregulated periplasmic glucans in Proteobacteria. FEMS Microbiol. Lett. 186:11-19. [DOI] [PubMed] [Google Scholar]
- 7.Bouchart, F., A. Delangle, J. Lemoine, J. P. Bohin, and J. M. Lacroix. 2007. Proteomic analysis of a nonvirulent mutant of the phytopathogenic bacterium Erwinia chrysanthemi deficient in osmoregulated periplasmic glucans: change in protein expression is not restricted to the envelope, but affects general metabolism. Microbiology 153:760-767. [DOI] [PubMed] [Google Scholar]
- 8.Burr, T., A. M. L. Barnard, M. J. Corbett, C. L. Pemberton, N. J. Simpson, and G. P. C. Salmond. 2006. Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, Erwinia carotovora: the VirR repressor. Mol. Microbiol. 59:113-125. [DOI] [PubMed] [Google Scholar]
- 9.Chandu, D., and D. Nandi. 2003. PepN is the major aminopeptidase in Escherichia coli: insights on substrate specificity and role during sodium-salicylate-induced stress. Microbiology 149:3437-3447. [DOI] [PubMed] [Google Scholar]
- 10.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015-32024. [DOI] [PubMed] [Google Scholar]
- 11.Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 104:4112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829-30837. [DOI] [PubMed] [Google Scholar]
- 13.Cogez, V., P. Talaga, J. Lemoine, and J. P. Bohin. 2001. Osmoregulated periplasmic glucans of Erwinia chrysanthemi. J. Bacteriol. 183:3127-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulthurst, S. J., A. M. Barnard, and G. P. Salmond. 2005. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat. Rev. Microbiol. 3:295-306. [DOI] [PubMed] [Google Scholar]
- 15.Coulthurst, S. J., C. L. Kurz, and G. P. Salmond. 2004. luxS mutants of Serratia defective in autoinducer-2-dependent “quorum sensing” show strain-dependent impacts on virulence and production of carbapenem and prodigiosin. Microbiology 150:1901-1910. [DOI] [PubMed] [Google Scholar]
- 16.Coulthurst, S. J., K. S. Lilley, and G. P. C. Salmond. 2006. Genetic and proteomic analysis of the role of luxS in the enteric phytopathogen, Erwinia carotovora. Mol. Plant Pathol. 7:31-45. [DOI] [PubMed] [Google Scholar]
- 17.Crow, M. A. 2001. Ph.D. thesis. The University of Cambridge, Cambridge, United Kingdom.
- 18.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fineran, P. C., L. Everson, H. Slater, and G. P. Salmond. 2005. A GntR family transcriptional regulator (PigT) controls gluconate-mediated repression and defines a new, independent pathway for regulation of the tripyrrole antibiotic, prodigiosin, in Serratia. Microbiology 151:3833-3845. [DOI] [PubMed] [Google Scholar]
- 20.Fineran, P. C., H. Slater, L. Everson, K. Hughes, and G. P. Salmond. 2005. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 56:1495-1517. [DOI] [PubMed] [Google Scholar]
- 21.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimont, P. A., and F. Grimont. 1978. The genus Serratia. Annu. Rev. Microbiol. 32:221-248. [DOI] [PubMed] [Google Scholar]
- 23.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, A. K., N. R. Williamson, H. Slater, A. Cox, S. Abbasi, I. Foulds, H. T. Simonsen, F. J. Leeper, and G. P. Salmond. 2004. The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150:3547-3560. [DOI] [PubMed] [Google Scholar]
- 25.Hejazi, A., and F. R. Falkiner. 1997. Serratia marcescens. J. Med. Microbiol. 46:903-912. [DOI] [PubMed] [Google Scholar]
- 26.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hisert, K. B., M. MacCoss, M. U. Shiloh, K. H. Darwin, S. Singh, R. A. Jones, S. Ehrt, Z. Zhang, B. L. Gaffney, S. Gandotra, D. W. Holden, D. Murray, and C. Nathan. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defense and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234-1245. [DOI] [PubMed] [Google Scholar]
- 28.Hoang, T. T., and H. P. Schweizer. 1999. Characterization of Pseudomonas aeruginosa enoyl-acyl carrier protein reductase (FabI): a target for the antimicrobial triclosan and its role in acylated homoserine lactone synthesis. J. Bacteriol. 181:5489-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 30.Hood, D. W., R. Heidstra, U. K. Swoboda, and D. A. Hodgson. 1992. Molecular genetic analysis of proline and tryptophan biosynthesis in Streptomyces coelicolor A3(2): interaction between primary and secondary metabolism—a review. Gene 115:5-12. [DOI] [PubMed] [Google Scholar]
- 31.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 32.Jenal, U., and J. Malone. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385-407. [DOI] [PubMed] [Google Scholar]
- 33.Kazmierczak, B. I., M. B. Lebron, and T. S. Murray. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 35.Mendez-Ortiz, M. M., M. Hyodo, Y. Hayakawa, and J. Membrillo-Hernandez. 2006. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J. Biol. Chem. 281:8090-8099. [DOI] [PubMed] [Google Scholar]
- 36.Mikkelsen, H., Z. Duck, K. S. Lilley, and M. Welch. 2007. Interrelationships between colonies, biofilms, and planktonic cells of Pseudomonas aeruginosa. J. Bacteriol. 189:2411-2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidhardt, F. C., J. L. Ingraham, K. B. Low, B. Magasanik, and M. Schaechter (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 1st ed. American Society for Microbiology, Washington, DC.
- 38.Page, F., S. Altabe, N. Hugouvieux-Cotte-Pattat, J. M. Lacroix, J. Robert-Baudouy, and J. P. Bohin. 2001. Osmoregulated periplasmic glucan synthesis is required for Erwinia chrysanthemi pathogenicity. J. Bacteriol. 183:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker, W. L., M. L. Rathnum, J. S. Wells, Jr., W. H. Trejo, P. A. Principe, and R. B. Sykes. 1982. SQ 27,860, a simple carbapenem produced by species of Serratia and Erwinia. J. Antibiot. (Tokyo) 35:653-660. [DOI] [PubMed] [Google Scholar]
- 40.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romling, U., and D. Amikam. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218-228. [DOI] [PubMed] [Google Scholar]
- 42.Ryan, R. P., Y. Fouhy, J. F. Lucey, and J. M. Dow. 2006. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 188:8327-8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ryjenkov, D. A., R. Simm, U. Romling, and M. Gomelsky. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310-30314. [DOI] [PubMed] [Google Scholar]
- 44.Ryjenkov, D. A., M. Tarutina, O. V. Moskvin, and M. Gomelsky. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schmidt, A. J., D. A. Ryjenkov, and M. Gomelsky. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774-4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slater, H., M. Crow, L. Everson, and G. P. Salmond. 2003. Phosphate availability regulates biosynthesis of two antibiotics, prodigiosin and carbapenem, in Serratia via both quorum-sensing-dependent and -independent pathways. Mol. Microbiol. 47:303-320. [DOI] [PubMed] [Google Scholar]
- 48.Stock, I., T. Grueger, and B. Wiedemann. 2003. Natural antibiotic susceptibility of strains of Serratia marcescens and the S. liquefaciens complex: S. liquefaciens sensu stricto, S. proteamaculans and S. grimesii. Int. J. Antimicrob. Agents 22:35-47. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki, K., P. Babitzke, S. R. Kushner, and T. Romeo. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20:2605-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson, N. R., M. A. Crow, S. J. McGowan, A. Cox, and G. P. Salmond. 2000. Biosynthesis of carbapenem antibiotic and prodigiosin pigment in Serratia is under quorum sensing control. Mol. Microbiol. 36:539-556. [DOI] [PubMed] [Google Scholar]
- 52.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wasserman, H. H., R. J. Skles, P. Peverada, C. K. Shaw, R. J. Cushley, and C. R. Lipsky. 1973. Biosynthesis of prodigiosin. Incorporation patterns of C-labeled alanine, proline, glycine, and serine elucidated by Fourier transform nuclear magnetic resonance. J. Am. Chem. Soc. 95:6874-6875. [DOI] [PubMed] [Google Scholar]
- 55.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 56.Williamson, N. R., P. C. Fineran, F. J. Leeper, and G. P. Salmond. 2006. The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4:887-899. [DOI] [PubMed] [Google Scholar]
- 57.Williamson, N. R., H. T. Simonsen, R. A. Ahmed, G. Goldet, H. Slater, L. Woodley, F. J. Leeper, and G. P. Salmond. 2005. Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56:971-989. [DOI] [PubMed] [Google Scholar]