Abstract
How alphaherpesvirus capsids acquire tegument proteins remains a key question in viral assembly. Using pseudorabies virus (PRV), we have previously shown that the 62 carboxy-terminal amino acids of the VP1/2 large tegument protein are essential for viral propagation and when transiently expressed as a fusion to green fluorescent protein relocalize to nuclear capsid assemblons following viral infection. Here, we show that localization of the VP1/2 capsid-binding domain (VP1/2cbd) into assemblons is conserved in herpes simplex virus type 1 (HSV-1) and that this recruitment is specifically on capsids. Using a mutant virus screen, we find that the protein product of the UL25 gene is essential for VP1/2cbd association with capsids. An interaction between UL25 and VP1/2 was corroborated by coimmunoprecipitation from cells transiently expressing either HSV-1 or PRV proteins. Taken together, these findings suggest that the essential function of the VP1/2 carboxy terminus is to anchor the VP1/2 tegument protein to capsids. Furthermore, UL25 encodes a multifunctional capsid protein involved in not only encapsidation, as previously described, but also tegumentation.
All herpesviruses share a common structure that includes a capsid core surrounded by a tegument layer, the latter consisting of viral and cellular proteins derived from the cytosol of the infected cell (10, 40). The capsid and tegument are enveloped by a lipid membrane derived from the cellular secretory pathway (reviewed in reference 39). Although the tegument layer was initially thought to lack defined structure, electron cryomicroscopy and tomography studies of the alphaherpesviruses have determined that the tegument interacts with the capsid surface exclusively at vertices where it adopts the underlying fivefold symmetry (22, 60). The occurrence of many specific interactions between tegument proteins, as well as between tegument proteins and membrane glycoproteins, suggests that the assembly of tegument components directs the production of virions by linking the capsid to emerging envelopes in the infected cell (9, 14, 16, 21, 26, 27, 32, 34, 42, 48, 59, 62). In accordance with these findings, a direct interaction between specific tegument and capsid proteins of the alphaherpesviruses should exist but has eluded discovery.
Here, we show that the carboxy terminus of the VP1/2 tegument protein is a capsid-binding domain (CBD) that is functionally conserved in herpes simplex virus type 1 (HSV-1) and pseudorabies virus (PRV). Using a screen to identify viral proteins required for assembly of VP1/2 onto capsids, we identify a minor capsid protein previously found to participate in DNA encapsidation (UL25) as the binding partner for VP1/2 (2, 37). The interaction between the UL25 capsid protein and the VP1/2 tegument protein defines new functions for both of these proteins and reveals a critical missing link in the morphogenesis of the alphaherpesviruses.
MATERIALS AND METHODS
Subcloning.
The green fluorescent protein (GFP) fusion to the carboxy-terminal VP1/2 conserved domain of PRV (pGS1163), previously referred to as GFP-VP1/2(aa3023-3084) (33), is referred to here as PRV GFP-VP1/2cbd (where cbd is capsid-binding domain). An mCherry-VP1/2cbd variant (pGS1775) was made by replacing the GFP open reading frame (ORF) with that of mCherry (46). The mCherry ORF provided in plasmid pRSET-B/mCherry (a generous gift of Roger Tsien) was first amplified by PCR and the subcloned into pEGFP-C1 (Clontech), such that the GFP ORF was replaced by the mCherry ORF. This produced pmCherry-C1, which was the source of the mCherry ORF (by BglII and NheI digestion) for pGS1775. The HSV-1 GFP-VP1/2cbd fusion was constructed by a polymerase elongation reaction using the following two primers that share overlapping 3′ ends: 5′-CCAGATCT]CCCGTATCGGCAAACGCAGTTCTGTCGCGACGCTACGTGCAACGCACCGGTCGTAGCGCCCTGGCGGTACTGATCCGCGCCTGTTACCGCCTACAACAGC and 5′-CCAAGCTTAGCCCAGTAACATGCGCACGTGATGTAGGCTGGTCAGCACGGCGTCGCTGTGATGAAGCAGCGCCCGGCGGGTCCGCTGTAACTGCTGTTGTAGGCGGTAA. The resulting product was subcloned in frame downstream of the GFP ORF in pEGFP-C1 by using the primer-encoded BglII and HindIII restriction sites (underlined). The resulting plasmid, pGS2033, expresses GFP fused to amino acids 3104 to 3164 of HSV-1 VP1/2.
A PRV mCherry-UL25 expression plasmid (pGS2243) was constructed by digesting pmCherry-C1 and pGS1962 (see below) with BglII and HindIII, followed by ligation. A PRV UL25-myc fusion (pGS2060) was made by PCR amplification of UL25 from the pBecker3 infectious clone using primers 5′-GGGGATCCATGGACCGCGCGTGGTTCGCCTTCG and 5′-GGTCTAGAGGCGGCGGCGAACTGCGGGATATAG (49). The resulting PCR product was subcloned into the pcDNA3.1/myc-His mammalian expression vector (Invitrogen) by using encoded BamHI and XbaI sites (underlined). The UL25 insert was confirmed by sequencing. An HSV-1 mCherry-UL25 expression plasmid (pGS2363) was constructed by PCR cloning of the UL25 ORF from an HSV-1 strain 17 infectious clone into pmCherry-C1 using BglII and HindIII(19).
Retroviral constructs used to produce complementing cell lines were made as follows. For the UL34-complementing cell line, a ∼0.9 kbp SalI-XhoI fragment of PRV genomic DNA encoding UL34 was subcloned into pLPCX (Clontech) from a previously isolated ∼4.5 kbp SalI fragment of PRV-Becker (51). The resulting clone, pGS1307, encodes the entire UL34 ORF and adjacent sequences (62 bp upstream and 101 bp downstream) behind the cytomegalovirus immediate-early promoter. For the UL25-complementing cell line, an NheI restriction site was inserted immediately downstream from the UL25 ORF of the pBecker3 infectious clone by RED-GAM mutagenesis (4). The UL25 ORF was subsequently subcloned using a modified RED-GAM protocol as described previously (36). Briefly, an R6K plasmid was amplified using the primers 5′ GCGGCGCGCGGCGGCGCGCGGCCCATGTCCCCCCGCGCCGCTAGCAAGCTTCCACATGTGGAATTCCCAT and 5′ GATCGCAGCGGCCTCGAAGGCGAACCACGCGCGGTCCATAGATCTGTCATCCATATCACCACG (underlined sequences encode NheI, HindIII, and BglII restriction sites). The resulting PCR product was inserted upstream of the UL25 start methionine codon based on homologies in the 5′ end of each primer, and the UL25 ORF was liberated by NheI digestion and self-ligation between the NheI sites engineered both upstream and downstream of the ORF. The HindIII and BglII sites were subsequently used for subcloning into pLPCX, resulting in plasmid pGS1962. The GFP-V1/2cbd construct was moved into pLPCX from pGS1163 as an SnaBI-HindIII fragment, resulting in plasmid pGS1262.
Virus construction.
All recombinant isolates of PRV were derived from the PRV-Becker infectious clone, pBecker3 (49). PRV encoding the VP26 capsid protein fused to either GFP (PRV-GS443) or monomeric red fluorescent protein (mRFP1; PRV-GS847) were previously described (51, 52). PRV-GS1214, encoding both mRFP1-VP26 and GFP-VP22, was previously described (35). Fluorescently tagged viruses carrying deletions in the UL35 (PRV-GS1205), UL36 (PRV-GS678), UL37 (PRV-GS993), and UL48 (PRV-GS1081) genes were previously described (4, 5, 36). Viruses encoding fluorescent capsids and carrying deletions in UL11 (PRV-GS718), UL25 (PRV-GS777), and UL34 (PRV-GS1248) were made by inserting a stop codon and a kanamycin marker flanked by Flp recombination target sites after the endogenous start codon by using RED-GAM recombination followed by Flp-mediated excision of the kanamycin marker, as described previously (4). This resulted in a TAA stop codon followed by a single 34-bp Flp recombination target site. For UL11, we deleted only codons 16 to 63 (of a total of 63), which kept the overlapping UL12 ORF intact. For UL25, codons 2 to 195 (of a total of 534) were deleted. The remaining UL25 ORF had no in-frame methionine codons and was left intact to prevent disruption of the downstream UL26 promoter. A second UL25-null virus was constructed in the RFP-capsid background. Briefly, a two-step recombination protocol was used to insert a stop codon, followed by deletion of codons 2 to 509 of the UL25 ORF (56).
To make a virus encoding deletions in both the US3 and UL13 protein kinase genes (PRV-GS1034), RecA-dependent homologous recombination was used to introduce a US3 deletion allele into a pBecker3 infectious clone already encoding a UL13 deletion. Both deletion alleles were previously described (4).
The recombinant HSV-1 strain encoding GFP-capsid fusions (HSV-GS1821) was derived from the HSV-1 strain KOS infectious clone, KOS-37 bacterial artificial chromosome (19). A two-step recombination protocol was used to place the GFP ORF between codons 5 and 6 of the UL35 ORF (56). Primers used for this RED recombination were 5′ ATATCGCTTCCCGACCTCCGGTCCCGATGGCCGTCCCGCACTCGACCATGGTGAGCAAGGGCGAGGAGC and 5′ AAGCGCCCGGACGCTATCGGTGGTAACGGTGCTGGGGCGGCTCGAGTTGTACAGCTCGTCCATGCCG. Linkers (underlined sequences) were placed at both fusion junctions to mimic a previously described GFP-UL35 fusion (12).
Virus and cells.
Pig kidney epithelial cells (PK15) were used for propagation of PRV stocks that did not require complementation. Recombinant PRV viruses were isolated following transfection of infectious clone plasmids into PK15 cells as previously described (50). PK15-UL36 and PK15-UL37 complementing cell lines required to propagate the PRV-GS678 (ΔUL36) and PRV-GS993 (ΔUL37) viruses were previously described (36). PK15-UL25 and PK15-UL34 complementing cell lines required to propagate PRV-GS777/GS2168 (ΔUL25) and PRV-GS1248 (ΔUL34), as well as PK15-GFPVP1/2cbd cells, were made by transduction of retroviral particles derived from pGS1307, pGS1962, and pGS1262 (see above) as previously described (36). All complementing cells were selected with 1 μg of puromycin/ml.
African green monkey kidney cells expressing Cre recombinase (CRE-Vero cells) were used to initially propagate HSV-1 KOS expressing the GFP-UL35 capsid fusion (HSV-GS1821) and were maintained as described previously (19). Vero cells were used for additional HSV propagation and subcellular localization studies of VP/2cbd and UL25 constructs. HEK-293 cells were used for cotransfection studies of viral proteins used in protein immunoprecipitation experiments due to the high transfection efficiency that could be attained.
Transfection and infection.
For fluorescence imaging, Vero cells were transfected with expression plasmids using polyethylenimine (catalog no. 23966; Polysciences) as previously described (33). In a subset of experiments, cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen) as follows: to 1 ml of Dulbecco's modified Eagle's medium, 30 μl of Lipofectamine 2000 and 30 μl of plasmid DNA were added, mixed, and incubated at room temperature for 20 min. The mixture was added to a 10-cm dish of subconfluent Vero cells and incubated for an additional 12 to 14 h. Transfected cells were passaged at a fivefold dilution onto 22- by 22-mm glass coverslips (no. 1.5), allowed to adhere for 2 h, and infected at an multiplicity of infection of 5. The samples were imaged between 5 to 8 h postinfection (hpi) (see below).
For experiments examining VP1/2cbd localization after infection with mutant viruses, either GFP-VP1/2cbd (pGS1163) or mCherry-VP1/2cbd (pGS1775) was transfected, depending upon the fluorescent protein fused to the VP26 capsid protein in the various mutant viruses examined (GFP-capsid virus mutants included ΔUL11, ΔUL25, and ΔUL36; RFP-capsid virus mutants included ΔUL13/ΔUS3, ΔUL34, ΔUL37, and ΔUL48).
For coimmunoprecipitation experiments, HEK-293 cells were transfected using 45 μl of polyethylenimine and 22.5 μl of each plasmid in 1 ml of Dulbecco's modified Eagle's medium.
Capsid isolation.
PK15 or PK15-GFPVP1/2cbd cells were grown in 850-cm2 roller bottles and infected with PRV-GS847, PRV-GS1214, or PRV-GS2168 at a multiplicity of infection of 3. Capsids were isolated at 24 hpi by freon extraction. The majority of capsids isolated by this method have been reported to be B and C capsids (50, 53).
Fluorescence microscopy.
Fluorescence imaging was performed on transfected and infected cells as previously described (33). To investigate GFP association with isolated RFP-capsids, a 1:10 dilution of isolated capsids was placed on a glass slide and then covered with a 22- by 22-mm glass coverslip (no. 1.5). Fluorescence emissions from isolated capsids were captured using a 60×objective with a numerical aperture of 1.4 (Nikon) and a Cascade 650 charged-coupled device (Roper Scientific) with 2-s RFP and 4-s GFP sequential exposures. Quantitation of RFP-GFP colocalization was performed using the Metamorph software package (Molecular Devices) and an automated algorithm that identified punctate emissions of RFP-capsid fluorescence and measured the amount GFP signal-over-background associated with each RFP spot.
Electron microscopy.
Capsids were placed onto carbon-coated copper electron microscopy grids (Ladd Research Industries) and allowed to adhere for 5 min at room temperature. Grids were washed two times with TNE (50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM EDTA) buffer, stained with 1% uranyl acetate for 15 s, and imaged with a Jeol 1220 electron microscope equipped with a megapixel-resolution digital camera (Kodak).
Immunoprecipitation, Western blot analysis, and antibodies.
HEK-293 cells transiently expressing combinations of VP1/2, UL25, and GFP were lysed 36 h posttransfection in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.5% NP-40 supplemented with 1 mM dithiothreitol, 1 mM NaF, 0.1 mM Na3VO4, 0.1 mM phenylmethylsulfonyl fluoride, and a protease inhibitor cocktail (P1860; Sigma). A total of 25 μg of anti-GFP antibody (Roche) was bound to 100 μl of a Sepharose A/G bead slurry (as provided by the manufacturer) (Amersham) for 2 h at 4°C. The beads were washed three times with lysis buffer to remove unbound antibody. Approximately 1 ml of precleared lysate was incubated with a 30-μl aliquot of the antibody-bound beads overnight at 4°C. The beads were washed three times with lysis buffer and then resuspended in 30 μl of 2× final sample buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100, 10% β-mercaptoethanol). Samples were boiled for 5 min, separated through an 8% sodium dodecyl sulfate-polyacrylamide gel, and transferred to a Hybond-P membrane (GE Healthcare). The membrane was incubated with a 1:1,000 dilution of an anti-myc antibody (Northwestern Monoclonal Antibody Facility) followed by a horseradish peroxidase-conjugated antibody used at a 1:10,000 dilution (Jackson ImmunoResearch). UL25-myc was visualized using a luminol-coumeric acid-H2O2 chemiluminescence solution, and exposed film was digitized with an EDAS 290 documentation system (Kodak). Densitometry analysis was performed on lysates and immunoprecipitations for each sample. A ratio of immunoprecipitate to lysate was calculated, and the ratio was plotted relative to the background sample (no GFP), as follows: (intensity of UL25-myc immunoprecipitation sample/intensity of UL25-myc lysate sample)/(background intensity of UL25-myc immunoprecipitation/background intensity of UL25-myc lysate). Three separate experiments were averaged for the final plot. Band intensities were quantified using ImageJ software (1).
HSV-1 GFP-VP1/2cbd (pGS2033) and mCherry-UL25 (pGS2343) constructs were transfected into HEK-293 cells, and immunoprecipitations were performed as stated above but with 20 μl of HSV-1 UL25 antiserum (a generous gift from Edouard Cantin) bound to 100 μl of beads (3). Bound HSV-1 GFP-VP1/2cbd was resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blotting with a 1:1,000 dilution of anti-GFP antibody (Santa Cruz).
For detection of capsid-associated proteins, 10 μl of PRV capsids isolated from infected PK15 cell nuclei was boiled for 5 min in an equal volume of 2× final sample buffer; samples were separated and blotted as stated above using either a 1:1,000 dilution of an anti-GFP (Santa Cruz) or a 1:1,000 dilution of an anti-VP5 antibody (a generous gift from Lynn Enquist).
RESULTS
The carboxy-terminal essential domain of VP1/2 is functionally conserved in HSV-1 and PRV.
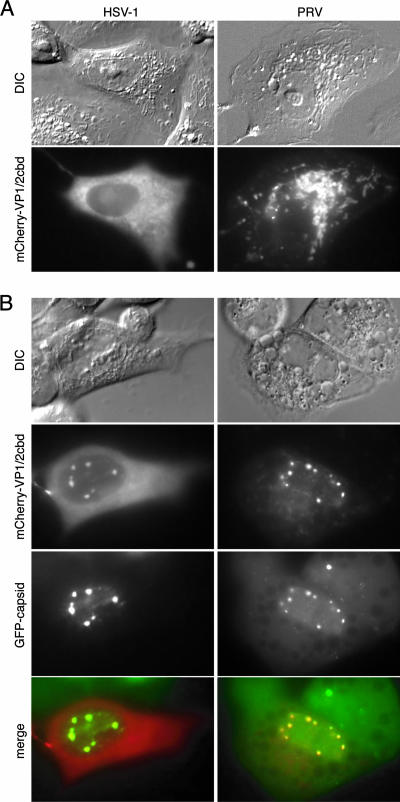
We have reported that expression of the carboxy-terminal domain of PRV VP1/2 fused to GFP is excluded from the nucleus of transfected Vero cells but upon infection with PRV becomes redistributed to nuclear capsid assemblons (33). Because the amino acid sequence of this region of VP1/2 is reasonably conserved throughout the alphaherpesviruses, we examined the equivalent region of HSV-1 VP1/2 for similar activity (33). The 61 carboxy-terminal codons of HSV-1 VP1/2 were synthetically made as two overlapping oligonucleotides that were elongated into a double-stranded DNA that was cloned in frame into an mCherry expression plasmid. Upon transfection into Vero cells, the fluorescence emissions from the mCherry fusion protein were predominantly cytoplasmic (Fig. 1A). Although the PRV and HSV-1 constructs showed different patterns of cytoplasmic fluorescence, upon infection with recombinants of either PRV (strain Becker) or HSV-1 (strain KOS) expressing GFP-tagged capsids, each fusion protein redistributed to nuclear capsid assemblons (Fig. 1B). Knowing that the association of the VP1/2 carboxy terminus with capsid assemblons was conserved between different alphaherpesviruses and was, therefore, likely an important feature of these viral proteins, we next further characterized this activity using the PRV model.
FIG. 1.
The VP1/2 CBD is conserved in HSV-1 and PRV. (A) Vero cells transiently expressing VP1/2cbd from either HSV-1 or PRV fused to the mCherry fluorescent protein. (B) mCherry-VP1/2cbd-expressing cells infected with HSV-1 or PRV encoding GFP-capsid fusions and imaged at 6 to 8 hpi. DIC, differential interference contrast.
UL25 is required for VP1/2 capsid association.
Although VP1/2 was previously stated to coimmunoprecipitate with the VP5 major capsid protein from infected cells, the nature of this interaction has not been resolved (38). VP1/2 is an inner tegument protein and, as such, associates with capsids in infected cells, but whether this interaction is directly with VP5 or through additional capsid and tegument proteins is not known (15). We tested the requirement of nine viral proteins in this interaction that were considered good candidates as participants in the VP1/2-capsid interaction (see Discussion). Viruses with a deletion of each candidate gene were used to infect PK15 cells transiently expressing either a GFP or mCherry fusion to VP1/2cbd, and relocalization of VP1/2cbd to nuclear capsid assemblons was monitored. The viral genes tested in this way were the UL11, UL13, UL25, UL34, UL35, UL48, UL36, UL37, and US3 genes.
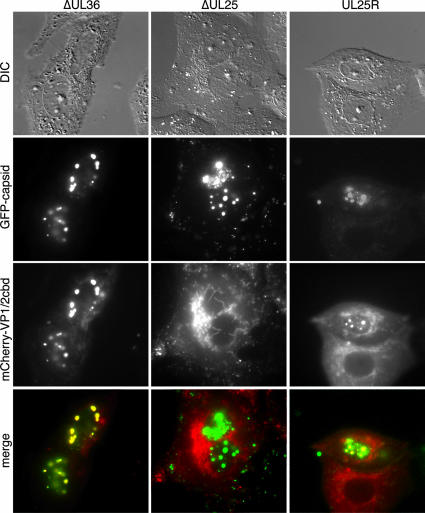
VP1/2cbd localization was imaged between 6 to 8 hpi, and cells were scored for the presence of VP1/2cbd in nuclear inclusions indicative of capsid assemblons (Table 1). Most mutant viruses induced VP1/2cbd relocalization equivalent to the wild type, including a virus lacking VP1/2 expression (ΔUL36), indicating that VP1/2cbd can function in the absence of full-length protein (Fig. 2). The notable exception was the ΔUL25 virus, which failed to induce relocalization of the VP1/2cbd in all instances. A revertant of the UL25-null virus was functionally restored (Fig. 2).
TABLE 1.
Dependence of VP1/2cbd localization at capsid assemblons
| Virus | Knocked out gene(s) | % of cells with VP1/2cbd at assemblons (n)a |
|---|---|---|
| PRV-GS847 | 100 (32) | |
| PRV-GS718 | ΔUL11 | 100 (38) |
| PRV-GS1034 | ΔUL13, ΔUS3 | 100 (25) |
| PRV-GS777 | ΔUL25 | 0 (28) |
| PRV-GS1248 | ΔUL34 | 91 (11) |
| PRV-GS1205 | ΔUL35 | 100 (25) |
| PRV-GS678 | ΔUL36 | 100 (22) |
| PRV-GS993 | ΔUL37 | 100 (12) |
| PRV-GS1081 | ΔUL48 | 100 (25) |
n, number of cells examined.
FIG. 2.
UL25 is required for VP1/2cbd localization to capsid assemblons. Vero cells were transfected with the PRV mCherry-VP1/2cbd construct and subsequently infected with PRV encoding GFP-capsid fusions and either ΔUL36, ΔUL25, or a revertant of ΔUL25 (UL25R). Cells were imaged at 6 to 8 hpi. DIC, differential interference contrast.
The VP1/2 carboxy terminus binds to capsids.
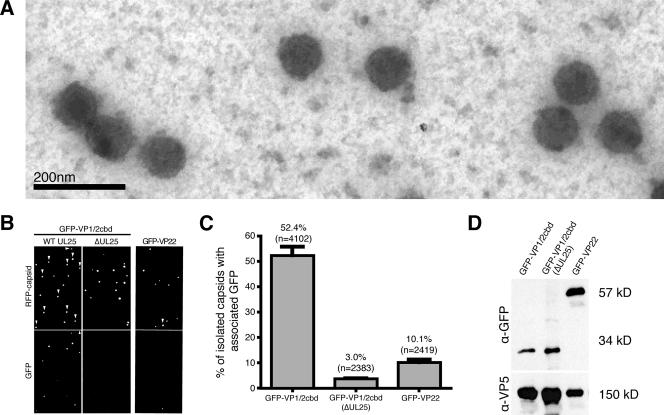
We next determined if the observed nuclear localization of the VP1/2 carboxy terminus was due to an association with capsids rather than another assemblon component. Fluorescent capsids were isolated from the nuclei of PK15 cells stably expressing the PRV GFP-VP1/2cbd construct, following infection with PRV-Becker encoding mRFP1-capsids. As controls, we isolated capsids from the nuclei of PK15 cells infected with either the UL25-null isolate of PRV or with a recombinant PRV-Becker expressing both mRFP1-capsids and a GFP-VP22 fusion (35). Although VP22 is present in the nuclei of infected cells, it is an outer-tegument protein that is thought to be added to capsids during cytoplasmic secondary envelopment and, as such, should not copurify with capsids isolated from the nucleus (8, 11, 15, 23). Capsids extracted from infected nuclei were spotted either on glass coverslips for fluorescence imaging or on electron microscopy grids. Capsids were confirmed to be present and structurally intact in the sample preparations by electron microscopy (Fig. 3A).
FIG. 3.
VP1/2cbd is associated with capsids isolated from PRV-infected cell nuclei. (A) Representative image of capsids isolated from the infected cell nuclei. (B) Fluorescence imaging of capsids isolated from PK15 cells stably expressing GFP-VP1/2cbd infected with wild-type RFP-capsid virus (left panels), ΔUL25 RFP-capsid virus (center panels), or PK15 cells infected with an RFP-capsid/GFP-VP22 virus (right panels). Arrowheads indicate capsids emitting detectable GFP fluorescence. (C) Percentage of RFP-capsid fusions emitting detectable GFP fluorescence, as shown in panel B. Error bars are standard deviations. (D) The presence of VP5 and GFP fusion proteins was detected in each capsid preparation by Western blot analysis. Predicted molecular sizes of proteins are indicated at right, which were consistent with molecular size markers. α, anti.
By fluorescence microscopy, 52.4% of capsids (n = 4,102) isolated from the nuclei of GFP-VP1/2cbd-expressing cells infected with a wild-type UL25 virus also emitted GFP fluorescence at various intensities, whereas only 3.0% of UL25-null capsids (n = 2,283) overlapped with GFP-VP1/2cbd fluorescence (Fig. 3B and C). UL25-dependent binding of GFP-VP1/2cbd was specific. Although there was abundant GFP-VP22 in our nuclear preparation, based on Western analysis (Fig. 3D), GFP-VP22 fluorescence was limited to 10.1% of capsids (n = 2,419) (Fig. 3B and C). Based on these findings, we conclude that the VP1/2 carboxy terminus binds capsids, and this binding is dependent upon the presence of UL25. The observed heterogeneity in VP1/2cbd-capsid binding may result from the differing amounts of UL25 present on A, B, and C capsids isolated from infected cell nuclei (41, 47, 55).
HSV-1 and PRV VP1/2cbd binds UL25.
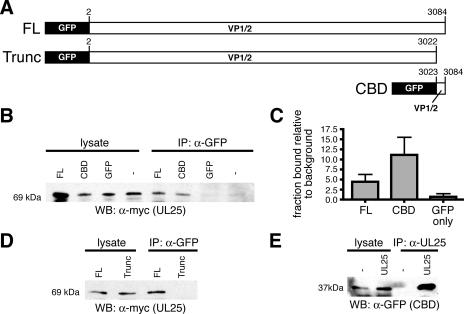
HEK-293 cells were transiently transfected with a PRV UL25-myc expression plasmid alone or together with a plasmid encoding either GFP fused to full-length PRV VP1/2 (GFP-VP1/2), GFP-VP1/2cbd, or GFP alone. The GFP constructs were immunoprecipitated with an anti-GFP antibody, and the presence of PRV UL25-myc in the immunoprecipitates was detected by Western analysis. Both full-length PRV VP1/2 and VP1/2cbd interacted with PRV UL25 (Fig. 4A and B). Surprisingly, full-length VP1/2 bound to UL25 less efficiently than VP1/2cbd alone, which may indicate that full-length VP1/2 is less stable than the CBD alone or that full-length VP1/2 requires posttranslational modification or processing to allow efficient capsid binding (Fig. 4C). In the absence of the CBD, a truncated VP1/2 failed to interact with UL25-myc, indicating that the VP1/2 carboxy terminus is the only domain of the protein sufficient to mediate UL25 binding and also confirming that the VP1/2-UL25 interaction was due to the carboxy-terminal domain in VP1/2 and not an artifact of having fused GFP to the VP1/2 protein constructs (Fig. 4D).
FIG. 4.
VP1/2cbd and UL25 interact in the absence of other viral proteins. (A) Illustration of GFP-VP1/2 fusion proteins. (B and D) PRV UL25-myc detection by Western blotting either directly from cell lysates or after immunoprecipitation from lysates with an anti-GFP antibody. HEK-293 cells were cotransfected with PRV UL25-myc and either PRV GFP-VP1/2 (FL, full-length), GFP-VP1/2Δcbd (Trunc, truncated), PRV GFP-VP1/2cbd (CBD), unfused GFP, or no additional construct. (C) Densitometry analysis of three duplicate experiments as documented in panel B. For each sample, the ratio of the amount of UL25-myc in the immunoprecipitate to the amount in the lysate is shown relative to the background (no GFP sample). Error bars are standard deviations. (E) HSV-1 VP1/2cbd detection by Western blotting from HEK-293 cells cotransfected with or without an HSV-1 UL25-mCherry fusion construct. Predicted molecular sizes of proteins are indicated at left, which were consistent with molecular size markers. α, anti; WB, Western blotting; IP, immunoprecipitation.
The VP1/2cbd-UL25 interaction was also evident in HSV-1. HSV-1 VP1/2cbd was efficiently immunoprecipitated from HEK-293 cells transiently transfected with both HSV-1 GFP-VP1/2cbd and UL25-mCherry expression plasmids, using an anti-UL25 antibody (Fig. 4E).
VP1/2cbd competes with full-length VP1/2 assembly into viral particles.
To test if the interaction observed between VP1/2cbd and UL25 in overexpressing cells was consistent with a role in tegument assembly during infection, PK15 cells stably expressing GFP-VP1/2cbd were infected with a recombinant of PRV expressing mRFP1 fused to the amino-terminus of full-length VP1/2 (mRFP1-VP1/2) (5). The resulting extracellular viral particles released from the infected cells were imaged for incorporation of green and red fluorescence as described previously (35). Both GFP-VP1/2cbd and mRFP-VP1/2 (full-length) were incorporated together in 34% of released viral particles in various amounts (n = 6,506), demonstrating that the carboxy-terminal CBD was sufficient for structural incorporation. In the remaining 66% of released viral particles examined, either GFP-VP1/2cbd (34%) or mRFP1-VP1/2 (32%) emissions were exclusively detected, indicating that the presence of VP1/2cbd can effectively compete for viral incorporation with the endogenous VP1/2 protein, which is normally present in 98% of released viral particles (35). The observation of a large proportion of extracellular viral particles lacking detectable full-length VP1/2 was unexpected, as VP1/2 is required for efficient viral egress from infected cells, and may indicate that VP1/2 was frequently incorporated at levels below our detection limit (13, 17). Nevertheless, these data support a critical role for the carboxy terminus of VP1/2 in capsid tegumentation. Consistent with this conclusion, VP1/2 with a deletion of the carboxy-terminal CBD fails to support viral propagation (7, 33).
UL25 binds capsids independently of VP1/2.
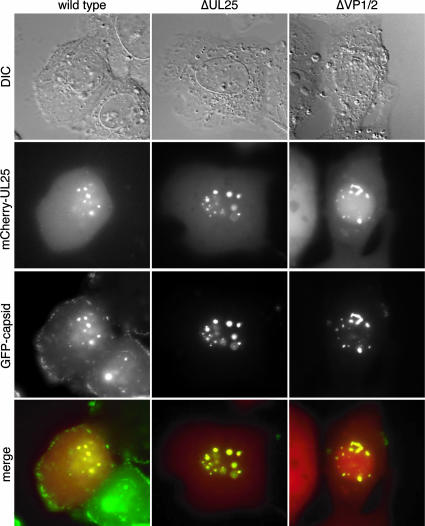
Transient expression of an mCherry-UL25 fusion construct resulted in only diffuse fluorescence in Vero cells, similar to expression of mCherry alone (data not shown). However, association of UL25 with nuclear capsid assemblons was consistently observed in cells infected with either wild-type, ΔUL25, or ΔVP1/2 recombinant PRV (Fig. 5). Similar UL25 nuclear localization was previously observed in cells infected with wild-type HSV-1 or PRV (24, 43). These observations are consistent with the finding that soluble UL25 binds to purified HSV-1 capsids in vitro and further demonstrate that this interaction is independent of VP1/2 binding to UL25 (41).
FIG. 5.
UL25 does not require VP1/2 to localize to capsid assemblons. Vero cells were transfected with a PRV mCherry-UL25 construct and subsequently infected with wild-type, ΔUL25, or ΔUL36 PRV encoding GFP-capsid fusions. Cells were imaged at 6 to 8 hpi.
DISCUSSION
The specific components of the capsid and tegument that interact to mediate tegumentation and ultimately produce infectious virions have been a missing link in the understanding of herpesvirus assembly. At least three alphaherpesvirus tegument proteins, VP1/2, UL37, and the US3 protein kinase, associate with capsids prior to membrane envelopment in the cytoplasm of infected cells (15, 20, 27). In contrast, other tegument proteins preferentially associate with cytoplasmic membranes prior to capsid envelopment (15, 31). As such, the tegument is considered to have an inner and outer layer, and reconstruction of the capsid-tegument interface from purified virions reveals that the inner tegument is bound to capsid vertices (22, 60). The VP1/2 tegument protein is considered the best candidate for serving as the bridge between the capsid and tegument of the alphaherpesviruses for a few reasons: (i) VP1/2 remains associated with capsids from purified virions after extraction procedures that remove other tegument proteins (18, 54); (ii) VP1/2 is found associated with cytosolic capsids in the absence of other tegument proteins, including the UL37 inner-tegument protein (27); and (iii) VP1/2 was observed to coimmunoprecipitate with the VP5 major capsid protein from infected cell lysates (38). None of these findings demonstrates a direct connection between VP1/2 and the capsid, however, as other viral proteins could tether VP1/2 to the capsid surface.
The carboxy terminus of VP1/2 is essential to the propagation of PRV in cell culture (7, 33). In addition, we found that the 62 carboxy-terminal amino acids of VP1/2 are sufficient to direct GFP into nuclear capsid assemblons in infected cells (33). This observation suggested that the VP1/2 carboxy terminus may be a long-sought link between capsids and tegument. We have confirmed here that this link is preserved in at least two alphaherpesviruses (HSV-1 and PRV) and that it represents a genuine tegument-capsid interaction.
Although there is no reason to expect that VP1/2cbd is either liberated or expressed independently of the full-length VP1/2 protein during infection, we rationalized that the phenomenon of capsid assemblon localization could be exploited to identify viral proteins required for the tegumentation of capsids and thus to define the precise linkage between capsids and the surrounding tegument.
Several recombinants of PRV with deletions of individual genes that seemed good candidates to participate in the initial steps of capsid tegumentation were immediately available in our laboratory (4, 36). Both UL37 and UL48 (VP16) encode tegument proteins that bind VP1/2 directly (27, 59). The VP26 capsid protein is present in 900 copies on the capsid surface, where it could influence tegumentation (57). Finally, a virus with a deletion of UL36 (VP1/2) was useful to rule out the possibility that the carboxy terminus was a multimerization domain that resulted in capsid association indirectly through interaction with full-length VP1/2. Because viruses with deletions of each of these genes recruited the VP1/2cbd to capsid assemblons, we next made viruses with deletions of the UL11, UL13, UL25, UL34, and US3 genes. We had previously observed a defect in nuclear egress of a virus with a VP1/2 gene deletion (36), and each of the genes we next targeted were previously reported to be important for primary envelopment of capsids in the nucleus (6, 25, 28-30, 37, 44, 45). A single virus with deletions of both the US3 and UL13 genes was made, as both encode protein kinases that could have functional redundancy. Of the viruses tested, only the UL25-null virus was incapable of recruiting the VP1/2cbd to capsid assemblons, and this failure was absolute.
UL25 is present on the capsid surface around the vertices, at five copies per vertex, where it binds to the capsid proteins VP19c, VP5, and UL17 (41, 43, 55, 58). In addition to these three interactions, we now show that UL25 also binds the VP1/2 carboxy terminus, thereby linking the capsid and tegument. The UL25-VP1/2 interaction was conserved in representative members of the herpes simplexviruses (HSV-1) and varicelloviruses (PRV), which together comprise the neuroinvasive groups of the alphaherpesviruses.
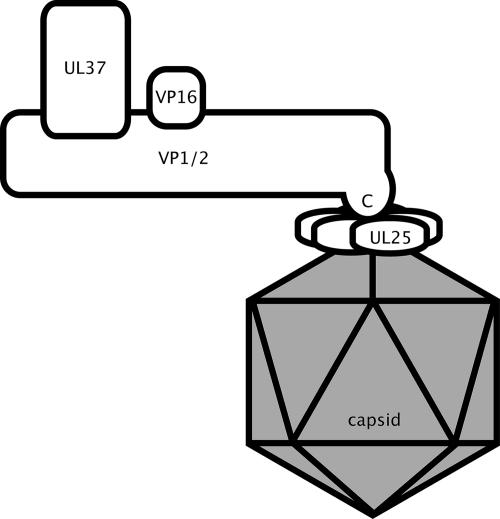
Because VP1/2 was not required for the UL25-capsid interaction and UL37 and VP16 were not required for the VP1/2cbd-UL25 interaction, a model of capsid-tegument interactions can be inferred (Fig. 6). We note that the orientation of VP1/2 in this model is opposite to that proposed for the homologue in the gammaherpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV) (61). Both UL25 and VP1/2 homologues are poorly conserved between alphaherpesviruses and KSHV, and it is therefore possible that these proteins function differently in more distantly related herpesviruses. We propose that this could be tested by transiently expressing the carboxy terminus of the KSHV VP1/2 homologue as a GFP fusion protein in KSHV-infected cells.
FIG. 6.
Illustration of capsid and tegument protein interactions. Protein interactions are based on the findings of this study and previous reports (27, 59). The juxtaposition of the protein interactions is based on the findings presented in this report that indicate that the VP1/2-binding proteins, UL37 and VP16, do not participate in the newly described VP1/2-UL25 interaction. N, amino terminus; C, carboxy terminus.
While defining these interactions lays the groundwork for understanding the steps that lead to capsid tegumentation, many critical questions remain: where in a cell does tegumentation begin, how is it regulated, and how are tegument assembly and egress coupled? Although VP1/2cbd bound to capsids in the nucleus in our current study, the nucleus may not be the native site of the VP1/2-capsid interaction. The small size of the CBD (62 amino acids) allowed for passive diffusion of the GFP fusion through nuclear pores and thereby provided a useful assay for identifying capsid-tegument interactions. Whether full-length VP1/2 enters the nucleus and binds to nuclear capsids during infection is controversial. While VP1/2 was found to copurify with capsids from the nuclei of HSV-1-infected cells in one recent study, no large proteins indicative of VP1/2 were observed in preparations of purified nuclear HSV-1 capsids from a subsequent study (8, 58). Similarly in three separate studies, VP1/2 was found to be either excluded from the nucleus, present at reduced levels in the nucleus relative to the cytoplasm, or evenly diffused throughout the nucleus and cytoplasm (27, 33, 38). While this study does not address the site of tegumentation during infection, a critical capsid-tegument interaction was identified. Determining when and where these interactions take place will clarify how these viruses are assembled and egress from cells.
Acknowledgments
We thank Hank Seifert and Allen Helm for help with negative staining and for their generous gift of electron microscopy grids. We also thank Caroline Freitag for help with fluorescence imaging, Sarah Antinone for production of the GFP-capsid HSV-1 recombinant virus, Gant Luxton for help with isolation of a stable cell line, Jenifer Klabis and Sarah Antinone for help with plasmid construction, and Sarah Haverlock-Moyns for help with viral mutagenesis. We are grateful to Patricia Spear, Richard Longnecker, and Hong Zhou for their insightful discussions of these data; David Leib for providing the KOS-37 bacterial artificial chromosome and CRE-Vero cells; Roger Tsien for the pRSET-B/mCherry construct; Edouard Cantin for anti-HSV-UL25 antibody; Lynn Enquist for anti-PRV-VP5 antibody; and Mindy Leelawong for her helpful comments on the manuscript.
This work was supported by NIH grant 1RO1AI056346 and an Investigators in Pathogenesis of Infectious Disease grant from the Burroughs Wellcome Fund (to G.A.S.). K.E.C. was supported by the training program in Immunology and Molecular Pathogenesis, NIHT32AI07476.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with Image J. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Addison, C., F. J. Rixon, J. W. Palfreyman, M. O'Hara, and V. G. Preston. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138:246-259. [DOI] [PubMed] [Google Scholar]
- 3.Ali, M. A., B. Forghani, and E. M. Cantin. 1996. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology 216:278-283. [DOI] [PubMed] [Google Scholar]
- 4.Antinone, S. E., G. T. Shubeita, K. E. Coller, J. I. Lee, S. Haverlock-Moyns, S. P. Gross, and G. A. Smith. 2006. The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J. Virol. 80:5494-5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antinone, S. E., and G. A. Smith. 2006. Two modes of herpesvirus trafficking in neurons: membrane acquisition directs motion. J. Virol. 80:11235-11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher, S., B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bucks, M. A., K. J. O'Regan, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 10.del Rio, T., C. J. DeCoste, and L. W. Enquist. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierasch, W. W., D. L. Zimmerman, S. L. Ward, T. K. Vanheyningen, J. D. Romine, and D. A. Leib. 2006. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods 135:197-206. [DOI] [PubMed] [Google Scholar]
- 20.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2004. The pseudorabies virus US3 protein is a component of primary and of mature virions. J. Virol. 78:1314-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1-12. [DOI] [PubMed] [Google Scholar]
- 22.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 23.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaelin, K., S. Dezelee, M. J. Masse, F. Bras, and A. Flamand. 2000. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J. Virol. 74:474-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishiyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp, B. G., S. Bottcher, H. Granzow, M. Kopp, and T. C. Mettenleiter. 2005. Complex formation between the UL16 and UL21 tegument proteins of pseudorabies virus. J. Virol. 79:1510-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 30.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam, N., and G. J. Letchworth. 2000. Bovine herpesvirus 1 UL3.5 interacts with bovine herpesvirus 1 α-transinducing factor. J. Virol. 74:2876-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loomis, J. S., R. J. Courtney, and J. W. Wills. 2003. Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 77:11417-11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luxton, G. W., J. I. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 39.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ng, T. I., W. O. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 43.Ogasawara, M., T. Suzutani, I. Yoshida, and M. Azuma. 2001. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J. Virol. 75:1427-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 47.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smibert, C. A., B. Popova, P. Xiao, J. P. Capone, and J. R. Smiley. 1994. Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68:2339-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 101:16034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sottong, P., R. Harris, B. Graham, B. Rupp, C. Bell, T. Conlon, and J. Klecker. 1976. Purification of herpesvirus nucleocapsids by fluorocarbon extraction. Microbios 16:105-110. [PubMed] [Google Scholar]
- 54.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40:191-197. [DOI] [PubMed] [Google Scholar]
- 57.Trus, B. L., F. L. Homa, F. P. Booy, W. W. Newcomb, D. R. Thomsen, N. Cheng, J. C. Brown, and A. C. Steven. 1995. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J. Virol. 69:7362-7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trus, B. L., W. W. Newcomb, N. Cheng, G. Cardone, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA-filled HSV-1 capsids. Mol. Cell 26:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu, Q., and R. J. Courtney. 1994. Chemical cross-linking of virion envelope and tegument proteins of herpes simplex virus type 1. Virology 204:590-599. [DOI] [PubMed] [Google Scholar]