Abstract
The recent identification of antiretroviral tripartite motif-bearing restriction factors that protect against retroviral infection has revealed a novel branch of innate immunity. The factors target the retroviral capsid and inhibit infectivity soon after the capsid has entered the cytoplasm by an incompletely characterized mechanism. Restriction is species specific. For example, TRIM5α from Old World monkeys, but not humans, restricts human immunodeficiency virus type 1 infection. Here, we identify an antiviral TRIM5 molecule in rabbits that is closely related to antiviral TRIM5 of both primates and cattle. We demonstrate that the rabbit TRIM5 protein is active against divergent retroviruses and leads to a strong block to viral DNA synthesis and infectivity. Furthermore, we show that antiviral activity is directed against the viral capsid and that human TRIM5 proteins are dominant negative to restriction in rabbit cells. We propose that the sequence and restriction characteristics conserved between restriction factors from primates, cattle, and rabbits indicate that these factors have evolved from a common ancestor with antiretroviral properties.
The study of host factors influencing replication of retroviruses has recently uncovered a novel branch of the innate immune system mediated by tripartite motif, or TRIM, proteins. The first TRIM protein to be unambiguously shown to have antiviral properties was rhesus macaque TRIM5α (51). Since this discovery, TRIM5 orthologues from a variety of primates, including humans, have also been shown to have species-specific antiretroviral properties (19, 27, 33, 42, 49, 64, 66). Importantly, an antiretroviral TRIM5-like protein from cattle has been described, indicating that TRIM-mediated restriction of retroviruses is not unique to primates (48, 67). The tripartite motif-bearing protein family is large, comprising around 70 members in humans and mice (reviewed by Nisole et al. [35]). The functions of the vast majority of TRIM proteins are unknown or, at best, poorly understood. How many TRIM proteins are involved in immunity remains unclear, although there are clear suggestions that several TRIM proteins are involved in diverse aspects of immunity, including TRIM19 (14, 55), TRIM25 (16), TRIM22 (9, 56), TRIM21 (28), and possibly TRIM20 (47; reviewed in reference 58). The tripartite motif comprises RING, B-Box, and coiled-coil domains and is otherwise known as an RBCC motif. Many TRIM proteins, including TRIM5α, additionally bear B30.2 domains, also known as PRY SPRY domains, at their C termini. The roles of the various domains in restriction of retroviral infection are not completely clear (12, 24, 30, 63). However, it is likely that all four domains have roles in restriction of infection in vivo. The mechanism of restriction remains incompletely characterized, but recruitment of incoming virions to the proteasome and either accelerated uncoating of the incoming retroviral capsids (CAs) or steric hindrance of the progression of the life cycle have been suggested (2, 41, 52, 58).
It is of great interest to consider how many TRIM5-like genes exist within mammalian genomes and to consider whether these genes have evolved from a common ancestor with antiretroviral activity or whether they have arisen independently, as has been recently suggested for the primate and bovine antiviral TRIM proteins (48). It is also interesting to consider whether the mechanism of restriction is conserved between antiviral TRIM proteins from different species. Here, we identify an antiviral TRIM protein from rabbits and characterize its antiviral activities against divergent retroviruses.
MATERIALS AND METHODS
Identification and molecular cloning of rabbit tripartite motif genes.
Cell lines SIRC (Statens Seruminstitut rabbit cornea), a gift from Yasuhiro Takeuchi, CRFK (Crandel Reese feline kidney), a gift from Yasuhiro Ikeda, and EREp (rabbit kidney) (ATCC) were grown in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal calf serum (Biosera). cDNA was prepared from the rabbit cell line SIRC as follows. Total RNA was purified using TRIzol (Invitrogen) according to the manufacturer's instructions, and this was used to make cDNA by reverse transcription using superscript (Invitrogen) according to the manufacturer's instructions. The cDNA was used as a template for PCR using two primers directed against central regions of TRIM5, forward primer TS1 (5′GGAGGAGGTGACCTGTCC3′) and reverse primer TS16 (5′CATAGTCTAGGAAAACTCCAACACG3′). The 5′ and 3′ ends of the cDNA were cloned by rapid amplification of cDNA ends (RACE) using a second-generation 5′/3′ RACE kit (Roche Applied Science) according to the instructions supplied. Forward internal primer TS2 (5′TGTGGCCACAGCTTCTGCCAAG3′) and nested primer TS6 (5′TCAGGGAAACATTACTGGG3′) were used for 3′ RACE, whereas reverse internal primer TS10 (5′GAGCAGGAGTTTCTCTCCATG3′) and nested primer TS8 (5′CTTGGCAGAAGCTGTGGCC3′) were used for 5′ RACE. The sequence of the rabbit TRIM5 gene has the GenBank accession number EU014879. A search of the rabbit genome, using BLAST (1), for genes similar to the rabbit TRIM5 gene that we had identified returned the rabbit TRIM5 gene as well as a closely related rabbit TRIM gene (rabbit TRIM6) in a single genomic contig (GenBank accession no. AC186253). For phylogenetic analyses, the putative rabbit TRIM6 open reading frame was assembled by stepwise alignment of each rabbit TRIM5 exon sequence with the genomic sequence and subsequent assembly (data not shown).
Phylogenetic analysis.
The nucleotide sequences for human TRIM genes 22 (RefSeq no. NM_006074), 34 (RefSeq no. NM_021616), 5α (RefSeq no. NM_033034), and 6 (RefSeq no. NM_058166); porcine TRIM5 (GenBank no. AY970971); African green monkey TRIM5α (GenBank no. AB210050); rhesus macaque TRIM5α (GenBank no. AY523632); gorilla TRIM5α (AY923178); murine TRIM genes 6 (RefSeq no. NM_001013616), 5 (RefSeq no. NM_175677), 12 (RefSeq no. NM_023835), 30 (RefSeq no. NM_009099), 34 (GenBank no. AF220139), and 21 (GenBank no. CT010336); bovine TRIM genes 5 (Lv1) (GenBank no. DQ380509), 5b (GenBank no. AY970972), and 5d (RefSeq no. XM_864679); and rabbit TRIM5 (GenBank no. EU014879) were manually aligned using the application Se-Al (44). A maximum-likelihood phylogenetic tree was then reconstructed under the general time-reversible model of nucleotide substitution with a proportion of invariable sites and gamma-distributed rate heterogeneity, using the program PAUP* (54). The robustness of the tree topology was assessed by bootstrap analysis with 1,000 replicates.
Viral vector preparation and infectivity assays.
Vesicular stomatitis virus G envelope protein (VSV-G)-pseudotyped, green fluorescent protein (GFP)-encoding retroviral vectors were prepared as previously described (6, 18, 26) by triple transfection of 293T cells with Fugene 6 (Invitrogen). Plasmids used to generate the human immunodeficiency virus type 1 (HIV-1) (4, 69), HIV-2 (17), simian immunodeficiency virus from rhesus macaques (SIVmac) (34), feline immunodeficiency virus (FIV) (43), equine infectious anemia virus (EIAV) (21), N-tropic murine leukemia virus (MLV-N), and MLV-B (8) vectors have been described previously. Virus infectivity was measured by titrating serially diluted virus onto 2 × 105 cells per well in six-well plates. Infected cells were enumerated 48 h later by measuring GFP expression by fluorescence-activated cell sorting (FACS) (BD Biosciences). The HIV-1 (H/SCA) vector was made by cloning the gag gene from H/SCA (37) into the HIV-1 gag-pol expression vector pCMVΔR8.9 (NotI) (22, 69) between the NotI and BglII sites. The SIV encoding HIV-1 CA-p2 was made by replacing the wild-type SIVmac CA-p2 in the SIVmac gag-pol expression vector SIV3+ (34) with the modified SIVmac gag fragment encoding HIV-1 CA-p2, from p239SpSp5′(HIV-1 CA) (13), between the PacI and DraIII sites at SIV3+ positions 1718 and 5460, respectively. The modified vectors were then prepared as described above.
Disruption of rabbit TRIM5 expression with shRNA.
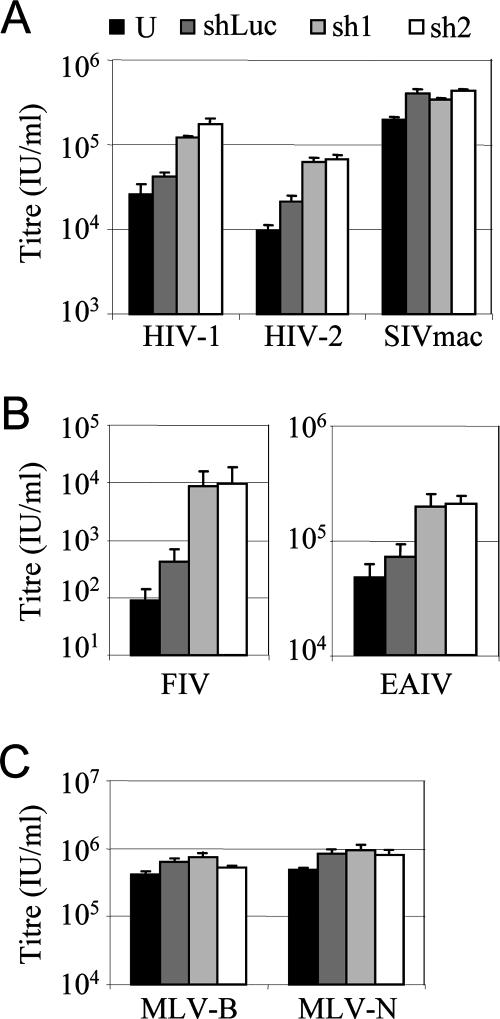
Two short hairpin RNA (shRNA) sequences directed against the rabbit TRIM5 mRNA were designed and cloned into the MLV hairpin RNA delivery vector pSIREN RetroQ (Clontech) according to the manufacturer's instructions. The target sequences were shR1 (5′AGTAGAACCCCTGAGCATA3′) and shR2 (5′CGGTTACCAACTTGAGAAC 3′). The pSIREN control vector bearing the shRNA directed against luciferase (14) was a kind gift from Roger Everett. The pSIREN vectors were packaged into VSV-G-pseudotyped Moloney MLV cores as described above and used to transduce rabbit SIRC cells with neat supernatant and at a high multiplicity of infection (>2). Forty-eight hours later, the cells were replated and their permissivity was tested by titrating serial dilutions of VSV-G-pseudotyped, GFP-encoding vectors derived from MLV-N, MLV-B, HIV-1, HIV-2, SIVmac, FIV, or EIAV on 2 × 105 cells per well in six-well plates. The percentage of infected cells was measured by FACS 48 h after GFP-encoding vector infection, and data are plotted as numbers of infectious units/ml in Fig. 2.
FIG. 2.
Introduction of two independent shRNA sequences directed against rabbit TRIM5 into SIRC cells increases their permissivity to restricted retroviruses. SIRC cells were transduced with a high multiplicity of infection (>2) with the MLV pSIREN RetroQ vectors, each bearing one of two independently designed shRNAs (sh1 and sh2) directed against rabbit TRIM5 or shRNA targeting luciferase (shLuc) as a control. Forty-eight hours later, the permissivity of these cells, as well as unmodified SIRC cells (U), was tested by titrating serial dilutions of VSV-G-pseudotyped, GFP-encoding vectors derived from the viruses shown. The percentages of infected cells were measured by FACS 48 h after GFP-encoding vector infection, and data are plotted as numbers of infectious units/ml. Errors are standard deviations for titers determined at three different multiplicities and are representative of data from two independent experiments.
QPCR.
TaqMan quantitative PCR (QPCR) for measurement of viral DNA synthesis was performed using primer/probe sequences specific to GFP as described previously (6). Cells (4 × 105) were infected in six-well plates in triplicate with equivalent doses of virus treated with DNase (70 units/ml for 2 h; Promega, United Kingdom). Six hours after infection, total DNA was extracted from two samples by using a QiaAmp DNA extraction kit (QIAGEN, United Kingdom). The third sample was subjected to FACS analysis 48 h after infection to enumerate infected cells. DNA (100 ng) was subjected to TaqMan QPCR as described previously (6, 60). Absolute numbers of GFP DNA per PCR were determined by reference to a standard curve. The numbers of GFP molecules per 100 ng of total DNA are plotted. As a negative control for plasmid contamination of the viral inoculum, cells were infected with virus that had been boiled for 5 min. QPCR was then performed as described above.
RESULTS
Identification of rabbit TRIM genes closely related to primate TRIM5.
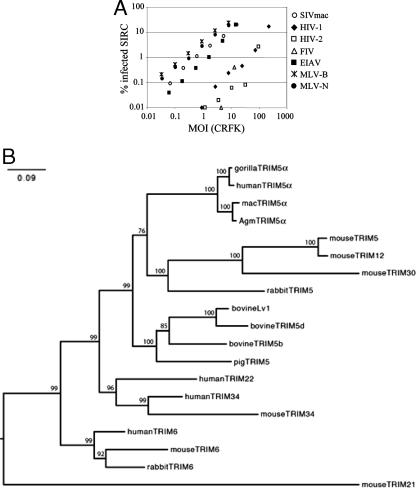
Several studies have demonstrated that rabbit cell lines are particularly resistant to infection with retroviral vectors, even when specific envelope/receptor requirements are obviated using the VSV-G (6, 11, 20, 31). We therefore determined the infectious titers of retroviral vectors derived from a variety of retroviruses on the rabbit cell line SIRC (Fig. 1A). We prepared VSV-G-pseudotyped, GFP-encoding retroviral vectors from MLV-N, MLV-B, HIV-1, HIV-2, EIAV, FIV, and SIVmac and determined their infectious titers on feline CRFK cells, as previously described (6, 18, 67). These cells have been shown to not restrict any of the retroviruses in this study (21, 45, 57, 66). MLV-N, MLV-B, and SIVmac have similarly high titers on CRFK cells and SIRC cells, whereas HIV-1, HIV-2, and FIV are significantly less infectious on SIRC cells than on CRFK cells. EIAV titer is slightly reduced on SIRC cells. We found a weaker but similar pattern of restriction in a second rabbit cell line, EREp (data not shown). We then cloned a TRIM gene closely related to primate TRIM5 genes from SIRC cDNA by PCR using primers specific to conserved central regions of TRIM5, followed by 5′ and 3′ RACE. A search of the rabbit genome sequence by BLAST (1) confirmed that we had cloned a rabbit cDNA. The sequence of the cDNA PCR product differed from the rabbit genome sequence in only two places, both of which were silent (data not shown). We also identified a second rabbit TRIM gene, closely related to primate TRIM6, by BLAST on the same rabbit contig sequence (GenBank accession number AC186253). We were unable to PCR amplify the TRIM6-like rabbit gene from SIRC cDNA, suggesting that this gene is not expressed in these cells (data not shown). The sequences of the rabbit TRIM genes, and a variety of closely related mammalian TRIM sequences, were aligned and assembled into a phylogenetic tree (Fig. 1B). The tree reveals that the rabbit TRIM gene that we cloned by PCR is closely related to primate, bovine, and murine TRIM5 genes. In fact, the antiretroviral TRIM5 genes from rabbit, primates, and cattle form a single cluster with the murine and porcine TRIM5 homologues. Importantly, the human TRIM genes most closely related to TRIM5 form independent clusters, one including orthologues of TRIM34 and TRIM22 and the other including orthologues of TRIM6. We therefore refer to the rabbit TRIM gene under consideration in this study as rabbit TRIM5.
FIG. 1.
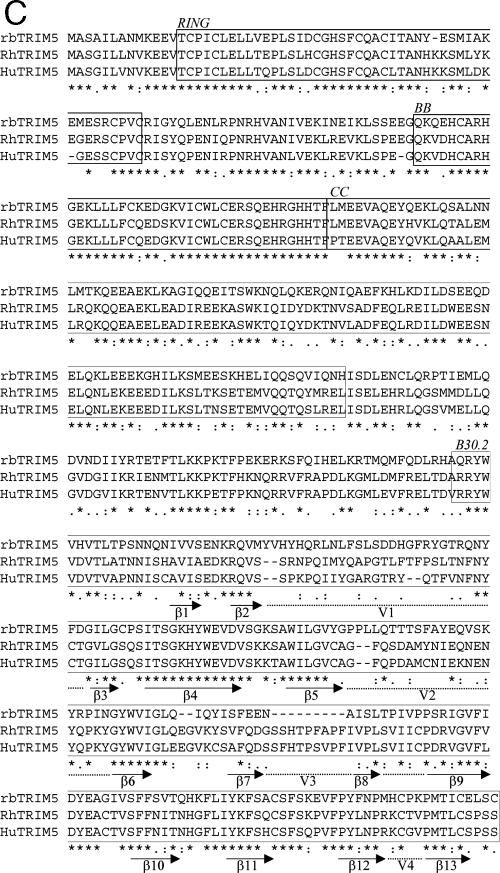
Identification of rabbit TRIM genes closely related to primate TRIM5. (A) VSV-G-pseudotyped, GFP-encoding vectors derived from the viruses shown were titrated on permissive feline CRFK cells and rabbit SIRC cells. The doses of virus are plotted as multiplicities of infection (MOI) on CRFK cells. Data are representative of three independent experiments. (B) A selection of mammalian TRIM nucleotide sequences was aligned and used to construct a maximum-likelihood phylogenetic tree to consider the relatedness of rabbit TRIM genes to mammalian TRIM sequences. Percent bootstrap support values for 1,000 replicates are shown on the branches. Abbreviations: mac, rhesus macaque; Agm, African green monkey. Branch lengths represent the number of substitutions per site. (C) Alignment of protein sequences of rabbit TRIM5 and rhesus macaque and human TRIM5α reveals strong conservation between RING and B-Box 2 (BB) domains and the beta sheet sequences at the core of the B30.2 domain (arrows) (23). The coiled-coil (CC) domain is also shown. Dotted lines mark variable regions V1 to V4. HuTRIM5, human TRIM5; rbTRIM5, rabbit TRIM5; asterisk, identical residue; colon, conserved substitution; period, semiconserved substitution; gap, no conservation.
Alignment of TRIM5 sequences from rabbit, human, and rhesus macaque indicates strong sequence conservation in the RBCC motif, particularly in the RING and B-Box 2 domains (Fig. 1C). There is also reasonably strong conservation in the B30.2 domain, particularly in the regions predicted to form the beta sheets that constitute the core of the B30.2 fold (23, 61). As expected, the sequences of the B30.2 V1, V2, and V3 loops thought to form the viral interaction surface and specificity determinant for TRIM5 restriction (33, 36, 40, 46, 53, 65) are not conserved (Fig. 1C).
Introduction of two independent shRNA sequences directed against rabbit TRIM5 into SIRC cells increases the permissivity of these cells to restricted retroviruses.
To test whether rabbit TRIM5 expression contributes to the poor permissivity of SIRC cells to retroviruses, we expressed two independent shRNAs directed against rabbit TRIM5 in SIRC cells. We then tested the permissivity of these cells to retroviral infection (Fig. 2). Expression of either shRNA specifically increased the titers of poorly infectious HIV-1 and HIV-2 on SIRC cells but did not significantly affect the titer of SIVmac, a virus that has a relatively high titer on these cells (Fig. 2A). Furthermore, the titers of both FIV and EIAV were increased by expression of shRNAs directed against rabbit TRIM5 (Fig. 2B), whereas the infectivities of MLV-N and MLV-B were not affected (Fig. 2C). While expression of the shRNAs in SIRC cells led to an only modest rescue of restricted infection, the rescue of infectivity was specific to viruses with low titers on SIRC cells, namely, HIV-1, HIV-2, EIAV, and FIV but not SIVmac or MLV. These data support the conclusion that rabbit TRIM5 specifically restricts the poorly infectious retroviruses. As a negative control, we expressed an shRNA directed against luciferase (14), which slightly increased the permissivity of SIRC cells in a nonspecific way, underlining the importance of this control.
Expression of rabbit TRIM5 in permissive feline cells confers the ability to restrict retroviral infection.
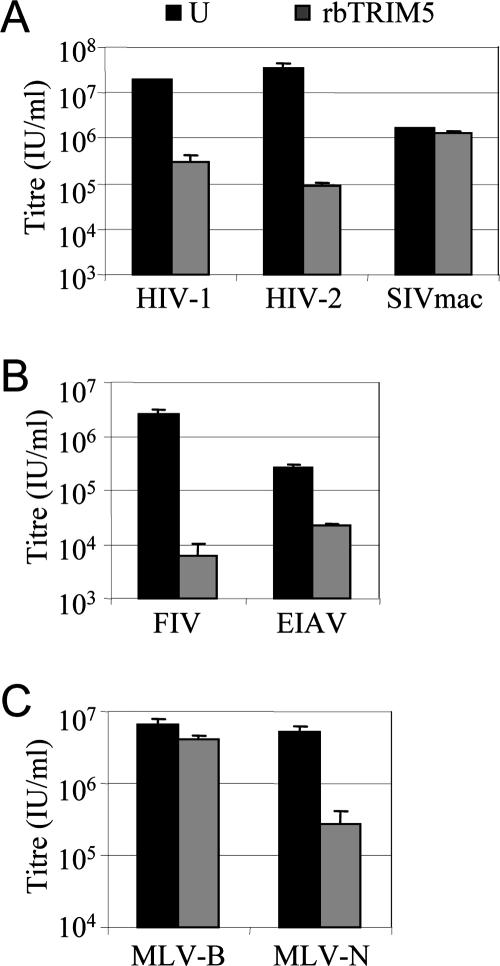
To confirm that rabbit TRIM5 is able to restrict retroviral infection, we expressed the protein in permissive feline CRFK cells and titrated retroviral vectors onto two clones of CRFK cells stably expressing rabbit TRIM5 and unmodified cells as a control (Fig. 3). Cells expressing rabbit TRIM5 were around approximately 2 orders of magnitude less permissive to HIV-1 and HIV-2 infection, whereas the infectivity of SIVmac was not affected (Fig. 3A). CRFK cells expressing rabbit TRIM5 were also less permissive to FIV and EIAV vectors, by 3 and 1 order of magnitude, respectively (Fig. 3B). Surprisingly, CRFK cells expressing rabbit TRIM5 were also 1 order of magnitude less permissive to infection by MLV-N but not by MLV-B, suggesting that overexpression of the TRIM protein broadens its antiviral specificity to MLV-N (Fig. 3C). A broadening of antiviral specificity following overexpression has been reported previously both for TRIM genes and for the gag-like restriction factor Fv1 (8, 39, 66). Introduction of the shRNAs directed against rabbit TRIM5, described in the legend to Fig. 2, into CRFK cells expressing rabbit TRIM5 specifically rescued the infectivities of restricted viruses to an extent similar to that in SIRC cells, confirming the abilities of the shRNAs to reduce rabbit TRIM5 expression levels (data not shown).
FIG. 3.
Expression of rabbit TRIM5 (rbTRIM5) in permissive feline cells confers the ability to restrict retroviral infection. CRFK cells were transduced with MLV retroviral vector encoding rabbit TRIM5 and dsRED express and cloned by dilution. They were then tested for their permissivity to VSV-G-pseudotyped, GFP-encoding retroviral vectors as shown. The data plotted are the titers of these viruses on CRFK cells expressing rabbit TRIM5 versus those on unmodified CRFK cells (U) as a control. Errors are standard deviations for titers determined at three different multiplicities of infection. Data shown are representative of experiments performed on two independent clones of cells.
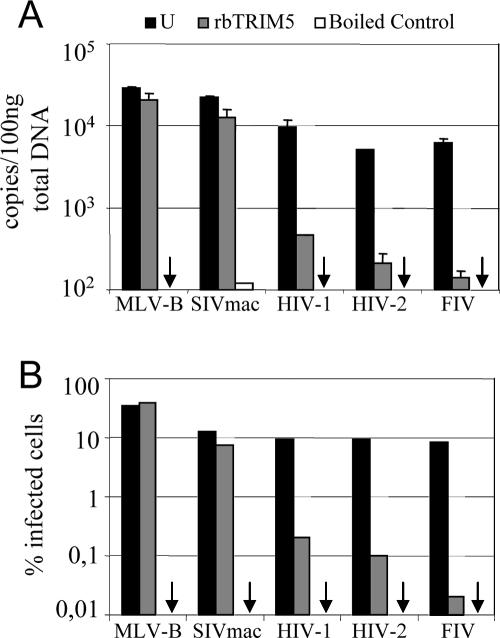
Rabbit TRIM5 blocks DNA synthesis by restricted viruses.
TRIM5α characteristically restricts retroviral infection early after virus entry into target cells, before significant viral reverse transcription occurs (6, 7, 10, 51, 57, 59). Other cases, such as restriction of SIVmac by the New World squirrel monkey TRIM5α protein or restriction of HIV-2 by bovine TRIM5, are exceptions, with strong blocks to infection but not reverse transcription (66, 67). To examine whether rabbit TRIM5 is able to block viral reverse transcription, we infected CRFK cells or CRFK cells expressing rabbit TRIM5 and measured viral DNA levels 6 hours after infection by TaqMan QPCR (Fig. 4). Cells were infected in triplicate, DNA was extracted from two samples for QPCR, and the third sample was subjected to FACS to enumerate infected cells (Fig. 4B). As a negative control, cells were exposed to virus that had been boiled for 5 min to ensure that the DNA signal was due to reverse transcription and not plasmid contamination in the inoculate. Data in Fig. 4A demonstrate that expression of rabbit TRIM5 led to a significant reduction in DNA synthesis by restricted HIV-1, HIV-2, and FIV, compared to that by unrestricted MLV-B and SIVmac (Fig. 4B).
FIG. 4.
Rabbit TRIM5 (rbTRIM5) blocks DNA synthesis by restricted virus. CRFK cells expressing TRIM5 were infected in triplicate with VSV-G-pseudotyped, GFP-encoding vectors derived from the viruses shown. (A) Two samples were extracted for DNA and viral DNA measured by QPCR using primers/probe specific to GFP. Data plotted are the numbers of viral DNA molecules per 100 ng of total DNA after infection of unmodified CRFK cells (U; black bars) or CRFK cells expressing rabbit TRIM5 (gray bars). (B) The third sample was subjected to FACS 48 h after infection to determine the percentage of infected cells. Results for negative-control infections of unmodified cells with boiled virus are shown (white bars or arrows for values below the limit of reliable detection). Errors are standard deviations for duplicate samples and are representative of experiments performed on two independent clones.
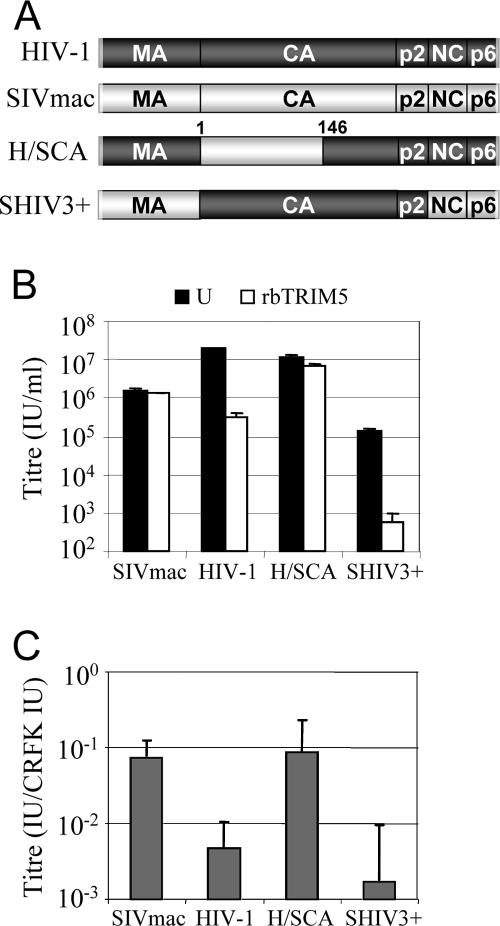
The viral determinant for restriction of HIV-1 by rabbit TRIM5 is the CA.
Previous studies have mapped the viral determinant of sensitivity to restriction by TRIM proteins to the CA protein (7, 10, 42, 57, 66). To test whether the CA determines sensitivity to rabbit TRIM5, we replaced amino acids 1 to 146 of the HIV-1 CA with the equivalent residues from the SIVmac CA to create H/SCA, as previously described (38) (Fig. 5A). We also replaced the entire CA-p2 region of SIVmac with that of HIV-1 to create SIV(HIV-1CA-p2), as previously described (Fig. 5A) (13). VSV-G-pseudotyped, GFP-encoding vectors were produced using the two simian-HIV (SHIV) gag-pol constructs, and their titers were determined on CRFK cells expressing rabbit TRIM5 and unmodified CRFK cells as a control (Fig. 5B). The titers of wild type HIV-1 and SIVmac vectors were also determined for comparison. The titer of the SHIV3+ vector is lower than its wild-type counterpart due to, we presume, a loss in fitness (Fig. 5B). We therefore equalized the doses of the SHIVs and wild-type vectors according to their titers in CRFK cells and measured their titers on SIRC cells. We then plotted the CRFK-adjusted SIRC titers (Fig. 4C). Together, these data demonstrate that the presence of an N-terminal CA domain from SIVmac in HIV-1 allows it to escape restriction both in CRFK cells expressing TRIM5 and in SIRC cells. Furthermore, the presence of an HIV-1 CA in SIVmac causes it to become restricted in both cell lines.
FIG. 5.
The viral determinant for restriction of HIV-1 by rabbit TRIM5 (rbTRIM5) is the CA. (A) Cartoons of HIV-1 and SIVmac gag are shown to illustrate the structure of the HIV-1/SIVmac SHIV hybrids used. H/SCA is HIV-1 with CA amino acids 1 to 146 from SIVmac. SHIV3+ is SIVmac with the CA-p2 domains of HIV-1. MA, matrix; NC, nucleocapsid. (B) Absolute titers of wild-type and SHIV vectors were determined on CRFK cells expressing rabbit TRIM5 (white bars) and unmodified CRFK cells (U; black bars) as a control. (C) Titers of the wild-type and SHIV vectors on rabbit SIRC cells. Doses are standardized on permissive feline CRFK cells. Errors are standard deviations for titers determined at different doses and are representative of experiments performed on two independent clones.
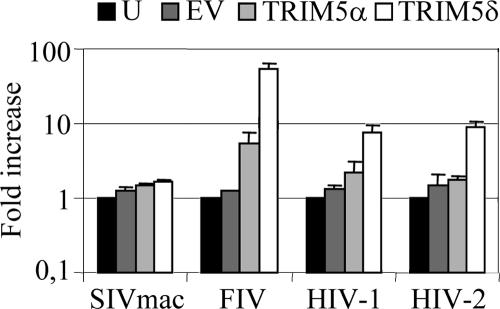
Human TRIM5 isoforms are dominant negatives against restriction of retroviruses in SIRC cells.
A feature of restriction by TRIM5α-related antiviral factors is that the expression of closely related molecules rescues infectivity of restricted viruses through dominant-negative activity. This is true for the splice variants δ and γ of the human TRIM5 gene (39, 51) as well as for closely related proteins from the same species, e.g., human TRIM34 (68) or even TRIM5α from different species (5). We therefore tested whether human TRIM5α or TRIM5δ is dominant negative against the restricting capabilities of SIRC cells by expressing them in SIRC and measuring the infectivities of SIVmac, HIV-1, HIV-2, and FIV (Fig. 6). Expression of human TRIM5α weakly rescued the infectivity of HIV-1 and more strongly rescued the infectivity of FIV. Expression of human TRIM5δ efficiently rescued the infectivities of HIV-1 and HIV-2 by an order of magnitude and that of FIV by close to 2 orders of magnitude. The infectivity of SIVmac was not affected by any of the TRIM5s expressed. As a negative control, we infected cells with empty vector and this did not affect the titer of any of the tester viruses. These data suggest that, like primate TRIM5α, rabbit TRIM5 acts in a multimeric complex (32), although the dominant-negative activity might also be mediated via saturation of a cofactor.
FIG. 6.
Human TRIM5 isoforms are dominant negatives against restriction of retroviruses in SIRC cells. Rabbit SIRC cells were left uninfected (U; black bars) or infected with empty vector (EV; dark gray bars), MLV vector encoding human TRIM5α (light gray bars), or human TRIM5δ (white bars). Forty-eight hours later, the cells were challenged with GFP-encoding SIVmac, FIV, HIV-1, or HIV-2. The increases (n-fold) in infectivity of the GFP-encoding virus are plotted. Errors are standard deviations for parallel infections. Results are representative of two independent experiments performed with independent virus stocks.
DISCUSSION
This study demonstrates that rabbits bear an antiretroviral TRIM gene which clusters in a phylogenetic tree with antiviral TRIM5s from primates and cattle. Moreover, the human TRIM genes most closely related to TRIM5, TRIM genes 6, 34, and 22, cluster independently with their orthologues from mouse, cattle, and rabbit. We therefore propose that the antiviral rabbit protein is a true TRIM5 orthologue, as is the antiretroviral bovine gene previously referred to as bovine Lv1 (48, 67). Orthology indicates a common ancestor for these TRIM5 genes, and we presume that it had antiretroviral properties that have since been selected by pathogenic retroviral infection, leading to the species-specific antiviral properties that we observe today.
Reduction of rabbit TRIM5 expression using shRNAs rescues infectivity with the appropriate specificity, increasing the infectivities of viruses that are poorly infectious in rabbit cells, namely, HIV-1, HIV-2, FIV, and EIAV, but not the relatively infectious viruses SIVmac and MLV (Fig. 2). This specificity strongly suggests that TRIM5 significantly contributes to the poor permissivity of rabbit cells to HIV-1, HIV-2, EIAV, and FIV. The abilities of two independent shRNA sequences to specifically increase infectivity supports this notion. The partial rescue of infectivity suggests that the reduction of rabbit TRIM5 expression in these experiments might be incomplete. Pools of drug-selected SIRC cells expressing shRNAs against rabbit TRIM5 grew poorly consistently, suggesting that strong reduction of this protein is not tolerated (data not shown). Furthermore, the transient assay employed is unlikely to deliver a high dose of shRNA-bearing vector to all the cells in the well, and this also might contribute to the incomplete reduction in rabbit TRIM5 expression. The modest effect of the shRNA could also be explained by the existence of further, as-yet-unidentified antiviral proteins in rabbit cells, and it is also possible that the rabbit cells might lack factors important for the high infectivity seen in feline CRFK cells.
A role for TRIM5 in restriction of retroviruses in rabbit cells is confirmed by expression in feline CRFK cells (Fig. 3). Although it is likely that CRFK cells also express feline TRIM molecules, feline cells have been shown to be very permissive to retroviral vector infection (21, 27, 45, 66). Expression of rabbit TRIM5 significantly reduces the titers of HIV-1, HIV-2, EIAV, and FIV in these cells. Importantly, it does not reduce the infectivity of SIVmac, a virus that is highly infectious in rabbit cells. Strikingly, MLV-N but not MLV-B infectivity is reduced by overexpression of rabbit TRIM5, despite the fact that SIRC cells do not restrict MLV-N in comparison to MLV-B (57) (Fig. 1 and 2). It is possible that the feline CRFK cells express a cofactor that expands the specificity of rabbit TRIM5 to MLV-N, but it is more likely that overexpression of TRIM5 leads to expanded specificity, as has been described for human TRIM5 (66). We imagine that low-affinity interactions between TRIM5 and, for example, the MLV-N CA do not lead to restriction at low expression levels, but when rabbit TRIM5 is highly expressed, this interaction leads to restriction, as shown in Fig. 3. Whether such broadening of specificity represents an artifact of overexpression is unclear, particularly given that TRIM5 expression is up-regulated by interferon (3).
Analysis of DNA synthesis by rabbit TRIM5-restricted viruses indicates strong reduction in reverse transcription by restricted viruses. As in most other cases of TRIM-mediated restriction, the block to viral DNA synthesis is weaker than the block to infectivity, suggesting that many viruses synthesize viral DNA but remain uninfectious (7, 66, 67). This observation has been investigated further by the members of the Hope laboratory, who have shown that inhibition of the proteasome rescues DNA synthesis by TRIM5α-restricted virus but not infectivity (2, 62). This suggests that TRIM5α blocks infection independently of the proteasome and that rapid recruitment to an active proteasome prevents reverse transcription.
The observation that the CA bears the determinant for sensitivity to rabbit TRIM5 is consistent with determinants of sensitivity to other TRIM5 molecules (7, 10, 42, 57, 66) and implies conservation in the mechanism of viral restriction. The dominant-negative activities of human TRIM5α and -δ against rabbit TRIM5 are also consistent with previous observations (5, 39, 51). The simplest explanation for these data is that the human TRIM5 proteins multimerize with rabbit TRIM5 and compromise its ability to restrict through titration of the virus binding B30.2 domains. The stronger activity of TRIM5δ may be due to this shorter molecule being more readily able to interact with a heterologous trimer. It may also be that without a B30.2 domain, it cannot contribute to antiviral activity (39, 51). The strength of the dominant-negative rescue of infectivity is also related to the strength of the restriction, with FIV being both restricted most strongly and rescued most effectively (Fig. 1 and 6).
A number of studies have proposed that rabbits might be developed into an animal model for HIV-1/AIDS. Indeed, several studies have suggested that HIV-1 will replicate in rabbits if they are inoculated with human cells infected with HIV-1 (15, 29). Unfortunately, these promising early results did not culminate in the development of rabbits as an animal model, perhaps because rabbit cells are generally poorly permissive to HIV-1 infection (6, 11, 20). Another study has shown that expression of an HIV-1 receptor (CD4) and a coreceptor (CCR5) on rabbit SIRC cells renders these cells permissive to HIV-1 infection (50). We imagine that these data reflect the fact that while SIRC cells express a restriction factor strongly active against HIV-1, they are not completely nonpermissive to HIV-1 infection (Fig. 1) (6, 11). It is therefore possible that a high-dose infection might lead to infected cells that can secrete HIV-1 into the supernatant, confounding the interpretation of this study. Our data do not rule out the development of rabbits for use as an animal model for HIV/AIDS but do suggest that a knockout of the rabbit TRIM5 gene will be an important component of this work. A recent study has suggested that rabbit cells might be nonpermissive to HIV-1 infection due to a recessive block caused by the lack of a host factor important for HIV-1 replication (11). This notion was based on the observation that heterokaryons between permissive human cells and rabbit SIRC cells were permissive to HIV-1 infection. These data might now be explained by the observation that human TRIM5α and -δ proteins act as dominant negatives to the anti-HIV-1 activity found in SIRC cells (Fig. 6).
The study of antiviral proteins is likely to be influenced by the narrow range of viruses employed. However, it is striking that primate, bovine, and rabbit TRIM5 proteins restrict multiple unrelated retroviruses, whereas the closely related human TRIM proteins 6, 22, and 34 do not (68). This implies that the antiretroviral activities of the TRIM5 orthologues have evolved from a common antiretroviral ancestor, whereas TRIM proteins 6, 22, and 34 have evolved independently and may have alternative functions. Importantly, restriction by the TRIM5 orthologues has common features, such as targeting the viral CA early after viral entry and sensitivity to dominant-negative activities of related TRIM proteins. These observations suggest a conserved antiviral mechanism and support the notion of orthology.
The most important finding in this study is that rabbits bear an active orthologue of TRIM5. This suggests that rabbits have been under selection pressure from pathogenic retroviruses similar to that for primates and cattle. The recent identification of rabbit endogenous lentivirus type K supports this notion (25). Moreover, the discovery of rabbit endogenous lentivirus type K extends the age of lentiviruses to approximately 7 million years and increases the likelihood that lentiviruses have contributed to the selection of TRIM5 antiviral activity. The phylogenetic tree in Fig. 1 also indicates that murine TRIM genes 5, 12, and 30 are homologues of primate TRIM5. It is difficult to designate the true murine TRIM5 orthologue, which evolved from a common TRIM5 ancestor, and TRIM5 paralogues, which were derived by duplication of the TRIM5 orthologue, particularly when their antiviral activities remain as yet uncharacterized. The names assigned to the murine TRIM genes predate our study, and we cannot be sure whether the murine TRIM5 or TRIM30 gene is the true orthologue of primate TRIM5. However, we note that murine TRIM12 is likely to be derived by duplication of murine TRIM5, as they are almost identical, except for the loss of the B30.2 domain in TRIM12 (data not shown).
Cattle also appear to have multiple TRIM5 genes, here named TRIM5b and -d. Pigs have a single sequence, although they may bear further, as-yet-unidentified TRIM5-like genes. It seems certain that the TRIM genes in the TRIM5 cluster (Fig. 1B) are homologues of TRIM5, many of which are likely to have antiretroviral properties. It will be interesting to test this further, although a negative result is difficult to interpret as it may be that the viruses tested simply do not reflect the viruses that have driven TRIM5 selection. However, the TRIM5 proteins identified thus far tend to restrict multiple divergent retroviruses. It is certainly clear that the continued study of TRIM5 and related molecules will help us understand complex host-retrovirus relationships, particularly those that concern species-specific replication and zoonosis. We hope that eventually this information will improve animal models and therapeutics for HIV/AIDS.
Acknowledgments
This work was funded by a senior biomedical research fellowship from the Wellcome Trust (G.J.T.), The European Commission (S.H.), and a Medical Research Council PhD studentship (T.S.).
We thank Roger Everett, Heinrich Gottlinger, Joe Sodroski, Didier Trono, Adrian Thrasher, Andrew Lever, Francois Loic Cosset, Kyriacos Mitrophanous, Eric Poeschla, Yasuhiro Takeuchi, and Yasuhiro Ikeda for reagents, Ben Webb for critical reading of the manuscript, and Ari Fassati for helpful discussion.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. L., E. M. Campbell, X. Wu, N. Vandegraaff, A. Engelman, and T. J. Hope. 2006. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 80:9754-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asaoka, K., K. Ikeda, T. Hishinuma, K. Horie-Inoue, S. Takeda, and S. Inoue. 2005. A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem. Biophys. Res. Commun. 338:1950-1956. [DOI] [PubMed] [Google Scholar]
- 4.Bainbridge, J. W., C. Stephens, K. Parsley, C. Demaison, A. Halfyard, A. J. Thrasher, and R. R. Ali. 2001. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8:1665-1668. [DOI] [PubMed] [Google Scholar]
- 5.Berthoux, L., S. Sebastian, D. M. Sayah, and J. Luban. 2005. Disruption of human TRIM5alpha antiviral activity by nonhuman primate orthologues. J. Virol. 79:7883-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besnier, C., L. Ylinen, B. Strange, A. Lister, Y. Takeuchi, S. P. Goff, and G. Towers. 2003. Characterization of murine leukemia virus restriction in mammals. J. Virol. 77:13403-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bock, M., K. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouazzaoui, A., M. Kreutz, V. Eisert, N. Dinauer, A. Heinzelmann, S. Hallenberger, J. Strayle, R. Walker, H. Rubsamen-Waigmann, R. Andreesen, and H. von Briesen. 2006. Stimulated trans-acting factor of 50 kDa (Staf50) inhibits HIV-1 replication in human monocyte-derived macrophages. Virology 356:79-94. [DOI] [PubMed] [Google Scholar]
- 10.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cutino-Moguel, T., and A. Fassati. 2006. A phenotypic recessive, post-entry block in rabbit cells that results in aberrant trafficking of HIV-1. Traffic 7:978-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Griffero, F., N. Vandegraaff, Y. Li, K. McGee-Estrada, M. Stremlau, S. Welikala, Z. Si, A. Engelman, and J. Sodroski. 2006. Requirements for capsid-binding and an effector function in TRIMCyp-mediated restriction of HIV-1. Virology 351:404-419. [DOI] [PubMed] [Google Scholar]
- 13.Dorfman, T., and H. G. Gottlinger. 1996. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70:5751-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filice, G., P. M. Cereda, and O. E. Varnier. 1988. Infection of rabbits with human immunodeficiency virus. Nature 335:366-369. [DOI] [PubMed] [Google Scholar]
- 16.Gack, M. U., Y. C. Shin, C. H. Joo, T. Urano, C. Liang, L. Sun, O. Takeuchi, S. Akira, Z. Chen, S. Inoue, and J. U. Jung. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916-920. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda, Y., M. K. Collins, P. A. Radcliffe, K. A. Mitrophanous, and Y. Takeuchi. 2002. Gene transduction efficiency in cells of different species by HIV and EIAV vectors. Gene Ther. 9:932-938. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda, Y., L. Ylinen, M. Kahar-Bador, and G. J. Towers. 2004. The influence of gag on HIV-1 species specific tropism. J. Virol. 78:11816-11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.James, L. C., A. H. Keeble, Z. Khan, D. A. Rhodes, and J. Trowsdale. 2007. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. USA 104:6200-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 25.Katzourakis, A., M. Tristem, O. G. Pybus, and R. J. Gifford. 2007. From the cover: discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. USA 104:6261-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keckesova, Z., L. Ylinen, and G. J. Towers. 2006. Cyclophilin A renders HIV-1 sensitive to old world monkey but not human TRIM5a antiviral activity. J. Virol. 80:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong, H. J., D. E. Anderson, C. H. Lee, M. K. Jang, T. Tamura, P. Tailor, H. K. Cho, J. Cheong, H. Xiong, H. C. Morse III, and K. Ozato. 2007. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J Immunol. 179:26-30. [DOI] [PubMed] [Google Scholar]
- 29.Kulaga, H., T. Folks, R. Rutledge, M. E. Truckenmiller, E. Gugel, and T. J. Kindt. 1989. Infection of rabbits with human immunodeficiency virus 1. A small animal model for acquired immunodeficiency syndrome. J. Exp. Med. 169:321-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, X., Y. Li, M. Stremlau, W. Yuan, B. Song, M. Perron, and J. Sodroski. 2006. Functional replacement of the RING, B-box 2, and coiled-coil domains of tripartite motif 5alpha (TRIM5alpha) by heterologous TRIM domains. J. Virol. 80:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKnight, A., P. R. Clapham, and R. A. Weiss. 1994. HIV-2 and SIV infection of nonprimate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology 201:8-18. [DOI] [PubMed] [Google Scholar]
- 32.Mische, C. C., H. Javanbakht, B. Song, F. Diaz-Griffero, M. Stremlau, B. Strack, Z. Si, and J. Sodroski. 2005. Retroviral restriction factor TRIM5alpha is a trimer. J. Virol. 79:14446-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakayama, E. E., H. Miyoshi, Y. Nagai, and T. Shioda. 2005. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5α determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79:8870-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Negre, D., P. E. Mangeot, G. Duisit, S. Blanchard, P. O. Vidalain, P. Leissner, A. J. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J. L. Darlix, and F. L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7:1613-1623. [DOI] [PubMed] [Google Scholar]
- 35.Nisole, S., J. P. Stoye, and A. Saib. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 36.Ohkura, S., M. W. Yap, T. Sheldon, and J. P. Stoye. 2006. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80:8554-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Owens, C. M., P. C. Yang, H. Gottlinger, and J. Sodroski. 2003. Human and simian immunodeficiency virus capsid proteins are major viral determinants of early, postentry replication blocks in simian cells. J. Virol. 77:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passerini, L. D., Z. Keckesova, and G. J. Towers. 2006. Retroviral restriction factors Fv1 and TRIM5α act independently and can compete for incoming virus before reverse transcription. J. Virol. 80:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perron, M. J., M. Stremlau, M. Lee, H. Javanbakht, B. Song, and J. Sodroski. 2007. The human TRIM5α restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 81:2138-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poeschla, E. M., F. Wong-Staal, and D. Looney. 1998. Efficient transduction of non dividing human cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 44.Rambaut, A. 1996, posting date. Se-Al: Sequence Alignment Editor. http://tree.bio.ed.ac.uk/software/seal/.
- 45.Saenz, D. T., W. Teo, J. C. Olsen, and E. M. Poeschla. 2005. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5α proteins. J. Virol. 79:15175-15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. USA 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaner, P., N. Richards, A. Wadhwa, I. Aksentijevich, D. Kastner, P. Tucker, and D. Gumucio. 2001. Episodic evolution of pyrin in primates: human mutations recapitulate ancestral amino acid states. Nat. Genet. 27:318-321. [DOI] [PubMed] [Google Scholar]
- 48.Si, Z., N. Vandegraaff, C. O'Huigin, B. Song, W. Yuan, C. Xu, M. Perron, X. Li, W. A. Marasco, A. Engelman, M. Dean, and J. Sodroski. 2006. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc. Natl. Acad. Sci. USA 103:7454-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song, B., H. Javanbakht, M. Perron, H. Park do, M. Stremlau, and J. Sodroski. 2005. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol. 79:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speck, R. F., M. L. Penn, J. Wimmer, U. Esser, B. F. Hague, T. J. Kindt, R. E. Atchison, and M. A. Goldsmith. 1998. Rabbit cells expressing human CD4 and human CCR5 are highly permissive for human immunodeficiency virus type 1 infection. J. Virol. 72:5728-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 52.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. USA 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stremlau, M., M. J. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (* and other methods), 4th ed. Sinauer Associates, Sunderland, MA.
- 55.Tavalai, N., P. Papior, S. Rechter, M. Leis, and T. Stamminger. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tissot, C., and N. Mechti. 1995. Molecular cloning of a new interferon-induced factor that represses human immunodeficiency virus type 1 long terminal repeat expression. J. Biol. Chem. 270:14891-14898. [DOI] [PubMed] [Google Scholar]
- 57.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Towers, G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 60.Towers, G. J., D. Stockholm, V. Labrousse-Najburg, F. Carlier, O. Danos, and J. C. Pages. 1999. One step screening of retroviral producer clones by real time quantitative PCR. J. Gene Med. 1:352-359. [DOI] [PubMed] [Google Scholar]
- 61.Woo, J. S., J. H. Imm, C. K. Min, K. J. Kim, S. S. Cha, and B. H. Oh. 2006. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 25:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, X., J. L. Anderson, E. M. Campbell, A. M. Joseph, and T. J. Hope. 2006. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. USA 103:7465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yap, M. W., G. B. Mortuza, I. A. Taylor, and J. P. Stoye. 2007. The design of artificial retroviral restriction factors. Virology 365:302-314. [DOI] [PubMed] [Google Scholar]
- 64.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 66.Ylinen, L., Z. Keckesova, S. J. Wilson, S. Ranasinghe, and G. J. Towers. 2005. Differential restriction of HIV-2 and SIVmac by TRIM5α alleles. J. Virol. 79:11580-11587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ylinen, L. M., Z. Keckesova, B. L. Webb, R. J. Gifford, T. P. Smith, and G. J. Towers. 2006. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J. Virol. 80:7332-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang, F., T. Hatziioannou, D. Perez-Caballero, D. Derse, and P. D. Bieniasz. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396-409. [DOI] [PubMed] [Google Scholar]
- 69.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotech. 15:871-875. [DOI] [PubMed] [Google Scholar]