Abstract
In this study, we investigated the cell entry characteristics of dengue virus (DENV) type 2 strain S1 on mosquito, BHK-15, and BS-C-1 cells. The concentration of virus particles measured by biochemical assays was found to be substantially higher than the number of infectious particles determined by infectivity assays, leading to an infectious unit-to-particle ratio of approximately 1:2,600 to 1:72,000, depending on the specific assays used. In order to explain this high ratio, we investigated the receptor binding and membrane fusion characteristics of single DENV particles in living cells using real-time fluorescence microscopy. For this purpose, DENV was labeled with the lipophilic fluorescent probe DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt). The surface density of the DiD dye in the viral membrane was sufficiently high to largely quench the fluorescence intensity but still allowed clear detection of single virus particles. Fusion of the viral membrane with the cell membrane was evident as fluorescence dequenching. It was observed that DENV binds very inefficiently to the cells used, explaining at least in part the high infectious unit-to-particle ratio. The particles that did bind to the cells showed different types of transport behavior leading to membrane fusion in both the periphery and perinuclear regions of the cell. Membrane fusion was observed in 1 out of 6 bound virus particles, indicating that a substantial fraction of the virus has the capacity to fuse. DiD dequenching was completely inhibited by ammonium chloride, demonstrating that fusion occurs exclusively from within acidic endosomes.
Dengue virus (DENV) is an enveloped, positive-strand RNA virus belonging to the family Flaviviridae, which also includes tick-borne encephalitis virus, yellow fever virus, and West Nile virus. Flavivirus virions contain three structural proteins: the C (capsid) protein, the M (membrane) protein, and the E (envelope) protein (12, 26). Multiple copies of the C protein associate with the viral RNA to form the nucleocapsid (26). The nucleocapsid is surrounded by a lipid bilayer in which the M and E glycoproteins are inserted. In the infected cell, the M protein is produced as a precursor protein called prM, which is believed to function as a chaperone during the folding and assembly of the E protein (2, 27). The E glycoproteins are assembled as homodimers on the surface of mature virions and mediate the infectious entry of flaviviruses into cells (14, 21). The crystal structure of the major external part of the E glycoprotein has been solved and reveals that the protein contains three distinct domains: domain I is the structurally central domain, domain II is the dimerization domain and contains the fusion peptide, and domain III has an immunoglobulin-like fold and mediates receptor binding (1, 6, 7, 21, 39).
The initial step in the viral life cycle of DENV is attachment of the virus to a cellular receptor. DENV has been proposed to bind to the glycosaminoglycan heparan sulfate, which is expressed on many cell types (5, 10, 17, 25). Additionally, dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin, a mannose-specific C-type lectin, has been identified as a DENV receptor on immature dendritic cells (33, 46). Recently, it has been suggested that both dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin and heparan sulfate act as attachment factors and that cell entry may be mediated by another as yet unidentified receptor (28, 35).
Following receptor binding, flaviviruses enter cells via receptor-mediated endocytosis through which the virus particles are transported to endosomes (11, 19). Acidification of the endosome lumen promotes fusion of the viral membrane with the endosomal membrane (14, 28). For flaviviruses, the following mechanism for membrane fusion is proposed (15, 30, 31). Upon exposure of the virions to low pH, the E homodimers dissociate, and the fusion peptide of the E monomers inserts into the endosomal membrane, which triggers the formation of E trimers. Then, domain III of the trimeric E protein folds back toward the fusion peptide, and the energy released by this conformational change results in the formation of a hemifusion intermediate. Subsequently, a fusion pore is formed, and the nucleocapsid is delivered into the cell cytoplasm.
Viral replication is initiated after uncoating of the nucleocapsid in the cytoplasm of the cell (26). Following RNA replication and translation of the viral structural proteins, immature virions are assembled by budding of newly formed nucleocapsids into the lumen of the endoplasmic reticulum. During this process the virus particles obtain their lipid bilayer envelope in which the prM-E heterodimeric complex is inserted. Subsequently, the particles mature by passing through the Golgi and trans-Golgi network (TGN). In the TGN or another post-TGN compartment, prM is cleaved by furin, resulting in the formation of mature particles containing the M and E proteins. Finally, progeny virus particles are released from the cell by exocytosis (26).
Several studies have shown that, for many viruses, the number of physical particles in a virus preparation is higher than the number of infectious units and also that the specific infectivity numbers vary substantially among different viruses. For example, for alphaviruses, such as Sindbis virus, a PFU-to-particle ratio of ∼1:10 has been observed (44), while for flaviviruses such as DENV and yellow fever virus, a ratio of ∼1:4,000 has been described (4, 19, 20, 40). The high ratios might be explained by a limited capacity of the virus to bind to cells, inefficient uptake of virions in cells, or defects in membrane fusion properties, viral replication, or virus assembly.
In this study, we dissected the early events involved in DENV type 2 (S1 strain) cell entry. It is demonstrated that the concentration of virus particles measured with biochemical methods is 3 to 4 orders of magnitude higher than the number of infectious particles determined by infectivity assays. Subsequently, real-time fluorescence microscopy was used to obtain a direct insight into the binding, entry, and membrane fusion properties of single DENV particles using virus labeled with DiD (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt). The results show that DENV particles do not bind efficiently to cells, which partially explains the low specific infectivity of the virus. Fusion occurs exclusively from within acidic endosomes. It is initiated on average 12 min after the onset of cell entry of the virus. Interestingly, we observed different types of transport dynamics of the virus particles inside cells, leading to membrane fusion in the cell periphery as well as in the perinuclear region. Viral fusion was observed in 17% of the DENV particles that bind to the cell, which shows that a substantial fraction of the virus has the capacity to fuse.
MATERIALS AND METHODS
Cells.
BHK-21 clone 15 cells (BHK-15) were maintained in minimal essential medium ([MEM] Life Technologies, Breda, The Netherlands) supplemented with 10% fetal bovine serum, 25 mM HEPES, 7.5% sodium bicarbonate, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C and 5% CO2. Aedes albopictus C6/36 cells were cultured at 30°C and 5% CO2 in the same medium as BHK-15 cells with the addition of 200 mM glutamine and 100 μM nonessential amino acids. BS-C-1 cells were maintained in MEM Eagle (American Type Culture Collection, Manassas, VA) with 10% fetal bovine serum at 37°C and 5% CO2.
Virus.
DENV serotype 2 strain PR159 S1 was produced by inoculating monolayers of C6/36 cells at a multiplicity of infection of 0.1. DENV particles released from the cells were harvested at 72 h postinfection and cleared from cell debris by low-speed centrifugation. Subsequently, virions were pelleted by ultracentrifugation at 4°C in a Beckman type 19 rotor for 15 h at 30,000 × g. Virus pellets were resuspended in HNE buffer (5 mM HEPES, 150 mM NaCl, 0.1 mM EDTA, pH 7.4) and further purified on 30% glycerol-potassium tartrate density gradients (10 to 35%, wt/vol) by ultracentrifugation in a Beckman SW41 rotor at 4°C for 2 h at 125,000 × g. The 20% to 25% potassium tartrate section was harvested, aliquoted, and stored at −80°C. Prior to experiments, the virus was concentrated and cleared from tartrate using 100-kDa filter devices (Millipore, Amsterdam, The Netherlands). The purity of DENV was evaluated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The identity of the viral proteins was confirmed with Western blotting analysis using antibody MAB8702 (Chemicon, Hampshire, United Kingdom) against DENV type 2. Protein concentration was determined with a Lowry protein determination (34). The distribution of mature and immature particles was determined by use of cryoelectron microscopy. Briefly, small aliquots of purified DENV were applied to perforated carbon-coated electron microscope grids, vitrified in liquid nitrogen, and analyzed as described before (21).
Viral infectivity.
The number of PFU was determined on BHK-15 cells at 34°C. Plaques were counted 7 days after infection. The number of infectious units (IU) was measured by an infectious center assay. DENV was titrated at 34°C for BHK-15 and BS-C-1 cells or at 30°C for C6/36 cells. At 24 h postinfection, cells were fixed to prevent a second round of infection (24) and stained by using antibody MAB8702 to measure infection. In control experiments, 60 μM nocodazole (Sigma, Zwijndrecht, The Netherlands) was added to the cells 30 min prior to the addition of virus. At 1 h postinfection, cells were washed, and medium containing 20 mM ammonium chloride was added to prevent further virus infection. The numbers of infected cells were counted in five random image areas and used for calculation of the titer with the following formula: number of IU/ml = N × (Swell/SImageArea) × dilution factor × 1/V, where N is the average number cells expressing the E protein within a field, Swell is the surface area of the cell culture dish, SImageArea is the surface of the image area, and V is the volume of inoculum. All titration analyses were carried out at least three times.
qPCR.
The number of genome-containing particles (GCPs) was determined by quantitative PCR (qPCR). Viral RNA was extracted from 4 μl of purified virus or 125 μl of unpurified virus by use of a QIAamp Viral RNA mini kit (QIAGEN, Venlo, The Netherlands). Next, cDNA was synthesized from viral RNA with reverse transcription-PCR (RT-PCR) using the forward primer 5′-ACAGGCTATGGCACTGTTACGAT-3′, the reverse primer 5′-TGCAGCAACACCATCTCATTG-3′, deoxynucleoside triphosphates, RNasin (Promega, Leiden, The Netherlands), and Omniscript (QIAGEN). For the qPCR, cDNA was mixed with the TaqMan probe (5′-FAM-AGTGCTCTCCAAGAACGGGCCTCG-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine) (Eurogentec, Maastricht, The Netherlands), the same primers as for the RT-PCR, deoxynucleoside triphosphates, and HotStarTaq DNA polymerase (QIAGEN). DNA was amplified for 40 cycles (15 s at 95°C and 60 s at 60°C) on a Smart-Cycler (Cepheid, Sunnyvale, CA) with the baseline set to 9 cycles. Determination of the number of genomic RNA copies was performed with a standard curve (correlation coefficient of >0.995) of a quantified DNA plasmid containing the DENV prM and E sequence (pcDNA3-prM/E), which was constructed using standard DNA techniques. Determination of the number of GCPs was carried out at least three times. In order to determine the efficiency of the RT-PCR and qPCR, a Sindbis virus expression vector was constructed in which the structural genes of Sindbis were replaced by the structural genes C, prM, and E of DENV using standard DNA techniques. Briefly, in vitro RNA transcripts of the linearized cDNA clone were generated, subjected to a DNase I (Fermentas, St. Leon-Rot, Germany) treatment, and used in the RT-PCR and qPCRs, as described above. In parallel, the RNA content was quantified with a NanoDrop (NanoDrop Technologies, Wilmington, DE). The efficiency of the RT-PCR was calculated by comparing the number of RNA molecules measured by qPCR to the RNA concentration determined by the NanoDrop.
DiD-labeling of DENV.
Approximately 5 × 109 GCPs were mixed with 2 nmol of DiD (Molecular Probes, Eugene, OR) dissolved in dimethyl sulfoxide in a total dimethyl sulfoxide concentration of less than 2.5%. After a 10-min incubation, the unincorporated dye was removed by gel filtration on a Sephadex G-50 column (Pharmacia, Uppsala, Sweden) in HNE buffer. DiD-labeled virus was stored at 4°C and used within 2 days. Emission scans were recorded at an excitation wavelength of 640 ± 4 nm in an AB2 fluorometer (SLM/Aminco, Urbana, IL) in the presence or absence of octaethyleneglycol monododecyl ether (C12E8; Fluka Chemie AG, Buchs, Switzerland). For the determination of the number of incorporated DiD molecules, the fluorescence emission of DiD-labeled virus was measured at 665 ± 4 nm in the presence of C12E8 and compared to a standard curve of DiD. We attempted to estimate the number of individual virus particles labeled with DiD by fluorescence microscopy. Briefly, DiD-labeled DENV was added to a glass coverslip and excited with a 633-nm helium-neon laser. Fluorescent emission was collected by an oil immersion objective with a numerical aperture of 1.45 and imaged through a 665-nm long-pass filter (Chroma, Rockingham, VT) onto a charge-coupled-device camera. Image analysis was performed as described previously (22). Briefly, background and noise were removed by convolving with a Gaussian spatial filter. Virus spots were detected by recursively integrating over bright regions connected to each local maximum. The numbers of viral spots were counted in five random image areas, and the concentration of DiD-labeled DENV was calculated according to the following formula: number of spots/ml = N × (SCoverSlip/SImageArea) × dilution factor × 1/V, where N is the average number of virus spots, SCoverSlip is the surface area of the glass coverslip, SImageArea is the surface of the image area, and V is the volume.
Binding assay.
BS-C-1 cells were cultured on glass coverslips (MatTek, Ashland, MA). To determine the number of cells per coverslip, cells of a control coverslip were trypsinized and counted in a Bürker counting chamber (VWR, Amsterdam, The Netherlands). Prior to the binding experiment, cells were washed with cold serum-free, phenol red-free MEM (MEM−, Life Technologies). Subsequently, DiD-labeled DENV (1.2 × 105 IU) was added to the cells and incubated for at least 15 min on ice. Unbound virus was removed and when indicated added to new cells to initiate a new binding experiment. After removal of unbound virus, cells were washed with MEM−, and the number of bound virus particles was determined with fluorescence microscopy, as described above.
Real-time fluorescence microscopy.
The tracking experiments were performed as described previously (22). Briefly, DiD-labeled DENV was added to BS-C-1 cells on ice under the same experimental conditions as described for the binding assay. In control experiments, cells were treated with 20 mM ammonium chloride (Sigma) before and after binding of virus to the cells. Subsequently, the temperature was rapidly elevated to 37°C and kept at 37°C throughout the tracking experiment using a thermostatted stage and objective heater. The moment of the temperature shift to 37°C is referred to as time point 0 min. DiD-labeled DENV was detected with a 633-nm helium-neon laser. Image series of the fluorescent emission were recorded with a charge-coupled-device camera at 1 frame per s for a total of 25 min. Before and after fluorescence imaging, the localization of the nucleus and plasma membrane of the cell was determined by use of differential interference contrast (DIC) optics. The cell shape and position did not change significantly during the fluorescence recording period. Image analysis was performed as described previously (22). Briefly, trajectories were generated by pairing peaks in each frame to previously established trajectories according to proximity and similarity in intensity. Only those virus particles that moved roughly within the focal plane and showed more than a fivefold increase in fluorescence intensity after membrane fusion were used for single-particle tracking analysis. The two types of transport behavior were differentiated in a subjective manner on the basis of the velocity and direction of the virus particles. The transport behavior of virus particles that showed a rapid movement with a speed of more than 0.5 μm/s toward the nucleus is classified as long-range three-stage transport, whereas particles that exhibited barely detectable movement (<0.2 μm/s) or moved slowly over short distances are referred to as stationary.
RESULTS
Characterization of DENV preparations.
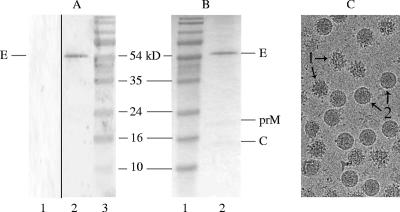
In this study, we investigated the cell entry properties of DENV using biochemical methods and real-time fluorescence microscopy. First, a method was developed to produce highly purified DENV. Briefly, DENV (S1 strain) was cultured on C6/36 mosquito cells, pelleted from the medium, purified on a continuous tartrate gradient, and filtered as described in Materials and Methods. Analysis by Western blotting identified the isolated material as DENV (Fig. 1A). SDS-PAGE analysis showed that virus preparations produced in this manner are pure, although it should be noted that small amounts of protein contamination might go unnoticed (Fig. 1B). Figure 1B shows the E, prM, and C proteins but not M, presumably due to its low abundance in combination with its small size (8.3 kDa). The presence of prM indicates the existence of immature particles in the preparation, which is characteristic for DENV type 2 (3, 13, 16, 32, 36, 38, 50). To further analyze the ratio of mature versus immature particles cryoelectron microscopy images were taken of a similar preparation of DENV S1 strain (21, 51). Figure 1C shows an example. Detailed analysis of multiple micrographs indicated that approximately 40% of the particles in the preparation contain prM.
FIG. 1.
Analysis of purified DENV grown on C6/36 cells. (A) Western blot analysis using MAB8702 antibody against DENV E glycoprotein. Lane 1, mock-infected cells; lane 2, purified DENV; lane 3, protein marker. (B) Gel stained with Coomassie blue. Lane 1, protein marker; lane 2, purified DENV. The positions of the bands for proteins E (55 kDa), prM (18.44 kDa), and C (12.55 kDa) and of the marker are indicated. The viral M protein (8.3 kDa) could not be detected, presumably due to its small size. (C) Cryoelectron micrograph of purified DENV S1. Immature (“spikey”) particles (1) and mature (“smooth”) particles (2) are indicated.
Subsequently, viral infectivity was evaluated by plaque assay on BHK-15 cells, and the concentration of physical virus particles was measured on the basis of a protein determination. A theoretical amount of 2.26 × 10−17 g of protein per virus particle was used for calculation. It was observed that the number of IU was considerably lower than the number of physical virus particles, with values of 8.0 × 107 PFU/ml and 8.6 × 1012 particles/ml, respectively (Table 1). Consequently, the PFU-to-particle ratio was approximately 1:100,000 under the conditions of these experiments.
TABLE 1.
Titer determinations of DENV and DiD-labeled DENV
| Preparation | Virus infectivitya
|
No. of GCPs/ml | No. of particles/ml | IU:GCP ratio | IU:particle ratio | |
|---|---|---|---|---|---|---|
| No. of PFU/ml | No. of IU/ml | |||||
| DENVb | 8.0 × 107 | 1.2 × 108 | 3.1 × 1011 | 8.6 × 1012 | 1:2,600 | 1:72,000 |
| Unpurified DENV | ND | 3.6 × 106 | 8.3 × 109 | ND | 1:2,300 | ND |
| DiD-labeled DENVc | ND | 7.5 × 106 | 1.3 × 1010 | ND | 1:1,800 | ND |
Average of at least two determinations. ND, not done.
Used for preparation of DiD-labeled virus.
Used for binding and tracking experiments.
Due to the fact that a plaque assay could underestimate viral infectivity, the number of IU was also measured by an infectious center assay on BHK-15 cells. In this assay, serial dilutions of virus were added to cells, incubated for 24 h, fixed, and stained for the E protein. The IU titer was 1.2 × 108 IU/ml, which is only slightly higher than the plaque titer (Table 1). Hence, the IU-to-particle ratio is 1:72,000 under the conditions of the experiments. Additionally, the infectious center assay was performed on BS-C-1 cells and C6/36 cells and revealed IU titers of 1.0 × 108 IU/ml and 7.6 × 107 IU/ml, respectively, similar to the titers determined on BHK-15 cells.
The high IU-to-particle ratio prompted us to characterize the DENV preparations in more detail. To this end, the number of GCPs was determined in the same virus preparation using qPCR. RNA was extracted from the virus, and cDNA was synthesized with RT-PCR and amplified with qPCR using a TaqMan probe. The concentration of GCPs was determined by use of a standard curve as described in Material and Methods. It was observed that the DENV preparation used in the above titration experiments contains 3.1 × 1011 GCPs/ml, which results in an IU-to-GCP ratio of 1:2,600 (Table 1).
To investigate whether the high ratio was caused by the purification procedure of the virus, the IU-to-GCP ratio of unpurified virus was also determined (Table 1). The results show that the IU-to-GCP ratio of unpurified virus was similar to that of purified virus, which demonstrates that the purification procedure does not damage the particles and therefore cannot explain the low specific infectivity of the virus.
Repeated analyses of different purified DENV preparations showed that the concentration of physical virus particles based on protein determination was consistently 10- to 25-fold higher than the concentration of GCPs (Table 2). A possible reason for this discrepancy may be that a substantial fraction of the virus particles does not contain an (complete) RNA genome (42). Another possibility is that the protein determination overestimates the concentration of physical virus particles in the preparation due to contaminations, while on the other hand the concentration of GCPs might represent an underestimation caused by incomplete RNA extraction or inefficient RT-PCR and/or qPCRs. In order to determine the efficiency of the RT-PCR and qPCRs, a known amount of in vitro transcribed RNA was subjected to the procedures described in Materials and Methods. The efficiency of the RT-PCR and qPCR was found to be significantly less than 100% (55% ± 7%; n = 8), which partially contributes to the discrepancy between the particle number based on protein quantification and GCPs. In addition, it is also possible that an incomplete RNA extraction contributes to an underestimation of the number of GCPs in the virus preparations.
TABLE 2.
Ratio between GCPs and physical virus particles in different preparations
| Preparation no. | No. of GCPs/mla | No. of particles/mla | GCP:particle ratio |
|---|---|---|---|
| 1 | 3.1 × 1011 | 8.6 × 1012 | 1:28 |
| 2 | 7.8 × 1011 | 7.0 × 1012 | 1:9 |
| 3 | 1.3 × 1012 | 1.4 × 1013 | 1:11 |
| 4 | 6.4 × 1011 | 1.6 × 1013 | 1:25 |
| 5 | 9.9 × 1011 | 1.1 × 1013 | 1:11 |
Values are the average of three determinations.
In conclusion, determination of the IU-to-particle ratio would suggest that under the conditions employed in our studies, the majority of the virus particles produced are not infectious in BHK-15, C6/36, and BS-C-1 cells, since the ratio ranges from 1:2,600 to 1:72,000, depending on the assay used.
Characterization of DiD-labeled DENV.
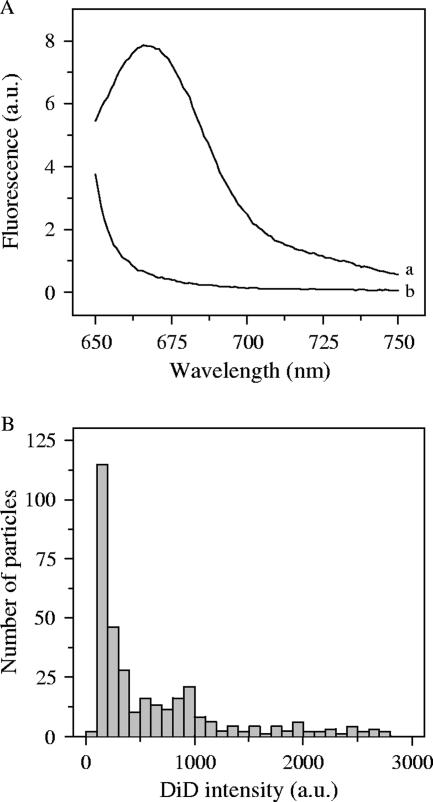
In an attempt to further explain the observed high IU-to-particle ratios, we dissected the receptor binding and membrane fusion properties of DENV using real-time fluorescence microscopy. For these experiments, DENV was labeled with a relatively high surface density of the fluorescent probe DiD in the viral membrane so that its fluorescence was largely quenched but still allowed single virus particles to be clearly detected. Membrane fusion of virus particles labeled in this manner is observed as fluorescence dequenching due to the dilution of the DiD probe into the target membrane. Optimal DiD labeling was observed when 1.9 × 106 IU (corresponding to 5 × 109 GCPs and 1.4 × 1011 physical particles) were mixed with 2 nmol of DiD. After gel filtration, on average 5.5 × 105 IU or 1 × 109 GCPs was recovered, which demonstrates that DiD labeling in itself has no effect on the specific infectivity of the virus (Table 1). To summarize, in this DiD-labeled DENV preparation, 1 IU equals 1,800 GCPs and a maximum of 50,000 physical particles. Under these labeling conditions, the average number of DiD molecules per virus particle (based on protein) was determined to be 550. This results in efficient quenching of the fluorescent probe (Fig. 2A, curve b), as a significant increase in DiD intensity was observed when the viral membrane was dissolved by the addition of C12E8 detergent (Fig. 2A, curve a).
FIG. 2.
Characterization of DiD-labeled DENV particles. (A) Fluorescence emission spectra of DiD-labeled DENV. DiD-labeled DENV was mixed with HNE buffer at 37°C. Emission scans were recorded at wavelengths 650 to 750 nm with excitation at 640 ± 4 nm. Curve a, DiD-labeled DENV in presence of C12E8; curve b, DiD-labeled DENV. (B) The intensity of DiD-labeled DENV was determined with fluorescence microscopy and plotted in a histogram, as described in Materials and Methods. Particles with low fluorescence intensity (0 to 500 AU) were selected for image analysis.
Subsequently, we attempted to estimate the number of DiD-labeled particles in the virus preparation. To this end, DiD-labeled DENV was added to a glass coverslip, and the numbers of fluorescent spots were counted in five random images in which one spot was taken as one virus particle. Three independent labeling experiments revealed that 20, 22, and 44% of the added number of GCPs and, thus, a minimum of 1% of the physical particles were detected. It should be noted, however, that the observed number of DiD-labeled virus particles most likely represents an underestimate, since clusters of virus particles present in the preparation or formed in the counting procedure were considered as a single particle. Indeed, about one-third of all the particles detected in our experiments had a variable high fluorescence intensity and presumably represented clusters of multiple particles (Fig. 2B). On the other hand, the majority of the fluorescent spots had a comparable low DiD fluorescence intensity, which suggests that all virus particles were labeled with DiD to a similar extent (Fig. 2B). In tracking experiments, only those particles that fell into the peak with the lowest fluorescence intensity (0 to 500 arbitrary units [AU]) were selected for data analysis since these most probably resemble single virus particles.
Binding and infectivity of DENV in cells.
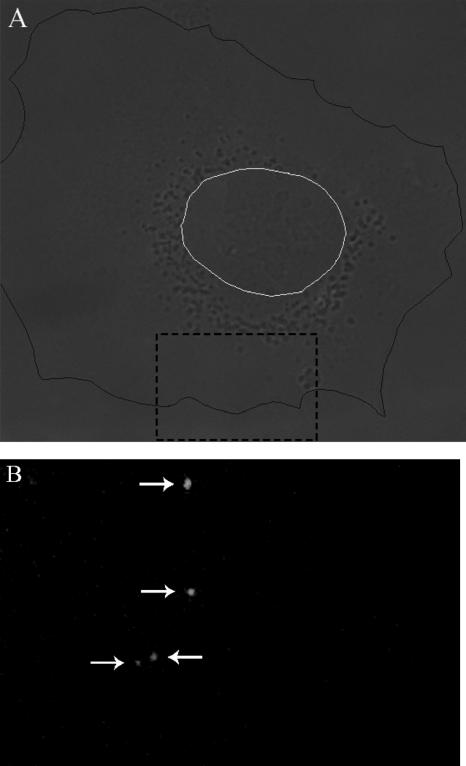
To determine the extent of virus binding, 1.2 × 105 IU of DiD-labeled virus particles was incubated with 200,000 BS-C-1 cells for 15 min at 4°C. This amount of virus equals 0.6 IU, 1,000 GCPs, and a maximum of 28,000 physical particles added per cell. After removal of unbound virus, on average 28 bound DiD-labeled virus particles were observed per cell (Table 3 and Fig. 3). Longer incubation periods (up to 2 h) did not result in a significantly higher extent of virus binding to the cells (data not shown). These observations show that the vast majority of the DENV particles did not bind to the cell surface under the conditions of the experiment.
TABLE 3.
Binding and fusion events of DENV in cellsa
| No. of virus spots per cell | No. of fusion events per cell | % Fusion |
|---|---|---|
| 37 | 3 | 8 |
| 36 | 3 | 8 |
| 29 | 1 | 3 |
| 24 | 5 | 21 |
| 36 | 8 | 22 |
| 19 | 2 | 11 |
| 9 | 3 | 33 |
| 17 | 3 | 18 |
| 14 | 3 | 21 |
| 41 | 8 | 20 |
| 45 | 9 | 20 |
| 24 | 4 | 17 |
| 30 | 4 | 13 |
Average values are as follows: number of virus spots per cell, 28; number of fusion events per cell, 4; percent fusion, 16%.
FIG. 3.
Binding of DiD-labeled DENV to cells. Virus binding was determined by fluorescence microscopy as described in Materials and Methods. (A) Cell image obtained with DIC optics. The plasma membrane is indicated in black, and the nucleus is in white. (B) Fluorescent image of a small portion of the cell shown in panel A (dashed black square) to clearly demonstrate the virus spots. In this image, four virus spots can be seen.
To determine whether these virus particles are simply not capable of binding to cells, we collected unbound virions from a previous binding experiment and added them to new cells to evaluate their binding properties. Interestingly, these particles still bound to the cell surface with the same efficiency as the original DENV preparation (data not shown). This observation suggests that the low binding efficiency cannot be entirely attributed to heterogeneity of the virus preparation, with the large majority of the virions being intrinsically unable to bind to the cells, but, rather, that it is the result of the low binding affinity of the virus particles involved.
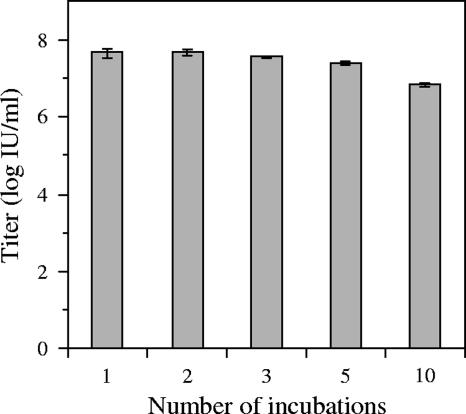
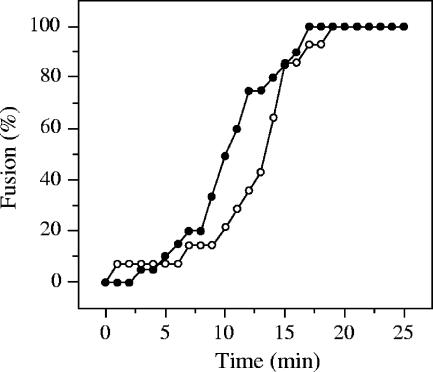
To further investigate whether the low specific infectivity of DENV is, indeed, primarily related to a low binding affinity of the virus for the cells, we determined the infectious properties of collected unbound virus particles. For this purpose, we performed an infectious center assay in which DENV particles were allowed to bind to BS-C-1 cells for 15 min at 4°C, after which unbound virions were collected and added to new cells. This procedure was repeated 10 times. In order to carefully determine the number of positive cells and limit the chance that multiple virus particles infect one cell, the virus concentration in this experiment was reduced by a factor 10 compared to the above binding experiment. Interestingly, the number of positive cells did not significantly change over three incubations, and even after 10 rounds of cell binding the titer of the virus was high (Fig. 4). This observation clearly demonstrates that the titration analyses severely underestimate the actual concentration of infectious virus particles in the preparation due to the low binding affinity of the virus for the cells involved and therefore at least partially explains the high IU-to-particle ratios.
FIG. 4.
Infectious properties of unbound DENV particles in cells. Virus binding was allowed for 15 min at 4°C on BS-C-1 cells, after which the unbound virus particles were collected and transferred to new cells. This procedure was repeated 10 times. Infection was analyzed by E protein expression in cells, and the titer was calculated as described in Material and Methods.
Tracking single DiD-labeled DENV particles in cells.
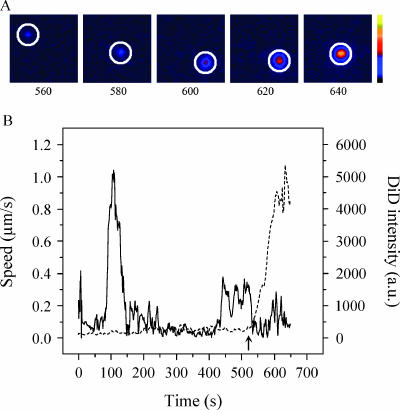
To investigate if virus particles that do bind to the cell surface are internalized and undergo membrane fusion within the cell, we tracked entry of single DENV particles into the cells using real-time fluorescence microscopy (22, 43). The tracking experiments were performed in BS-C-1 cells, since these cells have a flat morphology and, therefore, the virus particles are likely to move in the focal plane of the microscope. Prior to the tracking experiments, DiD-labeled DENV was incubated with BS-C-1 cells for 15 min at 4°C to synchronize infection. After removal of unbound virus, the temperature was rapidly elevated to 37°C to initiate virus cell entry. Subsequently, fluorescence images were recorded at 1 frame per s for 25 min. A typical example of a trajectory is shown in Fig. 5. Fluorescent images of one DiD-labeled particle are depicted at different time points after initiation of virus cell entry to demonstrate the increase of fluorescence intensity upon membrane fusion (Fig. 5A). The DiD intensity of this virus particle starts to increase at 580 s, and after dilution of the probe is completed, the intensity is more than 20-fold of its original value (Fig. 5B, dashed curve). For a real-time movie of single-virus-particle entry and membrane fusion in cells, see the supplemental material.
FIG. 5.
Trajectory of a single DENV particle in a cell. (A) Fluorescence images show one DiD-labeled DENV particle (surrounded by a white circle) at 560, 580, 600, 620, and 640 s after the temperature shift to 37°C. The colored bar indicates the DiD fluorescence intensity from low (black) to high (yellow) intensity. (B) Time trajectories of DiD fluorescence intensity (dashed line) and velocity (solid line) of the same virus particle. Membrane fusion of the virus particle with the endosomal membrane is detected as an increase in DiD fluorescence intensity. The arrow indicates the time point of membrane fusion. a.u., arbitrary units.
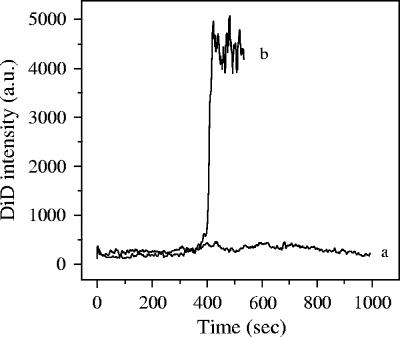
When the cells were treated with ammonium chloride, the DiD signal of all 114 observed virus particles remained constant (Fig. 6, curve a), which demonstrates that the observed increase in DiD intensity in untreated cells (Fig. 6, curve b) represents membrane fusion of DENV particles from within acidic endosomes.
FIG. 6.
DENV entry in cells (ammonium chloride treated). Tracking analysis was performed as described in Materials and Methods. Curve a, time trajectory of the fluorescence intensity of DiD-labeled DENV in ammonium chloride-treated cells; curve b, untreated cells.
Transport dynamics and membrane fusion of DENV in cells.
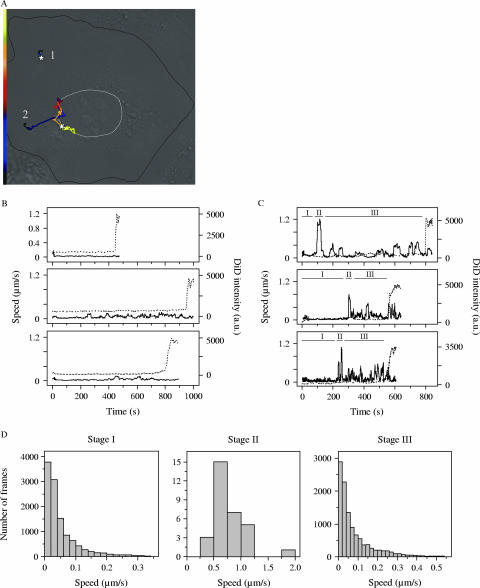
Tracking of single virus particles allowed us to investigate the detailed dynamics of viral transport. Lakadamyali and coworkers found a clear three-stage transport pattern in Chinese hamster ovary cells for influenza virus (22, 23). First, the virus particles move slowly in the cell periphery (stage I). Then, the virus travels rapidly on microtubules to the perinuclear region (stage II), where the particles move intermittently and often in a bidirectional manner (stage III) before fusing with an endosome (22). In BS-C-1 cells, among the influenza virus particles that eventually fused with the endosomes, about 90% showed this type of long-range three-stage movement and fused in the perinuclear region. The remaining 10% of the particles exhibited only stage I movement and fused in the cell periphery (M. Rust and X. Zhuang, unpublished data). This typical behavior of influenza virus prompted us to also examine the details of DENV entry. Analysis of 44 virus trajectories of viruses that fused with endosome membranes showed that DENV also exhibits these two types of transport behavior (Fig. 7A), but the fraction of virus particles that fused in the cell periphery was significantly larger. Approximately two-thirds of DENV particles exhibited the long-range three-stage transport behavior. Quantitative analysis revealed that these DENV particles moved with a velocity of less than 0.2 μm/s in the cell periphery (stage I), followed by a rapid transport with a speed of more than 0.5 μm/s toward the nucleus in stage II (Fig. 7A, C, and D). Subsequently, the particles moved with typically lower velocity in the perinuclear region in an intermittent manner and often appeared to travel back and forth along the same track in stage III before they finally fused with the endosome membrane (Fig. 7A, C, and D). The remaining one-third of DENV particles were relatively stationary (<0.2 μm/s) and exhibited barely detectable movement or moved slowly over short distances, after which they fused in the cell periphery (Fig. 7A and B).
FIG. 7.
Distinct transport behavior of DENV particles in cells. (A) Two virus trajectories in a cell. The morphology of the cell was visualized with DIC optics as described in the legend of Fig. 3. The trajectories are color coded for time to indicate the velocity of the virus particle. The white stars represent the fusion sites. Trajectory 1, virus transport in the cell periphery with time axis from 0 s (black) to 450 s (yellow); trajectory 2, long-range three-stage movement toward the nucleus with time axis from 0 s (black) to 800 s (yellow). (B and C) Three time trajectories of DiD fluorescence intensity (dashed line) and speed (solid line) of stationary and long-range three-stage transport behavior, respectively. The upper graphs in both panels are trajectories 1 and 2 shown in panel A, respectively. In panel C the stages in the long-range three-stage movement are indicated (I, II, and III). (D) Quantitative analysis of the velocity in stages I, II, and III. For every frame within each stage, the velocities of all 30 virus particles that showed long-range three-stage movement are plotted.
To investigate if both transport behaviors lead to infection, the effect of nocodazole, which prevents microtubule-dependent stage II movement, on viral infectivity was measured. The infectious titer dropped to 30% ± 3%, which suggests that microtubule-dependent movement is important but not essential in order to induce viral infection.
Next, we analyzed how much time a DENV particle needs to reach membrane fusion after cell entry is initiated. Here, cellular entry is triggered by raising the temperature rapidly from 4°C to 37°C. The time point of membrane fusion was defined as the moment at which the DiD intensity starts to increase (Fig. 5B, arrow). Figure 8 shows the cumulative number of fusion events in the cell periphery and in the perinuclear region. Virus particles that exhibited long-range three-stage transport fused significantly earlier than particles that remained in the cell periphery (P = 0.002, Mann-Whitney U test), the average time being 10 and 13 min, respectively. The vast majority of all DENV particles had fused within 17 min. Similar fusion kinetics were detected with a twofold higher laser intensity, indicating that the results are not affected by photobleaching of DiD in our experiments.
FIG. 8.
Kinetics of DENV fusion in cells. The time point of membrane fusion was defined as described in the legend of Fig. 5. In total, 14 membrane fusion events in the cell periphery (open circles) were detected, and 30 were detected in the perinuclear region (closed circles). Time point 0 min is the moment of the temperature shift to 37°C. The fraction of fused virus particles was plotted as a function of time.
Finally, we determined how many virus particles that bind to the cell surface eventually undergo membrane fusion from within acidic endosomes. This was found to be, on average, 1 out of 6 particles (Table 3). The particles that did not produce a membrane fusion event possibly failed to enter cells, lacked the capacity to fuse, or may have entered a pathway that does not support membrane fusion.
DISCUSSION
The mechanisms involved in the cell entry process of DENV are poorly understood. In this study, we have used biochemical methods, titration assays, and real-time fluorescence microscopy to investigate the cell entry properties of DENV (S1 strain) in cells. This approach allowed us to determine the binding properties of DENV to cells and to address in real time the transport dynamics of single DENV particles and their membrane fusion characteristics in cells.
DENV is propagated on C6/36 cells, and the titer of virus particles is generally determined by plaque titration or infectious center assays on BHK-15 cells (8, 24, 41). The results presented in this paper show that a typical purified DENV preparation contains 8.0 × 107 PFU and 1.2 × 108 IU per ml. Similar virus titers were observed in C6/36 and BS-C-1 cells. Subsequent determination of the protein and GCP content revealed concentrations of 8.6 × 1012 and 3 × 1011 per ml, respectively, suggesting that there would be a substantial fraction of noninfectious virus particles in the preparation. Also, there appeared to be a consistent discrepancy, by an approximate order of magnitude, between the numbers of virus particles based on protein determination and on genome content. The results show that the number of GCPs is underestimated in our experiments by at least a factor of 2 due to inefficient RT-PCR and qPCRs. The remaining discrepancy may be explained by incomplete RNA extraction and/or protein contamination. Another possibility is that our DENV preparations contain subviral particles lacking a genome. It is well established that flavivirus infection of cells may result in the production of subviral particles (42). These particles are usually smaller than native virus and, unlike native virus, exhibit a T=1 symmetry (9). Since a DENV (S1 strain) preparation very similar to our current virus has been used for structural studies using cryoelectron microscopy image reconstruction (21), it is unlikely that the majority of the particles in the preparation are subviral particles, since this would not have gone unnoticed in the structural analyses. In summary, under the conditions of our experiments the IU-to-GCP or IU-to-particle ratio ranges from approximately 1:2,600 to 1:72,000. Similar PFU-to-GCP ratios have been observed for DENV type 2 strains 16681 (20) and JAM1409 (40) and yellow fever virus strain 17D (4). Irrespective of whether the IU-to-GCP or IU-to-particle ratio is considered, these large numbers indicate that the majority of the produced DENV particles did not initiate an infection under the conditions of commonly used titration assays.
The high IU-to-particle ratio was found to be related to the low efficiency of virus binding to cells. Interestingly, similar binding and infectivity properties were observed when unbound virus particles from a previous experiment were transferred to new cells, indicating that most of the particles in the preparation that are inherently infectious do not, in fact, initiate a productive infection in our titration experiments. This, in turn, implies that the virus preparations used in our studies may, in fact, be more homogeneous than the low specific infectivity numbers would suggest. This conclusion is consistent with the observation that most virus particles are also largely identical in terms of structure since the DENV S1 strain has been used for the solution of a high-resolution density map of the virus (21). Taking these results together, we argue that our DENV preparations are intrinsically more infectious than the high particle-to-IU ratios suggest but that the majority of the infectious virions go unnoticed in the commonly used titration assays due to the limited capacity of the virus to bind to the cells used in these assays. It is very interesting that in a recent paper, Thomas et al. (48) arrive at an almost identical conclusion with regard to low specific infectivity values commonly seen for human immunodeficiency virus type 1 preparations. The authors demonstrate that the observed high ratios of physical particles to infectious titers are primarily related to the limited binding affinity of the virions for cells and that many virus particles that do not initiate a productive infection are not inherently defective but, rather, simply do not get the chance to bind to and infect target cells under the conditions used in titration experiments.
A substantial fraction (17%) of the virus particles that bound to the cell surface eventually underwent membrane fusion. This result is comparable to the degree of fusion described for other viruses, for example, influenza virus and Semliki Forest virus (22, 45, 49). It remains to be determined to what extent these membrane fusion events result in a productive infection. The DENV S1 strain used in this study is highly attenuated and therefore might have a relatively large fraction of particles with a defective genome such that not all particles that fuse initiate infection. Another, more intriguing, possibility is that in some circumstances, after fusion of the viral envelope, the nucleocapsid is not delivered to the proper site for efficient replication (29).
Tracking the entry of single virus particles into cells allowed us to gain a detailed insight into the dynamics of DENV internalization, transport, and the cellular localization of membrane fusion. Roughly two-thirds of the particles showed a three-stage transport pattern. First, the virus particles enter the cell and move slowly in the cell periphery with a speed of less than 0.2 μm/s. Then, the particles move in a unidirectional manner toward the nucleus with a velocity of more than 0.5 μm/s, after which the particles move intermittently and often bidirectionally in the perinuclear region before fusing with the endosomal membrane. The remaining one-third of the fused DENV particles was nearly stationary (<0.2 μm/s) and stayed in the cell periphery where membrane fusion was observed. This behavior was similar to that described for influenza virus (22), albeit that the fraction of influenza virions that were nearly stationary and fused in the cell periphery was substantially smaller.
The above results suggest that DENV particles may possibly be targeted to different transport routes. In this respect it is interesting that DENV particles purified from cell culture contain a mixture of mature and immature particles (3, 13, 36, 37, 50) in which immature particles often predominate (3, 13, 36). Functional analysis has shown that immature DENV particles do have the capacity to bind to cells and are able to initiate viral infection (18, 38). Currently, it is not clear whether DENV particles have evolved in such a way that cleavage of prM to M is not required for viral infectivity or that immature particles are infectious due to cleavage by furin upon cell entry (47). SDS-PAGE analysis of our DENV S1 preparation showed a protein band corresponding to the molecular weight of prM, indicating the existence of immature particles. Cryoelectron microscopy analysis showed that, on average, 40% of the DENV S1 particles are released by the cell in their immature form. Therefore, one possible explanation for our findings might be that immature particles, since they are structurally very different from mature particles (51), may exhibit a different trafficking behavior from the mature particles.
The different types of transport dynamics lead to membrane fusion at the periphery and perinuclear regions of the cell. Interestingly, we observed that particles that show long-range three-stage transport fused significantly earlier than particles fused in the cell periphery. Future studies are required to investigate whether the two types of transport behavior reflect entry via distinct endocytic pathways. In both cases, membrane fusion activity of DENV is strictly dependent on exposure of the virus to low pH since the addition of weak bases during cell entry completely inhibited membrane fusion. The observation that exposure of the virus to low pH is an obligatory step in the infectious entry of DENV is in agreement with Randolph and Stollar, who observed that the threshold for fusion of DENV with the plasma membrane is pH 6.5 (37).
This is the first study describing the binding, transport behavior, and membrane fusion characteristics of single DENV virus particles in living cells. Future research will be required to elucidate which membrane fusion events lead to a productive DENV infection. We expect that this technique will dissect important aspects of the cell entry properties of DENV and, thus, ultimately the pathogenesis of the disease.
Supplementary Material
Acknowledgments
We thank Izabela Zybert and Melike Lakadamyali for many stimulating discussions, Chen Chen for his help in counting the number of DiD-labeled virions, and Tjarko Meijerhof for virus growth and purification.
This work was supported by the Pediatric Dengue Vaccine Initiative, by The Netherlands Organization for Scientific Research under the auspices of the Foundation of Medical Sciences, by the University of Groningen, by the Ter Meulen fund of the Royal Netherlands Academy of Arts and Sciences (travel grant to H.M.S.), by the National Institutes of Health (National Institute of General Medical Sciences), and by a Packard Science and Engineering Fellowship (to X.Z.). X.Z. is a Howard Hughes Medical Institute investigator.
Footnotes
Published ahead of print on 29 August 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, and F. X. Heinz. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, R., S. Wang, C. Osiowy, and A. C. Issekutz. 1997. Activation of endothelial cells via antibody-enhanced dengue virus infection of peripheral blood monocytes. J. Virol. 71:4226-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, H. G., A. Nitsche, A. Teichmann, S. S. Biel, and M. Niedrig. 2003. Detection of yellow fever virus: a comparison of quantitative real-time PCR and plaque assay. J. Virol. Methods 110:185-191. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3:866-871. [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., T. Maguire, and R. M. Marks. 1996. Demonstration of binding of dengue virus envelope protein to target cells. J. Virol. 70:8765-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond, M. S., D. Edgil, T. G. Roberts, B. Lu, and E. Harris. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 10.Germi, R., J. M. Crance, D. Garin, J. Guimet, H. Lortat-Jacob, R. W. Ruigrok, J. P. Zarski, and E. Drouet. 2002. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292:162-168. [DOI] [PubMed] [Google Scholar]
- 11.Gollins, S. W., and J. S. Porterfield. 1984. Flavivirus infection enhancement in macrophages: radioactive and biological studies on the effect of antibody on viral fate. J. Gen. Virol. 65:1261-1272. [DOI] [PubMed] [Google Scholar]
- 12.Hahn, Y. S., R. Galler, T. Hunkapiller, J. M. Dalrymple, J. H. Strauss, and E. G. Strauss. 1988. Nucleotide sequence of dengue 2 RNA and comparison of the encoded proteins with those of other flaviviruses. Virology 162:167-180. [DOI] [PubMed] [Google Scholar]
- 13.He, R. T., B. L. Innis, A. Nisalak, W. Usawattanakul, S. Wang, S. Kalayanarooj, and R. Anderson. 1995. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J. Med. Virol. 45:451-461. [DOI] [PubMed] [Google Scholar]
- 14.Heinz, F. X., K. Stiasny, and S. L. Allison. 2004. The entry machinery of flaviviruses. Arch. Virol. Suppl. 2004:133-137. [DOI] [PubMed] [Google Scholar]
- 15.Heinz, F. X., K. Stiasny, G. Puschner-Auer, H. Holzmann, S. L. Allison, C. W. Mandl, and C. Kunz. 1994. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology 198:109-117. [DOI] [PubMed] [Google Scholar]
- 16.Henchal, E. A., J. M. McCown, D. S. Burke, M. C. Seguin, and W. E. Brandt. 1985. Epitopic analysis of antigenic determinants on the surface of dengue-2 virions using monoclonal antibodies. Am. J. Trop. Med. Hyg. 34:162-169. [DOI] [PubMed] [Google Scholar]
- 17.Hilgard, P., and R. Stockert. 2000. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology 32:1069-1077. [DOI] [PubMed] [Google Scholar]
- 18.Huang, K. J., Y. C. Yang, Y. S. Lin, J. H. Huang, H. S. Liu, T. M. Yeh, S. H. Chen, C. C. Liu, and H. Y. Lei. 2006. The dual-specific binding of dengue virus and target cells for the antibody-dependent enhancement of dengue virus infection. J. Immunol. 176:2825-2832. [DOI] [PubMed] [Google Scholar]
- 19.Ishak, R., D. G. Tovey, and C. R. Howard. 1988. Morphogenesis of yellow fever virus 17D in infected cell cultures. J. Gen. Virol. 69:325-335. [DOI] [PubMed] [Google Scholar]
- 20.Keelapang, P., R. Sriburi, S. Supasa, N. Panyadee, A. Songjaeng, A. Jairungsri, C. Puttikhunt, W. Kasinrerk, P. Malasit, and N. Sittisombut. 2004. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J. Virol. 78:2367-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lakadamyali, M., M. J. Rust, H. P. Babcock, and X. Zhuang. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100:9280-9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lakadamyali, M., M. J. Rust, and X. Zhuang. 2006. Ligands for clathrin-mediated endocytosis are differentially sorted into distinct populations of early endosomes. Cell 124:997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambeth, C. R., L. J. White, R. E. Johnston, and A. M. de Silva. 2005. Flow cytometry-based assay for titrating dengue virus. J. Clin. Microbiol. 43:3267-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, E., M. Pavy, N. Young, C. Freeman, and M. Lobigs. 2006. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antivir. Res. 69:31-38. [DOI] [PubMed] [Google Scholar]
- 26.Lindenbach, D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 27.Lorenz, I. C., S. L. Allison, F. X. Heinz, and A. Helenius. 2002. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 76:5480-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozach, P. Y., L. Burleigh, I. Staropoli, E. Navarro-Sanchez, J. Harriague, J. L. Virelizier, F. A. Rey, P. Despres, F. Arenzana-Seisdedos, and A. Amara. 2005. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J. Biol. Chem. 280:23698-23708. [DOI] [PubMed] [Google Scholar]
- 29.Marsh, M., and R. Bron. 1997. SFV infection in CHO cells: cell-type specific restrictions to productive virus entry at the cell surface. J. Cell Sci. 110:95-103. [DOI] [PubMed] [Google Scholar]
- 30.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313-319. [DOI] [PubMed] [Google Scholar]
- 31.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13-22. [DOI] [PubMed] [Google Scholar]
- 32.Murray, J. M., J. G. Aaskov, and P. J. Wright. 1993. Processing of the dengue virus type 2 proteins prM and C-prM. J. Gen. Virol. 74:175-182. [DOI] [PubMed] [Google Scholar]
- 33.Navarro-Sanchez, E., R. Altmeyer, A. Amara, O. Schwartz, F. Fieschi, J. L. Virelizier, F. Arenzana-Seisdedos, and P. Despres. 2003. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4:723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson, G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83:346-356. [DOI] [PubMed] [Google Scholar]
- 35.Pokidysheva, E., Y. Zhang, A. J. Battisti, C. M. Bator-Kelly, P. R. Chipman, C. Xiao, G. G. Gregorio, W. A. Hendrickson, R. J. Kuhn, and M. G. Rossmann. 2006. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell 124:485-493. [DOI] [PubMed] [Google Scholar]
- 36.Putnak, R., D. A. Barvir, J. M. Burrous, D. R. Dubois, V. M. D'Andrea, C. H. Hoke, J. C. Sadoff, and K. H. Eckels. 1996. Development of a purified, inactivated, dengue-2 virus vaccine prototype in Vero cells: immunogenicity and protection in mice and rhesus monkeys. J. Infect. Dis. 174:1176-1184. [DOI] [PubMed] [Google Scholar]
- 37.Randolph, V. B., and V. Stollar. 1990. Low pH-induced cell fusion in flavivirus-infected Aedes albopictus cell cultures. J. Gen. Virol. 71:1845-1850. [DOI] [PubMed] [Google Scholar]
- 38.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174:450-458. [DOI] [PubMed] [Google Scholar]
- 39.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 40.Richardson, J., A. Molina-Cruz, M. I. Salazar, and W. Black. 2006. Quantitative analysis of dengue-2 virus RNA during the extrinsic incubation period in individual Aedes aegypti. Am. J. Trop. Med. Hyg. 74:132-141. [PubMed] [Google Scholar]
- 41.Roche, R. R., M. Alvarez, M. G. Guzman, L. Morier, and G. Kouri. 2000. Comparison of rapid centrifugation assay with conventional tissue culture method for isolation of dengue 2 virus in C6/36-HT cells. J. Clin. Microbiol. 38:3508-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Russell, P. K., W. E. Brandt, and J. M. Dalrymple. 1980. Chemical and antigenic structure of flaviviruses, p. 503-529. In R. W. Schlesinger (ed.), The togaviruses: biology, structure, replication. Academic Press, New York, NY.
- 43.Sieczkarski, S. B., and G. R. Whittaker. 2002. Dissecting virus entry via endocytosis. J. Gen. Virol. 83:1535-1545. [DOI] [PubMed] [Google Scholar]
- 44.Smit, J. M., R. Bittman, and J. Wilschut. 1999. Low-pH-dependent fusion of Sindbis virus with receptor-free cholesterol- and sphingolipid-containing liposomes. J. Virol. 73:8476-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stegmann, T., H. W. Morselt, J. Scholma, and J. Wilschut. 1987. Fusion of influenza virus in an intracellular acidic compartment measured by fluorescence dequenching. Biochim. Biophys. Acta 904:165-170. [DOI] [PubMed] [Google Scholar]
- 46.Tassaneetrithep, B., T. H. Burgess, A. Granelli-Piperno, C. Trumpfheller, J. Finke, W. Sun, M. A. Eller, K. Pattanapanyasat, S. Sarasombath, D. L. Birx, R. M. Steinman, S. Schlesinger, and M. A. Marovich. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas, G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell. Biol. 3:753-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, J. A., D. E. Ott, and R. J. Gorelick. 2007. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J. Virol. 81:4367-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vonderheit, A., and A. Helenius. 2005. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLOS Biol. 3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 73:2547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.