Abstract
Previous analysis of hepatitis B virus (HBV) indicated base pairing between two cis-acting sequences, the 5′ half of the upper stem of ɛ and φ, contributes to the synthesis of minus-strand DNA. Our goal was to identify other cis-acting sequences on the pregenomic RNA (pgRNA) involved in the synthesis of minus-strand DNA. We found that large portions of the pgRNA could be deleted or substituted without an appreciable decrease in the level of minus-strand DNA synthesized, indicating that most of the pgRNA is dispensable and that a specific size of the pgRNA is not required for this process. Our results indicated that the cis-acting sequences for the synthesis of minus-strand DNA are present near the 5′ and 3′ ends of the pgRNA. In addition, we found that the first-strand template switch could be directed to a new location when a 72-nucleotide (nt) fragment, which contained the cis-acting sequences present near the 3′ end of the pgRNA, was introduced at that location. Within this 72-nt region, we uncovered two new cis-acting sequences, which flank the acceptor site. We show that one of these sequences, named ω and located 3′ of the acceptor site, base pairs with φ to contribute to the synthesis of minus-strand DNA. Thus, base pairing between three cis-acting elements (5′ half of the upper stem of ɛ, φ, and ω) are necessary for the synthesis of HBV minus-strand DNA. We propose that this topology of pgRNA facilitates first-strand template switch and/or the initiation of synthesis of minus-strand DNA.
Hepatitis B virus (HBV), the prototype member of the Hepadnaviridae family, is a liver-tropic virus with a small, double-stranded DNA genome. HBV replicates by reverse transcription of an RNA intermediate termed pregenomic RNA (pgRNA) (17). The pgRNA has several roles during virus replication. It is the mRNA for the translation of two viral replication proteins: the polymerase (P) and the capsid subunit (C). In the cytoplasm of the hepatocyte, pgRNA is selectively encapsidated along with P protein into capsid particles (2, 8). It is within these viral capsids that pgRNA serves as the template for reverse transcription.
RNA encapsidation requires that P protein recognize and bind an encapsidation signal (ɛ), a stem-loop within the 5′ end of pgRNA (2, 15). P protein acts as primer and reverse transcriptase to initiate the synthesis of minus-strand DNA, using a bulge within ɛ as a template (20, 21). Synthesis of minus-strand DNA pauses after 4 nucleotides (nt). At this point, nascent minus-strand DNA, which is covalently bound to P protein (6, 16, 18, 19), switches templates and anneals to a complementary 4-nt acceptor site (AS) within the 3′ copy of DR1 (Fig. 1) (16, 18, 19). Synthesis of minus-strand DNA resumes from this location.
FIG. 1.
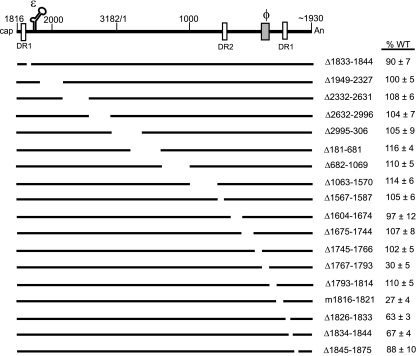
Schematic representation of HBV WT pgRNA and variants. The black lines represent either full-length pgRNA or pgRNAs expressed from variants. The white vertical boxes represent the direct repeat (DR) sequences, DR1:1822-1832 and DR2:1588-1598. The 5′ end of pgRNA is at nt 1816. The poly(A) tail begins at approximately nt 1930. The encapsidation signal, ɛ (nt 1845 to 1905), and a previously identified cis element, φ (nt 1771 to 1789), are indicated. The names of the deletion variants reflect the deleted nucleotides on the HBV pgRNA (inclusive). The m1816-1821 variant changes 6 nt to the respective Watson-Crick partners. On the right are the mean values and standard deviations of the relative levels of minus-strand DNA compared to the WT reference. The ability of a virus to synthesize minus-strand DNA was determined as the level of minus-strand DNA divided by the sum of pgRNA and minus-strand DNA levels. The mean values represent analysis from at least six independent transfections of each variant. The representations of the deleted sequences are not drawn to scale.
ɛ plays a crucial role in at least two processes: pgRNA encapsidation and initiation of minus-strand DNA synthesis (2, 4, 15). More recently, we (1) and others (13) demonstrated that the 5′ half of the upper stem of ɛ base pairs with another sequence, φ, which is near the 3′ end of pgRNA, to make an important contribution to the synthesis of minus-strand DNA. For the 5′ half of the upper stem of ɛ and φ to base pair, a conformational reorganization of pgRNA takes place. Specifically, the upper stem of ɛ is disrupted and φ anneals to the 5′ half of the upper stem of ɛ. The mechanism of this reorganization is not understood but could require other cis-acting sequences on the pgRNA and/or trans-acting factors.
For the present study we wanted to know whether additional cis-acting sequences contribute to the synthesis of minus-strand DNA. A set of deletion and substitution variants that collectively represent ∼95% of the HBV genome were analyzed. We found that most of the pgRNA is dispensable for the synthesis of minus-strand DNA. We removed large portions of the pgRNA without any appreciable effect on the level of minus-strand DNA, indicating that synthesis of minus-strand DNA does not depend on the specific size of the template. Also, substitutions of heterologous sequence into pgRNA could be copied into DNA. Our analysis revealed two new cis-acting sequences: a 6-nt region immediately upstream and a 19-nt region downstream of the AS. In addition, a 72-nt fragment containing the cis-acting sequences around the AS could direct first-strand template switch to a new location when inserted elsewhere on the pgRNA.
Furthermore, we learned that the cis-acting sequence (ω) present downstream of the AS functions by base pairing with a subregion of φ. Previous studies showed that base pairing between the adjacent portion of φ and the 5′ half of the upper stem of ɛ was important for function (1, 13). Thus, φ appears to act as a scaffold by base pairing with two regions: ω and the 5′ half of the upper stem of ɛ. We propose that base pairing juxtaposes the nascent minus-strand DNA and the AS to facilitate first-strand template switch, although contributions to either initiation of minus-strand DNA synthesis or elongation cannot be excluded.
MATERIALS AND METHODS
Molecular clones.
All molecular clones of HBV were derived from the subtype ayw (GenBank accession number V01460). The C of the unique EcoRI site (GAATTC) was designated nucleotide position 1. The names of mutants indicate the first and last nucleotide altered. Deletion variants are indicated by the “Δ” prefix and substitution variants by the “m” prefix.
The wild-type (WT) HBV reference virus, NL84, is null for P, C, X, and surface proteins and has been described previously (1). LJ96 provided the HBV replication proteins in trans and has been described previously (9).
Variants Δ1833-1844, Δ2332-2631, Δ2632-2996, Δ2996-304, Δ1069-1570, Δ1604-1674, and Δ1675-1744 have been described previously (9). Variants Δ1744-1766, Δ1767-1793, and Δ1793-1814 have been described previously (1). Variants Δ1949-2327, Δ181-681, Δ682-1069, and Δ1567-1588 were derived from NL84 by removing restriction fragments or by oligonucleotide-directed mutagenesis. The variant m1816-1821 has 6 nt mutated at the 3′ copy of the sequence. Variants Δ1826-1833, Δ1834-1844, Δ1845-1875, and m1816-1821 were made in the NL84 background. To prevent the potential expression of a truncated form of the precore protein from the partial core gene found in the 3′ end of the pgRNA, these variants had a mutation that changed the start codon (T-to-C change at nt 1813).
HBV/LacZ and HBV/DHBV chimeras were made in the NL84 background. LacZ sequence was amplified from plasmid pON249 (5). DHBV was amplified from a plasmid containing DHBV3 sequence. Both LacZ and DHBV oligonucleotides had restriction sites of HBV appended to the 5′ ends. The PCR-amplified fragment was cloned into the analogous restriction sites in NL84.
In the analysis to introduce a new AS into pgRNA, clones were made by amplifying the relevant sequences by PCR using HBV-derived oligonucleotides. Variant mφ1767-1913 contained the changes A1776T and G1777C within the introduced fragment. The resulting fragments were cloned after nt 181, 824, or 1575 into NL84 using restriction endonucleases. To clone after nt 181, 824, and 1575, restriction enzymes AvrII, BstZ17I, and RsrII, respectively, were used.
Cell culture, transfection, and isolation of encapsidated nucleic acid.
The human hepatoma cell line Huh7 was used in all experiments. Cells were cultured and transfected as previously described (1). Viral nucleic acid was harvested from cytoplasmic capsids as previously described (1).
Analysis of viral nucleic acid.
Primer extension was performed on encapsidated minus-strand DNA and pgRNA by using the two end-labeled oligonucleotides 1661+ and 1948− as described previously (1). For Δ1604-1674 and Δ1675-1744, oligonucleotide 1556+ was used to detect minus-strand DNA instead of oligonucleotide 1661+. Oligonucleotide 1556+ has a sequence complementary to nt 1556 to 1573 on the HBV ayw genome. Oligonucleotide 1556+ anneals at nt 1556 on the minus-strand DNA and extends to its 5′ end, resulting in 200- and 201-nt products for Δ1604-1674 and Δ1675-1744, respectively.
For Southern blotting analysis, viral DNA was first heat denatured at 95°C for 3 min and then placed on ice. Samples were electrophoresed through a 1.25% Tris-borate-EDTA agarose gel at 2 V/cm for 18 h. Southern blotting was done as previously described (10). A minus-strand-specific RNA probe spanning nt 3096 to 264 was used to detect minus-strand DNA. This probe does not detect DNA synthesized from encapsidated spliced RNA. The membrane was exposed in a phosphorimaging cassette and scanned with a Molecular Dynamics Typhoon apparatus (model 8600).
Primer extension with two primers in a single reaction was performed on viral minus-strand DNA isolated from variants where sequences were introduced after nt 824. Two end-labeled oligonucleotides, 1661+ and 749+, were used. Oligonucleotide 1661+ anneals at position 1661 on minus-strand DNA and extends to the 5′ end (normal AS), resulting in a 165-nt product. This oligonucleotide had sequence complementary to nt 1661 to 1685 on the HBV ayw genome. Oligonucleotide 749+ anneals at position 749 on minus-strand DNA and extends to the 5′ end (introduced AS), resulting in a 136-nt product. This oligonucleotide had sequence complementary to nt 749 to 769 on the HBV ayw genome. The primer extension reaction mixtures contained 1× ThermoPol buffer [10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100 (New England Biolabs)], 0.2 mM (each) deoxynucleoside triphosphates, 2 U of Vent exo-DNA polymerase (New England Biolabs), and 0.8 pmol of 32P-end-labeled primer. Viral DNA from one-tenth of a 60-mm plate was analyzed. Each reaction was in a final volume of 10 μl. ins@824 plasmid was digested with HphI restriction enzyme, and primer extension was performed with both primers. Primer extension reactions on plasmid DNA produced extension products from both primers. The ratio of the amount of product from each primer was used to correct for the efficiency with which each primer was end labeled and the ability with which each primer was able to extend on its template. Sequencing ladders were generated by using individual oligonucleotides and HBV plasmid DNA as a template as described previously (1). Primer extension products were electrophoresed on a 5% denaturing polyacrylamide gel. The gel was dried, exposed in a phosphorimaging cassette, and scanned by using a Molecular Dynamics Typhoon (model 8600).
RESULTS
Most of the pgRNA is dispensable for the synthesis of minus-strand DNA.
The variants illustrated in Fig. 1 were analyzed for their ability to make minus-strand DNA. Huh7 cells were cotransfected with plasmids that expressed derivatives of pgRNA and a plasmid that provided the replication proteins in trans, as described previously (9). Encapsidated nucleic acid was isolated from cells 4 days after transfection. Two oligonucleotides were used simultaneously in a primer extension reaction to measure the levels of pgRNA and minus-strand DNA (1). The ability of a variant to synthesize minus-strand DNA was defined as the level of minus-stand DNA divided by the sum of minus-strand DNA and pgRNA and then compared to the WT reference.
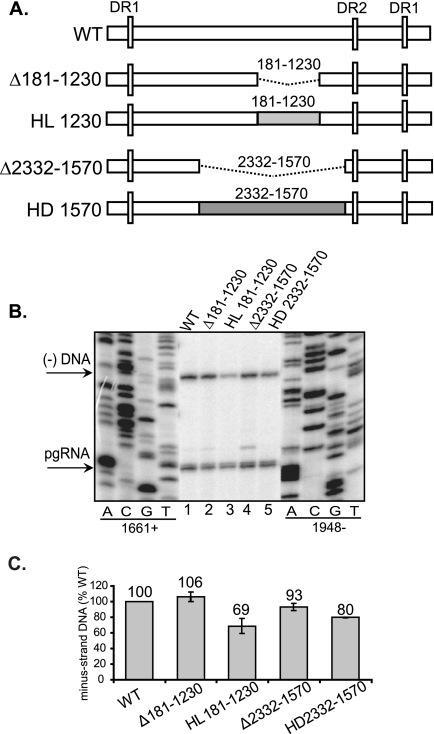
We found that most of the variants synthesized minus-strand DNA at levels similar to that of the WT reference, except for Δ1767-1793, m1816-1821, Δ1826-1833, and Δ1834-1844 (Fig. 1 and 2). This result indicates that most of the pgRNA does not contain cis-acting sequences for the synthesis of minus-strand DNA. Given that the majority of deletions (Fig. 1 and 2) do not affect this process, we wanted to determine whether short derivatives of pgRNA could support normal synthesis of minus-strand DNA. Thus, we deleted either 1.0 kb (Δ181-1230) or 2.4 kb (Δ2332-1570) from the pgRNA (Fig. 3A). As indicated in Fig. 3B and C, variants Δ181-1230 and Δ2332-1570 synthesized minus-strand DNA at levels similar to the WT reference. This result indicates that the size of the pgRNA template does not influence the synthesis of minus-strand DNA.
FIG. 2.
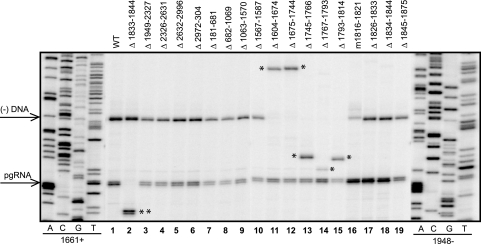
Representative primer extension analysis of variants. Replicative intermediates of WT reference and variants were isolated from cytoplasmic capsids from Huh7 cells and analyzed by primer extension to measure the levels of pgRNA and minus-strand DNA. The oligonucleotides 1661+ and 1948− were used. The positions of the 5′ ends of pgRNA and minus-strand DNA are indicated by arrows on the left. The single asterisk indicates the altered positions at which minus-strand DNA migrates reflecting the size of the deletion. The double asterisks indicate the altered position at which pgRNA migrates, reflecting the size of the deletion. Oligonucleotide 1556+ was used instead of 1661+ to measure levels of minus-strand DNA for Δ1604-1674 and Δ1675-1744 (lanes 11 and 12).
FIG. 3.
Large deletions and heterologous substitutions of the pgRNA are tolerated during the synthesis of minus-strand DNA. (A) WT HBV and HBV/LacZ (HL) or HBV/DHBV (HD) chimeric variants. The white rectangles represent HBV sequences. The dotted lines represent deletions. The coordinates of the deletions are inclusive. The light and dark gray boxes represent LacZ and DHBV sequences, respectively, that were substituted for HBV sequences. The vertical white rectangles represent the DR sequences. (B) Primer extension analysis of replicative intermediates of WT and deletion and substitution variants harvested from cytoplasmic capsids from Huh7 cells. Oligonucleotides 1661+ and 1948− were used to measure minus-strand DNA and pgRNA, respectively. The 5′ ends of minus-strand DNA and pgRNA are indicated on the left. (C) Proportions of minus-strand DNA relative to the WT reference. The level of synthesis of minus-strand DNA is defined as the amount of minus-strand DNA divided by the sum of pgRNA and minus-strand DNA. The values for all variants are normalized to the WT reference. The mean values are from at least six independent transfections of each variant. The error bars indicate standard deviations.
Our findings led us to predict that substitutions of heterologous sequences would not affect the synthesis of minus-strand DNA. To test this hypothesis, LacZ or DHBV sequences were substituted instead of HBV sequence (Fig. 3A). Substitution variants HL181-1230 and HD2332-1570 synthesized minus-strand DNA at 69 and 80% the level of the WT reference (Fig. 3C). Since HD2332-1570 encompasses the region substituted by HL181-1230, the lower levels of minus-strand DNA could be attributed to the sequence of the substitution. Even though substitution variants showed a slight decrease in levels of minus-strand DNA, this analysis shows that foreign sequences can be reverse transcribed into DNA. In summary, these results corroborate findings from the deletion analysis, where most of the HBV genome is not necessary for the synthesis of minus-strand DNA.
Although most of the genome did not make cis-acting contributions to minus-strand DNA synthesis, we identified two new regions that did. These two cis-acting sequences flank the 4-nt AS. Changing 6 nt (m1816-1821) immediately upstream of the AS reduced the level of minus-strand DNA to 27% of the level of the WT reference (Fig. 1 and 2). Deleting 19 nt (Δ1826-1833 and Δ1834-1844) downstream of the AS also reduced levels of minus-strand DNA (63 and 67%, respectively, compared to the level of the WT reference) (Fig. 1 and 2).
In summary, this analysis of the HBV genome demonstrates that most of the pgRNA does not play an active role in the synthesis of minus-strand DNA. Our results indicate that the cis-acting sequences that contribute to the synthesis of minus-strand DNA are located near the 5′ and 3′ ends of the pgRNA.
The sequence between nt 1767 and 1838 is sufficient to direct first-strand template switch to a new location.
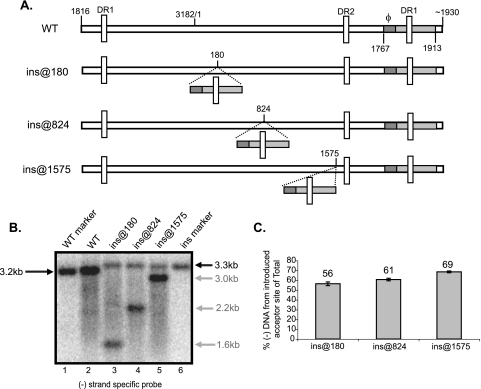
With the exception of ɛ, the cis-acting sequences for the synthesis of minus-strand DNA are located within the 3′ end of pgRNA, i.e., φ, the two new sequences identified in Fig. 1 and the 4-nt AS. We sought to determine whether this region was sufficient to direct the first-strand template switch to a new location on the pgRNA. We inserted a 147-nt segment (nt 1767 to 1913) in different locations within the pgRNA (Fig. 4A). Viral DNA was isolated from cytoplasmic capsids from transfected Huh7 cells. The DNA was denatured, and Southern blotting was performed. The membranes were probed (see Materials and Methods) such that DNAs derived from spliced mRNAs (7) were not detected. Southern blotting showed two bands consistent in size with minus-strand DNA elongated from the normal and the introduced AS (Fig. 4B, lanes 3, 4, and 5). Primer extension mapped the 5′ end of the new minus-strand DNA species to the AS within the introduced fragment (data not shown). Also, the new minus-strand DNA species were covalently linked to the P protein (data not shown). Overall, we found that the region between nt 1767 and 1913 was sufficient to direct the first-strand template switch to new locations on the pgRNA.
FIG. 4.
Nucleotides 1767 to 1913 direct the first-strand template switch to a new AS when introduced at different locations on the pgRNA. (A) Representation of WT HBV and insertion variants. The gray rectangle represents nt 1767 to 1913 (inclusive) and contains φ (dark gray). For variants ins@180, ins@824, and ins@1575, nt 1767 to 1913 have been inserted after nt 180, 824, and 1575, respectively. The location of the direct repeat (DR) sequences are indicated by vertical white rectangles. The nucleotide coordinates of the 5′ and 3′ ends of pgRNA are indicated. (B) Southern blot of WT and insertion variants. DNA replicative intermediates harvested from Huh7 cells were denatured prior to electrophoresis. A minus-strand specific RNA probe spanning nt 3096 to 264 was used to detect minus-strand DNA. This probe does not detect DNA synthesized from encapsidated spliced RNA. WT and insertion genome length markers were used to indicate the size of single-stranded DNA arising from the AS (lanes 1 and 6). The sizes of minus-strand DNA arising from normal and introduced AS are indicated in black and gray, respectively. (C) For each insertion variant, gray bars represent minus-strand DNA arising from introduced AS as a proportion of total minus-strand DNA. Total minus-strand DNA is the sum of completely elongated minus-strand DNA from normal and introduced AS. Minus-strand DNA derived from spliced RNA are not detected in this analysis due to probe choice. The mean values represent analysis from at least six independent transfections of each variant. The error bars indicate standard deviations.
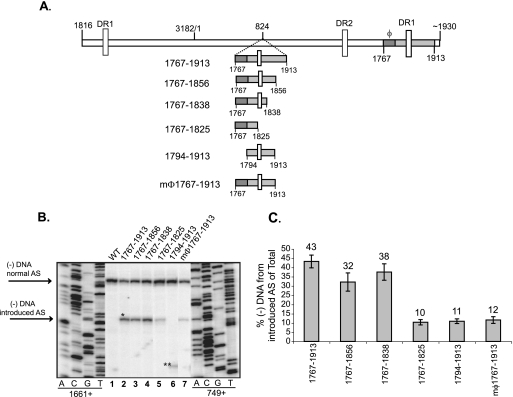
In order to determine the smallest contiguous region that was sufficient to direct the first-strand template switch to a new location, truncations were made from the 5′ and 3′ ends of the region from nt 1767 to 1913 (Fig. 5A). Primer extension analysis was performed with two primers in the same reaction to detect the 5′ ends of minus-strand DNA from the normal and introduced AS. When φ was removed (variant 1794-1913), only ∼10% of minus-strand DNA compared to the WT reference was made from the introduced AS (Fig. 5B, lane 6, and 5C). In addition, mutating φ at the introduced site, such that base pairing with the 5′ half of the upper stem of ɛ is disrupted (mφ1767-1913), also reduced the levels of minus-strand DNA (Fig. 5B, lane 7, and 5C). Inserting nt 1767 to 1838 resulted in minus-strand DNA levels similar to levels resulting from inserting nt 1767 to 1913, whereas inserting nt 1767 to 1825 resulted in a reduction in levels of minus-strand DNA (Fig. 5B, lanes 2, 4, and 5, and 5C). Based on these findings, we have identified the smallest contiguous region, nt 1767 to 1838, that can direct the template switch to a new location. We conclude that sequences surrounding the normal AS play crucial roles in accurately placing the nascent minus-strand DNA at the normal AS.
FIG. 5.
Nucleotides 1767 to 1838 are sufficient to direct first-strand template switch to a new AS when introduced at another location on the pgRNA. (A) WT HBV and insertion variants. The gray rectangle represents the sequence introduced after nt 824. The dark gray rectangle represents φ. The 5′ and 3′ ends of the introduced sequence are indicated. The coordinates are inclusive of region inserted after nt 824. The locations of the DR sequences are indicated by vertical white rectangles. The 5′ and 3′ ends of the pgRNA are also indicated. (B) Primer extension analysis of WT and insertion variants. Oligonucleotides 1661+ and 749+ were used to measure minus-strand DNA arising from the normal and introduced AS, respectively. The 5′ ends of minus-strand DNA arising from normal and introduced AS are indicated. The single and double asterisks indicate the positions of minus-strand DNA arising from the introduced AS. The double asterisks indicate the altered position of minus-strand DNA reflecting the 27-nt difference between the oligonucleotide 749+ annealing site and the introduced AS. (C) For each insertion variant, the gray bars represent minus-strand DNA arising from the introduced AS as a proportion of total minus-strand DNA. Total minus-strand DNA is determined as the sum of minus-strand DNA from the normal and introduced AS. Primer extension detects all minus-strand DNAs within the capsid, i.e., incompletely elongated minus-strand DNAs and minus-strand DNA derived from pgRNA and spliced RNAs. This could account for the difference in levels of minus-strand DNA from the introduced AS for ins@824 in the analysis in Fig. 4C. The mean values represent analysis from at least six independent transfections. The error bars represent the standard deviation.
Novel cis-acting sequence downstream of the AS functions by base pairing with part of φ.
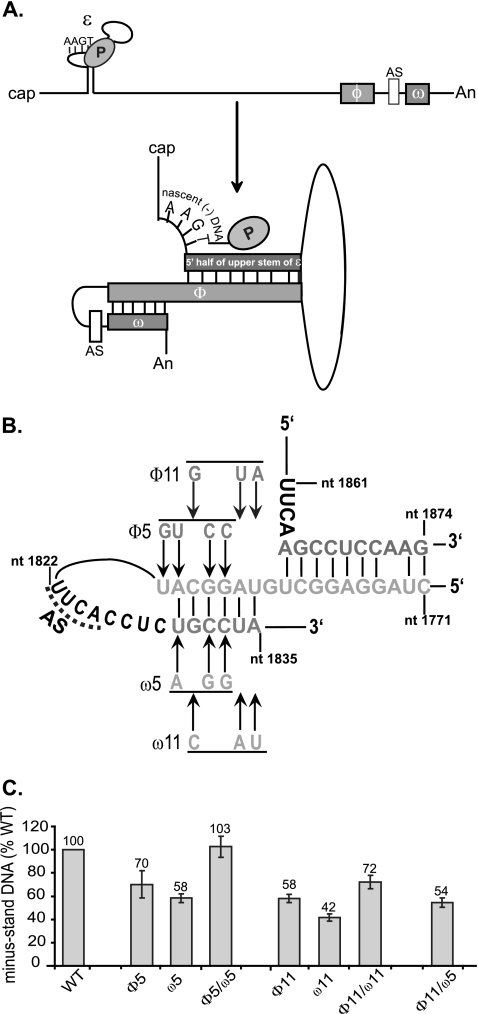
We identified potential base pairing between nt 1782 and 1787 (part of φ) and nt 1830 and 1835 (contained within the newly identified cis-acting sequence downstream of the AS). We named this 6-nt region ω (Fig. 6A). We sought to determine whether disrupting putative base pairs decreased the level of minus-strand DNA and whether restoring these base pairs, albeit with mutant sequences, restored minus-strand DNA levels. We made two sets of variants (sets 5 and 11). We found that the single variants that disrupted putative base pairs synthesized lower levels of minus-strand DNA (φ5, ω5, φ11, and ω11) compared to the WT reference (Fig. 6B and C). More important, variants that restored putative base pairs (φ5/ω5 and φ11/ω11) made higher levels of minus-strand DNA than the constitutive single variants (Fig. 6B and C). The φ5/ω5 variant showed complete restoration (103% of the WT reference), whereas φ11/ω11 showed a partial restoration (72% of the WT reference). In addition, we found that variant φ11/ω5, which would not restore putative base pairing, did not restore the levels of minus-strand DNA. These results indicate that base pairing between φ and ω contribute to the synthesis of minus-strand DNA. Furthermore, the partial restoration of φ11/ω11 may indicate a requirement for a specific sequence.
FIG. 6.
Base pairing between ω and a portion of φ contributes to the synthesis of minus-strand DNA. (A) Schematic representation of the dynamic conformation of pgRNA. The 5′ half of upper stem of ɛ, φ, and ω are shown as gray rectangles. The nascent minus-strand DNA attached to P protein is shown on the bulge of ɛ. The 4-nt AS upstream of ω is shown as a white rectangle. The figure is not drawn to scale. (B) Putative base pairing between the 5′ half of the upper stem of ɛ, φ, and ω. Substitutions designed to disrupt base pairing between φ and ω are shown. Variants that restore base pairing combine the corresponding φ and ω substitutions. P, P protein; AS, AS for minus-strand DNA synthesis. (C) Proportion of minus-strand DNA compared to WT reference. The level of synthesis of minus-strand DNA is defined as the amount of minus-strand DNA divided by the sum of pgRNA and minus-strand DNA. The mean values represent analysis from at least six independent transfections of each variant. The error bars represent the standard deviation.
Our analysis shows that φ base pairs with two regions: one portion of φ base pairs with the 5′ half of the upper stem of ɛ, and the adjacent portion of φ base pairs with ω, as depicted in Fig. 6A. Thus, base pairing between at least three cis-acting sequences is necessary for normal synthesis of minus-strand DNA.
DISCUSSION
This analysis provides a comprehensive understanding of the sequence requirements for the synthesis of minus-strand DNA in HBV. The present study introduces several new findings and concepts into the field. (i) The majority of pgRNA does not make cis-acting contributions to minus-strand DNA synthesis. (ii) Short (∼700-nt) derivatives of pgRNA can be reverse transcribed efficiently, demonstrating that a specific size of pgRNA is not required. (iii) Base pairing between three cis-acting sequences promotes minus-strand DNA synthesis, indicating a complex topology for pgRNA.
Our studies show that most of the pgRNA does not make cis-acting contributions to the synthesis of minus-strand DNA (Fig. 1 and 2). We were able to remove most of the pgRNA sequence without affecting its ability to be an efficient template for this process (Fig. 3). Furthermore, foreign sequences substituted for HBV sequence could be reverse transcribed into DNA (Fig. 3). Taken together, these results demonstrate that neither the specific size nor the sequence identity of most of the pgRNA is a determinant for the efficient synthesis of minus-strand DNA. Thus, the majority of the pgRNA is not necessary to maintain a conformation that is most favorable for the synthesis of minus-strand DNA.
In addition, we identified a region downstream of the AS that contributes to the synthesis of minus-strand DNA. Within this region, we found that 6 nt (ω) base pair with a portion of φ (Fig. 6B and C). In our previous study, we could not ascribe a mechanism to this portion of φ (1). In light of our current findings, we revise our model of the conformation of pgRNA for the synthesis of minus-strand DNA. We propose that φ base pairs with two sequences, the 5′ half of the upper stem of ɛ and ω, which are present near the 5′ and 3′ ends of the pgRNA, respectively. Although, our analysis shows that base pairing between ɛ, φ, and ω are necessary for efficient synthesis of minus-strand DNA, it needs to be determined how this conformation of the pgRNA contributes to the synthesis of minus-strand DNA. The proposed conformation of pgRNA could be contributing to minus-strand DNA synthesis at several junctures. One idea is that this base pairing facilitates the first-strand template switch by juxtaposing the donor (bugle of ɛ) and the acceptor (4-nt) sites. In addition, the conformation of pgRNA at the 3′ end could help to maintain the AS sequence in a single-stranded loop, such that it is accessible for base pairing with the 4-nt nascent minus-strand DNA. Another possibility worth considering is that the conformation of pgRNA is the template for initiation of minus-strand DNA, rather than the ɛ stem-loop structure.
The conformation of pgRNA (Fig. 6A) is reminiscent of the proposed conformation of minus-strand DNA that promotes the synthesis of plus-strand DNA for duck HBV (DHBV). For DHBV, base pairing between the middle (M region) and the 5′ and 3′ ends (5E and 3E regions) of the minus-strand DNA was shown to contribute to template switches during plus-strand DNA synthesis (11). This similarity between HBV and DHBV illustrates a more general theme: dynamic conformations or topologies of replication templates guide hepadnavirus replication.
We found a 6-nt sequence immediately upstream of the AS that contributes to minus-strand DNA synthesis. When mutated (m1816-1821), the level of minus-strand DNA is reduced to 27% of the level of the WT reference (Fig. 1 and 2). Interestingly, additional 5′ ends of minus-strand DNA were seen for this variant (Fig. 2, lane 16). We speculate that the new 5′ ends probably arise as a result of either imprecise priming of nascent minus-strand DNA or inaccurate first-strand template switch. At present, the mechanism through which this sequence functions is not known. One possibility is that it interacts with another cis-acting sequence via base pairing. However, we have been unable to identify such interactions. Alternatively, this sequence may interact with a trans-acting factor to facilitate synthesis of minus-strand DNA.
Although complementarity between the nascent minus-strand DNA and the pgRNA is important for the first-strand template switch (12), it is not the only determinant for defining the AS. Two lines of evidence lend credence to this idea. First, the authentic AS is used almost exclusively, despite the presence of numerous (21 for subtype ayw) potential acceptor sequences on the pgRNA. Second, the introduction of tandem acceptor sequences adjacent to the authentic AS still showed a strong preference for the authentic site (data not shown). We found that introducing the region between nt 1767 and 1838 (contains all of the cis-acting sequences near the 3′ end of pgRNA) at another location on the pgRNA is sufficient to direct first-strand template switch to this location (Fig. 5). Our results help to explain why complementarity alone does not drive the template switch and how additional cis-acting sequences contribute to define the normal AS.
ɛ plays multiple roles during viral replication. First, it is crucial for the selective encapsidation of pgRNA into capsids (2, 4, 14, 15). Second, the bulge of ɛ is the site of initiation of nascent minus-strand DNA (19). Third, the 5′ half of the upper stem of ɛ has been shown to base pair with φ to contribute to the synthesis of minus-strand DNA (1, 13). In order for the 5′ half of the upper stem of ɛ and φ to base pair, the upper stem of ɛ has to be disrupted. Although the mechanism of this change is not understood, we can look to the DHBV model for insights. For DHBV, when P protein binds to the ɛ stem-loop, the upper stem of ɛ melts (3). This conformational change is thought to be necessary for the initiation of minus-strand DNA synthesis (3). By analogy, for HBV, if P binding to the ɛ stem-loop melts the upper stem of ɛ, then the 5′ half of the upper stem of ɛ is free to base pair with φ to facilitate the synthesis of minus-strand DNA. Thus, the impetus for the conformational change could be provided by the interaction between P protein and the ɛ stem-loop. Although a conformational change wrought by HBV P binding to the ɛ stem-loop is an attractive idea, it is possible that additional trans-acting factors and/or cis-acting sequences also play a role. The inability to reconstitute protein priming in vitro has impeded understanding early steps in viral replication for HBV.
Our findings show that the cis-acting sequences necessary for the synthesis of minus-strand DNA are the bulge of ɛ, the 5′ half of the upper stem of ɛ, φ, the 6-nt region upstream of the AS, the AS, and ω. We have not determined the mechanism by which the sequence upstream of the AS functions. However, we have shown that ω base pairs with a portion of φ. These results led us to propose the model in Fig. 6A. In this model, base pairing with φ brings the ends of the pgRNA together to facilitate first-strand template switch and/or initiation of minus-strand DNA synthesis.
Acknowledgments
We thank Katy Haines, Eric Lewellyn, Thomas Lentz, and Megan Maguire for many helpful discussions and critical review of the manuscript.
This study was supported by National Institutes of Health grant CA22443.
Footnotes
Published ahead of print on 15 August 2007.
REFERENCES
- 1.Abraham, T. M., and D. D. Loeb. 2006. Base Pairing between the 5′ half of ɛ and a cis-acting sequence, φ, makes a contribution to the synthesis of minus-strand DNA for human hepatitis B virus. J. Virol. 80:4380-4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and H. Schaller. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11:3413-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck, J., and M. Nassal. 1998. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 18:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fallows, D., and S. Goff. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geballe, A. P., R. R. Spaete, and E. S. Mocarski. 1986. A cis-acting element within the 5′ leader of a cytomegalovirus β transcript determines kinetic class. Cell 46:865-872. [DOI] [PubMed] [Google Scholar]
- 6.Gerlich, W., and W. Robinson. 1980. Hepatitis B Virus contains protein attached to the 5′ terminus of its complete DNA strand. Cell 21:801-809. [DOI] [PubMed] [Google Scholar]
- 7.Gunther, S., G. Sommer, A. Iwanska, and H. Will. 1997. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238:363-371. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch, R., J. Lavine, L. Chang, H. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B virus are required for genomic RNA packaging as well as for reverse transcription. Nature 334:552-555. [DOI] [PubMed] [Google Scholar]
- 9.Liu, N., L. Ji, M. L. Maguire, and D. D. Loeb. 2004. cis-Acting sequences that contribute to the synthesis of relaxed-circular DNA of human hepatitis B virus. J. Virol. 78:642-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu, N., K. M. Ostrow, and D. D. Loeb. 2002. Identification and characterization of a novel replicative intermediate of heron hepatitis B virus. Virology 295:348-359. [DOI] [PubMed] [Google Scholar]
- 11.Liu, N., R. Tian, and D. D. Loeb. 2003. Base pairing among three cis-acting sequences contributes to template switching during hepadnavirus reverse transcription. Proc. Natl. Acad. Sci. USA 100:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nassal, M., and A. Rieger. 1996. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70:2764-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oropeza, C. E., and A. McLachlan. 2007. Complementarity between epsilon and phi sequences in pregenomic RNA influences hepatitis B virus replication efficiency. Virology 359:371-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollack, J., and D. Ganem. 1993. An RNA stem-loop structure directs Hepatitis B virus genomic RNA encapsidation. J. Virol. 67:3254-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollack, J., and D. Ganem. 1994. Site-specific RNA binding by a Hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68:5579-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rieger, A., and M. Nassal. 1996. Specific Hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J. Virol. 70:585-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 18.Tavis, J., S. Perri, and D. Ganem. 1994. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, G., and C. Seeger. 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67:6507-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, G., and C. Seeger. 1992. The reverse transcriptase of the hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 21.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]