Abstract
Herpes simplex virus mutants with single substitutions that decrease DNA binding by the DNA polymerase processivity subunit UL42 are only modestly impaired for viral replication. In this study, recombinant viruses harboring two or four of these mutations were constructed. The more substitutions, the more severe the defects in viral replication and DNA synthesis, suggesting that DNA binding by UL42 is important for these processes.
Herpes simplex virus DNA polymerase (Pol) consists of a heterodimer with a catalytic-subunit Pol and an accessory subunit, UL42. UL42 is a processivity factor that stimulates long-chain DNA synthesis in vitro and is essential for viral replication (3, 7, 9, 10). Unlike the sliding clamp processivity factors that interact only topologically with DNA (reviewed in reference 4), UL42 interacts with DNA directly with high affinity as a monomer (11) and increases the binding affinity of the Pol to the primer/template (2, 15) by decreasing the dissociation rate (2). Several results indicate that the DNA binding activity of UL42 is crucial for processive DNA synthesis by the viral Pol. Insertion mutations of UL42 that result in undetectable DNA binding impair long-chain DNA synthesis and prevent complementation of a UL42 null mutant (1). UL42 mutants containing alanine substitutions for any of four conserved arginine residues (at positions 113, 182, 279, and 280), which reside on the positively charged surface of UL42, decrease DNA binding without affecting binding to the Pol C terminus and decrease the stimulation of long-chain DNA synthesis (12). However, when incorporated into herpes simplex virus, these single substitutions have only modest effects on viral replication and DNA synthesis (5). Because combining two or four substitutions resulted in more-severe effects on DNA binding in vitro and the quadruple substitution impaired complementation of a UL42 null mutant (12), we wished to investigate the effects of such multiple substitutions on viral replication and DNA replication when they were incorporated into the virus.
We constructed two plasmids, pHC-R113/182A and pHC-R279/280A, each of which contains two arginine-to-alanine substitutions, as previously described using pHC700 (5) and plasmids based on pMBP-ppΔ340 containing the corresponding mutations (12). Each plasmid was sequenced to confirm the presence of the desired mutations and no others. These two plasmids were tested for their ability to complement the replication of CgalΔ42, a UL42 null mutant, in Vero cells as described previously (5, 12). As expected from the previous result that a plasmid containing all four substitutions was able to complement CgalΔ42 replication, albeit with reduced efficiency, the two new plasmids were able to complement the null mutant. The complementation efficiencies of the two double-mutation plasmids were intermediate between those of the quadruple-substitution mutant and a wild-type control plasmid (data not shown).
Recombinant viruses containing UL42 double and quadruple mutations were then constructed and purified as described previously (5). Two independent isolates of each mutant were derived from separate transfections, and the presence of the UL42 mutations was confirmed by sequencing. Plaque assays were performed on Vero cells and on V9 cells, which express wild-type UL42 protein (6), and images of at least 20 plaques were recorded using a SPOT camera on a model TE300 inverted microscope (Nikon). Plaque sizes were measured using Photoshop software (Adobe). The data presented in Fig. 1 were from a single experiment using equal densities of cells. While all recombinants formed plaques of similar sizes on V9 cells, mutants with double and quadruple mutations formed significantly smaller plaques on Vero cells than did two single-substitution mutants that were tested or a control virus constructed in the same manner as the mutants but encoding wild-type UL42 (Fig. 1). The quadruple mutant was particularly impaired in plaque size, with a mean area more than 20-fold less than that of the control virus.
FIG. 1.
UL42 mutants form smaller plaques on Vero cells than the wild type (wt). Plaques formed on Vero and V9 cells were analyzed as described in the text. Numbers shown below the plaques are average sizes (mm2) of at least 20 plaques ± standard deviations. *, the mutant with quadruple mutations formed only tiny plaques, which could be observed clearly only under a high magnification (×200). The plaques formed by other viruses were observed under a magnification of ×100 at 3 days postinfection.
The double- and quadruple-substitution mutants along with two single-substitution mutants and the control virus were then tested in single-cycle growth assays on Vero cells, performed as previously described (5). The titers of all viruses peaked at 36 h postinfection, with the exception of that of the quadruple mutant, which peaked at 48 h postinfection. Burst sizes were then determined as the total yield of virus at the time of peak titer divided by the number of infected cells (Table 1). Consistently with a previous report (5), the single-substitution R182A and R279A mutants exhibited two- and fourfold-reduced virus yields, respectively, relative to the yield of the control virus. The double-substitution R113/182A and R279/280A mutants had 10- and 30-fold-decreased virus yields, respectively, while the quadruple mutant with the R113/182/279/280A mutations yielded nearly 100-fold less virus. Thus, the double and quadruple mutants were substantially impaired for viral replication.
TABLE 1.
Burst sizes and DNA copy numbers per PFU of UL42 mutants
| Virus (recombinant)c | Burst sizea (PFU/cell) | No. of DNA copies/PFUb |
|---|---|---|
| Wild-type virus (A) | 30 ± 6.1 | 190 ± 10 |
| R182A mutant (A) | 18 ± 1.8 | 490 ± 10 |
| R279A mutant (A) | 7.9 ± 1.1 | 1,200 ± 160 |
| R113/182A mutant (A) | 3.6 ± 0.1 | 2,000 ± 70 |
| R113/182A mutant (B) | 2.6 ± 0.2 | 2,900 ± 40 |
| R279/280A mutant (A) | 1.1 ± 0.0 | 4,800 ± 1,400 |
| R279/280A mutant (B) | 1.1 ± 0.2 | 5,200 ± 1,900 |
| R113/182/279/280A mutant (A) | 0.4 ± 0.0 | 4,800 ± 80 |
| R113/182/279/280A mutant (B) | 0.4 ± 0.0 | 3,200 ± 1,000 |
Burst size was calculated as the ratio of the peak titer (at 48 h postinfection for the quadruple mutants and at 36 h postinfection for all other recombinants) to the number of cells. Data are averages ± standard errors of the means from the same two experiments of single-cycle growth curves, and the amounts of viral DNA were synthesized as indicated for Fig. 2.
Numbers of DNA copies per PFU were calculated by determining the ratios of total DNA copy numbers synthesized to peak titers (at 48 h for the quadruple mutant and at 36 h postinfection for other recombinants).
Wild-type virus refers to the control virus with wild-type UL42. The A and B recombinants were two independent constructs. All recombinations were performed with the control virus and the indicated mutant.
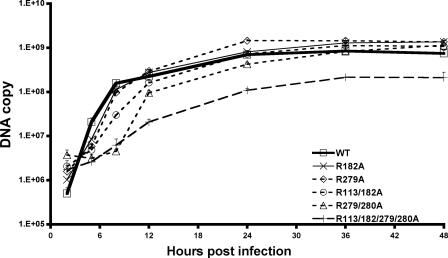
To examine the kinetics of viral DNA synthesis, a quantitative real-time PCR assay was applied, as described previously (5). Figure 2 shows the amounts of viral DNA synthesized at 2, 5, 8, 12, 24, 36, and 48 h postinfection by the various viruses. Similarly to what was observed previously with single-substitution mutants (5), the two double-substitution mutants replicated DNA with slower kinetics during the early phase of infection than that of the control virus with wild-type UL42. Interestingly, the R113/182A mutant synthesized less DNA than the single mutants during the early phase of infection (P < 0.05 and P = 0.003 at 5 and 8 h postinfection, respectively; Student's t test), and the R279/280A mutant replicated DNA even more slowly during this phase (P < 0.05 and P < 0.001 at 5 and 8 h postinfection, respectively). These double mutants, however, synthesized amounts of DNA similar to that of the wild-type UL42 control virus by 36 h after infection. The quadruple mutant, on the other hand, was substantially impaired for viral DNA synthesis at all time points (P was ≤0.001 except at 48 h postinfection, at which time, P was 0.003); it synthesized fourfold-less DNA than the control virus with wild-type UL42 even at late times. Even so, the double and quadruple mutants exhibited significantly higher ratios of DNA copy numbers to numbers of PFU relative to those of viruses with wild-type UL42 or a single-substitution mutation (Table 1).
FIG. 2.
Viral DNA synthesized by UL42 recombinants. Vero cells (1 × 105) were inoculated with UL42 recombinants at a multiplicity of infection of 3. Infected cells were harvested at different times postinfection. DNA was isolated from aliquots of both infected cells and medium containing cell-free virus. Purified DNA was serially diluted and quantified by real-time PCR as described in reference 5 to determine the relative copy numbers of viral DNA. Data shown are averages from two independent experiments. Since the independent isolates of each virus exhibited similar kinetics, only the results for one recombinant (recombinants A in Table 1) are shown for clarity. WT, wild type.
The increasing severity of replication and DNA synthesis phenotypes with an increased number of substitutions and their increasingly severe effects on DNA binding (11) are consistent with the interpretation that reduced DNA binding by UL42 results in a reduction in viral DNA synthesis and thus viral replication. It must be cautioned that in the study of the effects of the double and quadruple mutations on DNA binding, we could not rule out an effect on Pol binding. Nevertheless, no single substitution affected the ability of UL42 to bind to Pol in vitro (12). In addition, we cannot exclude the possibility that these substitutions might also influence other properties of UL42, such as interactions with other proteins. Regardless, the simplest interpretation of the results is that DNA binding by UL42 is important for viral replication and DNA synthesis.
Given this interpretation, one could ask why these mutants synthesize any viral DNA since the mutant proteins were so highly impaired in DNA binding (12). It should be noted, however, that not every arginine and lysine on the positively charged surface of UL42 is replaced by alanine in the quadruple mutant, so the mutant likely retains some affinity for DNA. Moreover, UL42 may interact not only with Pol but also with UL9 (8, 14), the origin binding protein, and UL29 (13), the single-stranded DNA binding protein. The interactions of these other proteins with DNA in infected cells may permit the mutant UL42s to function, albeit less efficiently, in supporting processive DNA synthesis, which may result in slower kinetics of viral DNA replication and the synthesis of less viral DNA during the early phase of infection.
An interesting property of all of the viral UL42 mutants containing arginine-to-alanine substitutions is that they synthesize relatively large amounts of DNA per PFU. This was true even for the quadruple-substitution mutant, which is substantially defective in viral DNA synthesis, even at late times postinfection. This mutant is 25-fold-more defective in viral yield than it is in viral DNA synthesis. It is possible that the high DNA copy number/PFU ratio is due in part to the mutagenic effect of the UL42 mutation (5; unpublished results). Indeed, examination of the copy numbers of cell-free virion DNA by real-time PCR, which mimicked the numbers of cell-free virus particles, and of the relative virus titers demonstrated that the quadruple-substitution mutant exhibited a 10-fold-higher ratio of particle numbers to PFU than the control virus with wild-type UL42. This suggests that the mutant synthesized more virions that contain lethal mutations and thus do not form plaques. Another possible explanation for the elevated DNA copy number/PFU ratio stems from the delayed kinetics of DNA replication exhibited by the mutants. It may be that much of the DNA made later in infection is not productively packaged into virions. An electron microscopy study of the distributions of different nucelocapsids formed in cells infected with viruses with mutant and wild-type UL42 should clarify this possibility. A third possibility is that UL42 has a role in viral replication other than DNA synthesis and that the mutations affect this function. Further studies of these and other UL42 mutants should resolve the possibility.
Acknowledgments
J.C.W.R. was a predoctoral fellow of the Howard Hughes Medical Institute. The study was supported by NIH grants AI056359 (C.B.C.H.) and AI19838 (D.M.C.).
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Chow, C. S., and D. M. Coen. 1995. Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J. Virol. 69:6965-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb, J., and M. D. Challberg. 1994. Interaction of herpes simplex virus type 1 DNA polymerase and the UL42 accessory protein with a model primer template. J. Virol. 68:4937-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottlieb, J., A. I. Marcy, D. M. Coen, and M. D. Challberg. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hingorani, M. M., and M. Donnell. 2000. Sliding clamps: a (tail)ored fit. Curr. Biol. 10:R25-R29. [DOI] [PubMed] [Google Scholar]
- 5.Jiang, C., Y. T. Hwang, J. C. W. Randell, D. M. Coen, and C. B. C. Hwang. 2007. Mutations that decrease DNA binding of the processivity factor of the herpes simplex DNA polymerase reduce viral yield, alter the kinetics of viral DNA replication, and decrease fidelity of DNA replication. J. Virol. 81:3495-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson, P. A., M. G. Best, T. Friedmann, and D. S. Parris. 1991. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J. Virol. 65:700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchetti, M. E., C. A. Smith, and P. A. Schaffer. 1988. A temperature-sensitive mutation in a herpes simplex virus type 1 gene required for viral DNA synthesis maps to coordinates 0.609 through 0.614 in UL. J. Virol. 62:715-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monahan, S. J., L. A. Grinstead, W. Olivieri, and D. S. Parris. 1998. Interaction between the herpes simplex virus type 1 origin-binding and DNA polymerase accessory proteins. Virology 241:122-130. [DOI] [PubMed] [Google Scholar]
- 9.Parris, D. S., A. Cross, L. Haarr, A. Orr, M. C. Frame, M. Murphy, D. J. McGeoch, and H. S. Marsden. 1988. Identification of the gene encoding the 65-kilodalton DNA-binding protein of herpes simplex virus type 1. J. Virol. 62:818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purifoy, D. J. M., R. B. Lewis, and K. L. Powell. 1977. Identification of the herpes simplex virus DNA polymerase gene. Nature 269:621-623. [DOI] [PubMed] [Google Scholar]
- 11.Randell, J. C., and D. M. Coen. 2004. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J. Mol. Biol. 335:409-413. [DOI] [PubMed] [Google Scholar]
- 12.Randell, J. C., G. Komazin, C. Jiang, C. B. Hwang, and D. M. Coen. 2005. Effects of substitutions of arginine residues on the basic surface of herpes simplex virus UL42 support a role for DNA binding in processive DNA synthesis. J. Virol. 79:12025-12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trego, K. S., and D. S. Parris. 2003. Functional interaction between the herpes simplex virus type 1 polymerase processivity factor and origin-binding proteins: enhancement of UL9 helicase activity. J. Virol. 77:12646-12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weisshart, K., C. S. Chow, and D. M. Coen. 1999. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol. 73:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]