Abstract
We are still inadequately prepared for an influenza pandemic due to the lack of a vaccine effective for subtypes to which the majority of the human population has no prior immunity and which could be produced rapidly in sufficient quantities. There is therefore an urgent need to investigate novel vaccination approaches. Using a combination of genomic and traditional tools, this study compares the protective efficacy in macaques of an intrarespiratory live influenza virus vaccine produced by truncating NS1 in the human influenza A/Texas/36/91 (H1N1) virus with that of a conventional vaccine based on formalin-killed whole virus. After homologous challenge, animals in the live-vaccine group had greatly reduced viral replication and pathology in lungs and reduced upper respiratory inflammation. They also had lesser induction of innate immune pathways in lungs and of interferon-sensitive genes in bronchial epithelium. This postchallenge response contrasted with that shortly after vaccination, when more expression of interferon-sensitive genes was observed in bronchial cells from the live-vaccine group. This suggested induction of a strong innate immune response shortly after vaccination with the NS1-truncated virus, followed by greater maturity of the postchallenge immune response, as demonstrated with robust influenza virus-specific CD4+ T-cell proliferation, immunoglobulin G production, and transcriptional induction of T- and B-cell pathways in lung tissue. In conclusion, a single respiratory tract inoculation with an NS1-truncated influenza virus was effective in protecting nonhuman primates from homologous challenge. This protection was achieved in the absence of significant or long-lasting adverse effects and through induction of a robust adaptive immune response.
In the event of an influenza pandemic, two doses of a killed vaccine would be needed to protect an immunologically naïve human population, but the delay associated with a prime-boost regimen would result in many casualties. Even if a single dose of an inactivated vaccine could be used, demand is likely to exceed production capabilities because of the large antigenic dose needed and the lack of infrastructure required to scale up vaccine production. Vaccination trials with unadjuvanted killed avian influenza virus have suggested that up to six times the normal antigen amount is needed to achieve effective protection, in part due to the lack of prior immunity to the avian subtypes (19). Trials using a killed-virus formulation with relatively small amounts of antigen combined with aluminum hydroxide adjuvant have been disappointing, as the most promising results still required two inoculations and up to twice the normal antigen dose (12, 33, 48). The predicted shortage may also be compounded by the low yield of modified H5N1 viruses in eggs (47).
Modified live-vaccine formulations, alternate routes of administration, and/or the use of novel adjuvants for influenza vaccination may therefore have to be pursued to meet the needs during an influenza pandemic. One advantage of live attenuated vaccines lies in their ability to replicate and provide exposure to large amounts of antigens despite a low starting dose; another advantage is the significant levels of mucosal immunity induced through local production of immunoglobulin A (IgA) at viral replication sites (17). Exposure to a modified live virus may also be partially protective across several strains or subtypes through cell-mediated and humoral responses, since internal proteins of influenza viruses share a high degree of conservation (3, 11, 36). Cold-adapted live attenuated vaccines, produced by reassortment of the genes encoding the surface proteins of the circulating strains with those encoding the internal proteins of a cold-adapted attenuated strain, have offered significantly better protection than killed vaccines in children (7). In elderly people, a combination of live and killed vaccines has proven more efficacious than killed vaccine alone (17). Although these cold-adapted vaccines have been shown to be safe and effective for a majority of people, they still require large amounts of viruses per vaccine dose, and improvements might be possible if novel vaccines that require lower antigenic doses to induce a more robust immune response are developed, particularly for the elderly.
New approaches to attenuate live viruses are currently being investigated. Nonstructural protein 1 (NS1) truncation to attenuate influenza viruses has suggested that critical and apparently species-specific functions of NS1 inhibit cellular interferon responses and promote viral replication through a variety of molecular interactions (1, 6, 21, 23, 24, 29, 32, 34, 39, 40, 45, 46, 49, 50, 52). NS1 also appears to negatively affect dendritic cell maturation, resulting in suppression of the adaptive immune response (15, 22, 30, 31) and implying that impairment of NS1 function may enhance adaptive immunity. The present study is the first in vivo demonstration of the potential of NS1-truncated influenza viruses as modified live vaccines in a macaque model. This influenza virus macaque model was shown in prior studies using the mildly pathogenic Texas strain to reproduce human pathology and immune responses very well (2, 4). Macaques, which are among the closest genetic relatives to humans after great apes, can be infected with human influenza virus without prior adaptation and share other key features with humans, such as lung physiology, size, anatomy, and posture. The availability of a macaque-specific oligoarray has also allowed detailed functional genomic studies in the past.
MATERIALS AND METHODS
Animals.
Eight female and four male adult pigtailed macaques (Macaca nemestrina), ranging in age from 3.3 to 18 years and in weight from 6.7 to 17.5 kg, were obtained for this study from the Washington National Primate Research Center. All preassignment screening, husbandry, animal handling, and biosafety procedures were performed as previously described (31, 32), in accordance with guidelines approved by the University of Washington Environmental Health and Safety Committee, the Occupational Health Administration, the Primate Center Research Review Committee, and the Institutional Animal Care and Use Committee.
Viruses and vaccine preparation.
The H1N1 human influenza viruses used in this study, a reconstructed wild-type influenza A/Texas/36/91 (wtTX91) virus and a modified live version of the same containing a truncated NS1 gene coding for the first 126 amino acids (TX91 NS1▴126), were generated from plasmid DNA, propagated, concentrated, and purified as previously described (45). The wtTX91 and TX91 NS1▴126 virus stocks had titers of 3 × 108 PFU/ml and 1 × 108 PFU/ml, respectively. Sequences of both viruses were confirmed by reverse transcription-PCR (RT-PCR) and sequence analysis.
For production of the formalin-inactivated wtTX91 vaccine, the wtTX91 virus was concentrated and purified by centrifugation of infected allantoic fluid through a 30% sucrose density gradient before resuspension in calcium borate buffer. The purified virus was adjusted to a protein concentration of 1 mg/ml and treated with 0.025% formalin at 4°C for 3 days. Protein concentrations were measured and standardized based on the optical density at 280 nm, Bio-Rad protein assay (Bio-Rad), and hemagglutinin (HA) units. The inactivated vaccine stock contained 45 μg of HA protein (6,000 HA units) per 150-μl dose. Hemagglutination titers were indicative of the presence of conformationally active antigen in the vaccine preparations. Additionally, inactivation of the killed virus was confirmed by the absence of viral growth following inoculation of embryonated eggs.
Animal protocol.
The animal protocol was adapted from the work of Rimmelzwaan et al. (42-44) and was based on our previous work (2, 4). Briefly, the 12 animals were assigned to four experimental groups and were matched for age, weight, and gender to the extent possible. One group (L; n = 6) was vaccinated with the TX91 NS1▴126 virus, one group (K; n = 4) was vaccinated with the formalin-killed wtTX91 virus, and one group (N; n = 2) was not vaccinated. Two of the “L” animals were never challenged and were sacrificed on day 4 postvaccination (referred to hereafter as day 4). All other animals were challenged on day 21 and sacrificed on day 25, as shown in Fig. 1. Vaccination with the TX91 NS1▴126 virus and challenge with the wtTX91 virus were done with 6 × 107 PFU intratracheally, on tonsils and conjunctivae (2, 4). Vaccination with the formalin-killed wtTX91 virus was done by intramuscular injection of 150 μl of preparation at 1 mg/ml viral proteins, corresponding to 45 μg of HA, which is three times the amount present in the commercial trivalent human vaccine for each virus component.
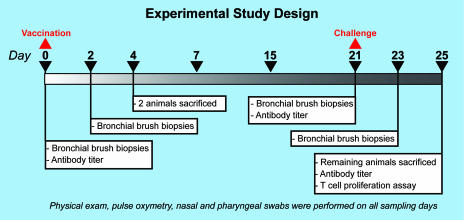
FIG. 1.
The study consisted of six animals receiving a live attenuated influenza virus vaccine (A/Texas/36/91; NS1 truncated), four animals receiving a killed influenza virus vaccine (formalin-killed wtTX91), and two animals receiving no vaccine on day 0. Two of the animals that received the live vaccine were sacrificed on day 4. The remainder of the animals were sacrificed on day 25, which was also 4 days after challenge with a 107 50% tissue culture infective dose of wild-type influenza A/Texas/36/91 virus.
Animals were monitored as previously described (2, 4, 51). Peripheral blood was collected on days 0, 21, and 25 for determination of antibody titers to wtTX91 or to TX91 NS1▴126 by hemagglutination inhibition and by IgG antibody enzyme-linked immunosorbent assay (ELISA) (26). Influenza virus-specific T cells in peripheral blood mononuclear cells were measured on day 25. This was achieved by culture of peripheral blood mononuclear cells with 107 PFU of wtTX91 per 2 million cells, followed by treatment with brefeldin A to prevent secretion of cytokines. Cells were then surface stained for CD3, CD4, and CD8 and subjected to intracellular staining for gamma interferon. Flow cytometric data were acquired on an LSR-II or FACScanto flow cytometer (BD Biosciences, San Jose, CA), and data analysis was done using Flo-Jo software (Tree Star, Inc., Ashland, OR).
Bronchial brush biopsies were taken by bronchoscopy on days 0, 2, 21, and 23 (2.00-mm bronchial cytology brush; Telemed Systems, Inc., Hudson, MA) and were flash frozen until being processed for expression arrays. Finally, the presence or absence of viral shedding was determined by viral isolation from nasal and pharyngeal swabs (Fig. 1).
Our previous studies (2, 4) demonstrated that acute lung pathology due to mildly pathogenic influenza virus infection peaks at or around day 4 and that by day 7 the lesions are resolving and the virus is largely cleared. This is also consistent with the kinetics of lung pathology induced by other respiratory viral infections in other animals (8). This knowledge formed the rationale for terminating the animals at the peak of the infection to allow for a direct comparison of viral loads in vaccinated versus nonvaccinated animals at this key point of the infection. At necropsy, all tissues were examined grossly and lung tissue was harvested, particularly from those areas within or near gross lesions. Other tissues harvested included tracheobronchial lymph nodes, tracheas, and tonsils. All samples not set aside for arrays were either snap frozen for viral isolation or fixed in 10% formalin for histology and immunohistochemistry (IHC). Methods for viral isolation, histopathology, and IHC for staining of influenza virus NP antigen were previously described (4). Staining for mature dendritic cells (CD83+) in lungs and tracheobronchial lymph nodes was done using a standardized protocol for formaldehyde-fixed tissues involving antigen retrieval, several blocking steps, and a 2-h incubation with a mouse monoclonal antibody specific for human CD83 (Vision Biosystems Inc., Norwell, MA), with visualization of bound antibody by the ABC method (9). All microscopy was performed on an Olympus BX41 light microscope. Photomicrographs were acquired with an Olympus Q-Color 3 camera and associated computer software.
Pathology, antigen, and dendritic cell scores were assigned to each animal according to standard methods. Statistical calculations were performed with SigmaStat for Windows, and figures were made with SigmaPlot for Windows (Systat Software, Inc.).
Tissue processing and analysis for qRT-PCR and macaque oligonucleotide arrays.
Tissue samples were processed and total RNA extracted and purified for quantitative real-time RT-PCR (qRT-PCR) and macaque oligonucleotide arrays as previously described (2, 4). Oligonucleotide array analyses (experiments) consisted of hybridization of individual lung samples from infected animals to pooled lung samples from mock-infected animals from a prior study (2). Prior to inclusion in the array experiments, samples from experimental animals were selected for evidence of active infection by qRT-PCR for viral HA mRNA of TX91 when any could be found in clinically and pathologically relevant samples. RNAs from bronchial brush biopsies from randomly selected animals in each group on days 2, 21, and 23 were hybridized against a pool from all animals on day 0.
In accordance with proposed standards (10), all data described in this report, including sample information, intensity measurements, gene lists, error analysis, microarray content, and slide hybridization conditions, are available through Expression Array Manager at http://expression.viromics.washington.edu.
RESULTS
The TX91 NS1▴126 influenza virus caused no lung pathology.
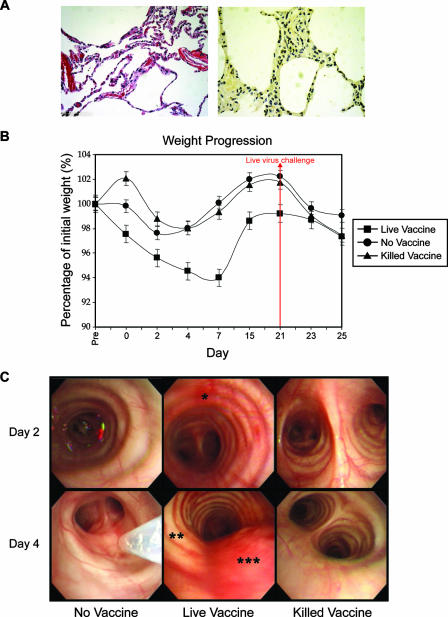
To evaluate NS1 truncation as a means of attenuating influenza viruses while maintaining immunogenicity, we vaccinated pigtailed macaques intratracheally with TX91 NS1▴126 virus and challenged the animals with the wild-type version 3 weeks later. To verify that the live attenuated virus caused little or no pathology or disease on its own in macaques, we sacrificed two animals on day 4, i.e., 4 days after vaccination. No significant macroscopic or microscopic lung pathology was seen in these animals (Fig. 2A, left panel), and influenza virus antigen staining was minimal (Fig. 2A, right panel). This is in stark contrast to the pathology and antigen staining results observed on day 4 postinfection and later for macaques infected with the wild-type virus in the present and past studies (2, 4). Animals vaccinated with the modified live vaccine had statistically significant but transient weight loss (Fig. 2B) which was not accompanied by anorexia, again in contrast to the findings during infection with the wild-type Texas virus (4). Animals vaccinated with the modified live vaccine had regained most of the weight by the time of challenge and went on to lose less weight after challenge than did animals in the other groups, but these differences were not statistically significant. No significant skewing of the data due to cycling of the sex skin in four of the seven females included in the study could be teased out of the statistical analysis. Bronchoscopic examination revealed mild inflammation in the upper respiratory tract from day 2 to day 4 (Fig. 2C), but no other upper or lower respiratory signs were observed, unlike during infection with the wild-type virus (2, 4). This inflammation resolved by day 15, i.e., 6 days prior to challenge. Collectively, these results suggest that vaccination with the TX91 NS1▴126 virus causes a transient local and systemic inflammatory response consistent with a nonproductive respiratory infection and no lung pathology (Fig. 2).
FIG. 2.
Lung histology (A, left panel), influenza virus nucleoprotein antigen staining (A, right panel), weight progression (B), and bronchoscopic examination (C) after vaccination. (A) Histology and IHC for influenza virus antigens revealed the lack of pathology and productive infection 4 days after vaccination (average pathology/IHC score, 0.93 out of 5). (B) Vaccination with the live attenuated virus was followed by significant but transient weight loss in that group (ANOVA on ranks; P ≤ 0.05). While all animals had signs of inflammation in the upper respiratory tract from manipulations and repeated lavages, these signs were very mild in the mock-vaccine and killed-vaccine groups. (C) Animals that received the live vaccine had mild to moderate inflammation, manifested as hyperemia (*), edema (**), and increased mucus production (***), shortly after vaccination. All signs of upper respiratory tract inflammation resolved by day 15.
Vaccination with live TX91 NS1▴126 influenza virus prevented replication of wtTX91 in lungs of challenged pigtailed macaques and decreased respiratory tract inflammation and pathology.
TX91 NS1▴126 virus-vaccinated animals were challenged on day 21 with wtTX91 virus. One group of animals that had received formalin-killed wtTX91 virus vaccine parenterally, at an antigenic dose equivalent to three times the trivalent commercial human vaccine dose, and one unvaccinated group were similarly challenged. The end point for the study was set at day 25, i.e., day 4 after challenge (Fig. 1).
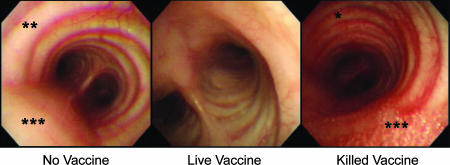
HA mRNA levels in lung lesions showed considerably lower levels of active viral replication postchallenge in animals that had received the live NS1-truncated virus than those in animals vaccinated with the formalin-killed virus or not vaccinated at all (Fig. 3A). Bronchoscopic examination showed less severe upper respiratory tract inflammation, defined as hyperemia, edema, and excessive serous or mucous secretions, in animals that had received the live-vaccine prototype (Fig. 4). As in previous experiments (2, 4), other clinical signs, such as weight loss and abnormal pulmonary auscultation findings, were subtle, without significant differences among groups. Active viral replication in nasopharyngeal passages and the lower respiratory tract was below the detection level of plaque assays, consistent with the mild nature of infection with wtTX91. Nevertheless, our previous work has shown that even minute amounts of viral mRNA have a pronounced effect on cellular gene expression in infected and adjacent tissues (2).
FIG. 3.
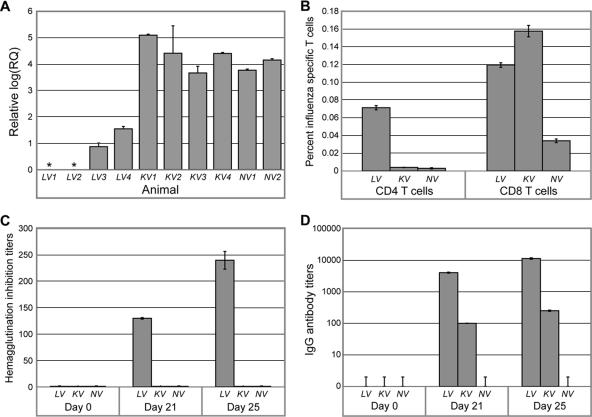
(A) Relative viral mRNA quantification by qRT-PCR 4 days after challenge, using lung tissue sampled from main gross lesions in individual animals (limit of detection = 0.3 on log scale). (B) Percentages of CD4+ and CD8+ T cells producing gamma interferon after an 8-hour in vitro stimulation with Texas influenza virus. These cells were isolated from blood samples on day 25 (4 days postchallenge). Hemagglutination inhibition (C) and enzyme-linked immunosorbent assay IgG (D) antibody titers to whole wtTX91 virus in sera of macaques after vaccination with TX91 NS1▴126 or formalin-killed wtTX91 virus are shown. LV, live attenuated vaccine; KV, killed vaccine; NV, no vaccine.
FIG. 4.
Representative bronchoscopic images for animals in all groups from day 23 to day 25, i.e., 2 to 4 days after challenge with the wild-type Texas virus. Animals in the group vaccinated with the live attenuated virus had less upper airway inflammation, as defined by hyperemia (*), edema (**), and production of serous or mucous respiratory fluids (***), and fewer other respiratory signs, such as rhinorrhea or abnormal auscultation findings, than did animals in the other challenged groups.
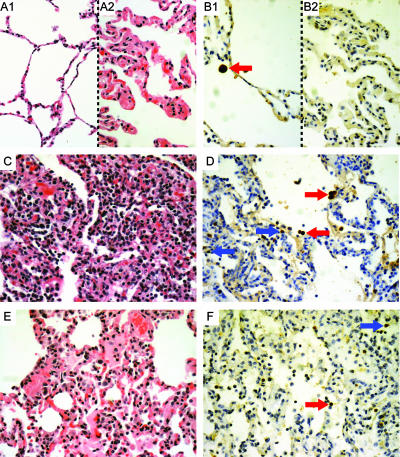
Lung lesions, although present in the experimental group that received the TX91 NS1▴126 virus, were less extensive than those in other challenged animals. Macro- and microscopic lesions were rather heterogeneous in that group, but pathology (Fig. 5A1 and A2) and influenza virus antigen staining (Fig. 5B1 and B2) scores were consistently and significantly lower than those for the killed-vaccine (Fig. 5C and D) and no-vaccine (Fig. 5E and F) groups, as demonstrated with a one-way analysis of variance (ANOVA) on ranks (P ≤ 0.01). These scores took into account whether the location of a positive influenza virus antigenic signal was in respiratory epithelial cells or in phagosomes of macrophages and therefore were augmented by evidence of an active infection at day 25 (4 days postchallenge) for the killed- and no-vaccine groups (Fig. 5D and F). A series of pairwise comparisons (Student-Newman-Keuls method; P ≤ 0.05) performed on influenza virus antigen staining scores also substantiated the difference between the live- and killed-vaccine groups after challenge, consistent with the qRT-PCR data, as well as the lack of significant differences between the killed-vaccine and the no-vaccine groups.
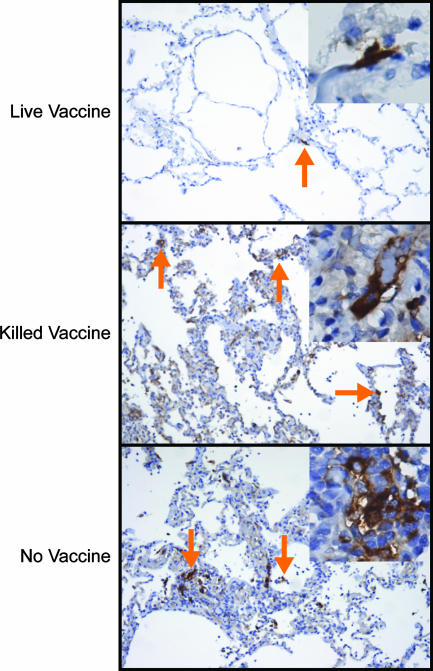
FIG. 5.
Vaccination with TX91 NS1▴126 resulted in significant protection of lower airways during challenge. The images show microscopic histopathological changes (A1, A2, C, and E) and influenza virus nucleoprotein (NP) antigen staining (B1, B2, D, and F) for the live-vaccine group (A1, A2, B1, and B2) (average pathology/IHC score, 1.86 out of 5), the killed-vaccine group (C and D) (average pathology/IHC score, 3.11 out of 5), and the no-vaccine group (E and F) (average pathology/IHC score, 3.3 out of 5) 4 days after challenge. Split images A1-A2 and B1-B2 illustrate the range of pathology seen in the live-vaccine group. Red arrows point to cells that were positive for the viral antigen inside phagocytes, whereas blue arrows point to positive epithelial cells, indicative of recent active infection. All photographs were taken at an optical magnification of ×20, with an additional digital magnification of ×2.
Vaccination with TX91 NS1▴126 virus resulted in cellular and humoral immunity.
Subtyping of influenza virus-specific T lymphocytes in peripheral blood on day 25 as well as pre- and postchallenge serology validated the clinical, pathology, and virology data and suggested an explanation for those observations. Four days after challenge with wtTX91, the percentage of influenza virus-specific CD4+ T lymphocytes was approximately seven times higher in the live-vaccine prototype group than in the other groups (Fig. 3B, left panel). The higher relative abundance of CD4+ T cells in the live-vaccine group was consistent with the much higher hemagglutination inhibition (Fig. 3C) and IgG (Fig. 3D) titers in sera from this group before challenge on day 21 and, even more so, 4 days after challenge on day 25. Interestingly, animals in the killed-vaccine group had slightly higher percentages of influenza virus-specific CD8+ T cells after challenge than did animals in the live-vaccine prototype group (Fig. 3B, right panel), perhaps reflecting differences in abundance of actively infected pneumocytes observed by influenza virus-specific antigen staining in lung tissues of the same animals. Unvaccinated animals had the lowest percentages of influenza virus-specific CD4+ and CD8+ T cells, most likely because they were unprimed and lacked the necessary time after challenge to mount an antigen-specific response. The percentages of CD4+ and CD8+ cells observed were consistent with previously reported data for humans exposed to influenza virus (18).
Lungs of unvaccinated or killed-virus-vaccinated animals showed a stronger induction of the innate immune response gene profile after challenge.
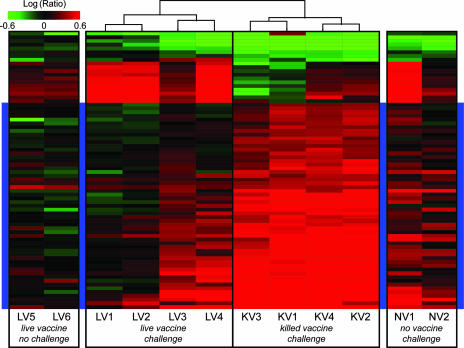
To highlight transcriptional differences between lungs of animals that had received the live attenuated vaccine prototype versus those that had received the killed vaccine, we performed a one-way ANOVA on the two sets of lung expression profiles obtained on day 25 (4 days after challenge). The results of this ANOVA were corrected using the Benjamini-Hochberg false discovery rate test for multiple testing, with a P value cutoff of 0.01, and genes were included in an unsupervised hierarchical cluster if they met the final criterion for inclusion, namely, a ≥2-fold change (P ≤ 0.01) in at least three of the eight experiments. The resulting set of genes was used to produce side clusters illustrating transcriptional regulation of the same genes in animals from other experimental groups (Fig. 6). This analysis revealed that the genes most strongly induced in the killed-vaccine group and in those animals that had intermediate pathology in the live attenuated vaccine group were either interferon dependent or relevant to the innate immune response (Table 1). There appeared to be a positive relationship between induction of these genes and the severity of lung pathology or relative abundance of influenza virus mRNA. Not surprisingly, an opposite relationship was found with hemagglutination inhibition titers at the same time point. Gene expression profiles for animals that received the live attenuated vaccine but were not challenged and for challenged animals that received no vaccine were in overall agreement with these observations.
FIG. 6.
Differential gene expression in lung tissue postchallenge in animals vaccinated with the live attenuated vaccine or the killed vaccine. The dendrogram at top center was obtained by performing a one-way ANOVA between the live attenuated vaccine group and the killed-vaccine group (P ≤ 0.01; Benjamini-Hochberg false discovery rate). The final criterion for inclusion in this cluster was a ≥2-fold change (P ≤ 0.01) in at least three of the experiments. The resulting set of genes was used to produce the side clusters illustrating transcriptional regulation in lungs of animals that received no vaccine and were challenged and in those of animals that received the live vaccine but were not challenged. Animals LV3 and LV4 had relatively more pathology than did LV1 and LV2, but less than all the animals in the killed-vaccine group. LV, live attenuated vaccine; KV, killed vaccine; NV, no vaccine. LV5 and LV6 were not challenged. The blue highlighted region, corresponding to higher induction in the killed-vaccine group, is comprised of ∼33% immune response and ∼18% interferon-induced genes.
TABLE 1.
Gene symbols and accession numbers corresponding to blue highlighted and nonhighlighted regions of clusters shown in Fig. 6
Vaccination with the TX91 NS1▴126 influenza virus is associated with the presence of fewer mature dendritic cells in lung tissue but with stronger induction of an adaptive immune response after challenge.
With the goal of better understanding the local response to the NS1-truncated virus, we stained lung sections in areas affected by the infection for the presence of CD83, a marker of dendritic cell maturation expressed as early as 4 h after internalization of foreign antigens for presentation to T cells either in situ or in local lymph nodes (13). We found that activated dendritic cells were relatively scarce on day 25 (4 days postchallenge) in lung tissue of animals that had received the NS1-truncated vaccine (Fig. 7). This presumably reflects the lack of active infection in lung tissue in these animals at the time of sacrifice (Fig. 3A). Conversely, activated dendritic cells were abundant in lung tissue and less consistently found in tracheobronchial lymph nodes in animals that received either no vaccine or the killed vaccine, a finding compatible with ongoing viral replication and antigen internalization by dendritic cells in these animals.
FIG. 7.
Antigen presentation in lung tissue on day 25 (4 days postchallenge). The images show staining of CD83 (mature or activated dendritic cells) in lung tissues harvested on day 25 (4 days postchallenge) from challenged animals in the live-vaccine group (average mature dendritic cell score, 1.62 out of 4), the killed-vaccine group (average mature dendritic cell score, 2.25 out of 4), and the no-vaccine group (average mature dendritic cell score, 2.5 out of 4). All photographs were taken at an optical magnification of ×10 (insets were taken at an optical magnification of ×20, with an additional digital magnification of ×2).
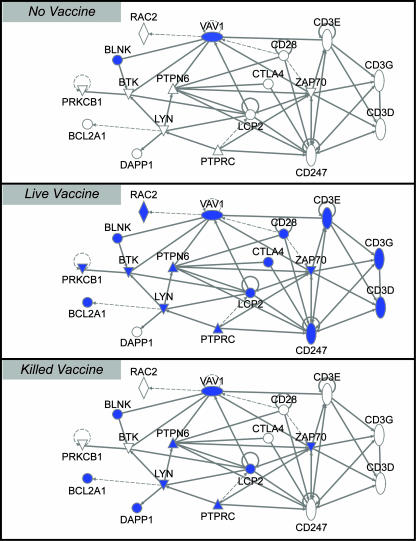
At the same time, we mined lung transcriptional profiles for all experimental groups, with a focus on genes associated with induction of adaptive T- and B-cell responses. This analysis showed that in lung tissue, the induction of adaptive immunity was clearly stronger at 4 days postchallenge in animals that had received the live vaccine (Fig. 8). Therefore, vaccination with the NS1-truncated virus not only produced a superior systemic neutralizing antibody response that reduced replication of the challenge virus but also induced a transcriptional response in situ consistent with stronger or earlier stimulation of a cellular immune response after challenge than that seen for the other groups. The timing of induction of genes with T- and B-cell stimulatory functions coincided with the early migration of dendritic cells away from the primary lesions.
FIG. 8.
Diagrams produced with Ingenuity Systems, Inc., software show induced genes (shown in blue and identified by their HUGO symbols) involved in T- and B-cell receptor activation for each experimental group on day 25 (4 days postchallenge). Genes were selected to be included on all diagrams if they were induced twofold or more (P ≤ 0.01) in half of any experimental group or, in the case of the live vaccine, any subgroup (for the purpose of gene selection, this group was subdivided into two subgroups reflecting the range of pathology observed). Genes on these diagrams are connected if the relationship between the resulting proteins is activation, expression, binding regulation, or inhibition. Solid lines denote direct relationships, and dotted lines denote indirect relationships.
Transcriptional regulation of tracheobronchial cells 2 days after vaccination with the TX91 NS1▴126 virus suggests strong induction of interferon pathways by the attenuated virus.
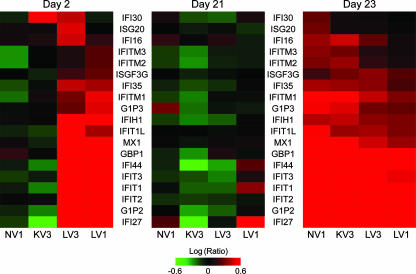
In order to investigate possible mechanisms for the stronger antibody responses pre- and postchallenge associated with the TX91 NS1▴126 virus, we looked at transcriptional regulation in bronchial brush biopsies 2 days after vaccination of the live-vaccine, killed-vaccine, and no-vaccine groups. Bronchial cells sampled at the level of the carina or from primary bronchi were exposed only very briefly to the live attenuated vaccine during inoculation. Consistently, only one of the four animals in that experimental group showed the slight presence of viral mRNA by qRT-PCR at day 2 postvaccination (data not shown). Yet we found striking differences in transcriptional induction of interferon-sensitive genes at that time between bronchial epithelial cells sampled by brush biopsy from animals which had received the TX91 NS1▴126 virus as a live attenuated vaccine and those sampled from the other groups (Fig. 9, left cluster). Since differences in these cells existed in the absence of active viral replication shortly after a brief exposure to the attenuated virus, it is possible that the diminished functionality of NS1 allowed a relatively exaggerated interferon response locally. Provided that what we observed in bronchial cells was representative of the events taking place at the same time in the lungs, a stronger interferon response during a brief instance of viral replication would have facilitated stimulation of dendritic cells, eventually contributing to greater production of influenza virus-specific antibodies (15, 22, 30, 31). By the time the animals were challenged, at day 21 postvaccination, differences in induction of interferon-sensitive genes among vaccine groups were no longer apparent (Fig. 9, middle cluster). As expected, challenge reactivated these genes, but it did so to a lesser extent in animals vaccinated with the live attenuated virus (Fig. 9, right cluster), which is consistent with our findings suggesting that these animals had an abbreviated and possibly attenuated innate immune response due to either the significantly reduced replication of the challenge virus (Fig. 3A), the more robust adaptive immune response (Fig. 3B, C, and D and 8), or both.
FIG. 9.
Differential expression in bronchial brush biopsies on days 2 (day 2 postvaccination), 21 (day of challenge), and 23 (day 2 postchallenge). The criterion for inclusion in the hierarchical cluster illustrating transcriptional regulation in bronchial cells from randomly selected animals from each experimental group 2 days after vaccination (left) was a fourfold change or more (P ≤ 0.01) in at least two of the experiments, followed by mining for genes relevant to the innate immune response. The same set of genes was used to produce the clusters showing transcriptional regulation on the day of challenge (middle) and 2 days after challenge (right). LV, live attenuated vaccine; KV, killed vaccine; NV, no vaccine.
DISCUSSION
Truncation of the nonstructural protein (NS1) of influenza virus to attenuate viruses and to produce live-vaccine prototypes has been explored in a series of in vitro (22, 40, 46, 52) and in vivo (20, 21, 41, 45) studies. The functions of NS1 as an inhibitor of the antiviral response and as a facilitator of viral replication in infected cells are well documented (5, 23, 29, 32, 35, 40). Although the consequences of truncating NS1 to its first 126 amino acids have yet to be studied systematically, it is presumed that the truncation affects NS1 at multiple levels, including its ability to bind double-stranded RNA, because the truncation appears to destabilize the homodimer, the functional unit of the protein. Prior studies have supported such assumptions; for example, swine were protected during homologous and heterologous challenge by an NS1-truncated H3N2 influenza virus (41) whose attenuation was previously verified, as was its limited ability to replicate in interferon-competent cells (45).
As with any live vaccine, the NS1-truncated virus was expected to result in stronger immune stimulation than the killed vaccine. Yet the low capacity of this virus to replicate, even locally, compared to other modified live influenza virus vaccines, along with the robust serum IgG response (17), which is usually more characteristic of formalin-inactivated vaccines, lead us to believe that factors beyond the live nature of the vaccine, and perhaps unique to NS1 truncation or to the route of vaccination, may provide a better explanation for what we observed. We noted evidence of a strong type I interferon transcriptional response in the upper respiratory tract shortly after vaccination with the NS1-truncated virus. This induction, which most likely also took place in lower airways exposed to the live vaccine, was comparable to that observed with the wild-type virus in other groups within the same time frame after challenge (Fig. 9). However, unlike after challenge, it took place in the absence of detectable viral replication in bronchial epithelial cells. While we have only indirect evidence that this could be the mechanism responsible for the observed differences in our study, previous experiments have shown a link between strong innate and adaptive immunity through interferon-induced acceleration of dendritic cell maturation and more efficient antigen presentation to T lymphocytes (22, 30, 31). There is also evidence that B lymphocytes may be stimulated directly by interferon (15), and our previous influenza virus experiments with macaques were in fact indicative of early B-cell stimulation following the virus-induced interferon response (2). The lungs are also known to be a Th2-biased organ, signifying that antigens of any kind delivered directly to the respiratory tract will be presented preferentially to Th2 CD4+ T cells and lead to the development of a strong humoral response (44, 45), so it is reasonable to assume that the route of inoculation of the live vaccine did play a role in the results that were seen. Indeed, a key finding of the present study was the much higher production of influenza virus-specific antibodies before and after challenge in animals that had received the live vaccine. Along with substantially greater percentages of influenza virus-specific CD4+ T cells in blood 4 days after challenge, this suggested a larger peripheral pool of memory CD4+ T cells induced by the live vaccine. These results were accentuated by the stronger induction of the adaptive immune response and relative repression of interferon and inflammatory pathways in lung tissue during challenge. This was a clinically important result considering the dysregulation of the innate immune response observed in mice and macaques infected with the fully reconstructed 1918 pandemic virus (25, 27) and the importance of humoral immunity in providing protection against this virus (28). Conversely, the relative immaturity of the postchallenge immune response after vaccination with the killed or mock vaccine was further supported by the abundance of CD83+ dendritic cells in lung tissue of these animals. Staining of tracheobronchial lymph nodes revealed similar but less dramatic differences (not shown), suggesting that animals in the killed-vaccine and no-vaccine groups were still in the early stages of antigen presentation 4 days after challenge.
The present study with pigtailed macaques demonstrates the protective potential of live influenza viruses attenuated through modification of the NS1 gene. The use of oligonucleotide arrays on lung and bronchial tissues served to validate the clinical findings and to further explore the effects of the truncation on transcriptional control after vaccination as well as the effects of the vaccines on transcriptional induction after challenge. Compared to the TX91 NS1▴126 virus, the formalin-killed virus, although administered at the same dose as the commercial vaccine, was poorly immunogenic. This is consistent with its performance in previously unvaccinated children, for whom two doses of the killed vaccine are needed for consistent protection (37, 38). On the other hand, a single vaccination with the truncated virus induced strong systemic immunity and also prevented inflammation in the upper respiratory tract following virus challenge, a finding in agreement with higher local IgA production following vaccination with live vaccines (17) and with the superior protective efficacy of live influenza virus vaccines, particularly for children (16).
Infections of mice with the fully reconstructed 1918 influenza virus have resulted in much stronger upregulation of interleukin-6 (IL-6) and IL-10 in lung tissue, both of which are normally secreted by activated dendritic cells, than in infections with less pathogenic viruses (25). They also resulted in strong upregulation of CXCL2 and IL-8, both of which are neutrophil chemoattractants secreted in response to dendritic cell activation (14). Vaccination with the TX91 NS1▴126 virus resulted in less induction of all of these cytokines after challenge than that in the other vaccine groups, in spite of the stronger antibody response, suggesting that an effective adaptive immune response is protective against overstimulation of dendritic cells in the lungs.
In conclusion, vaccination of pigtailed macaques with a human influenza virus attenuated by truncation of the NS1 gene resulted in significant protection of the animals against viral replication, local inflammation, and lung pathology during homologous challenge. This protection was achieved through strong induction of the humoral and cellular immune responses subsequent to vaccination, likely as a result of an early innate immune response despite the absence of detectable replication, resulting in more efficient maturation of dendritic cells. The intrarespiratory route of immunization also very likely contributed to enhancing the humoral response to the modified live vaccine. Therefore, NS1 truncation constitutes a promising means of attenuating influenza viruses for the purpose of devising live vaccines. Further modifications to genes encoding surface proteins of H5N1 viruses would be required to ensure the safety of the vaccine, and these vaccines may not be appropriate for all segments of the human population, but they have the potential to be protective after a single inoculation, they can be produced with far fewer eggs than those required for killed vaccines, and they do not require the use of internal genes from a cold-adapted strain, thereby decreasing the odds of recombination with circulating human viruses.
Acknowledgments
This work was partially supported by NIH grants R01AI46954, P01AI58113, and U01AI070469 to A.G.-S. and R24 RR16354-04, P51 RR00166-45, and R01 AI022646-20A1 to M.G.K.
We thank Richard Cadagan, Kay Larsen, and Leon Flanary for their expert technical assistance.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Aragon, T., S. de la Luna, I. Novoa, L. Carrasco, J. Ortin, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baas, T., C. R. Baskin, D. L. Diamond, A. Garcia-Sastre, H. Bielefeldt-Ohmann, T. M. Tumpey, M. J. Thomas, V. S. Carter, T. H. Teal, N. Van Hoeven, S. Proll, J. M. Jacobs, Z. R. Caldwell, M. A. Gritsenko, R. R. Hukkanen, D. G. Camp II, R. D. Smith, and M. G. Katze. 2006. An integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 80:10813-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bardiya, N., and J. H. Bae. 2005. Influenza vaccines: recent advances in production technologies. Appl. Microbiol. Biotechnol. 67:299-305. [DOI] [PubMed] [Google Scholar]
- 4.Baskin, C. R., A. Garcia-Sastre, T. M. Tumpey, H. Bielefeldt-Ohmann, V. S. Carter, E. Nistal-Villan, and M. G. Katze. 2004. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina). J. Virol. 78:10420-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., and A. Garcia-Sastre. 2002. Viruses and the type I interferon antiviral system: induction and evasion. Int. Rev. Immunol. 21:305-337. [DOI] [PubMed] [Google Scholar]
- 6.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belshe, R. B., P. M. Mendelman, J. Treanor, J. King, W. C. Gruber, P. Piedra, D. I. Bernstein, F. G. Hayden, K. Kotloff, K. Zangwill, D. Iacuzio, and M. Wolff. 1998. The efficacy of live attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine in children. N. Engl. J. Med. 338:1405-1412. [DOI] [PubMed] [Google Scholar]
- 8.Bielefeldt-Ohmann, H., L. A. Babiuk, and R. Harland. 1991. Cytokine synergy with viral cytopathic effects and bacterial products during the pathogenesis of respiratory tract infection. Clin. Immunol. Immunopathol. 60:153-170. [DOI] [PubMed] [Google Scholar]
- 9.Bielefeldt-Ohmann, H., D. H. Barouch, A. M. Bakke, A. G. Bruce, M. Durning, R. Grant, N. L. Letvin, J. T. Ryan, A. Schmidt, M. E. Thouless, and T. M. Rose. 2005. Intestinal stromal tumors in a simian immunodeficiency virus-infected, simian retrovirus-2 negative rhesus macaque (Macaca mulatta). Vet. Pathol. 42:391-396. [DOI] [PubMed] [Google Scholar]
- 10.Brazma, A., T. Freeman, K. Gardiner, S. Weissman, T. Werner, and B. Korn. 2004. Report on the thirteenth international workshop on the identification and functional, evolutionary and expression analysis of transcribed sequences: comparative and functional genomics workshop. Cytogenet. Genome Res. 105:11-17. [DOI] [PubMed] [Google Scholar]
- 11.Breathnach, C. C., H. J. Clark, R. C. Clark, C. W. Olsen, H. G. Townsend, and D. P. Lunn. 2006. Immunization with recombinant modified vaccinia Ankara (rMVA) constructs encoding the HA or NP gene protects ponies from equine influenza virus challenge. Vaccine 24:1180-1190. [DOI] [PubMed] [Google Scholar]
- 12.Bresson, J. L., C. Perronne, O. Launay, C. Gerdil, M. Saville, J. Wood, K. Hoschler, and M. C. Zambon. 2006. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet 367:1657-1664. [DOI] [PubMed] [Google Scholar]
- 13.Cao, W., S. H. Lee, and J. Lu. 2005. CD83 is preformed inside monocytes, macrophages and dendritic cells, but it is only stably expressed on activated dendritic cells. Biochem. J. 385:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu, B. C., C. M. Freeman, V. R. Stolberg, J. S. Hu, E. Komuniecki, and S. W. Chensue. 2004. The innate pulmonary granuloma: characterization and demonstration of dendritic cell recruitment and function. Am. J. Pathol. 164:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coro, E. S., W. L. Chang, and N. Baumgarth. 2006. Type I IFN receptor signals directly stimulate local B cells early following influenza virus infection. J. Immunol. 176:4343-4351. [DOI] [PubMed] [Google Scholar]
- 16.Cox, N. J., and C. B. Bridges. 2007. Inactivated and live attenuated influenza vaccines in young children—how do they compare? N. Engl. J. Med. 356:729-731. [DOI] [PubMed] [Google Scholar]
- 17.Cox, R. J., K. A. Brokstad, and P. Ogra. 2004. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand. J. Immunol. 59:1-15. [DOI] [PubMed] [Google Scholar]
- 18.de Bree, G. J., E. M. van Leeuwen, T. A. Out, H. M. Jansen, R. E. Jonkers, and R. A. van Lier. 2005. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J. Exp. Med. 202:1433-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enserink, M. 2005. Avian influenza. ‘Pandemic vaccine’ appears to protect only at high doses. Science 309:996. [DOI] [PubMed] [Google Scholar]
- 20.Falcon, A. M., A. Fernandez-Sesma, Y. Nakaya, T. M. Moran, J. Ortin, and A. Garcia-Sastre. 2005. Attenuation and immunogenicity in mice of temperature-sensitive influenza viruses expressing truncated NS1 proteins. J. Gen. Virol. 86:2817-2821. [DOI] [PubMed] [Google Scholar]
- 21.Ferko, B., J. Stasakova, J. Romanova, C. Kittel, S. Sereinig, H. Katinger, and A. Egorov. 2004. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 78:13037-13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Sesma, A., S. Marukian, B. J. Ebersole, D. Kaminski, M. S. Park, T. Yuen, S. C. Sealfon, A. Garcia-Sastre, and T. M. Moran. 2006. Influenza virus evades innate and adaptive immunity via the NS1 protein. J. Virol. 80:6295-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 24.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. Garcia-Sastre. 2002. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kash, J. C., T. M. Tumpey, S. C. Proll, V. Carter, O. Perwitasari, M. J. Thomas, C. F. Basler, P. Palese, J. K. Taubenberger, A. Garcia-Sastre, D. E. Swayne, and M. G. Katze. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443:578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz, J. M., X. Lu, S. A. Young, and J. C. Galphin. 1997. Adjuvant activity of the heat-labile enterotoxin from enterotoxigenic Escherichia coli for oral administration of inactivated influenza virus vaccine. J. Infect. Dis. 175:352-363. [DOI] [PubMed] [Google Scholar]
- 27.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, H. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319-323. [DOI] [PubMed] [Google Scholar]
- 28.Kong, W. P., C. Hood, Z. Y. Yang, C. J. Wei, L. Xu, A. Garcia-Sastre, T. M. Tumpey, and G. J. Nabel. 2006. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc. Natl. Acad. Sci. USA 103:15987-15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 30.Le Bon, A., G. Schiavoni, G. D'Agostino, I. Gresser, F. Belardelli, and D. F. Tough. 2001. Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 14:461-470. [DOI] [PubMed] [Google Scholar]
- 31.Le Bon, A., and D. F. Tough. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14:432-436. [DOI] [PubMed] [Google Scholar]
- 32.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 33.Lin, J., J. Zhang, X. Dong, H. Fang, J. Chen, N. Su, Q. Gao, Z. Zhang, Y. Liu, Z. Wang, M. Yang, R. Sun, C. Li, S. Lin, M. Ji, X. Wang, J. Wood, Z. Feng, Y. Wang, and W. Yin. 2006. Safety and immunogenicity of an inactivated adjuvanted whole-virion influenza A (H5N1) vaccine: a phase I randomised controlled trial. Lancet 368:991-997. [DOI] [PubMed] [Google Scholar]
- 34.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neirynck, S., T. Deroo, X. Saelens, P. Vanlandschoot, W. M. Jou, and W. Fiers. 1999. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat. Med. 5:1157-1163. [DOI] [PubMed] [Google Scholar]
- 37.Neuzil, K. M., and J. A. Englund. 2006. Influenza vaccine for young children: two doses are better than one. J. Pediatr. 149:737-738. [DOI] [PubMed] [Google Scholar]
- 38.Neuzil, K. M., L. A. Jackson, J. Nelson, A. Klimov, N. Cox, C. B. Bridges, J. Dunn, F. DeStefano, and D. Shay. 2006. Immunogenicity and reactogenicity of 1 versus 2 doses of trivalent inactivated influenza vaccine in vaccine-naive 5-8-year-old children. J. Infect. Dis. 194:1032-1039. [DOI] [PubMed] [Google Scholar]
- 39.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 40.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 79:8431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richt, J. A., P. Lekcharoensuk, K. M. Lager, A. L. Vincent, C. M. Loiacono, B. H. Janke, W. H. Wu, K. J. Yoon, R. J. Webby, A. Solorzano, and A. Garcia-Sastre. 2006. Vaccination of pigs against swine influenza viruses using an NS1-truncated modified live-virus vaccine. J. Virol. 80:11009-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rimmelzwaan, G. F., M. Baars, R. van Beek, G. van Amerongen, K. Lovgren-Bengtsson, E. C. Claas, and A. D. Osterhaus. 1997. Induction of protective immunity against influenza virus in a macaque model: comparison of conventional and iscom vaccines. J. Gen. Virol. 78:757-765. [DOI] [PubMed] [Google Scholar]
- 43.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2001. Pathogenesis of influenza A (H5N1) virus infection in a primate model. J. Virol. 75:6687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rimmelzwaan, G. F., T. Kuiken, G. van Amerongen, T. M. Bestebroer, R. A. Fouchier, and A. D. Osterhaus. 2003. A primate model to study the pathogenesis of influenza A (H5N1) virus infection. Avian Dis. 47:931-933. [DOI] [PubMed] [Google Scholar]
- 45.Solorzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. Garcia-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 79:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stasakova, J., B. Ferko, C. Kittel, S. Sereinig, J. Romanova, H. Katinger, and A. Egorov. 2005. Influenza A mutant viruses with altered NS1 protein function provoke caspase-1 activation in primary human macrophages, resulting in fast apoptosis and release of high levels of interleukins 1beta and 18. J. Gen. Virol. 86:185-195. [DOI] [PubMed] [Google Scholar]
- 47.Stephenson, I. 2006. H5N1 vaccines: how prepared are we for a pandemic? Lancet 368:965-966. [DOI] [PubMed] [Google Scholar]
- 48.Stohr, K., M. P. Kieny, and D. Wood. 2006. Influenza pandemic vaccines: how to ensure a low-cost, low-dose option. Nat. Rev. Microbiol. 4:565-566. [DOI] [PubMed] [Google Scholar]
- 49.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas, M. J., L. R. Flanary, B. A. Brown, M. G. Katze, and C. R. Baskin. 2006. Use of human nasal cannulas during bronchoscopy procedures as a simple method for maintaining adequate oxygen saturation in pigtailed macaques (Macaca nemestrina). J. Am. Assoc. Lab. Anim. Sci. 45:44-48. [PubMed] [Google Scholar]
- 52.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]