Abstract
The host cell protein cyclophilin A (CypA) binds to CA of human immunodeficiency virus type 1 (HIV-1) and promotes HIV-1 infection of target cells. Disruption of the CypA-CA interaction, either by mutation of the CA residue at G89 or P90 or with the immunosuppressive drug cyclosporine (CsA), reduces HIV-1 infection. Two CA mutants, A92E and G94D, previously were identified by selection for growth of wild-type HIV-1 in cultures of CD4+ HeLa cell cultures containing CsA. Interestingly, infection of some cell lines by these mutants is enhanced in the presence of CsA, while in other cell lines these mutants are minimally affected by the drug. Little is known about this cell-dependent phenotype of the A92E and G94D mutants, except that it is not dependent on expression of the host factor TRIM5α. Here, we show that infection by the A92E and G94D mutants is restricted at an early postentry stage of the HIV-1 life cycle. Analysis of heterokaryons between CsA-dependent HeLa-P4 cells and CsA-independent 293T cells indicated that the CsA-dependent infection by A92E and G94D mutants is due to a dominant cellular restriction. We also show that addition of CsA to target cells inhibits infection by wild-type HIV-1 prior to reverse transcription. Collectively, these results support the existence of a cell-specific human cellular factor capable of restricting HIV-1 at an early postentry step by a CypA-dependent mechanism.
Human immunodeficiency virus type 1 (HIV-1) encodes precursor polyproteins necessary for particle assembly and budding from infected host cells (26). Following the release of virions as immature particles, Gag and Gag-Pol polyprotein precursors undergo proteolytic cleavage by the viral protease into mature proteins, including CA (61). Maturation results in the formation of the condensed conical viral capsid required for HIV-1 particles to be infectious (64). Upon fusion of the viral and cellular membranes, the viral core, consisting of the genomic RNA and associated proteins within the viral capsid, then is released into the cytoplasm. Partial or complete disassembly of the viral capsid occurs in a reaction referred to as uncoating (reviewed in references 3 and 41). Mutations in CA that affect the capsid stability in either the positive or negative direction result in marked reduction of HIV-1 infectivity. The mutations are frequently associated with either impaired reverse transcription or nuclear targeting and integration of the preintegration complex in target cells (17, 21, 22).
Using a yeast two-hybrid screening system, it was found that the Gag polyprotein binds to the host cell protein cyclophilin A (CypA) (38). Subsequent in vitro and in vivo studies further demonstrated the direct interaction between CypA and the CA domain in the Gag protein as well as specific incorporation of CypA into HIV-1 virions (10, 24, 38, 57). The binding site for CypA mapped to the loop between helix 4 and helix 5 on the N-terminal domain of CA (27). It also has been shown, through the use of X-ray and nuclear magnetic resonance analyses, that CA residues A88, G89, and P90 were critical for interaction with the hydrophobic pocket of CypA (18, 23, 33, 57, 59, 70, 71).
CypA is the most abundant member of a family of peptidyl-prolyl isomerase enzymes and was first identified by virtue of its high affinity for the immunosuppressive drug cyclosporine (CsA) (20, 28, 56). The importance of the CA-CypA interaction for efficient HIV-1 replication was demonstrated by disrupting the interaction by CA mutations (10, 24, 57) and with a human CD4+ T-cell line with a homozygous deletion of the CypA gene (12). HIV-1 replication also is inhibited by blocking of the interaction between CA and CypA by CsA or related compounds that form a high-affinity complex with CypA (5, 9, 23, 44, 62). Previously, it has been shown that HIV-1 particles released from cells cultured in the presence of CsA exhibit reduced infectivity in single-cycle assays, suggesting that incorporation of CypA into HIV-1 particles is important for the particles to be infectious (4, 24, 57). The poor infectivity of CypA-deficient virions was traced to a failure of the virus to undergo reverse transcription in target cells (11, 12, 57). Despite the initial focus on producer-cell-dependent effects of CypA, recent studies have revealed that engagement of the viral capsid by CypA in the target cell represents the biologically relevant interaction (30, 37, 52). These studies also demonstrated that the reduced infectivity observed with virions in the presence of CsA or mutations in CA is independent of encapsidated CypA and that producer cell CypA is not necessary for the production of fully infectious HIV-1 virions (2, 30, 52, 69).
Recent studies have suggested that the requirement of the CA-CypA interaction may be related to the effects of CA-specific endogenous host restriction activities, originally termed Lv1 and Ref1 (7, 8, 16, 29, 31, 36, 37). The responsible cellular factors recently were identified as the α-spliced isoforms of TRIM5, which target the CA of a wide variety of retroviruses (36, 42, 55, 67). The fact that disruption of the HIV-1 CA-CypA interaction renders HIV-1 poorly infectious in human cells indicates that the binding of CypA to the incoming HIV-1 core may protect HIV-1 from an endogenous host restriction. By contrast, CypA binding to CA promotes restriction of HIV-1 by TRIM5α in Old World monkey cells (6, 35) and cells of owl monkeys (58). Additional studies identified a novel restriction factor, TRIM-Cyp, in owl monkey cells that originated from retrotransposition of a complete CypA cDNA into the TRIM5 gene (40, 49). Collectively, these studies demonstrate that binding of CypA to CA sensitizes HIV-1 to restriction in cells from a variety of nonhuman primates.
Adaptation of wild-type HIV-1 for replication in cultures of human cells containing CsA resulted in the identification of two CA mutations, A92E and G94D, which confer resistance to CsA (1). Interestingly, these mutants retain the ability of CA to bind CypA (10), suggesting that they are no longer sensitive to the putative restriction in human cells that targets HIV-1 in the absence of CypA. The phenotype of the A92E and G94D mutants depends on the human cell line in which viral infection is tested: addition of CsA is required for replication of A92E or G94D mutant virus in some human cell lines, such as CD4+ HeLa or H9 cells (1, 30, 52, 69), while in Jurkat, HOS, or 293T cells, the mutations confer CsA resistance but not drug dependence (10, 30, 52). These results suggest that in the human cell lines that are nonpermissive for the A92E and G94D mutants, the binding of CypA to CA has a detrimental effect on replication. In single-cycle infection assays, CsA promotes infection of these nonpermissive human cell types when added at the time of virus inoculation, indicating that the negative effect of CypA occurs in the target cell (30, 52).
Based on these findings, it was proposed that the binding of CypA to CA renders A92E and G94D susceptible to a host-cell-dependent restriction (52). A prediction of this model is that the nonpermissive cell lines express an activity that dominantly restricts A92E and G94D mutants in a CypA-dependent manner. In the present study, we tested this hypothesis by studying the phenotype of these mutants in heterokaryons generated between human cells that are permissive and nonpermissive for infection by the mutant viruses.
MATERIALS AND METHODS
Plasmids.
Capsid mutations (A92E and G94D) were transferred from the HIV-1 NL4-3 clone to the previously described HIV-1 vector expressing enhanced green fluorescent protein (HIV-GFP) (32) by transfer of the corresponding BssHII-SalI DNA fragments. The presence of those mutations in the CA in the final HIV-1-GFP construct was confirmed by DNA sequencing. The pHCMV-G plasmid encodes a vesicular stomatitis virus G (VSV-G) protein (68). The plasmid pMM310 expresses the β-lactamase-Vpr (BlaM-Vpr) fusion protein (obtained from Mike Miller, Merck Research Laboratories). The pTMT plasmid encodes the Mason-Pfizer monkey virus glycoprotein (M-PMV Env) under the transcriptional control of the myeloproliferative sarcoma virus promoter (53).
Cells lines and viruses.
293T and HeLa-P4 cells (15) were cultured at 37°C in a humidified incubator with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Cellgro) supplemented with 10% fetal bovine serum (HyClone), penicillin (50 IU/ml), and streptomycin (50 μg/ml). Wild-type and mutant GFP reporter virus stocks were prepared by cotransfection of 293T cells with HIV-GFP and pHCMV-G plasmids using polyethylenimine (PEI; Polysciences) as described previously (19). Briefly, viruses were produced by transient cotransfection of about 70% confluent 293T cells using 10 μg of HIV-1-GFP and 5 μg pHCMV-G plasmid DNAs with 60 μl of PEI reagent (1 mg/ml). Two days after transfection, virus-containing supernatant was collected, clarified by filtration through 0.45-μm-pore-size filters, and frozen into aliquots at −80°C. The CA content of the virus stocks was quantified by a p24-specific enzyme-linked immunosorbent assay utilizing monoclonal antibody 183-H12-5C for capture (63).
Infection assay.
Adherent target cells were plated at 20,000 per well in 24-well plates the day before infection. Cells were inoculated with fivefold serially diluted VSV-G-pseudotyped GFP reporter virus stocks in the presence of Polybrene (8 μg/ml). In experiments with CsA, the drug (5 μM final concentration) was added to the medium at the time of infection. Viruses and CsA were removed 24 h later, and the cells were cultured in fresh medium. Cells were harvested, and infection was measured at 48 h postinfection by quantifying GFP+ cells with a FACSCalibur instrument (Becton Dickinson). Ten thousand cells were analyzed per sample. For heterokaryon experiments, one million cells were plated in six-well plates, and one-half million cells per sample were analyzed with an LSRII flow cytometer (Becton Dickinson) 48 h after virus infection. The resulting data were analyzed using FlowJo software (Tree Star).
Virus-cell fusion assay.
The BlaM-Vpr HIV-1 fusion assay was performed as previously described (14, 34, 65). Briefly, reporter viruses were produced by cotransfection of proviral HIV-1 DNA (10 μg) with pHCMV-G (3 μg) and pMM310 (7 μg), which encodes a fusion protein consisting of the Escherichia coli BlaM fused to the amino terminus of HIV-1 Vpr. Quantities of wild-type and mutant reporter viruses were normalized by p24 and used to inoculate HeLa-P4 cells (20, 000) for 2 h at 37°C. Cells then were loaded with the CCF2-AM fluorogenic substrate (20 μM; PanVera Corp.) overnight at room temperature in the dark. The supernatant was replaced with 100 μl of phosphate-buffered saline (PBS), and fluorescence was measured from the bottom at 450 and 520 nm after excitation at 410 nm in a BMG FLUOstar fluorometer. Because cleavage of CCF2-AM by BlaM results in an emission spectral shift from 520 to 450 nm, the extent of fusion was calculated as the emission signal at 450 nm divided by that at 520 nm, after subtraction of the appropriate background values.
Quantitative analysis of HIV-1 reverse transcription in HeLa-P4 cells.
One day prior to infection, 293T and HeLa-P4 cells (100,000) were plated into each well of 12-well plates. Viruses were treated with 20 μg/ml of DNase I and 10 mM MgCl2 at 37°C for 1 h to remove contaminating plasmid DNA. 293T and HeLa-P4 cells were inoculated with VSV-G-pseudotyped viruses normalized by p24 content to 30 ng/well, along with 8 μg/ml of Polybrene. As a negative control for reverse transcription, wild-type HIV-1 was inoculated in the presence of a nonnucleoside reverse transcriptase inhibitor (Efavirenz; 5 μM). At 6 and 12 h postinfection, infected cells were washed with 1 ml of PBS and then treated with 500 μl of trypsin. Trypsin was inactivated by addition of 750 μl of DMEM containing 10% fetal bovine serum, and cells were collected and washed once with 500 μl of PBS. Cell pellets were resuspended in 200 μl of PBS, and DNA was isolated using a DNeasy kit (QIAGEN) by following the manufacturer's instructions. Viral DNA was quantified by real-time PCR using an MX-3000p thermocycler (Stratagene) utilizing TaqMan chemistry (ABI). Early reverse transcription products (minus-strand strong-stop DNA) were amplified with the primers ERT-SS-F (5′-GCTAACTAGGGAACCCACTGCTT-3′) and ERT-SS-R (5′-ACAACAGACGGGCACACACTAC-3′) and were detected with probe ERT-SS (5′-6-carboxyfluorescein-AGCCTCAATAAAGCTTGCCTTGAGTGCTTC-6-carboxytetramethylrhodamine-3′). Late reverse transcription products (U5-Gag) were amplified with primers MH531 (5′-TGTGTGCCCGTCTGTTGTGT-3′) and MH532 (5′-GAGTCCTGCGTCGAGAGAGC-3′) and were detected with the probe LTR-P (5′-6-carboxyfluorescein-CAGTGGCGCCCGAACAGGGA-6-carboxytetramethylrhodamine-3′) as previously described (13, 17). Thermal cycling conditions were 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s.
Generation and analysis of human cell heterokaryons.
293T cells were transfected with either pTMT plasmid or empty control plasmid (10 μg) using the PEI method described above. After 24 h posttransfection, the cells were washed once with PBS and then labeled with either CellTrace Far Red DDAO-SE (15 μM) or CellTracker Orange CMTMR (5 μM) fluorescent probe (both dyes from Molecular Probes) for 45 min at 37°C in DMEM. HeLa-P4 cells also were labeled with the alternate dye. After incubation, cells were washed twice with PBS, and then complete media were added. After another 1-h incubation at 37°C in complete media, trypsin was added to detach the cells. Equal numbers of 293T and HeLa-P4 cells were mixed, and 500,000 cells were plated into two sets of six-well plates to assay infection as well as fusion efficiency. Cells also were plated on coverslips for fluorescent microscopic examination. The cells were washed with PBS and then fixed with 3.7% paraformaldehyde (PFA) in PBS for 15 min at room temperature. The fixed cells were observed by an LSM 510 META inverted confocal microscope. Twenty-four hours after the plating of 500,000 cells in six-well plates, one set of cells was harvested, fixed with 2% PFA, and then analyzed with an LSR II instrument to test the efficiency of heterokaryon formation. Populations of cells positive for both DDAO and CMTMR fluorescent probes were identified as heterokaryons. Remaining cultures were inoculated with wild-type as well as mutant viruses with a predetermined amount of virus sufficient to infect 5% of HeLa-P4 cells with the A92E mutant. Forty-eight hours postinfection, 500,000 cells were analyzed with the LSR II instrument by first gating the population of cells positive for both DDAO and CMTMR and then analyzing this population for GFP expression.
Immunoblot analysis.
Cells were lysed in a solution of 50 mM Tris-HCl, pH 7.4, 0.5% NP-40, 150 mM NaCl, 1 mM EDTA, and cell debris and nuclei were removed by centrifugation. Equivalent amounts of protein, as determined by bicinchoninic acid assay (Pierce Chemical Co.), were subjected to sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis followed by transfer of proteins to nitrocellulose membranes (0.45-μm pore size; Perkin-Elmer). The membranes were blocked for 30 min at room temperature with 5% nonfat milk in PBS and were probed with a 1:10,000 dilution of rabbit anti-CypA antibody (Biomol) for CypA and mouse anti-β-tubulin (8.7 ng/ml; Sigma Chemical Co.) overnight at 4°C. The membranes were washed with PBS containing 0.1% Tween 20 four times for 10 min and were probed with the appropriate species-specific Alexa fluor 680-conjugated secondary antibody. Protein bands were visualized using the Odyssey infrared imaging system (LI-COR) and quantified using the instrument's software.
RESULTS
CsA enhances infection of HeLa-P4 cells by the A92E and G94D HIV-1 mutants.
Previously, it has been reported that infection by A92E and G94D mutants is enhanced by CsA in H9 and HeLa cells but not in Jurkat, HOS, and 293T cells (30, 52). To verify these reports, HIV-GFP vectors expressing GFP in place of Nef were engineered to encode either the A92E or G94D mutation. VSV-G-pseudotyped virions were produced by transfection of 293T cells with the engineered HIV-GFP CA variants as well as with the wild type. The resulting pseudotyped reporter viruses were used to infect the HeLa-P4 or 293T cell line in the presence or absence of CsA, and cultures were analyzed by flow cytometry for GFP expression to determine the percentage of infected cells. When CsA was present during inoculation of 293T cells with the wild-type virus, the level of infection was reduced by approximately two-thirds (Fig. 1A). Infection levels of these cells by the A92E and G94D mutants also were reduced by one-third to one-half with CsA treatment (Fig. 1C and E). Similar effects of CsA were obtained when either HOS or Jurkat cells were used as target cells instead of 293T cells (data not shown).
FIG. 1.
Cell-type-specific effects of CsA on HIV-1 infection. VSV-G-pseudotyped HIV-GFP viruses, either with wild-type (WT) or CA mutant (A92E and G94D) vectors, were produced by transfection of 293T cells, normalized by p24, and used to infect 293T cells (A, C, and E) or HeLa-P4 cells (B, D, and F) in the presence or absence of CsA (5 μM) during infection. Ten thousand live cells were analyzed per sample. The percentage of GFP-positive cells is plotted against an input viral dose (in nanograms of p24).
In contrast to its effects on 293T cells, CsA exhibited different effects on infection of HeLa-P4 cells. In the absence of CsA, the A92E and G94D mutants exhibited six- to eightfold reduced infectivity relative to that of the wild-type virus. Infection by the wild-type virus was reduced by half when CsA was present during inoculation (Fig. 1B). However, infection by the A92E and G94D mutants was stimulated 8- to 11-fold by CsA (Fig. 1D and F). Taken together, these results confirmed that infection by A92E and G94D mutants is restricted in HeLa-P4 cells but not in 293T cells and that the restriction is prevented by CsA. Additionally, infection of 293T cells by the wild-type virus was somewhat more sensitive to inhibition by CsA in 293T cells than in HeLa-P4 cells.
CsA does not inhibit fusion of A92E or G94D mutant HIV-1 particles with target cells.
Although CsA is known to facilitate an early step in infection of HeLa cells by A92E and G94D viruses, it has not been determined whether this enhancement occurs during fusion or during a postentry step. A previous study reported that CsA inhibits attachment of the wild-type virus to target cells, suggesting the possibility that virion-associated CypA promotes HIV-1 fusion (46, 47). To determine whether the increased infectivity of the A92E and G94D mutants in HeLa-P4 cells in the presence of CsA results from enhanced viral entry, we tested the fusion capacity of the wild type and the A92E and G94D mutants with HeLa-P4 cells. The assay employed utilizes BlaM as a fusion protein with the viral accessory protein Vpr, which is incorporated specifically into HIV-1 particles via an interaction with Gag during particle assembly (14). HeLa-P4 target cells were inoculated with VSV-G-pseudotyped HIV-1 particles containing BlaM in the presence and absence of CsA. Cultures subsequently were loaded with CCF2-AM, a cell-permeable fluorescent substrate for BlaM. The emission spectrum of CCF2 shifts from green to blue upon cleavage by BlaM, and the extent of CCF2-AM cleavage was quantified by spectrofluorimetry.
Inoculation with wild-type HIV-1 particles resulted in a dose-dependent increase in the blue/green fluorescent ratio (Fig. 2A). The addition of CsA during the fusion assay showed no effect on the blue/green fluorescent ratio in the wild-type cells compared to that of the cells not treated with CsA. The A92E and G94D mutants also exhibited a linear dose response of fusion regardless of CsA treatment (Fig. 2B and C). The observed fusion levels for the mutants were similar to that observed for the wild-type virus. HIV-1 particles lacking VSV-G protein exhibited only background signals (data not shown). These results indicate that addition of CsA to cells at the time of virus inoculation does not affect the fusion of the wild type or A92E and G94D mutants with HeLa-P4 cells when entry is mediated via VSV-G.
FIG. 2.
CsA does not affect HIV-1 fusion with target cells. VSV-G-pseudotyped HIV-1-GFP virus encoding wild-type (WT) (A), A92E mutant (B), and G94D mutant (C) CA were produced by cotransfection of 293T cells with a BlaM-Vpr expression plasmid and were subsequently titrated on HeLa-P4 cells. Fusion was quantified as the ratio of blue-to-green fluorescence resulting from cleavage of the cell-permeable BlaM substrate. Results are shown as the mean values of duplicate determinations plotted against the input viral dose (in nanograms of p24) in the presence or absence of CsA (5 μM). The results are representative of at least two independent experiments.
CsA inhibits HIV-1 reverse transcription in 293T cells.
The equivalent fusion observed for A92E and G94D HIV-1 particles in the presence and absence of CsA indicated that CsA-enhanced infection by these mutants results from the enhancement of early postentry events of the HIV-1 life cycle. To further identify the affected step, we employed quantitative real-time PCR (qPCR) to measure the amount of early and late products of reverse transcription. Wild-type and mutant viruses were used to infect 293T and HeLa-P4 target cells. At 6 and 12 h postinfection, infected cells were harvested and the DNA was purified. To measure early products, qPCR was performed using primers and probes specific for minus-strand strong-stop DNA using DNA harvested 6 h postinfection. For late products, primers and a probe specific for full-length plus-strand DNA were employed in qPCR with total DNA purified 12 h postinfection as a template.
When 293T cells were infected with wild-type and mutant viruses, the levels of early reverse transcription products for wild-type, A92E, and G94D viruses were reduced by approximately 20 to 60% with CsA treatment (Fig. 3A). The effect of CsA treatment on the synthesis of late reverse transcription products was somewhat more pronounced than that of early reverse transcription products. The addition of CsA during infection resulted in the reduction of late products by 70 to 90% compared to the levels of reverse transcription in the absence of CsA (Fig. 3B). We also tested viral infectivity of wild-type and mutant viruses with the same amount of virions used for the reverse transcription assay. As expected, the infectivity of the wild-type virions was reduced by approximately 80% (Fig. 3C, WT), and that of mutant viruses also was reduced by 20% (Fig. 3C, A92E) to 60% (Fig. 3C, G94D) with CsA treatment. Minimal signals were observed for both early and late reverse transcription products for infectivity of wild-type virus inoculated in the presence of a reverse transcriptase inhibitor (Efavirenz), indicating that the signals detected were dependent on reverse transcriptase activity and, thus, not from contaminating plasmid DNA. Taken together, these results indicate that the addition of CsA during infection decreases reverse transcription in 293T cells.
FIG. 3.
Effect of CsA treatment of target cells on reverse transcription (RT) of wild-type and mutant HIV-1 particles. VSV-G-pseudotyped HIV-1-GFP viruses were produced by transfection of 293T cells, normalized for p24, and used to inoculate 293T (A, B, and C) and HeLa-P4 (D, E, and F) cells in the presence or absence of 5 μM CsA. DNA was isolated from target cells at 6 and 12 h postinfection and assayed for early (A and D) and late (B and E) products of reverse transcription by quantitative PCR. As a control, the wild-type virus (WT) was inoculated in the presence of the reverse transcriptase inhibitor Efavirenz (WT/E). (C and F) Parallel cultures were analyzed 2 days postinoculation for GFP expression by flow cytometry. Both assays were performed with a high dose of virus to maximize the likelihood of detection of viral DNA in 293T and HeLa-P4 cells. Results are representative of two independent experiments.
CsA has different effects on wild-type and mutant HIV-1 reverse transcription in HeLa-P4 cells.
Wild-type virus produced one-half the level of early reverse transcription product with CsA treatment (Fig. 3D, WT). By contrast, the presence of CsA during infection by A92E and G94D mutants resulted in a 60 to 100% increase in levels of early reverse transcription products (Fig. 3D, A92E and G94D). Synthesis of late reverse transcripts by the wild-type virus was reduced by two-thirds with CsA addition (Fig. 3E, WT). The addition of CsA during inoculation with A92E and G94D mutant viruses resulted in a 60 to 100% increase in the levels of late reverse transcription products (Fig. 3E, A92E and G94D). In parallel, we also assayed infection of HeLa-P4 cells with the same doses of viruses to determine the effects of CsA on infection under these conditions. As expected, treatment of the wild-type virus with CsA reduced the viral infectivity by one-third, whereas CsA increased infection with the A92E and G94D mutants by about two- to threefold (Fig. 3F). Taken together, these results suggest that the enhancement of infection of the A92E and G94D mutants by CsA is a result of enhanced reverse transcription in HeLa-P4 cells.
HIV-1 mutants A92E and G94D are restricted by a dominant activity in HeLa-P4 cells.
The cell-specific phenotype of the A92E and G94D mutants suggests that an inhibitor of the mutant viruses is expressed in some cell types but not others. Alternatively, a cellular activity may facilitate infection by the mutants in cells that are permissive for the mutant viruses. To discriminate between these possibilities, we generated heterokaryons by fusion of permissive 293T and nonpermissive HeLa-P4 cells. Phenotypic analysis of heterokaryons previously has been employed to study the cell-specific requirements for Vif (51) and Vpu (60), as well as retroviral restriction in monkey and rabbit cell lines (16, 39).
In initial studies using polyethylene glycol-mediated fusion, we found the efficiency of heterokaryon formation (1% or lower) to be a significant experimental limitation. To circumvent this, we developed an approach involving cell fusion mediated by M-PMV Env. M-PMV Env efficiently induces fusion among a wide range of human cell types to produce syncytia with two to four nuclei per syncytium (53, 54). We exploited the fusion activity of M-PMV Env to generate heterokaryons between 293T and HeLa-P4 cells. 293T cells were transfected either with empty control DNA or with the pTMT, M-PMV Env expression vector. The transfected 293T cells also were labeled with the amine-reactive fluorescent probe DDAO, and HeLa-P4 cells were labeled with CMTMR. These probes have distinct spectral properties, allowing the labeled cells to be discriminated by flow cytometry. After being labeled, cells were detached, and equal numbers of labeled 293T and HeLa-P4 cells were cocultured to allow syncytium formation between these cells. Cultures subsequently were analyzed by flow cytometry (Fig. 4A) and by confocal microscopy (Fig. 4B) to determine the efficiency of fusion between 293T and HeLa-P4 cells.
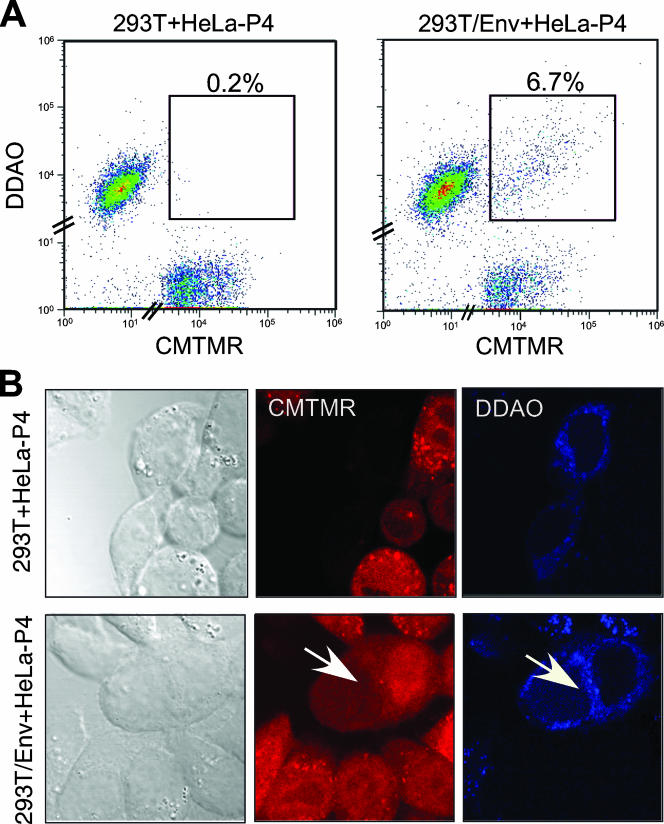
FIG. 4.
Generation of heterokaryons between 293T and HeLa-P4 cells by expression of the M-PMV Env glycoprotein. 293T cells were transfected with the empty control plasmid (293T) or the M-PMV Env expression vector (293T/Env), labeled with the DDAO fluorescence probe, and mixed with HeLa-P4 that had been labeled with CMTMR probe. (A) Twenty-four hours after coculture, 10,000 cells were analyzed by flow cytometry to detect heterokaryon formation. Ten thousand cells were analyzed per sample. (B) Twenty-four hours after coculture, cultures were analyzed for both probes by confocal microscopy. The arrow indicates a heterokaryon.
When DDAO-labeled control vector-transfected 293T cells were cocultured with CMTMR-labeled HeLa-P4 cells, double-positive cells were detected at levels corresponding to 0.2% of the total cell population. These cells exhibited low levels of fluorescence and did not form a well-organized and distinct subpopulation (Fig. 4A, left panel). By contrast, when MPMV Env-expressing 293T cells were cocultured with HeLa-P4 cells, syncytia were apparent as a discrete population (6.7%) of cells positive for both DDAO-SE and CMTMR labels (Fig. 4A, right panel).
Upon being labeled with CMTMR, the entire cytoplasm and nuclei were stained by this fluorescent probe (Fig. 4B, CMTMR); in contrast, cells labeled with DDAO probe displayed a distinctive staining pattern in the cytoplasm and no labeling of nuclei (Fig. 4B, DDAO). Using confocal microscopy, we observed double-positive cells labeled with both DDAO and CMTMR only in coculture in which 293T cells expressed the M-PMV Env protein, indicating that this protein was required for efficient formation of 293T-HeLa-P4 heterokaryons (Fig. 4B, arrow).
To determine the permissiveness of the heterokaryons to infection, cultures were inoculated with VSV-G-pseudotyped HIV-GFP viruses 24 h after coculture of DDAO-labeled M-PMV Env-expressing 293T cells with CMTMR-labeled HeLa-P4 cells. For these experiments, the viral inocula were chosen so as to give a 10-fold increase of infectivity by A92E and G94D viruses in the presence of CsA in HeLa-P4 cultures. Even 72 h after coculture of DDAO-labeled 293T and CMTMR-labeled HeLa-P4 cells, the labeled cells maintained a discrete cell population in heterokaryon-containing cultures (Fig. 5A and B, 293T and HeLa-P4), indicating that the labeling with the fluorescent dyes was stable and that the labels were not spontaneously transferred between the two cell types. Cells that were doubly positive for both fluorescent probes maintained a distinctive subpopulation of about 3% of the total cell population (Fig. 5B).
FIG. 5.
Effect of CsA on HIV-1 infection of 293T-HeLa-P4 heterokaryons. Heterokaryons were produced by coculture of DDAO-stained 293T cells expressing the M-PMV Env glycoprotein with CMTMR-stained HeLa-P4 cells. Cultures were inoculated with the indicated GFP reporter viruses in the presence or absence of CsA. One-half million cells from each culture were analyzed by three-color flow cytometry for GFP, DDAO, and CMTMR. (A) Analysis of control uninfected cultures lacking M-PMV Env expression. (B) Analysis of heterokaryon-containing cultures. (C to F) Analysis of GFP expression in 293T cells (C), HeLa-P4 cells (D), and heterokaryon cells (E) in the presence or absence of CsA. For panel F, cultures of heterokaryons in which the DDAO and CMTMR staining was reversed were analyzed. Results are representative of two independent experiments. WT, wild type.
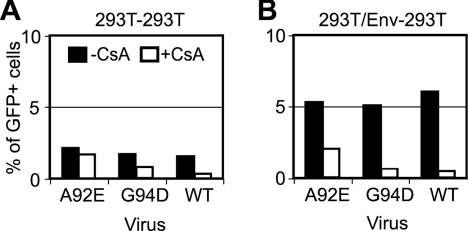
Following inoculation with the GFP reporter viruses, unfused 293T cells within the cocultures using the A92E and G94 mutants remained sensitive to inhibition by CsA. Treatment of these cells with CsA reduced viral infectivity by about fourfold for the wild type and by about 20 to 50% for A92E and G94D virions (Fig. 5C). In contrast, infection of HeLa-P4 cells in the cocultures by A92E and G94D mutant viruses was markedly enhanced by the addition of CsA, while the infectivity of wild-type virions was little changed with the addition of CsA (Fig. 5D). These results indicated that the labeling of the cells with the fluorescent dyes did not significantly alter the effect of CsA on the infection of either cell type.
Analysis of heterokaryons in these cultures revealed that CsA slightly reduced the level of infection by wild-type virus (Fig. 5E, WT). In contrast, infection by the A92E and G94D virions was increased by about sixfold in the presence of CsA (Fig. 5E, A92E and G94D). Thus, infection of the heterokaryons by the A92E and G94D mutants was markedly enhanced by addition of CsA during virus inoculation.
We next asked whether the fluorescence probes used to label the 293T or HeLa-P4 cells might alter the CsA-dependent infection of heterokaryons by A92E and G94D mutant viruses. To test this, the labeling of the 293T and HeLa-P4 cells was reversed, and heterokaryons were generated by coculture of control and M-PMV Env-expressing cells with HeLa-P4 cells. The resulting cultures then were challenged with wild-type and mutant viruses. The results were nearly identical to those obtained with the heterokaryon subpopulation that had 293T cells labeled with DDAO and HeLa-P4 cells labeled with CMTMR probes, indicating that the probes themselves had no effect on the CsA-dependent phenotype (Fig. 5F). Interestingly, heterokaryons were consistently more permissive than original 293T or HeLa-P4 cells regardless of CsA treatment and virus (Fig. 5, compare E and F to C and D). Taken together, these data indicate that infection of 293T-HeLa-P4 heterokaryons by the A92E and G94D mutant viruses is enhanced by CsA.
293T-293T hybrids do not exhibit CsA-dependent infection by A92E and G94D HIV-1 mutants.
It was possible that the process of cell-cell fusion itself or M-PMV Env expression resulted in CsA-dependent infection by A92E or G94D mutant virions, thus confounding interpretation of the results for the 293T-HeLa-P4 heterokaryons. To test this, we generated syncytia by fusing DDAO-labeled 293T with CMTMR-labeled 293T cells. 293T cells were transfected with the empty control vector or with M-PMV Env vector. Following transfection, 293T cells were labeled with CMTMR, and untransfected 293T cells were labeled with DDAO. Equal numbers of labeled cells were cocultured and subsequently challenged by wild-type and mutant viruses in the presence and absence of CsA.
In control cocultures lacking expression of M-PMV Env, we observed no distinctive cell population positive for both fluorescence probes (data not shown). The permissiveness of these control cells exhibited a CsA-independent pattern identical to that of untransfected, unlabeled 293T cells (Fig. 6A). When 293T cells were mixed with M-PMV Env-expressing 293T cells, about 4% of cells comprised a subpopulation positive for both fluorescent probes (data not shown). Analysis of double-positive cells for GFP expression revealed that wild-type and G94D viruses were markedly inhibited by CsA; infection by A92E was inhibited by approximately twofold (Fig. 6B). As previously observed for the heterokaryons between HeLa-P4 and 293T, the fused cells were about three times more permissive than unfused 293T cells with no CsA treatment. Thus, fusion between 293T cells did not result in CsA-dependent enhancement of infection by A92E and G94D viruses.
FIG. 6.
Effect of CsA on HIV-1 infection of syncytia between 293T cells. 293T cells were transfected with control (A) and M-PMV Env (B) plasmids. Twenty-four hours posttransfection, 293T cultures were labeled with DDAO and cocultured with an equal number of untransfected 293T cells labeled with CMTMR. After another 24 h, cultures were challenged with wild-type (WT) or mutant (A92E and G94D) HIV-1-GFP reporter virions in the presence or absence of 5 μM CsA. Two days later, cultures were analyzed by flow cytometry for GFP, DDAO, and CMTMR. (A) Infectivity frequency of unfused 293T cells (293T-293T). In these samples, no gating was applied to discriminate DDAO or CMTMR. (B) Infection frequency of syncytia positive for both fluorescence dyes (293T/Env-293T). The results shown are representative of at least two independent experiments.
Overexpression of CypA in 293T cells does not result in CsA-dependent infection by the A92E and G94D HIV-1 mutants.
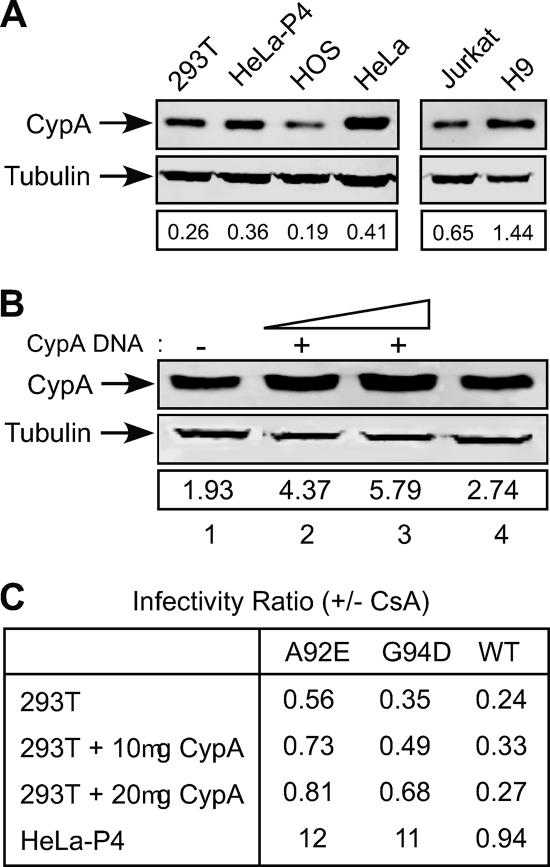
It has been suggested that the phenotypic consequences of mutations in the CypA-binding loop of CA may differ in cells as a result of differences in levels of CypA expression (2, 69). Specifically, H9 cells, in which replication of A92E was stimulated by CsA, contained higher levels of CypA than did Jurkat cells, in which CsA did not enhance A92E replication. To determine whether elevated levels of CypA contribute to the CsA-dependent infection for A92E and G94D mutants, we first performed immunoblotting to quantify the relative expression levels of endogenous CypA among cell lines in which infection by A92E is promoted by CsA (HeLa-P4, HeLa, and H9) and cells in which it is not (293T, HOS, and Jurkat) (Fig. 7A). To control for loading, the blots were reprobed with an antibody to tubulin, and the band intensities were quantified. Overall, CsA-dependent cells for those mutants displayed slightly higher levels of CypA expression than CsA-independent cells. HeLa and HeLa-P4 cells contained about twice the levels of CypA that HOS cells had and approximately 1.5 times the level in 293T cells. These results conflict somewhat with those of a previous report that the CypA level in HeLa cells is not significantly elevated relative to that in 293T cells (52). H9 cells also exhibited approximately twice the level of CypA exhibited by Jurkat cells, as previously reported (69). Collectively, these results are consistent with the possibility that the enhancement of A92E infection by CsA observed in HeLa and H9 cells is due to elevated CypA levels in these cells.
FIG. 7.
Overexpression of CypA in 293T cells does not result in CsA-dependent infection. (A) Analysis of CypA levels in lysates of 293T, HeLa-P4, HOS, HeLa, Jurkat, and H9 cells. Equal quantities of protein (20 μg) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were probed with anti-CypA and anti-tubulin antibodies. Protein bands were visualized with a LI-COR Odyssey infrared imaging system, and CypA levels were quantified by the instrument software, with tubulin as a control. Shown under each lane is the ratio of the CypA intensity to the tubulin intensity. (B) 293T cells were transfected with 10 and 20 μg of CypA expression vector (pCMX-CypA), and cells were analyzed 48 h posttransfection as described for panel A. Lane 1, 293T cells; lane 2, 293T cells transfected with 10 μg of pCMX-CypA; lane 3, 293T cells transfected with 20 μg of pCMX-CypA; lane 4, HeLa-P4 cells. (C) Analysis of the effects of CsA on infection of transfected 293T cells. HeLa-P4 and transfected 293T cells were plated onto 24-well plates 24 h posttransfection. After another 24 h, cultures were challenged with wild-type (WT) or mutant (A92E and G94D) HIV-GFP reporter virions in the presence or absence of 5 μM CsA. Two days later, cultures were analyzed by flow cytometry for GFP expression. Infectivity ratios were calculated by dividing the percentage of infected (i.e., GFP-positive) cells for CsA-treated cultures by the corresponding value for the parallel infections performed in the absence of CsA. The infectivity data shown are representative of three independent experiments.
To further test the relationship between CypA expression levels and cell-specific enhancement of infection by CsA, we overexpressed CypA in 293T cells by transient transfection with a CypA expression vector. Forty-eight hours later, the cells were challenged with wild-type or mutant HIV-1 in the presence and absence of CsA. Additionally, uninfected cultures were harvested and cellular proteins were extracted to quantify CypA levels. Immunoblot analysis revealed a two- to threefold higher level of CypA expression relative to that of control 293T cells. In contrast, HeLa-P4 cells exhibited a 40% higher level of CypA than that of 293T cells (Fig. 7B). Thus, the expression level of CypA in transfected 293T cells was approximately 1.5- to 2-fold greater than that observed in HeLa-P4 cells.
When transfected 293T and HeLa-P4 cells were analyzed after infection with wild-type or mutant virions, we found that CypA-transfected 293T cells displayed slightly enhanced levels of infectivity compared to that of mock-transfected 293T cells (Fig. 7C). Infection of HeLa-P4 cells by A92E and G94D mutant virions was enhanced by about 10-fold with CsA treatment. However, infection of CypA-overexpressing 293T cells was not enhanced by CsA, though the overall infection rate was slightly greater than that of control 293T cells. Thus, forced overexpression of CypA in 293T cells to a level exceeding that observed in HeLa cells did not result in CsA-dependent enhancement of A92E or G94D mutant. These data indicate that the CsA-stimulated infection by the mutant viruses is not due solely to the higher CypA level in HeLa cells relative to that in 293T cells.
DISCUSSION
The molecular mechanism by which CsA inhibits infection by wild-type HIV-1 and stimulates infection by some HIV-1 mutants is unknown. In this study, we showed that CsA does not promote fusion of CsA-dependent HIV-1 mutants with target cells, suggesting that the enhanced infection of HeLa-P4 cells by A92E and G94D HIV-1 mutants in cultures containing CsA is not due to altered viral entry (Fig. 2). CsA increased the level of synthesis of HIV-1 reverse transcripts, indicating that CsA facilitates an early postentry step necessary for reverse transcription of the HIV-1 genome (Fig. 3). Events in this phase of the virus life cycle that may be affected by CypA include uncoating (3, 41), proteasome- and lysosome-dependent degradation (25, 43, 50), and trafficking of the viral core to an intracellular compartment that is conducive to reverse transcription. We previously demonstrated that subtle changes in the stability of the capsid can lead to impaired reverse transcription (22). Because CsA prevents binding of CypA to CA, it is plausible that binding of CypA to the A92E and G94D viral capsids has a negative effect on uncoating of the mutant cores. Another possibility is that CsA treatment may protect the incoming viral core from proteasomal degradation, which can limit HIV-1 infection of human cells. However, treatment of HeLa-P4 cells with the proteasome inhibitor MG132 enhanced infection by A92E and G94D mutants to an extent similar to that of wild-type HIV-1 (our unpublished results), arguing against a necessary role of the proteasome in restriction of the mutants.
To determine whether CsA-dependent infection by the A92E or G94D mutant is due to expression of a dominant restriction factor, we analyzed heterokaryons generated with fluorescent-probe-labeled HeLa-P4 and 293T cells that were produced by expression of M-PMV glycoprotein and identified by flow cytometry. This approach eliminated the requirement for purification of heterokaryons, which we found to be difficult with the large adherent cells used in this study. The syncytia generated between HeLa-P4 and 293T cells (Fig. 5E and F) or between 293T cells (Fig. 6B) generally were more permissive to HIV-1 infection than either parental cell type regardless of CsA treatment (Fig. 5). Thus, it seems possible that the larger surface area of the syncytia, or the presence of M-PMV glycoprotein on the cell surface, facilitates viral entry.
Infection of the 293T-HeLa-P4 heterokaryons by the A92E and G94D mutants was stimulated by CsA to an extent that was intermediate between that observed in HeLa-P4 cells and in 293T cells (Fig. 5E). We observed similar results when the fluorescent staining of the cell lines was reversed. These results indicate that a dominant activity restricts infection by the A92E and G94D mutants in HeLa-P4 cells and that addition of CsA prevents this restriction. Infection of heterokaryons with the wild-type virus was not significantly affected by CsA, similar to what was observed in HeLa-P4 cells (Fig. 5E and F, WT). Thus, the 293T-HeLa-P4 heterokaryons resembled HeLa-P4 cells in their response to CsA for both wild-type and CsA-dependent mutant viruses. Several observations indicate that CypA protects the HIV-1 capsid from endogenous human restriction factors in target cells. Cross-saturation studies suggested that human cells restrict N-tropic murine leukemia virus and a CypA-binding-defective HIV-1 mutant (G89V) by a common activity (58). However, restriction of the HIV-1 G89V mutant in human cells did not require TRIM5α expression. Interestingly, Sayah and Luban also have reported that selection of human TE671 cells for loss of N-tropic murine leukemia virus restriction was correlated with a decrease in the dependence of HIV-1 infection on CypA binding (48). It is conceivable that a common restriction activity is responsible for inhibiting infection by A92E and G94D in a CypA-dependent manner in HeLa cells and inhibiting the wild-type virus in 293T cells when CypA binding to CA is prevented. However, the cell type specificity of these viral phenotypes implies the existence of multiple factors that determine the fate of the viral core in target cells. We propose that CypA binding to the A92E and G94D mutants leads to the formation of a specific binding site for an unknown host restriction factor. In agreement with this hypothesis, recent studies in our laboratory have identified a secondary CA mutation that restores infectivity to the A92E mutant in HeLa-P4 cells (66).
It was suggested previously that the CsA-dependent infection by the A92E and G94D mutants results from the increased levels of CypA expression in H9 cells compared to that in CsA-independent Jurkat cells (68), and we observed an overall positive correlation between cellular CypA levels and CsA-dependent infection of various cell lines (Fig. 7A). However, overexpression of CypA in 293T cells did not result in enhancement of infection by A92E and G94D mutant virions upon CsA treatment (Fig. 7B and C). We conclude that differences in levels of CypA do not fully account for the cell-specific differences in the A92E and G94D response to CsA. Thus, CypA, while not directly responsible for the restriction of these mutants in HeLa cells, is necessary for it.
CypA significantly enhances HIV-1 replication in cell lines (12, 24, 57) and primary T lymphocytes (45), yet the precise role of CypA in the virus life cycle remains unknown. Our results suggest that CsA-dependent HIV-1 infection of human cells is due to the expression of an unknown host factor that specifically restricts infection at an early postentry step of infection. Future identification of the putative cellular restriction factor will provide a better understanding of the consequences of CypA binding to the HIV-1 core and of cellular restriction mechanisms in human cells.
Acknowledgments
We thank Dana Gabuzda for the HIV-GFP plasmid, Jiong Shi for technical assistance, and David Hout for critical reading of the manuscript. The following reagent was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: monoclonal antibody 183-H12-5C from Bruce Chesebro.
This work was supported in part by a grant from the National Institutes of Health (AI050423). C.S. is the recipient of a fellowship award from the American Foundation for AIDS Research.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Aberham, C., S. Weber, and W. Phares. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 70:3536-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerson, B., O. Rey, J. Canon, and P. Krogstad. 1998. Cells with high cyclophilin A content support replication of human immunodeficiency virus type 1 Gag mutants with decreased ability to incorporate cyclophilin A. J. Virol. 72:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiken, C. 2006. Viral and cellular factors that regulate HIV-1 uncoating. Curr. Opin. HIV AIDS 1:194-199. [DOI] [PubMed] [Google Scholar]
- 4.Bachmann, B., J. Knuver-Hopf, B. Lambrecht, and H. Mohr. 1995. Target structures for HIV-1 inactivation by methylene blue and light. J. Med. Virol. 47:172-178. [DOI] [PubMed] [Google Scholar]
- 5.Bartz, S. R., E. Hohenwalter, M. K. Hu, D. H. Rich, and M. Malkovsky. 1995. Inhibition of human immunodeficiency virus replication by nonimmunosuppressive analogs of cyclosporin A. Proc. Natl. Acad. Sci. USA 92:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2005. Cyclophilin A is required for TRIM5α-mediated resistance to HIV-1 in Old World monkey cells. Proc. Natl. Acad. Sci. USA 102:14849-14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2004. Lv1 inhibition of human immunodeficiency virus type 1 is counteracted by factors that stimulate synthesis or nuclear translocation of viral cDNA. J. Virol. 78:11739-11750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. USA 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billich, A., F. Hammerschmid, P. Peichl, R. Wenger, G. Zenke, V. Quesniaux, and B. Rosenwirth. 1995. Mode of action of SDZ NIM 811, a nonimmunosuppressive cyclosporin A analog with activity against human immunodeficiency virus (HIV) type 1: interference with HIV protein-cyclophilin A interactions. J. Virol. 69:2451-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 70:5170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braaten, D., and J. Luban. 2001. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 20:1300-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 14.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 15.Clavel, F., and P. Charneau. 1994. Fusion from without directed by human immunodeficiency virus particles. J. Virol. 68:1179-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan, S., T. Hatziioannou, T. Cunningham, M. A. Muesing, H. G. Gottlinger, and P. D. Bieniasz. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc. Natl. Acad. Sci. USA 99:11914-11919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dismuke, D. J., and C. Aiken. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 80:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorfman, T., A. Weimann, A. Borsetti, C. T. Walsh, and H. G. Gottlinger. 1997. Active-site residues of cyclophilin A are crucial for its incorporation into human immunodeficiency virus type 1 virions. J. Virol. 71:7110-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer, G., B. Wittmann-Liebold, K. Lang, T. Kiefhaber, and F. X. Schmid. 1989. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337:476-478. [DOI] [PubMed] [Google Scholar]
- 21.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 22.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franke, E. K., and J. Luban. 1996. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology 222:279-282. [DOI] [PubMed] [Google Scholar]
- 24.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 25.Fredericksen, B. L., B. L. Wei, J. Yao, T. Luo, and J. V. Garcia. 2002. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J. Virol. 76:11440-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 27.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 28.Handschumacher, R. E., M. W. Harding, J. Rice, R. J. Drugge, and D. W. Speicher. 1984. Cyclophilin: a specific cytosolic binding protein for cyclosporin A. Science 226:544-547. [DOI] [PubMed] [Google Scholar]
- 29.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatziioannou, T., D. Perez-Caballero, S. Cowan, and P. D. Bieniasz. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatziioannou, T., D. Perez-Caballero, A. Yang, S. Cowan, and P. D. Bieniasz. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. USA 101:10774-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, J., Y. Chen, M. Farzan, H. Choe, A. Ohagen, S. Gartner, J. Busciglio, X. Yang, W. Hofmann, W. Newman, C. R. Mackay, J. Sodroski, and D. Gabuzda. 1997. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature 385:645-649. [DOI] [PubMed] [Google Scholar]
- 33.Howard, B. R., F. F. Vajdos, S. Li, W. I. Sundquist, and C. P. Hill. 2003. Structural insights into the catalytic mechanism of cyclophilin A. Nat. Struct. Biol. 10:475-481. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, J., and C. Aiken. 2006. Maturation of the viral core enhances the fusion of HIV-1 particles with primary human T cells and monocyte-derived macrophages. Virology 346:460-468. [DOI] [PubMed] [Google Scholar]
- 35.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2006. Cyclophilin A renders human immunodeficiency virus type 1 sensitive to Old World monkey but not human TRIM5α antiviral activity. J. Virol. 80:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2004. The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. USA 101:10780-10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kootstra, N. A., C. Munk, N. Tonnu, N. R. Landau, and I. M. Verma. 2003. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 39.Münk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nisole, S., C. Lynch, J. P. Stoye, and M. W. Yap. 2004. A Trim5-cyclophilin A fusion protein found in owl monkey kidney cells can restrict HIV-1. Proc. Natl. Acad. Sci. USA 101:13324-13328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nisole, S., and A. Saib. 2004. Early steps of retrovirus replicative cycle. Retrovirology 1:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5α mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. USA 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi, M., and C. Aiken. 2007. Selective restriction of Nef-defective human immunodeficiency virus type 1 by a proteasome-dependent mechanism. J. Virol. 81:1534-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenwirth, B., A. Billich, R. Datema, P. Donatsch, F. Hammerschmid, R. Harrison, P. Hiestand, H. Jaksche, P. Mayer, P. Peichl, et al. 1994. Inhibition of human immunodeficiency virus type 1 replication by SDZ NIM 811, a nonimmunosuppressive cyclosporine analog. Antimicrob. Agents Chemother. 38:1763-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saini, M., and M. J. Potash. 2006. Novel activities of cyclophilin A and cyclosporin A during HIV-1 infection of primary lymphocytes and macrophages. J. Immunol. 177:443-449. [DOI] [PubMed] [Google Scholar]
- 46.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 2002. Cyclophilin a plays distinct roles in human immunodeficiency virus type 1 entry and postentry events, as revealed by spinoculation. J. Virol. 76:4671-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayah, D. M., and J. Luban. 2004. Selection for loss of Ref1 activity in human cells releases human immunodeficiency virus type 1 from cyclophilin A dependence during infection. J. Virol. 78:12066-12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569-573. [DOI] [PubMed] [Google Scholar]
- 50.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 52.Sokolskaja, E., D. M. Sayah, and J. Luban. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 78:12800-12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Song, C., and E. Hunter. 2003. Variable sensitivity to substitutions in the N-terminal heptad repeat of Mason-Pfizer monkey virus transmembrane protein. J. Virol. 77:7779-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song, C., K. Micoli, and E. Hunter. 2005. Activity of the Mason-Pfizer monkey virus fusion protein is modulated by single amino acids in the cytoplasmic tail. J. Virol. 79:11569-11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 56.Takahashi, N., T. Hayano, and M. Suzuki. 1989. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature 337:473-475. [DOI] [PubMed] [Google Scholar]
- 57.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 58.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 9:1138-1143. [DOI] [PubMed] [Google Scholar]
- 59.Vajdos, F. F., S. Yoo, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1997. Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein. Protein Sci. 6:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varthakavi, V., R. M. Smith, S. P. Bour, K. Strebel, and P. Spearman. 2003. Viral protein U counteracts a human host cell restriction that inhibits HIV-1 particle production. Proc. Natl. Acad. Sci. USA 100:15154-15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 62.Wainberg, M. A., A. Dascal, N. Blain, L. Fitz-Gibbon, F. Boulerice, K. Numazaki, and M. Tremblay. 1988. The effect of cyclosporine A on infection of susceptible cells by human immunodeficiency virus type 1. Blood 72:1904-1910. [PubMed] [Google Scholar]
- 63.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288-293. [DOI] [PubMed] [Google Scholar]
- 64.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H.-G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyma, D. J., J. Jiang, J. Shi, J. Zhou, J. E. Lineberger, M. D. Miller, and C. Aiken. 2004. Coupling of human immunodeficiency virus type 1 fusion to virion maturation: a novel role of the gp41 cytoplasmic tail. J. Virol. 78:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, R., and C. Aiken. 2007. A mutation in alpha helix 3 of CA renders human immunodeficiency virus type 1 cyclosporine-resistant and dependent: rescue by a second-site substitution in a distal region of CA. J. Virol. 81:3749-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 101:10786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 43:99-112. [DOI] [PubMed] [Google Scholar]
- 69.Yin, L., D. Braaten, and J. Luban. 1998. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J. Virol. 72:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yoo, S., D. G. Myszka, C. Yeh, M. McMurray, C. P. Hill, and W. I. Sundquist. 1997. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J. Mol. Biol. 269:780-795. [DOI] [PubMed] [Google Scholar]
- 71.Zhao, Y., Y. Chen, M. Schutkowski, G. Fischer, and H. Ke. 1997. Cyclophilin A complexed with a fragment of HIV-1 gag protein: insights into HIV-1 infectious activity. Structure 5:139-146. [DOI] [PubMed] [Google Scholar]