Abstract
Gammaherpesviruses establish lifelong, latent infections in host lymphocytes, during which a limited subset of viral gene products facilitates maintenance of the viral episome. Among the gamma-2-herpesvirus (rhadinovirus) subfamily, this includes expression of the conserved ORF73-encoded LANA proteins. We previously demonstrated by loss-of-function mutagenesis that the murine gammaherpesvirus 68 (MHV68) ORF73 gene product, mLANA, is required for the establishment of latency following intranasal inoculation of mice (N. J. Moorman, D. O. Willer, and S. H. Speck, J. Virol. 77:10295-10303, 2003). mLANA-deficient viruses also exhibited a defect in acute virus replication in the lungs of infected mice. The latter observation led us to examine the role of mLANA in productive viral replication. We assessed the capacity of mLANA-deficient virus (73.Stop) to replicate in cell culture at low multiplicities of infection (MOIs) and found that 73.Stop growth was impaired in murine fibroblasts but not in Vero cells. A recombinant virus expressing an mLANA-green fluorescent protein (GFP) fusion revealed that mLANA is expressed throughout the virus replication cycle. In addition, 73.Stop infection of murine fibroblasts at high MOIs was substantially more cytotoxic than infection with a genetically repaired marker rescue virus (73.MR), a phenotype that correlated with enhanced kinetics of viral gene expression and increased activation of p53. Notably, augmented cell death, viral gene expression, and p53 induction were independent of viral DNA replication. Expression of a mLANA-GFP fusion protein in fibroblasts correlated with both reduced p53 stabilization and reduced cell death following treatment with p53-inducing agonists. In agreement, accentuated cell death associated with 73.Stop infection was reduced in p53-deficient murine embryonic fibroblasts. Additionally, replication of 73.Stop in p53-deficient cells was restored to levels comparable to those of 73.MR. More remarkably, the absence of p53 led to an overall delay in replication for both 73.Stop and 73.MR viruses, which correlated with delayed viral gene expression, indicating a role for p53 in MHV68 replication. Consistent with these findings, the expression of replication-promoting viral genes was positively influenced by p53 overexpression or treatment with the p53 agonist etoposide. Overall, these data demonstrate the importance of mLANA in MHV68 replication and suggest that LANA proteins limit the induction of cellular stress responses to regulate the viral gene expression cascade and limit host cell injury.
Murine gammaherpesvirus 68 (MHV68) is a member of the Gammaherpesvirinae subfamily of viruses that naturally infects rodents. Following infection of inbred mice by various routes, MHV68 undergoes acute replication at the primary site of inoculation, a process that facilitates the seeding of several cell types, including macrophages, dendritic cells, epithelial cells, and B cells, for latent or persistent infection (24, 64, 67, 78, 80). In latent infections, the viral genomes are maintained in a quiescent state for the lifetime of the host but retain the capacity to reactivate the lytic replication cycle and produce progeny virions. Like all gammaherpesviruses, MHV68 is defined by its capacity to establish latent infections in host lymphocytes.
During latent gammaherpesvirus infections, viral transcription is restricted, and only those genes proposed to influence maintenance and immune evasion are transcribed. For MHV68, the latent transcription program varies by cell type (45), likely reflecting cell-type-specific needs for viral maintenance and demonstrating the dynamic nature of gammaherpesvirus latency. Of the viral transcripts expressed during latent infections, mLANA (a homologue of the Kaposi's sarcoma-associated herpesvirus [KSHV] latency-associated nuclear antigen [LANA]) is critically required, as mLANA-null viruses are unable to establish and maintain latent infections following intranasal inoculation of mice (26, 48). This is consistent with a proposed role for mLANA in episome maintenance during latency. However, mLANA influences more than just the capacity of MHAV68 to persist during latency, as we and others observed acute replication deficits in the lungs of mice infected with mLANA-null mutants (48, 63).
Although a replication deficit was not originally borne out in cell culture growth assays, several lines of evidence suggest a role for mLANA in facilitating acute-phase viral replication in addition to the demonstrated role in regulating viral latency. First, in global and gene-specific MHV68 transcription analyses, orf73 was detected at very early times after infections in culture, and orf73 transcription occurred in the presence of cycloheximide, classifying orf73 as an immediate-early transcript (2, 15, 46). Further, herpesvirus saimiri (HVS) and rhesus rhadinovirus (RRV) LANAs suppress the progression of lytic replication in culture by inhibiting transcription of the major lytic transactivator protein, RTA (19, 58). KSHV LANA can either repress activity of the promoter for RTA (37) or, conversely, contribute to its activation via HIF-1α under hypoxic conditions (9, 10).
In addition to regulating transcription of viral lytic genes, KSHV LANA inhibits the activity of several host cell cycle regulatory proteins. The tumor suppressor protein p53 functions as a transcriptional regulator of cell cycle checkpoints and apoptosis in response to numerous cellular stressors, including DNA damage and viral infection (32, 38, 75). Consistent with these roles, loss of p53 function is a major contributor to cellular transformation and cancer (75). Biochemical analyses demonstrated that p53, as well as the tumor suppressor pRb, are targets of KSHV LANA- and HVS LANA-mediated repression (7, 27, 53), which may contribute to LANA inhibition of p16INK4a-induced cell cycle arrest (4). Moreover, KSHV LANA contributes to H-ras-induced transformation of rat fibroblasts and tumor growth in SCID mice (53), and KSHV LANA expressed from its cognate promoter induces lymphoproliferative disease in transgenic mice (22). Together with the observations that LANA is highly expressed in KS and KSHV-derived tumor cell lines (34), these data demonstrate the capacity of LANA proteins to promote oncogenesis by deregulating cellular tumor suppressors, in addition to their proposed role in maintaining gamma-2-herpesvirus episomes during latency. However, LANA-expressing KSHV-derived cell lines are sensitive to p53 agonistic chemotherapeutic agents (17, 52), perhaps illustrating the complexity of extending the results of isolated biochemical analyses to the setting of virus infection. Moreover, a role for LANA proteins in regulating cellular stress responses, such as p53 activation or cell cycle arrest, during lytic viral replication has not been analyzed.
In this report, we examined the role of MHV68 mLANA during productive virus replication. We found that mLANA is expressed with diverse localization patterns throughout the MHV68 replication cycle and is required for efficient replication, especially at low multiplicities of infection (MOIs), in murine fibroblasts. Infection with mLANA-null (73.Stop) MHV68 more readily induced cell death than marker rescue virus (73.MR) at high MOIs. Death occurred with p53 activation and enhanced kinetics of viral gene expression, even in the presence of inhibitors of viral DNA replication. Stable mLANA expression correlated with reductions in both p53 stabilization and cell death upon treatment with p53-inducing stimuli, and p53-deficient murine embryonic fibroblasts (MEFs) were modestly protected from 73.Stop-induced death at early times after infection. The absence of p53 equilibrated 73.Stop and 73.MR replication but led to an overall delay in the progression of viral gene expression and replication compared to that of p53-expressing cells. Consistent with these findings, lytic-cycle viral genes were inducible by p53 overexpression or treatment with p53-inducing drugs. These data provide the first evidence that LANA proteins regulate p53 induction during productive gammaherpesvirus replication to control the viral gene expression cascade. Furthermore, we demonstrate that p53 expression actively influences the gammaherpesvirus life cycle.
MATERIALS AND METHODS
Cell culture and viruses.
NIH 3T12, NIH 3T3, and 293T cells were purchased from the ATCC. BOSC 23 cells were a gift from Joshy Jacob (Emory University, Atlanta, GA). Vero cells used in this report express Cre recombinase and are commonly used by our laboratory and others to propagate bacterial artificial chromosome (BAC)-derived recombinant MHV68 (48). Vero-cre cells were a gift from David Leib (Washington University, St. Louis, MO). C57BL/6 MEFs were prepared from day-13.5 embryos as described previously (77). Trp53−/− (p53−/−) MEFs were a gift from Vera Tarakanova and Herbert W. Virgin IV (Washington University, St. Louis, MO). All cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine (cMEM). NIH 3T3 cells transduced with empty murine stem cell virus (MSCV) or mLANA-green fluorescent protein (GFP) MSCV or p53−/− MEFs transduced with DsRed MSCV or p53-DsRed MSCV were selected with 1 μg/ml puromycin (Sigma). Wild-type (WT) MHV68 was strain WUMS (ATCC VR1465). The derivation of 50.Stop, 73.Stop, and 73.MR viruses was previously described (48, 51). Passage 2 stocks propagated on Vero-cre cells were used for all experiments in this paper. Recombinant mLANA-GFP MHV68 was derived by allelic exchange in MHV68 BAC as previously described (1, 48). To derive the targeting construct, GFP from pIRES2-EGFP (Clontech) was fused to the carboxyl terminus of mLANA by overlap extension PCR using the following oligonucleotides: 73GFPFUS P1, 5′-CGAGATCTGGAGAGACAGACGATCCC-3′; 73GFPFUS P2, 5′-CTCGCCCTTGCTCACTGTCTGAGACCCTTG-3′; 73GFPFUS P3, 5′-GTGAGCAAGGGCGAGGAGCTGTTCACCGGG-3′; 73GFPFUS P4, 5′-ACCCTTACACTAAATTTACTTGTACAGCTC-3′; 73GFPFUS P5, 5′-ATTTAGTGTAAGGGTCTTATATAGGACCCG-3′; and 73GFPFUS P6, 5′-CGGCGGCCGCCACGCGTCGATAAAATT-3′. The overlap extension PCR product was cloned into pCR-blunt (Invitrogen), and the fidelity of cloning was verified by automated sequencing. The construct was subcloned into pGS284, and allelic exchange was performed as described previously (1, 48). Appropriate recombination was determined by colony PCR. Recombinant mLANA-GFP MHV68 virus was derived by BAC transfection into Vero-cre cells.
Vectors, transfections, and antibodies.
The −310 orf50 promoter and −800 v-cyclin promoter constructs were previously described (3, 40). The p53 expression vector pFC-p53 was purchased from Stratagene. mLANA-GFP was cloned via PCR from recombinant mLANA-GFP MHV68 with primers 73-1_Bam (5′-GATCGGATCCCTTGACCCACACCCTTCCTGTGC-3′ [restriction endonuclease recognition sequences are underlined 1]) and 73-3_Eco (5′-GATCGAATTCCATGCCCTGGCGAAGGTGTTG-3′) and restriction digested with BamHI and EcoRI for transfer into the BglII and EcoRI sites in pMSCV-puro (Becton Dickinson). Fidelity of cloning was verified by automated sequencing. DsRed was cloned from pDsRed Monomer-N1 (Clontech) by PCR with primers DSRED_US (5′-GATCGGATCCGCCACCATGGACAACACCGAG-3′) and DSRED_DS (5′-GATCGAATTCCTACTGGGAGCCGGAGTGGCGG-3′). To derive P53-DsRed, DsRed from pDsRed Monomer-N1 was fused to the carboxyl terminus of WT p53 from pFC-p53 by overlap extension PCR using the following oligonucleotides: P53_US, 5′-GATCGGATCCCTGCCATGGAGGAGCCGCAG-3′; P53-DSRED_comp, 5′-GTTGTCCATGGTGGCGTCTGAGTCAGGCCCTTCTGTC-3′; P53-DSRED, 5′-GGGCCTGACTCAGACGCCACCATGGACAACACCGAG-3′; and DSRED_DS (described above). Products were digested with BamHI and EcoRI for transfer into the BglII and EcoRI sites in pMSCV-puro. Retroviruses were generated by transfecting pMSCV constructs into BOSC 23 cells. Transfections were performed using Lipofectamine (Invitrogen). The antibodies used in this study include chicken anti-ORF59 (70), rabbit anti-v-cyclin (71), anti-poly(ADP-ribose) polymerase (PARP) no. 9542, anti-p53 no. 2524, anti-phospho-S15 p53 no. 9284S (Cell Signaling Technology), goat polyclonal and mouse monoclonal anti-GFP (Rockland Immunochemicals), and anti-β-actin (A-5316; Sigma).
Viral infections.
Viral stocks were diluted in cMEM and adsorbed to monolayers of cells plated the previous day. The time of adsorption was considered time zero. For an MOI of 2 to 20 infections, cells in 6-well plates were adsorbed in 200 μl inocula with rocking every 10 to 15 min for 1 h. Inocula were removed for single-step growth curves. After adsorption, cells were incubated in a normal culture volume of cMEM for the indicated times at 37°C. Cells were harvested by direct lysis for immunoblotting or by freezing at −80°C for titer determinations. Progeny virions were liberated by freeze-thaw lysis, lysates were serially diluted, and titers were determined by MHV68 plaque assay as described previously (69).
Drug and UV treatments.
Cidofovir (Vistide; Gilead Sciences, Inc.) was diluted to 20 ng/ml in cMEM. Viral stocks were diluted in cidofovir-containing medium for adsorption, and incubations were carried out at 37°C for the indicated times in the presence of cidofovir. Drug efficacy was verified by parallel growth for 24 h and subsequent plaque assay. Etoposide (Sigma) was dissolved in dimethylsulfoxide (DMSO) and diluted to the appropriate concentration in cMEM. Cells were treated with DMSO as a negative control or with etoposide for the indicated times. UV treatments were performed on cells plated to ∼70% confluence in 6-well plates. Culture medium was removed and replaced with 400 μl warm phosphate-buffered saline (PBS). Cells were exposed to 100 J/m2 UV light in a Stratalinker 2400 (Stratagene). PBS was removed and replaced with cMEM, and cells were incubated at 37°C for the indicated times. Mock treatments used identically manipulated plates without UV exposure.
Imaging and immunofluorescence.
Fixed and crystal-violet-stained cells (described below) for mock, 50.Stop, 73.Stop, and 73.MR infections were viewed by light microscopy, and mLANA-GFP-transfected 293T cells were imaged by fluorescence microscopy on a Leica DMRB microscope equipped with a Retiga EXi digital camera. For infections with recombinant mLANA-GFP MHV68, cells were plated on glass coverslips the day before infections. Cells were infected at an MOI of 10 PFU/cell. Cells were fixed with 10% formalin, washed with PBS, permeabilized, and blocked with 0.1% Triton X-100 (TX100) in PBS containing 5% bovine serum albumin (blocking buffer). Primary antibodies (anti-GFP [to increase the sensitivity of detection for mLANA-GFP], anti-ORF59, or polyclonal MHV68 antiserum) were diluted in blocking buffer with 1% normal donkey serum and were incubated for 1 h at 37°C. For mLANA-GFP detection, staining specificity was verified by colocalizing true GFP fluorescence with red (Alexa 568) anti-GFP immunofluorescence. Cells were washed three times with 0.1% TX100 in PBS (wash buffer) and incubated with species-appropriate Alexa fluor-conjugated secondary antibodies (488 for mLANA-GFP, 568 for ORF59, and 350 for MHV68 antiserum; Molecular Probes/Invitrogen) diluted in blocking buffer at 37°C for 45 min. Cells were washed three times with wash buffer and once with PBS and were mounted on slides using ProLong Anti-Fade Gold reagent (Molecular Probes/Invitrogen). DNA was detected with Hoechst 33342 (Molecular Probes/Invitrogen) or with 4′,6′-diamidino-2-phenylindole (DAPI) in the mounting medium. MSCV- or mLANA-GFP-transduced cells for p53 induction were plated on glass coverslips the day before treatments. Cells were fixed with 10% formalin for 15 to 30 min, washed with PBS, and then permeabilized with 0.5% TX100-PBS. Samples were incubated in blocking buffer. Samples were incubated overnight at 4°C with primary antibody (anti-p53; described above) diluted 1:2,000 in blocking buffer containing 1 μg/ml normal goat immunoglobulin G (Santa Cruz Biotechnology). Cells were washed three times in wash buffer and then were incubated for 1 h at room temperature with secondary antibody (Alexa fluor 568 goat anti-mouse) diluted in blocking buffer. After being washed three times with wash buffer and once with PBS, samples were mounted on slides using DAPI-containing ProLong Anti-Fade Gold reagent. Images were captured using a Zeiss Axio A1 Imager fluorescent microscope.
Immunoblot analyses.
Cells were lysed with alternative radioimmunoprecipitation assay buffer (150 mM NaCl, 20 mM Tris, 2 mM EDTA, 1% NP-40, 0.25% deoxycholate, 1 mM NaF, and 1 mM Na3VO4 supplemented with complete mini-EDTA-free protease inhibitors [Roche]) and quantitated using the Bio-Rad DC protein assay prior to resuspension in Laemmli sample buffer, or else equivalent numbers of cells (1 × 105 to 2 × 105) were directly lysed with 100 μl Laemmli sample buffer (36). Samples were heated to 100°C for 10 min and were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Resolved proteins were transferred to nitrocellulose and were identified with the indicated antibodies. Immobilized antigen and antibody were detected with horseradish peroxidase-conjugated secondary antibodies and ECL reagents (Amersham/GE Healthcare) and were exposed to film. Densitometric analysis was performed by scanning developed films using a Typhoon scanner (GE Healthcare) and were analyzed using Image Quant software (Amersham/GE Healthcare).
Reporter assays.
NIH 3T3 cells were transfected with the indicated plasmids and were incubated for 24 h at 37°C. A GFP reporter plasmid was included at a 1:10 dilution as a transfection control. Parallel assays using a pp53-TA-luc reporter verified the function of the p53 expression construct. Cells were scraped, pelleted by centrifugation, and lysed using Promega passive lysis buffer. Lysates were clarified, and luciferase activity was determined using luciferase reporter assay kits (Promega). Luciferase activity was measured using a Turner Designs 2000 luminometer.
RNA isolation and quantitative RT-PCR.
NIH 3T3 cells were infected at an MOI of 10 PFU/cell with MHV68 ORF50-deficient virus (50.Stop) (51). Ninety minutes postinfection, cells were treated with either DMSO or 100 μM etoposide. All treatments were performed in duplicate. Cells were harvested in TRIzol (Invitrogen) at 4 and 8 h after drug treatment. Total RNA was extracted according to the TRIzol RNA extraction protocol and was quantified with an ND-1000 (NanoDrop Technology). Following extraction, 2 μg of RNA was treated for 22 min with amplification-grade DNase I (1 U; Invitrogen) in a 10-μl reaction mixture containing 20 mM Tris-HCl (pH 8.4), 2 mM MgCl2, 50 mM KCl. Reactions were quenched by adding 1 μl 25 mM EDTA and heating for 10 min at 70°C. One microgram of treated RNA was reverse transcribed with SuperScript II (Invitrogen) in a total volume of 20 μl containing 180 ng random hexamers. No-reverse-transcription (RT) controls were carried out in parallel. Resulting cDNAs were diluted 1:3 in water.
Quantitative PCR was carried out on the resulting cDNA using a Bio-Rad iCycler. Each reaction mixture contained 25 μl iQ SYBR green supermix (Bio-Rad), 10 pmol each primer, 2 μl cDNA, and water to a final volume of 50 μl, and each reaction was performed in triplicate in optical 96-well plates. Primers for the reactions were as follows: orf73, 5′-AAGGGTTGTCTTGGCCTAC TGTG-3′ and 5′-AGAGATGCTGTGGGACCATGTTG-5′; orf50, 5′-GGCCGCAGACATTTAATGAC-3′ and 5′-GCCTCAACTTCTCTGGATATGCC-3′; GAPDH, 5′-CCTGCACCACCAACTGCTTAG-3′ and 5′-GTGGATGCAGGGATGATGTTC-5′; and murine p21 (13). Cycling conditions were as follows: 95°C for 5 min and then 40 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s.
Data were analyzed using the ΔΔCT method (41), where ΔΔCT = [CT(gene) − CT(GAPDH)]treated − [CT(gene) − CT(GAPDH)]mock treated. The data are presented as the evaluation of 2−ΔΔCT for each sample, representing the change (n-fold) in transcript level of each gene in the treated samples, normalized to the level for GAPDH and relative to the normalized level for that gene in the untreated control.
Cell viability assays.
For trypan blue exclusion, cell culture supernatants, which included nonadherent dead cells, were collected, and the remaining cells were detached by trypsinization. Supernatants and detached cells were combined and pelleted by centrifugation. Cells were resuspended in PBS and kept on ice until quantitation. Trypan blue solution (Sigma) was added to the cells at a final concentration of 0.1%, and positive and negative cells were identified by light microscopy. A modification of a previously described protocol was used for the imaging of infected cells (28, 29). Media and detached cells were removed by aspiration and then washed twice with PBS. Adherent cells were fixed for 15 to 30 min in 10% formalin. Cells were washed and stained with 0.1% crystal violet solution for 1 to 2 h. Stained cells were washed three times in deionized H20 and allowed to dry. Cells were imaged, or retained crystal violet was extracted with 10% acetic acid, transferred to microtiter plates, and quantified by spectrophotometry at 630 nm using a plate reader (Biotek). For p53-deficient MEFs, cells were fixed with 10% formalin and washed with PBS. Cells were permeabilized with 0.1% TX100-PBS, and nuclei were stained with 1 μg/ml ethidium bromide. Nuclei visualized by fluorescence microscopy were quantitated from three random fields of vision. For etoposide-treated cells, cell viability was determined using the CellTiter-Glo assay (Promega) according to the manufacturer's instructions.
RESULTS
MHV68 LANA is required for efficient viral replication in primary MEFs at low MOIs.
We previously demonstrated growth deficiencies for 73.Stop virus in the lungs of mice infected by intranasal inoculation compared to the growth for WT and 73.MR viruses. However, in our original analysis of 73.Stop replication in tissue culture, we did not appreciate a replication defect, despite modestly reduced titers at late time points in multistep viral growth curves in NIH 3T12 fibroblasts (48). To more thoroughly assess the role of mLANA in MHV68 replication in culture, we compared the capacities of 73.Stop and 73.MR viruses to replicate over time in Vero cells or in primary MEFs at an MOI of 0.05 (Fig. 1A and B). While 73.Stop replicates efficiently in Vero cells (replication kinetics and yields were enhanced compared to those of 73.MR at the times analyzed), 73.Stop replication was sevenfold reduced relative to that of 73.MR in MEFs 96 h after infection at an MOI of 0.05 PFU/cell. We further assessed the nature of the 73.Stop replication defect by infecting MEFs and NIH 3T3 fibroblasts (the cell line used by Song et al. [63]) at a very low MOI (0.001 PFU/cell) and observed an even greater diminution of 73.Stop replication, with 73.Stop yielding average reductions of 1.8 and 1.5 logs, respectively, relative to the levels of replication of 73.MR at time points corresponding to peak output titers (Fig. 1C and D). These results demonstrate a role for mLANA in virus replication, especially following low-multiplicity infection of both primary and immortalized murine fibroblasts. This correlates with the observed growth defect of the 73.Stop mutant in vivo.
FIG. 1.
mLANA promotes efficient viral replication in primary murine fibroblasts. Vero cells (A), MEFs (B and C), or NIH 3T3 fibroblasts (D) were infected with 73.Stop or 73.MR virus at an MOI of 0.05 (A and B) or 0.001 PFU/cell (C and D). Cells were harvested at the indicated times postinfection, and viral titers were determined by plaque assay. Results are the means of triplicate samples. Error bars represent standard deviations.
MHV68 LANA is a nuclear/cytoplasmic protein expressed during productive virus replication.
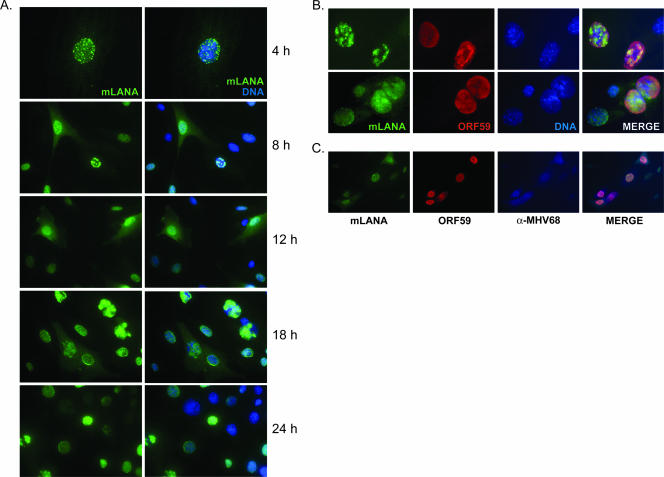
LANA proteins of KSHV, HVS, and RRV localize to the nuclei of host cells, where they regulate a variety of processes, from modulating host and viral transcription to facilitating episome maintenance (16). The requirement of MHV68 LANA for efficient viral replication, paired with data demonstrating that orf73 is an immediate-early transcript present throughout lytic replication (2, 15, 46), suggests an important role in the initial stages of viral replication. However, the localization of mLANA protein or its expression during productive replication has not been examined. To determine the expression and localization of mLANA during infection, we generated a recombinant mLANA-GFP fusion-expressing MHV68 (MHV68-73GFP). NIH 3T3 fibroblasts were infected with MHV68-73GFP and were observed over time by fluorescence microscopy. These experiments revealed mLANA-GFP expression throughout the course of infection with diverse and time-dependent patterns of localization (Fig. 2A). Expression was restricted to the nucleus early during infection and was largely diffuse, with focal areas of intensity. By 8 h postinfection, globular staining was detected in some cells. mLANA-GFP localization by 12 h postinfection was predominately diffuse, exhibiting nuclear and some cytoplasmic staining. By 18 and 24 h, mLANA localization varied dramatically. An interesting feature at these times was mLANA-GFP staining patterns that resembled those of herpesvirus replication foci (see especially the 18-h time point in Fig. 2A) (81, 82), a pattern also seen for KSHV LANA in productively infected endothelial cells (35). Expression and localization of mLANA relative to those of the viral DNA polymerase processivity factor encoded by ORF59 are shown at high magnification in Fig. 2B, as well as in cells that also were stained with MHV68 antiserum (Fig. 2C). These analyses depict mLANA localization during various stages of viral replication and provide further evidence that mLANA is expressed during active MHV68 replication. Taken together, these data demonstrate the unexpected diversity of mLANA localization during the course of a productive MHV68 infection, and its presence very early in infection suggests a role in modulating the viral replication process.
FIG. 2.
mLANA is a nuclear/cytoplasmic protein expressed during lytic replication. NIH 3T3 fibroblasts were infected with recombinant MHV68-73GFP at an MOI of 10 PFU/cell. (A) Cells were fixed at the indicated times postinfection and were stained with GFP-directed antiserum to visualize mLANA-GFP subcellular localization by fluorescence microscopy. (B and C) Cells were stained with antibodies to GFP or ORF59 18 h postinfection (B) or GFP, ORF59, and MHV68 antiserum 24 h postinfection (C). DNA was stained with DAPI (A) or Hoechst 33342 (B). The images in panel A at 4 h and in panel B were captured at ×100 magnification. All other images were captured at ×40 magnification.
mLANA-deficient MHV68 exhibits increased kinetics of cell death during viral replication in tissue culture.
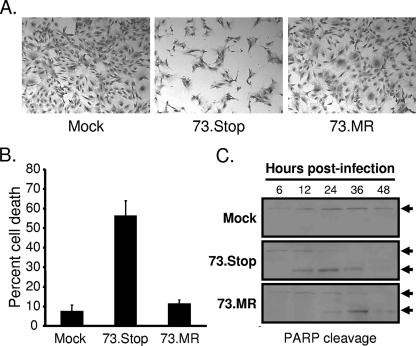
In contrast to the low-MOI replication deficit observed for 73.Stop (Fig. 1), high-MOI 73.Stop infections of MEFs did not reveal an overt replication defect (48). However, 73.Stop-infected cultures display cytopathic effect (CPE) much more rapidly than WT- or 73.MR-infected cells following high-MOI infections. Representative images of this phenotype at an MOI of 2 PFU/cell shown in Fig. 3A are from 24 h after infection of early-passage NIH 3T3 fibroblasts. The uniformity of CPE exhibited by NIH 3T3 fibroblasts and reduced basal levels of apoptosis made them more amenable than MEFs to these studies, although MEFs also exhibit increased CPE following high-MOI 73.Stop infections (data not shown). We quantitated the viability of infected cells by trypan blue exclusion and found that >50% of 73.Stop-infected cultures were nonviable 24 h after infection, whereas approximately 10% of cultures infected with 73.MR at equivalent MOIs were nonviable 24 h after infection (Fig. 3B). Moreover, infection by 73.MR at a higher MOI of 5.0 PFU/cell still resulted in less than 30% cell death at this time point (data not shown).
FIG. 3.
mLANA-deficient virus exhibits increased kinetics of cell death and PARP cleavage. (A and B) NIH 3T3 fibroblasts were mock infected or were infected with 73.Stop or 73.MR virus at an MOI of 2 PFU/cell and were harvested 24 h postinfection. (A) Representative images of cells that were fixed and stained with crystal violet for imaging by light microscopy. (B) Cell viability of triplicate samples determined by trypan blue exclusion. A minimum of 200 cells was counted per sample. Results are the means of triplicate samples. Error bars represent standard deviations. (C) NIH 3T3 fibroblasts were mock infected or were infected with 73.Stop or 73.MR virus at an MOI of 2 PFU/cell and were harvested at the indicated times postinfection. Equivalent amounts of total protein were resolved by SDS-PAGE, and the integrity of PARP was determined by immunoblotting. Arrows denote intact (upper) or cleaved (lower) PARP.
To complement the viability experiments, we examined the integrity of PARP over time following infection with 73.Stop or 73.MR at equivalent MOIs (Fig. 3C). During apoptosis and necrosis, PARP is specifically cleaved to smaller, faster-migrating products that are readily detected by immunoblotting and thus serves as a biochemical marker for cell death (21, 62). These experiments demonstrated that 73.Stop infection results in very rapid PARP cleavage, beginning between 6 and 12 h postinfection, with complete cleavage by 24 h postinfection. Conversely, 73.MR-infected cells exhibited PARP cleavage predominantly between 24 and 36 h postinfection at time points corresponding to peak output titers (Fig. 3C). Full-length PARP was detected throughout the time course for mock-infected cells. Intriguingly, we did not detect evidence of activated caspase-3, the effector caspase in the apoptotic cascade, nor did treatment with the pan-caspase inhibitor z-vad-fmk ameliorate cell death or rescue replication following 73.Stop infection (data not shown). These data suggest a nonapoptotic death pathway or reflect the functional activity of the viral bcl-2 orthologue (42, 74, 76) in MHV68-infected cells. Taken together, these data demonstrate that 73.Stop infection leads to increased cell death relative to that caused by 73.MR infection.
To assess the potential contribution of virus replication to 73.Stop-induced cell death, we analyzed the replication kinetics of 73.Stop and 73.MR in MEFs in a comprehensive time course analysis during a single replication cycle following infection at an MOI of 2 PFU/cell (Fig. 4A). This analysis revealed that 73.Stop replication proceeds with increased kinetics compared to those of 73.MR, with 73.Stop virus yields being 0.5 log higher than those of 73.MR by 12 h postinfection. With time, however, the virus yields from 73.MR-infected cells surpass those of 73.Stop, with maximal virus production from 73.Stop-infected cells occurring by ca. 18 to 24 h postinfection, while titers from 73.MR-infected cells continued to increase until ca. 30 h postinfection. Consistent with these data, we also detected more abundant cell-associated lytic antigens in 73.Stop-infected cells (data not shown), although it is unclear if this reflects retention due to cell death or truly correlates with increased early titers. Importantly, with the exception of the 18-h time point, these results are statistically significant and were consistent in multiple experiments. It is interesting that we observed a reduced eclipse phase in MEFs during 73.Stop infection at an MOI of 5 in our original study (48), which further supports the observations reported here. These data reveal the mLANA-dependent single-cycle replicative differences in murine fibroblasts and predict that differences in viral titers following low-MOI infections (Fig. 1) are the product of multiple inefficient rounds of 73.Stop replication. These data also suggest that in the absence of mLANA, either deregulation of the lytic gene expression cascade occurs, leading to increased replication kinetics and subsequent cell death, or mLANA is required to control an infection-related stress response that otherwise influences the course of the viral replication cycle.
FIG. 4.
73.Stop virus exhibits increased replication, viral antigen expression, and p53 induction. (A) MEFs were infected with 73.Stop or 73.MR virus at an MOI of 2 PFU/cell, and cells were harvested at the indicated times postinfection. Viral yields were determined by plaque assay. Results are the means from triplicate samples. Error bars represent standard deviations. P values were determined using a two-tailed paired Student's t test. (B) NIH 3T3 fibroblasts were infected with 73.Stop or 73.MR virus at an MOI of 2 PFU/cell. Equivalent numbers of cells were harvested directly by addition of Laemmli sample buffer at the indicated times postinfection. Total cell lysates were resolved by SDS-PAGE and were analyzed by immunoblotting with antibodies to the indicated proteins. Untreated and UV-exposed cells (100 J/m2) serve as negative and positive controls, respectively, for p53 stabilization and activation. The blot shown is representative of three independent experiments.
73.Stop infection is characterized by deregulated viral gene expression and activation of p53.
Given the increased cell death and expedited replication kinetics of 73.Stop, we asked if viral early gene expression is more rapidly detected during 73.Stop infection in culture. For these experiments, lysates of NIH 3T3 fibroblasts infected with 73.Stop or 73.MR at equivalent MOIs were analyzed by immunoblotting for the presence of v-cyclin and the viral processivity factor ORF59. Relative to the levels of 73.MR, infection by 73.Stop resulted in more rapid and more robust expression of both v-cyclin and ORF59 (Fig. 4B), suggesting that normal viral gene expression is dysregulated in the absence of mLANA.
KSHV LANA and HVS LANA previously were demonstrated to inhibit activation of the tumor suppressor p53 (7, 27), which led us to hypothesize that p53 is activated by MHV68 in the absence of mLANA. Activation of p53 leads to its stabilization, which is mediated by a variety of modifications, including acetylation, sumoylation, and phosphorylation, on a variety of serine residues (reviewed in references 32 and 38). To assess and compare the activation status of p53 during the course of infection with either 73.Stop or 73.MR virus, we performed immunoblots for total p53 and p53 phosphorylated on serine 18 (serine 15 in human p53), a residue commonly phosphorylated in response to DNA damage stimuli and observed with other herpesvirus infections (8, 30, 33, 38, 60). Untreated and UV-exposed cells served as negative and positive controls, respectively, for p53 induction. Increased stabilization and more robust phosphorylation of p53 on serine 18 was observed for 73.Stop-infected cells compared to the levels for 73.MR infections (Fig. 4B), suggesting that MHV68 infection is recognized by the host cell as an agonist of p53 and, further, that mLANA is involved in regulating p53 activation. Moreover, to our knowledge, these data provide the first evidence of a potential role for LANA proteins in regulating p53 during any aspect of gammaherpesvirus replication.
Increased cell death is independent of viral replication.
To help determine whether expedited virus replication and/or cell stress is the cause of 73.Stop-induced cell death, we tested the capacity of a pharmacologic inhibitor of MHV68 DNA replication to inhibit 73.Stop-induced cell death. Cidofovir is a nucleoside analog that potently inhibits viral DNA synthesis at concentrations that have no observable toxic effects on cultured cells (50; also data not shown) (Fig. 5A). NIH 3T3 fibroblasts were mock infected or were infected with 73.Stop (MOI = 2) or 73.MR (MOI = 2 to 20) in the presence of 20 ng/ml cidofovir, a concentration previously demonstrated and verified in these experiments (see Materials and Methods) to efficiently block MHV68 replication (50), and cell viability was assessed 24 to 26 h postinfection. Cidofovir treatment did not inhibit 73.Stop-induced CPE (Fig. 5A), nor did treatment inhibit the infection-associated loss of cell viability (Fig. 5B), while mock- and 73.MR-infected cells displayed no observable CPE or loss of viability. Importantly, infection with 73.MR in the presence of cidofovir at MOIs of up to 20 PFU/cell, or infection with a replication-incompetent ORF50-deficient virus (50.Stop; the titer was determined by plaque assay on complementing cells), exhibits much less CPE than 73.Stop at an MOI of 2 (Fig. 5A), providing confidence that 73.Stop-related cellular death is not simply the result of a particle effect due to an increased particle/PFU ratio. In addition, these data indicate that cell death associated with 73.Stop infection is not a direct result of virus replication (i.e., generation of progeny virus) and further suggest that mLANA inhibits an infection-related cellular stress response that would otherwise promote cell death.
FIG. 5.
73.Stop-induced cell death, p53 phosphorylation, and dysregulated early gene expression occur independently of viral replication. (A to C) NIH 3T3 fibroblasts were mock infected, infected with 73.Stop or 73.MR virus at MOIs of 2 to 20 PFU/cell in the presence of 20 ng/ml cidofovir (A and B), or infected with 50.Stop virus at MOIs of 2 to 20 PFU/cell (C). (A) Representative images of cells that were fixed and stained with crystal violet 24 h after infection for imaging by light microscopy. (B) Cell viability determined by trypan blue exclusion 26 h after infection. A minimum of 200 cells per sample was counted. Results are the means from triplicate samples. Error bars represent standard deviations. (C) NIH 3T3 fibroblasts were mock infected or were infected with 73.Stop or 73.MR virus in the presence or absence of 20 ng/ml cidofovir at an MOI of 2 PFU/cell. Equivalent numbers of cells were harvested directly by addition of Laemmli sample buffer 6 h postinfection. Total cell lysates were resolved by SDS-PAGE and were analyzed by immunoblotting with antibodies to the indicated proteins. UV-exposed cells (100 J/m2) serve as a positive control for p53 activation. The blot shown is representative of three independent experiments.
We also tested the capacity of cidofovir to inhibit the enhanced expression of viral early genes and activation of p53 observed following 73.Stop infection as demonstrated in Fig. 4. Lysates of NIH 3T3 fibroblasts infected with 73.Stop or 73.MR in the presence or absence of cidofovir were analyzed by immunoblotting. Notably, even in the presence of cidofovir, infection with 73.Stop virus resulted in robust expression of v-cyclin and ORF59, as well as eliciting p53 phosphorylation on serine 18, by 6 h postinfection (Fig. 5C). These data indicate that viral replication is not the cause of death and further suggest that the observed dysregulation of viral gene expression in the absence of mLANA is a direct consequence of very early events in MHV68 infection that trigger a cell stress response leading to p53 activation, a process that is modulated or inhibited by expression of mLANA.
Expression of mLANA correlates with decreased p53 stabilization and cell death following etoposide treatment.
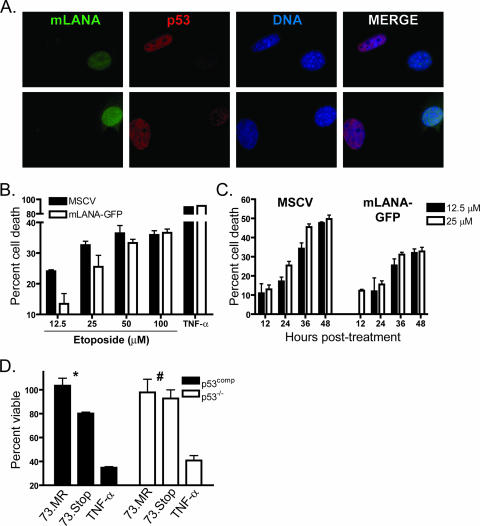
KSHV LANA was shown to prevent cell death triggered by a variety of p53-inducing agonists (27). The increased activation of p53, and the correlative induction of viral gene expression described above, suggested a role for mLANA in limiting p53 induction to facilitate survival of an infected cell. To analyze the capacity of mLANA to limit p53 induction, NIH 3T3 fibroblasts stably transduced with either empty vector (MSCV) or mLANA-GFP-expressing retroviruses were treated with the potent p53-inducing radio-mimetic drug etoposide, and p53 stabilization was examined by immunofluorescence (Fig. 6A). In these experiments, cells with the highest level of expression of mLANA-GFP correlated with the lowest levels of p53 accumulation in the nucleus, which coupled mLANA expression to p53 destabilization. As a functional complement to the immunofluorescence experiments, we examined the capacity of NIH 3T3 fibroblasts expressing mLANA-GFP to resist death relative to that of empty-vector-transduced control cells following treatment with increasing concentrations of etoposide (Fig. 6B). The presence of mLANA-GFP provided modest protection from death induced by the p53-inducing stimulus. Of note, the greatest protection from etoposide-induced cell death occurred with the lowest concentrations tested (≤25 μM). We also observed mLANA-related protection from death following UV exposure and in NIH 3T12 fibroblasts expressing untagged mLANA (data not shown). To provide a more detailed analysis of this phenomenon, we extended our experiments to a time course analysis of cell viability following etoposide treatment at concentrations of 12.5 and 25 μM (Fig. 6C). In support of the dose-response experiments, mLANA-GFP provided increased protection from etoposide-induced cell death, with the protection becoming more evident throughout the time course. These data provide functional evidence that MHV68 mLANA singularly has the capacity to increase the threshold for toxicity following treatment with classic p53 agonists and suggest that this is accomplished by limiting p53 induction.
FIG. 6.
mLANA inhibits p53 induction and reduces etoposide-induced cell death. (A) mLANA-GFP-transduced NIH 3T3 fibroblasts were treated with etoposide (100 μM). Cells were fixed and stained for p53 4 h posttreatment. Two representative images are shown at ×100 magnification. (B and C) Empty vector (MSCV) or mLANA-GFP-transduced NIH 3T3 fibroblasts were treated with etoposide at the indicated concentrations. Cell viability was analyzed 28 h posttreatment (B) or during a time course (C). Data are presented as the percentages of cell death with treatment relative to that for mock-treated controls. Results are the means from triplicate samples. Error bars represent standard deviations. (D) p53comp or p53−/− MEFs were infected with 73.Stop or 73.MR virus at an MOI of 2 PFU/cell in the presence of 20 ng/ml cidofovir. Cell viability was determined 12 h postinfection. Data represent the percentages of cell death relative to that for mock-infected samples. Results are the means from triplicate samples. Error bars represent standard errors of the means. Statistical analyses of differences for 73.MR- and 73.Stop-infected cells were performed using a two-tailed paired Student's t test (*, P = 0.05; #, P = 0.66). (B and D) Tumor necrosis factor alpha (TNF-α) samples were treated with 25 ng/ml TNF-α and 10 μg/ml cycloheximide as a positive control for cell death induction.
To gain additional insight into a role for mLANA in limiting p53 induction and death during MHV68 infection, we sought to determine the effects of 73.Stop infection in p53-deficient cells. In preliminary experiments, the differences in cellular morphology and growth rate made direct comparisons of cell death rates for WT and p53−/− MEFs difficult. To normalize the system, p53−/− MEFs were transduced with retroviruses encoding DsRed as a negative control (p53−/−) or a p53-DsRed fusion protein (p53comp) to reintroduce p53 expression. The function of p53 fusion proteins has been described previously (11, 23, 57; also data not shown). Immediately following puromycin selection and expansion, cells were infected with 73.Stop or 73.MR at an MOI of 2 PFU/cell. Infections were performed in the presence of cidofovir to block the contribution of virus replication to cell death. Viability determination 12 h postinfection demonstrated a reduction in cell death associated with 73.Stop infection in p53−/− cells relative to that in p53comp cells (Fig. 6D). The difference, however, was completely lost by 24 h postinfection (data not shown), suggesting that the contribution of p53 to 73.Stop-induced death occurs early during infection. These data additionally suggest roles for the unchecked viral gene expression or a currently undefined cellular stress in 73.Stop-related cellular injury.
p53 plays a general role in virus replication that influences the efficiency of 73.Stop replication.
The capacity of mLANA to regulate p53 induction paired with activation of p53 by 73.Stop virus suggests that p53 induction contributes to the replication deficit observed for 73.Stop following low-MOI infection. To understand the role of p53 activation in 73.Stop replication, we performed multistep growth analyses using the p53−/− or p53comp cells described above (see Fig. 6). Consistent with experiments with WT MEFs or NIH 3T3 cells (Fig. 1), viral titers for p53comp MEFs for 73.Stop peaked at a 1.4-log deficit relative to titers for 73.MR (Fig. 7A). Conversely, although 73.Stop displayed a delay in the onset of productive replication in p53−/− MEFs, by 168 h postinfection titers for both 73.Stop and 73.MR were virtually identical (73.Stop, ( 6.7 ± 0.4) × 106 PFU/ml; 73.MR, (7.1 ± 0.5) × 106 PFU/ml) (Fig. 7B). Remarkably, this equilibration was not due to an overt increase in 73.Stop replication. Rather, it represented an eightfold reduction in 73.MR output ([7.9 ± 0.1] × 106 PFU/ml in p53comp cells). As further evidence that p53 directly impacts MHV68 replication, while titers began to increase between 24 and 48 h postinfection in p53comp cells, the eclipse phase was prolonged a further 24 h in p53−/− MEFs for both 73.Stop and 73.MR in these assays. These data indicate that p53 regulates the timing and efficiency of MHV68 replication.
FIG. 7.
p53 regulates MHV68 replication. p53comp MEFs (A) or p53−/− MEFs (B) were infected with 73.Stop or 73.MR virus at an MOI of 0.001 PFU/cell. Cells were harvested at the indicated times postinfection, and viral titers were determined by plaque assay. Results are the means from triplicate samples. Error bars represent standard deviations. (C) WT or p53−/− MEFs were infected with WT, 73.Stop, or 73.MR MHV68 at an MOI of 2 PFU/cell. Equivalent numbers of cells were harvested directly by addition of Laemmli sample buffer at the indicated times postinfection. Total cell lysates were resolved by SDS-PAGE and were analyzed by immunoblotting with antibodies to the indicated proteins.
p53 promotes viral gene expression.
The data presented above, in conjunction with the correlation between p53 induction and increased viral gene expression following 73.Stop infection, suggested the capacity of p53 to promote the expression of viral genes. To more precisely define a role for p53 in augmenting MHV68 replication, we compared viral antigen production over time between WT and p53-deficient MEFs infected at an MOI of 2 PFU/cell with WT MHV68, 73.Stop, or 73.MR (Fig. 7C). In this assay, while ORF59 and v-cyclin were readily detectable 6 to 8 h after infection with WT MHV68 and 73.MR in WT MEFs, signal was not evident for these antigens in p53−/− MEFs. Identical blottings performed 14 h postinfection demonstrated that the p53−/− MEFs supported viral gene expression from the WT and MR viruses in these experiments (data available upon request). Although 73.Stop still displayed the increased viral gene expression portrayed in Fig. 4 and 5, expression of ORF59 and v-cyclin similarly exhibited relative delays in the absence of p53, which is in agreement with the early function of p53 in 73.Stop-induced death depicted in Fig. 6D. Together, these data support a role for p53 in enhancing MHV68 replication and potentially define the basis for increased viral gene expression and advanced replication kinetics at high MOIs for mLANA-deficient virus.
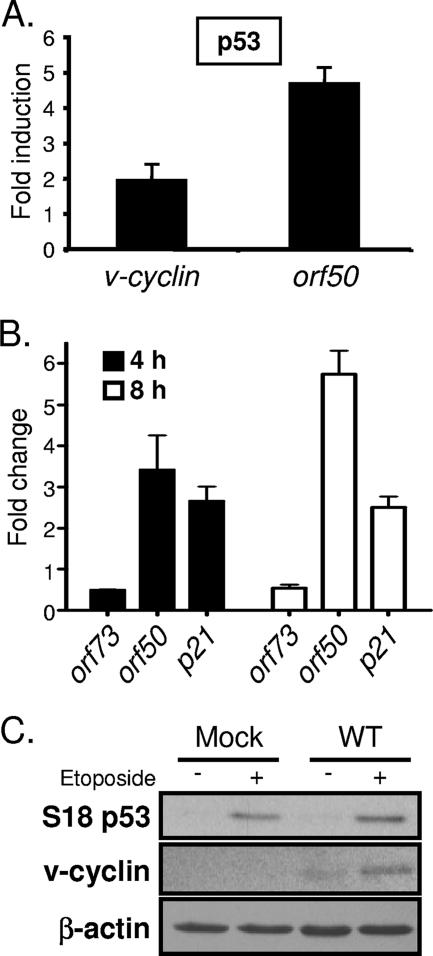
To more specifically determine a mechanism for p53-related induction of viral gene expression and replication, we tested the capacity of p53 and p53-inducing agonists to drive viral gene expression. Since v-cyclin and ORF59 were more rapidly and more abundantly expressed and p53 was activated during infection with mLANA-deficient virus, we hypothesized that p53 expression could induce activity of the proximal v-cyclin lytic-cycle-associated promoter (a promoter active during MHV68 replication in permissive fibroblasts [3]) in the absence of any viral gene products. Since this promoter is responsive to the immediate-early transactivator RTA, we further hypothesized that induction of early viral gene expression alternatively could be triggered through p53-mediated induction of orf50, leading to RTA expression and subsequent induction of viral early genes. To test these hypotheses, NIH 3T3 fibroblasts were transfected with v-cyclin or orf50 promoter-driven reporter constructs in the presence or absence of a p53 expression vector. Transfection of the p53 expression vector slightly increased v-cyclin promoter activity (twofold) while eliciting an approximate fivefold induction of the orf50 promoter (Fig. 8A). These data suggest an RTA-mediated positive feedback loop for increased v-cyclin expression in response to p53. Transfection efficiencies in these analyses were similar, as monitored by cotransfection of a GFP expression vector, and complementary experiments with a p53-responsive reporter vector verified the functionality of the p53 expression construct (data not shown). To gain additional insight into a potential role for p53 in MHV68 infection, we asked whether an exogenous p53-inducing stressor could influence viral gene expression by treating NIH 3T3 fibroblasts with etoposide following infection with the RTA-null 50.Stop MHV68 mutant (51) (Fig. 8B). 50.Stop virus was used to minimize potential influences of RTA-responsive genes. Analysis by quantitative RT-PCR demonstrated substantially increased orf50 transcription that accumulated over time following etoposide treatment. Induction of the p53-responsive cyclin inhibitor p21 served as a positive control for drug treatment (13) (Fig. 8B). Interestingly, we observed a twofold decrease in the level of orf73 transcripts following etoposide treatment, perhaps suggesting a negative feedback loop that facilitates the progression of the lytic gene expression cascade. Further, etoposide treatment of cells infected with WT MHV68 promoted a reproducible twofold increase in v-cyclin protein expression relative to that of vehicle-treated samples as assessed by densitometry (Fig. 8C). Together, these data provide strong evidence that MHV68 is poised to respond to either p53 itself or p53-activating stimuli and further implicate p53 as a factor that promotes viral gene expression toward enhanced lytic replication. Moreover, these findings suggest that mLANA critically regulates this process in a manner that limits host cell injury to permit efficient viral replication.
FIG. 8.
Overexpression of p53 and etoposide treatment induce viral gene expression. (A) NIH 3T3 fibroblasts were transfected with luciferase reporter promoter constructs for v-cyclin or orf50 in conjunction with either empty vector or p53 expression vector. Cells were harvested 24 h after transfection, and luciferase activity was determined. The data represent the difference (n-fold) between vector-transfected and p53-transfected cells. Results are the means from triplicate samples. Error bars represent standard deviations. (B) NIH 3T3 fibroblasts were infected with ORF50-deficient (50.Stop) MHV68 at an MOI of 10 PFU/cell. Ninety minutes postadsorption, cells were treated with DMSO or with etoposide (100 μM). Cells were harvested at 4 and 8 h posttreatment. Quantitative RT-PCR was performed for the indicated genes. Data represent the change (n-fold) in transcript levels normalized to the levels for GAPDH, as determined using the ΔΔCT method for etoposide-treated samples relative to DMSO-treated samples. Data are the averages from duplicate samples. Error bars represent the range of data. (C) NIH 3T3 fibroblasts were mock infected or were infected with WT MHV68 at an MOI of 2 PFU/cell. One hour postadsorption, cells were treated with either DMSO or etoposide (100 μM). Cells were harvested 12 h postinfection, and equivalent amounts of protein were resolved by SDS-PAGE. Samples were examined by immunoblot analysis with antibodies to the indicated proteins. The blot shown is representative of four independent experiments harvested between 8 and 12 h.
DISCUSSION
In this paper, we have examined the role of the MHV68 LANA homologue mLANA in virus replication. We found that mLANA-null virus is attenuated in primary and immortalized fibroblasts at low MOIs and that mLANA is a nuclear/cytoplasmic protein expressed during virus replication in permissive fibroblasts. At higher MOIs, 73.Stop resulted in increased kinetics of cell death with apparently protracted entry into the productive replication phase but an overall deficit in viral yield. Consistent with these findings, we detected increased viral gene expression in the absence of mLANA and determined that 73.Stop-induced cell death occurred in the presence of cidofovir, demonstrating that the induction of cell death was independent of viral replication. We further observed that infection with 73.Stop resulted in more robust activation of p53 than that with 73.MR, and that p53 activation also was independent of virus replication. In agreement with a role for mLANA in modulating p53 induction, expression of an mLANA-GFP fusion protein in fibroblasts provided modest protection from etoposide-induced death, characterized at the cellular level by diminished p53 stabilization. Moreover, p53-deficient cells were less susceptible than p53-competent cells to 73.Stop-induced death at early times postinfection. We found that 73-null virus replicated to the same level as 73.MR in p53-deficient cells but, more remarkably, that p53 generally promoted MHV68 replication. In a related determination, we found that p53 overexpression or treatment with the potent p53-inducing drug etoposide could increase promoter activity, transcription, and expression of viral genes that promote lytic replication. Together, these data further define the replication defect previously demonstrated for mLANA-null virus in vivo, reveal a previously unappreciated role for p53 in the MHV68 life cycle, and illuminate a role for mLANA in managing a cellular stress response to viral infection to prevent cellular injury and coordinate the MHV68 lytic-gene expression cascade.
Based on these studies, we propose a model for the roles played by mLANA to regulate MHV68 replication (Fig. 9). In this model, the immediate-early mLANA limits the induction of cellular stress responses that otherwise would activate p53 and currently undefined pathways that drive viral gene expression. We envision mLANA reducing the stressors directly or indirectly by counteracting feedback loops. We hypothesize that mLANA in this capacity orchestrates the cell and virus during infection in a manner that limits cellular injury and maximizes viral yield. We think it likely that unregulated cell stress and viral gene expression form a self-perpetuating loop that drives the host cell to death. We also illustrate the newly defined role for p53 in promoting lytic viral replication and that mLANA may alter p53 function by preventing activation, blocking downstream functions, or both. Our experiments did not delineate these possibilities, nor did we pinpoint an exact role for p53 in 73.Stop-induced cell death, although experiments with mLANA-transduced cells suggest an attenuation in p53-related death pathways (Fig. 6). Aside from a role for mLANA in episomal tethering to facilitate viral genome maintenance during latency, our data support the hypothesis that mLANA critically influences host cell survival and/or viral gene expression not only to allow efficient replication but also to promote establishment and maintenance of latent MHV68 infection.
FIG. 9.
Model of predicted mLANA functions to regulate host cell stress and promote efficient viral replication. MHV68 infection elicits the activation of cellular stress pathways that may promote p53 activation. Cell stress and/or p53 activity enhances expression of viral lytic genes, and likely cellular targets, in a manner that is deleterious to both the virus and host cell. We hypothesize that mLANA functions to limit propagation of the p53-inducing stressor or directly alters the activity of p53 to regulate viral gene expression and limit host cell injury, allowing for efficient viral replication.
MHV68 LANA and viral replication.
In previous studies we demonstrated that ORF73 mutant viruses exhibit a replication defect in the lungs of infected mice, and Song and colleagues reported a transposon mutant in ORF73 that was attenuated in NIH 3T3 fibroblasts (63). Moreover, the immediate-early transcription pattern described for orf73 (15) suggests an important role in facilitating MHV68 replication. The experiments reported here extend the previous findings by demonstrating an MOI-dependent requirement for mLANA in viral replication in fibroblasts (Fig. 1) but do not offer an immediate mechanism for potential cell-type-specific differences in 73.Stop replication. There are numerous possible explanations for 73.Stop replicating efficiently in Vero (Fig. 1) or BHK-21 (26) cells, including, but not limited to, species and tissue of origin, lack of expression of host inhibitory factors, and length of cell cycle. The data we present, particularly complementation of 73.Stop replication at low MOIs in p53-deficient MEFs, indicate the importance of p53 in limiting 73.Stop replication. It is interesting that the NIH 3T12 immortalization regimen for MEFs can promote mutations in p53 (55), which perhaps offers an explanation for why we did not originally appreciate a replication deficiency for 73.Stop in culture (48).
Unlike herpesvirus immediate-early genes with transactivator functions that are absolutely required for lytic replication, such as herpes simplex virus type 1 (HSV-1) ICP4 (18), human cytomegalovirus (HCMV) IE2 (44), and MHV68 RTA (ORF50) (51), a second class of immediate-early genes, such as ICP0 of HSV-1 (31) or HCMV IE1 (47), displays their defects only at low MOIs. Perhaps this reveals requirements for these genes in regulating cellular stresses very early in the infection process in the absence of high-MOI virus particle effects or in the face of an active cellular or host immune response. Detection of mLANA in the nucleus very early in infected cells (as early as 2 h postinfection in separate experiments; J. C. Forrest and S. H. Speck, unpublished data) suggests the importance of mLANA in modulating viral and host gene expression or limiting activation of nuclear stress pathways, such as a DNA damage response (see below), to provide an ideal cellular environment for viral replication.
Role of p53 in MHV68 replication.
Activation of p53 occurs in response to numerous cellular stresses, ranging from contact inhibition to nutrient deprivation to viral infection, and is central to maintaining metabolic homeostasis and genetic integrity (32, 38). In its role as a regulator of cell stress responses, p53 enforces cell cycle checkpoints to allow recovery from the sensed stress, but p53 also can direct initiation of apoptosis by promoting expression of apoptosis-initiating genes, repressing expression of anti-apoptotic genes, or directly insulting mitochondrial integrity (5, 13, 32). Because p53 is critically involved in recognizing and responding to cellular events that could elicit nutrient crisis or genomic instability, it stands to reason that nuclear invasion and exponential synthesis of large, foreign DNAs would elicit a p53 response. Conversely, the rationale for viral inactivation or deregulation of such a gatekeeper also is quite clear. Indeed, the characterization of DNA tumor virus replication (e.g., adenovirus, papillomavirus, and simian virus 40) helped define the regulation of several p53-mediated tumor-suppression-related pathways through the analysis of early gene functions (E1B, E6, and large T antigen, respectively) that either inhibit p53 activation or promote p53 degradation (49). Importantly, adenovirus and simian virus 40, with mutations in E1B and large T antigen, respectively, exhibit attenuated replication and increased cell death (65, 66).
In the case of herpesvirus replication, the role of p53 is less clear. Both HCMV (30, 33) and HSV-1 (8, 60) induce p53 during infection. During HSV-1 infection, p53 is sequestered into viral replication complexes (79, 82), although p53 status does not appear to regulate HSV-1 replication (8). The functional presence of p53 positively influences the kinetics and productivity of HCMV replication (11) in a manner analogous to that of MHV68 (Fig. 7). This remarkable functional conservation across herpesvirus families suggests similar mechanisms of p53 regulation and utilization during productive viral replication. Similarly to HSV-1, HCMV recruits p53 into viral replication compartments (25, 56), presumably in a manner that requires the IE2 viral protein and inactivates p53 function (12, 68). Although the mechanism whereby mLANA inhibits p53 activation is not apparent from our studies, it is tempting to speculate that gammaherpesvirus LANA proteins directly recruit p53 to viral replication compartments to inhibit p53 transcriptional regulation of host genes. This hypothesis is consistent with data demonstrating that KSHV LANA and HVS LANA engage p53 to inhibit its function (7, 27). KSHV LANA also limits p53 expression and activation of a p53 promoter (61), a finding consistent with the decrease in levels of nuclear p53 we observed in mLANA-GFP-expressing cells following UV (data not shown) or etoposide treatment (Fig. 6B). Alternatively, mLANA may limit p53 induction by altering one or more other host stress pathways, many of which have been described for KSHV LANA (16). A more refined interpretation will involve the temporal determination of viral and host proteins and genes targeted by LANA during productive rhadinovirus replication.
Our findings that mLANA limits p53 activation during productive MHV68 replication but that p53 promotes viral gene expression and replication are likely reflections of the tenuous relationship between herpesviruses and the host cell. Cassavant and colleagues suggested that p53 positively influences HCMV gene expression and subsequent replication as a transcriptional activator for viral replication and structural genes (11). In this case, sequestration of p53 into replication complexes would facilitate viral gene transactivation while limiting induction of p53-responsive host genes.
In light of mLANA limiting p53 activation during MHV68 replication, we propose additional hypotheses for p53 in the herpesvirus life cycle. LANA proteins of KSHV, HVS, and RRV inhibit activation of promoters for the lytic transactivator protein RTA (19, 37, 58). As a corollary for HVS in particular, mLANA induction limits the progression of lytic replication in culture (58). We did not detect mLANA-mediated inhibition of an orf50 promoter (data available upon request), but we did demonstrate p53-related orf50 induction (Fig. 8). In addition, the absence of mLANA promoted increased p53 activation and viral gene expression (Fig. 4 and 5). Together, these findings suggest that LANA proteins limit p53-dependent activation of lytic-phase viral genes and viral replication, thereby depicting the importance of mLANA in regulating the p53 response to facilitate efficient viral replication. Moreover, considering the delay in these events in cells lacking p53 (Fig. 7), these data predict that p53 serves as a catalyst for viral replication, perhaps even dictating the switch from latency to lytic replication.
Cell death and v-cyclin expression.
The mechanism of cell death induction by 73.Stop infection is not clear. The results shown in Fig. 6 suggest that p53 contributes to the cell death phenotype, especially early during infection, and to this end mLANA can protect cells from death caused by p53-inducing treatments. However, the absence of p53 is not protective to 73.Stop-infected cells at later time points (data not shown) even when viral replication is pharmacologically inhibited, indicating the execution of p53-independent death pathways. These results are not necessarily surprising and probably reflect the delay, but not total shutdown, in viral gene expression observed in p53-deficient MEFs (Fig. 7). To be sure, the DNA damage caused by UV light and etoposide induce numerous stress pathways that inhibit cell cycle progression and mediate repair or cell death, even in the absence of p53 (for example, see reference 54). Perhaps insight into other stress pathways altered by LANA functions can be gathered from studies of p53-deficient cells. Furthermore, as p53 mutations are very common in human cancer (75) and p53-independent cell death pathways thus form the basis for many cancer treatment regimens, understanding how gammaherpesvirus-infected cells limit death in the absence of p53 may offer new insights into treatments for gammaherpesvirus-related tumors.
Given the antigrowth and cell-cycle-inhibitory properties of p53, it is remarkable that v-cyclin expression is so dramatically increased early during 73.Stop infection (Fig. 4 and 5). MHV68 v-cyclin expression promotes tumor development in transgenic mice (71) and interacts with CDK1 and CDK2 to positively support pRb phosphorylation in vitro and cell cycle progression in serum-starved infected cells (70). Since p53-dependent transcription of the CDK inhibitor p21 potently blocks cellular cyclin-dependent pRb phosphorylation and progression from G1 to S phase (73, 75), we speculate that the robust v-cyclin expression observed in 73.Stop-infected cells is a compensation mechanism that facilitates replication when the cell cycle is arrested by p53 activation in the absence of mLANA expression. Indeed, despite p53 induction, we detect more rapid pRb hyperphosphorylation (which promotes E2F activity and cellular DNA synthesis [14]) in 73.Stop-infected cells than in those infected with 73.MR (Forrest and Speck, unpublished), which is consistent with increased v-cyclin expression. These ideas support our data demonstrating orf50 and v-cyclin induction by p53-related cellular stresses (Fig. 8). Since v-cyclin is a requirement for MHV68 reactivation following ex vivo plating in some settings (69, 72), these data further suggest that cell cycle arrest and/or p53 activation contribute to viral reactivation from latency.
Similarly to mLANA mutant viruses, v-cyclin mutants exhibit replication deficits in the lungs of infected mice, although the effect is dose dependent for v-cyclin mutants (69). It will be interesting to determine if mLANA similarly compensates for v-cyclin mutant MHV68 by increasing expression. If our hypotheses regarding compensatory functional mechanisms between v-cyclin and mLANA are correct, we would further speculate that a virus lacking both genes will be severely attenuated, perhaps even replication incompetent in some experimental settings. Moreover, as oncogene expression or deregulated expression of cell cycle control proteins, including cyclin E, can trigger p53 activation and cell cycle arrest (6, 20, 39, 43, 59), it is tempting to speculate that, while partially rescuing 73.Stop replication, v-cyclin overexpression during 73.Stop infection actually contributes to the infection-related cellular injury, thereby promoting increased cell death and an ensuing defect in virus replication. Indeed, we observed striking v-cyclin expression and death following 73.Stop infection, even in the presence of cidofovir (Fig. 5). Such a precarious balance between survival and death may exemplify the tightly regulated nature of herpesvirus gene expression.
Acknowledgments
This work was supported by NIH R01 grant CA52004. J.C.F. is supported by a Leukemia and Lymphoma Society postdoctoral fellowship. S.H.S. is supported by NIH R01 grants CA43147, CA52004, CA58524, CA95318, and AI05807.
We thank Rolf Renne for suggesting that we investigate p53 activation as a contributor to the 73.Stop death phenotype. We thank members of the Mocarski and Speck laboratories for helpful comments on this research.
Footnotes
Published ahead of print on 15 August 2007.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. W., K. L. Powell, P. Kellam, and D. G. Alber. 2002. Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol. 76:6244-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, R. D., III, M. N. Dezalia, and S. H. Speck. 2007. Identification of an Rta responsive promoter involved in driving gammaHV68 v-cyclin expression during virus replication. Virology 365:250-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An, F. Q., N. Compitello, E. Horwitz, M. Sramkoski, E. S. Knudsen, and R. Renne. 2005. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280:3862-3874. [DOI] [PubMed] [Google Scholar]
- 5.Baptiste, N., and C. Prives. 2004. p53 in the cytoplasm: a question of overkill? Cell 116:487-489. [DOI] [PubMed] [Google Scholar]
- 6.Bartkova, J., N. Rezaei, M. Liontos, P. Karakaidos, D. Kletsas, N. Issaeva, L. V. Vassiliou, E. Kolettas, K. Niforou, V. C. Zoumpourlis, M. Takaoka, H. Nakagawa, F. Tort, K. Fugger, F. Johansson, M. Sehested, C. L. Andersen, L. Dyrskjot, T. Orntoft, J. Lukas, C. Kittas, T. Helleday, T. D. Halazonetis, J. Bartek, and V. G. Gorgoulis. 2006. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444:633-637. [DOI] [PubMed] [Google Scholar]
- 7.Borah, S., S. C. Verma, and E. S. Robertson. 2004. ORF73 of herpesvirus saimiri, a viral homolog of Kaposi's sarcoma-associated herpesvirus, modulates the two cellular tumor suppressor proteins p53 and pRb. J. Virol. 78:10336-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutell, C., and R. D. Everett. 2004. Herpes simplex virus type 1 infection induces the stabilization of p53 in a USP7- and ATM-independent manner. J. Virol. 78:8068-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai, Q., K. Lan, S. C. Verma, H. Si, D. Lin, and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus latent protein LANA interacts with HIF-1α to upregulate RTA expression during hypoxia: latency control under low-oxygen conditions. J. Virol. 80:7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, P. A., H. L. Kenerson, R. S. Yeung, and M. Lagunoff. 2006. Latent Kaposi's sarcoma-associated herpesvirus infection of endothelial cells activates hypoxia-induced factors. J. Virol. 80:10802-10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casavant, N. C., M. H. Luo, K. Rosenke, T. Winegardner, A. Zurawska, and E. A. Fortunato. 2006. Potential role for p53 in the permissive life cycle of human cytomegalovirus. J. Virol. 80:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo, J. P., and T. F. Kowalik. 2002. Human cytomegalovirus immediate early proteins and cell growth control. Gene 290:19-34. [DOI] [PubMed] [Google Scholar]
- 13.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010-1014. [DOI] [PubMed] [Google Scholar]
- 14.Cobrinik, D. 2005. Pocket proteins and cell cycle control. Oncogene 24:2796-2809. [DOI] [PubMed] [Google Scholar]
- 15.Coleman, H. M., S. Efstathiou, and P. G. Stevenson. 2005. Transcription of the murine gammaherpesvirus 68 ORF73 from promoters in the viral terminal repeats. J. Gen. Virol. 86:561-574. [DOI] [PubMed] [Google Scholar]
- 16.Collins, C., and P. Medveczky. 2007. The multifunctional latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus, p. 141-153. In J. Minarovits, E. Gonczol, and T. Valyi-Nagy (ed.), Latency strategies of herpesviruses. Springer Science+Business Media, LLC, New York, NY.
- 17.Curreli, F., A. E. Friedman-Kien, and O. Flore. 2005. Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes. J. Clin. Investig. 115:642-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeWire, S. M., and B. Damania. 2005. The latency-associated nuclear antigen of rhesus monkey rhadinovirus inhibits viral replication through repression of Orf50/Rta transcriptional activation. J. Virol. 79:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Micco, R., M. Fumagalli, A. Cicalese, S. Piccinin, P. Gasparini, C. Luise, C. Schurra, M. Garre, P. G. Nuciforo, A. Bensimon, R. Maestro, P. G. Pelicci, and F. d'Adda di Fagagna. 2006. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444:638-642. [DOI] [PubMed] [Google Scholar]
- 21.Duriez, P. J., and G. M. Shah. 1997. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem. Cell Biol. 75:337-349. [PubMed] [Google Scholar]
- 22.Fakhari, F. D., J. H. Jeong, Y. Kanan, and D. P. Dittmer. 2006. The latency-associated nuclear antigen of Kaposi sarcoma-associated herpesvirus induces B cell hyperplasia and lymphoma. J. Clin. Investig. 116:735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 24.Flaño, E., S. M. Husain, J. T. Sample, D. L. Woodland, and M. A. Blackman. 2000. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J. Immunol. 165:1074-1081. [DOI] [PubMed] [Google Scholar]
- 25.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 84:3405-3416. [DOI] [PubMed] [Google Scholar]
- 27.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 28.García, M. A., M. Collado, C. Munoz-Fontela, A. Matheu, L. Marcos-Villar, J. Arroyo, M. Esteban, M. Serrano, and C. Rivas. 2006. Antiviral action of the tumor suppressor ARF. EMBO J. 25:4284-4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.García, M. A., S. Guerra, J. Gil, V. Jimenez, and M. Esteban. 2002. Anti-apoptotic and oncogenic properties of the dsRNA-binding protein of vaccinia virus, E3L. Oncogene 21:8379-8387. [DOI] [PubMed] [Google Scholar]
- 30.Gaspar, M., and T. Shenk. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. USA 103:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris, S. L., and A. J. Levine. 2005. The p53 pathway: positive and negative feedback loops. Oncogene 24:2899-2908. [DOI] [PubMed] [Google Scholar]
- 33.Jault, F. M., J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krug, L. T., V. P. Pozharskaya, Y. Yu, N. Inoue, and M. K. Offermann. 2004. Inhibition of infection and replication of human herpesvirus 8 in microvascular endothelial cells by alpha interferon and phosphonoformic acid. J. Virol. 78:8359-8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 37.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lavin, M. F., and N. Gueven. 2006. The complexity of p53 stabilization and activation. Cell Death Differ. 13:941-950. [DOI] [PubMed] [Google Scholar]
- 39.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, S., I. V. Pavlova, H. W. T. Virgin, and S. H. Speck. 2000. Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 42.Loh, J., Q. Huang, A. M. Petros, D. Nettesheim, L. F. van Dyk, L. Labrada, S. H. Speck, B. Levine, E. T. Olejniczak, and H. W. T. Virgin. 2005. A surface groove essential for viral Bcl-2 function during chronic infection in vivo. PLoS Pathogens. 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe, S. W., and H. E. Ruley. 1993. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 7:535-545. [DOI] [PubMed] [Google Scholar]
- 44.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marques, S., S. Efstathiou, K. G. Smith, M. Haury, and J. P. Simas. 2003. Selective gene expression of latent murine gammaherpesvirus 68 in B lymphocytes. J. Virol. 77:7308-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Guzman, D., T. Rickabaugh, T. T. Wu, H. Brown, S. Cole, M. J. Song, L. Tong, and R. Sun. 2003. Transcription program of murine gammaherpesvirus 68. J. Virol. 77:10488-10503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moorman, N. J., D. O. Willer, and S. H. Speck. 2003. The gammaherpesvirus 68 latency-associated nuclear antigen homolog is critical for the establishment of splenic latency. J. Virol. 77:10295-10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nevins, J. R.. 2001. Cell Transformation by viruses, p. 245-283. In B. N. Fields and D. M. Knipe (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 50.Neyts, J., and E. De Clercq. 1998. In vitro and in vivo inhibition of murine gamma herpesvirus 68 replication by selected antiviral agents. Antimicrob. Agents Chemother. 42:170-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pavlova, I. V., H. W. T. Virgin, and S. H. Speck. 2003. Disruption of gammaherpesvirus 68 gene 50 demonstrates that Rta is essential for virus replication. J. Virol. 77:5731-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petre, C. E., S. H. Sin, and D. P. Dittmer. 2007. Functional p53 signaling in Kaposi's sarcoma-associated herpesvirus lymphomas: implications for therapy. J. Virol. 81:1912-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 54.Reinhardt, H. C., A. S. Aslanian, J. A. Lees, and M. B. Yaffe. 2007. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11:175-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rittling, S. R., and D. T. Denhardt. 1992. p53 mutations in spontaneously immortalized 3T12 but not 3T3 mouse embryo cells. Oncogene 7:935-942. [PubMed] [Google Scholar]
- 56.Rosenke, K., M. A. Samuel, E. T. McDowell, M. A. Toerne, and E. A. Fortunato. 2006. An intact sequence-specific DNA-binding domain is required for human cytomegalovirus-mediated sequestration of p53 and may promote in vivo binding to the viral genome during infection. Virology 348:19-34. [DOI] [PubMed] [Google Scholar]
- 57.Rowland, B. D., S. G. Denissov, S. Douma, H. G. Stunnenberg, R. Bernards, and D. S. Peeper. 2002. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2:55-65. [DOI] [PubMed] [Google Scholar]
- 58.Schäfer, A., D. Lengenfelder, C. Grillhosl, C. Wieser, B. Fleckenstein, and A. Ensser. 2003. The latency-associated nuclear antigen homolog of herpesvirus saimiri inhibits lytic virus replication. J. Virol. 77:5911-5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 60.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336-30341. [DOI] [PubMed] [Google Scholar]
- 61.Si, H., and E. S. Robertson. 2006. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen induces chromosomal instability through inhibition of p53 function. J. Virol. 80:697-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soldani, C., and A. I. Scovassi. 2002. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis 7:321-328. [DOI] [PubMed] [Google Scholar]
- 63.Song, M. J., S. Hwang, W. H. Wong, T. T. Wu, S. Lee, H. I. Liao, and R. Sun. 2005. Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc. Natl. Acad. Sci. USA 102:3805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stewart, J. P., E. J. Usherwood, A. Ross, H. Dyson, and T. Nash. 1998. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J. Exp. Med. 187:1941-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stillman, B. 1986. Functions of the adenovirus E1B tumour antigens. Cancer Surveys 5:389-404. [PubMed] [Google Scholar]
- 66.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66:179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 68.Tsai, H. L., G. H. Kou, S. C. Chen, C. W. Wu, and Y. S. Lin. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J. Biol. Chem. 271:3534-3540. [PubMed] [Google Scholar]
- 69.Upton, J. W., and S. H. Speck. 2006. Evidence for CDK-dependent and CDK-independent functions of the murine gammaherpesvirus 68 v-cyclin. J. Virol. 80:11946-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Upton, J. W., L. F. van Dyk, and S. H. Speck. 2005. Characterization of murine gammaherpesvirus 68 v-cyclin interactions with cellular cdks. Virology 341:271-283. [DOI] [PubMed] [Google Scholar]
- 71.van Dyk, L. F., J. L. Hess, J. D. Katz, M. Jacoby, S. H. Speck, and H. I. Virgin. 1999. The murine gammaherpesvirus 68 v-cyclin gene is an oncogene that promotes cell cycle progression in primary lymphocytes. J. Virol. 73:5110-5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Dyk, L. F., H. W. T. Virgin, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vidal, A., and A. Koff. 2000. Cell-cycle inhibitors: three families united by a common cause. Gene 247:1-15. [DOI] [PubMed] [Google Scholar]
- 74.Virgin, H. W. T., P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 76.Wang, G. H., T. L. Garvey, and J. I. Cohen. 1999. The murine gammaherpesvirus-68 M11 protein inhibits Fas- and TNF-induced apoptosis. J. Gen. Virol. 80:2737-2740. [DOI] [PubMed] [Google Scholar]
- 77.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. I. Virgin. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weck, K. E., S. S. Kim, H. I. Virgin, and S. H. Speck. 1999. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J. Virol. 73:3273-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429-431. [DOI] [PubMed] [Google Scholar]
- 80.Willer, D. O., and S. H. Speck. 2003. Long-term latent murine gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J. Virol. 77:8310-8321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong, L., and G. S. Hayward. 1997. Assembly of complete, functionally active herpes simplex virus DNA replication compartments and recruitment of associated viral and cellular proteins in transient cotransfection assays. J. Virol. 71:3146-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]