Abstract
Foot-and-mouth disease (FMD) is a highly contagious viral disease of cloven-hoofed ungulates that can lead to severe losses in the livestock production and export industries. Although vaccines have been extensively used to control FMD, there is no antiviral therapy available to treat ongoing infections with FMD virus (FMDV). Six peptide-conjugated morpholino oligomers (PPMOs) with sequences complementary to various 21-nucleotide segments of the 5′ and 3′ untranslated regions (UTRs) of the FMDV genome (strain A24 Cruzeiro/Brazil/1955 [A24Cru]) were evaluated in cell cultures. Three of the PPMOs, targeting domain 5 of the internal ribosome entry site (5D PPMO), and the two translation start codon regions (AUG1 and AUG2 PPMOs), showed high levels of anti-FMDV activity. A dose-dependent and sequence-specific reduction in viral titers of greater than 5 log10, with a concomitant reduction of viral protein and RNA expression, was achieved at low micromolar concentrations. Under identical conditions, three other PPMOs targeting the 5′-terminal region of the genome, the cis-acting replication element in the 5′ UTR, and the 3′ “ab” stem-loop showed less dramatic titer reductions of 1.5 log10 to 2 log10. Treatment with 5D PPMO reduced the titers of FMDV strains representing five different serotypes by 2 log10 to 4 log10 compared to those of the controls. A24Cru-infected BHK-21 cells treated repeatedly with 5D or AUG2 PPMO generated resistant viruses for which phenotypic and genotypic properties were defined. Notably, three passages with low concentrations of the AUG1 PPMO extinguished all traces of detectable virus. The results indicate that PPMOs have potential for treating FMDV infections and that they also represent useful tools for studying picornaviral translation and evolution.
Foot-and-mouth disease (FMD) is a highly contagious disease caused by the FMD virus (FMDV) that affects cloven-hoofed ungulates. FMD can be transmitted directly by contact between infected and uninfected animals or indirectly through people, contaminated fomites, or aerosol and can spread rapidly among susceptible animals. The host range of FMDV includes domestic animals such as cattle, swine, sheep, and goats and more than 30 species of wild ruminants. Although mortality associated with FMD is usually low, the disease decreases livestock productivity, and affected countries cannot participate in international trade of animals or animal products. FMDV is the first disease for which the Office International des Epizooties (now World Organisation for Animal Health, OIE) established an official list of pathogen-free zones and countries. In the last decade, costly FMD outbreaks have occurred in Europe (United Kingdom and The Netherlands, 2001), Asia (India and Philippines, 1996; Taiwan, 1997; Korea, 2000 and 2002), South Africa (1999 and 2000), and South America (Brazil and Argentina, 2001), resulting in the slaughter of a large number of animals as well as implementation of quarantine and decontamination procedures (38), with associated socioeconomic ramifications.
FMDV belongs to the Picornaviridae family and is the prototype species of the genus Aphthovirus. Seven distinct FMDV serotypes have been identified worldwide (A, C, O, SAT1, SAT2, SAT3, and Asia1), with multiple subtypes within each serotype. As there is no cross-protection between serotypes, an animal which has been infected with or vaccinated against a virus of one serotype will not be protected against viruses from other serotypes (reviewed in reference 4). Vaccination with vaccines consisting of chemically inactivated whole virus is the primary method of control and eradication used in regions where FMD is endemic. However, vaccination does not protect an individual animal until at least 7 days after administration (15), presenting a practical problem in the event of an outbreak. It is widely agreed that an effective therapeutic agent useful for intervention early in the course of FMD infection of an individual or a herd would be a welcome development.
FMDV possesses a positive-sense, single-stranded RNA genome that contains a single long open reading frame encoding a polypeptide, the polyprotein, flanked by two untranslated regions (UTRs). The 5′ UTR is long, over 1,000 nucleotides (nt), and contains, from 5′ to 3′, a large stem-loop (S fragment), a poly(C) tract, three (or four) repeated pseudoknots, a stem-loop cis-acting replication element (cre), and a type II internal ribosome entry site (IRES) which is involved in cap-independent translation initiation of the viral polyprotein. Either of two AUG start codons may be used for translation initiation of the viral RNA, although the second (AUG2), which is located about 80 nt downstream of AUG1, has been shown to be preferentially utilized (6). The 3′ UTR is short, under 100 nt, but thought to contain cis-acting elements required for efficient genome replication (2).
At least three different nucleic-acid based strategies to intervene in FMDV infections have been described. Sense and antisense RNAs (3, 17, 18, 34), antisense DNA (18), and small interfering RNA (8, 10, 19) have generated various levels of antiviral activity. All of these strategies apparently function by interfering with expression of the viral RNA genome. Phosphorodiamidate morpholino oligomers (PMOs) are a single-stranded-DNA-like antisense structural type containing purine or pyrimidine bases attached to a backbone composed of six-member morpholine rings joined by phosphorodiamidate intersubunit linkages (37). PMOs are water soluble and nuclease resistant and are typically synthesized to about 20 to 25 subunits in length. They duplex with RNA by Watson-Crick base pairing and can interfere with gene expression by sterically blocking cRNA sequence (36). It has been demonstrated that PMO efficacy in a cell-free system (27), in cell cultures (24, 26), and in vivo (13) can be increased considerably by conjugation of arginine-rich peptides to the PMO 5′ terminus (creating peptide-conjugated PMOs [PPMOs]). PPMOs have demonstrated effective and specific suppression of several RNA viruses in cell cultures (9, 13, 14, 20, 28, 40) and against Ebola virus (13) and mouse hepatitis virus (5), both in cell cultures and in vivo. Recently, Yuan et al. (42) found that a PPMO designed against IRES sequence reduced coxsackievirus B3 production in cell cultures and in the heart tissues of infected mice.
This study evaluated the efficacy and specificity of PPMOs in inhibiting the productive replication of FMDV in BHK-21 cells. Six PPMOs were designed to duplex with sequence elements thought to be important in viral RNA synthesis and translation initiation. Three of the PPMOs, targeting sequences in the 3′ portion of the IRES and the two AUG translation initiation regions, produced potent antiviral activities. One of these PPMOs (targeting the IRES) was highly active against multiple FMDV serotypes. Serial exposure of cells to a low concentration of the PPMO directed against the AUG1 region was able to eliminate all traces of the virus.
MATERIALS AND METHODS
Viruses and cell line.
FMDV type A12 strains 119ab (GenBank accession number L11360), A24 Cruzeiro/Brazil/1955 (A24Cru, GenBank accession number AY593768), and SAT-2/Zim/7/83 were derived from infectious cDNA clones pRMC35 (31), pA24Cru (32), and pSAT-2 (41), respectively. FMDV isolates O1 Campos (GenBank accession number AJ320488), O/Taiwan/2/99 (O1 Taiwan, GenBank accession number AJ539136), C3 Resende (GenBank accession number AY593807), Asia1-1pak 54 (GenBank accession number AY593795), and bovine enterovirus type 1 (BEV-1, GenBank accession number D00214) were obtained from Marvin Grubman and Peter Mason. SAT1/SAR/9/81 virus (GenBank accession number AF056511) was provided by Francois Maree (Onderstepoort Veterinary Institute, Exotic Diseases Division, Onderstepoort, South Africa). Baby hamster kidney cells of strain 21 and the clone 13 cell line (BHK-21) were maintained in Eagle's basal medium (BME; Life Technologies, Gaithersburg, MD) supplemented with 10% bovine calf serum (HyClone, Logan, UT), 10% tryptose phosphate broth, and an antibiotic/antimycotic. The cells were grown at 37°C in a humidified 5% CO2 atmosphere.
PPMO design.
PMOs were synthesized at AVI BioPharma, Inc., (Corvallis, OR) by methods previously described (37). All PMOs were covalently conjugated at the 5′ end to the arginine-rich peptide NH2-RRRRRRRRRFFC-CONH2 (R9F2). The methods used for conjugation, purification, and analysis of PPMO compounds were similar to those previously described (26). Six PPMOs, each 21 bases in length, were designed to be complementary to sequences in the FMDV A24Cru genome. The exact PPMO sequences are presented in Table 1, and their relative target locations in the FMDV genome are shown in Fig. 1A. Two PPMOs with non-FMDV sequences were also prepared to control for PPMO-induced non-sequence-specific activities. AUG2scr has the same base composition as PPMO AUG2, but it is rearranged in random order, and DSscr is a random sequence with 50% G/C content. All PPMO sequences were screened by BLAST (NCBI) to detect mammalian mRNA sequences to preclude unintentional gene-silencing effects.
TABLE 1.
PPMO sequences and target locations in the FMDV genomea
| PPMO | PPMO sequence (5′-3′) | Location of PPMO target sequence in FMDV A24Crua | PPMO target region in FMDV |
|---|---|---|---|
| 5+ | AACCCTAGCGCCCCCTTTCAA | 1-21 | 5′ Terminus of genome |
| CRE | CTTAGATCGTGTTTGTACAAG | 569-589 | cis-Acting replication element in 5′ UTR |
| 5D | TTAAAAGAAAGGTGCCGGCCT | 1015-1035 | Domain 5 of the IRES |
| AUG1 | GTGTTCATAAGTCCAGTGTAA | 1036-1056 | First AUG of polyprotein gene |
| AUG2 | GAATTCCATCCTTCCTGTGGC | 1121-1142 | Second AUG of polyprotein gene |
| 3′SL | ACTCCTACGGCGTCGCGCGCC | 8085-8105 | Near end of 3′ UTR |
| AUG2scr | CTCAGCTGTCGTCAGTCTACT | AUG2 scrambled sequence control | |
| DSscr | AGTCTCGACTTGCTACCTCA | Nonsense sequence control |
GenBank accession number AY593768.
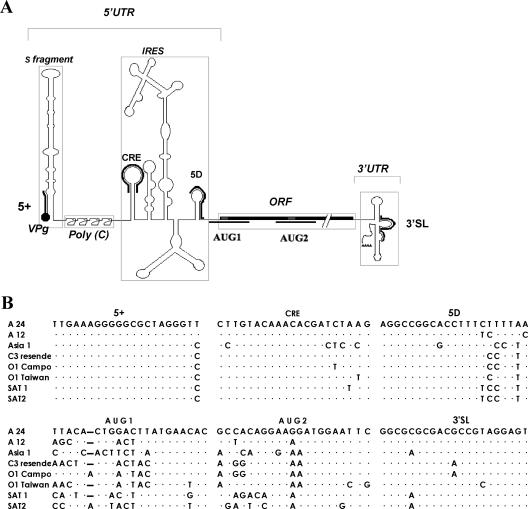
FIG. 1.
(A) Schematic representation of the FMDV genome and relative target locations of the PPMOs used in this study. PPMO names are nonitalicized, and PPMO locations are indicated by lines bordering the FMDV genome. Various FMDV genome regions are boxed and names are italicized. (B) Alignment of PPMO-targeted sequence regions of FMDV. Sequences from six FMDV serotypes (A, O, C, Asia, SAT1, and SAT2) were aligned 5′ to 3′, using Clustal W software. The sequence of WT A24Cru is included as a reference sequence. Dots indicate conserved nucleotides. For the GenBank accession numbers of the various strains, see Materials and Methods.
Cytotoxicity assays.
The viability of the cells after PPMO treatment was determined by using a Vybrant MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] cell proliferation assay kit (Molecular Probes, Inc., Eugene, OR) according to the manufacturer's instructions. Briefly, the MTT assay was performed on uninfected cell monolayers in 12-mm dishes mock treated or treated with PPMO at specified concentrations in BME media (Life Technologies, Grand Island, NY), without serum, for 24 h at 37°C. The cells were rinsed three times with phosphate-buffered saline (PBS), and 10 μl of MTT stock solution was added to each well, followed by 3 h of incubation at 37°C. To dissolve the blue formazan crystals, 50 μl of dimethyl sulfoxide was added to each well, and the optical density was then measured at 570 nm in a BioTek microplate reader. The absorbance values of the PPMO-treated samples were converted to percentages by comparing them to the average value of the mock-treated samples, which was set at 100% survival. An additional measurement of toxicity was performed in parallel with the MTT tests by counting trypan blue-excluding cells with a Neubauer hematocytometer.
Viral growth, inhibition of virus yield, and plaque assays.
To assess the effects of the PPMO on virus replication in BHK-21 cells, monolayers were seeded in triplicate in 12-well plates with 5 × 105 cells/well in BME supplemented with 10% bovine calf serum and grown to ∼90% confluence. Monolayer cells were rinsed three times with PBS before being treated with specified concentrations of PPMO diluted in serum-free BME. After being PPMO- or mock-treated (with BME vehicle) for 3 h, the cells were rinsed and infected with FMDV at a multiplicity of infection (MOI) of 0.5 PFU/cell (unless otherwise indicated) for 1 h, after which the inoculum was removed and the same concentrations of the BME/PPMO mixture used during preinfection were added to the cells. After 24 h at 37°C, the plates were frozen for the subsequent determination of virus titers. For virus growth curves, BHK-21 monolayers were treated with PPMO as described above and then infected with A24Cru at an MOI of 5 to 10 PFU/cell. After 1 h of adsorption at 37°C, the monolayers were rinsed with 25 mM HEPES (N-2-hydroxyethylpiperazine-NÍ-2- ethanesulfonic acid) buffer (pH 5.5) and then twice with PBS, followed by the addition of fresh BME containing no serum along with PPMO at specified concentrations. At various times postinfection (p.i.), viral titers were determined by plaque assays (31), using a 1% gum tragacanth overlay, and the mixture was incubated for 24 h at 37°C. The plates were fixed and stained with crystal violet (0.3% in HistoChoice; AMRESCO, Solon, OH), and the plaques were counted. All assays were performed in triplicate.
Selection of drug-resistant viruses.
BHK-21 cells in six-well culture plates were pretreated with PPMO as described above, the virus inoculum was adsorbed to cells for 1 h at 37°C and then removed, and the cells were rinsed twice with 25 mM HEPES buffer (pH 5.5) and twice with PBS, followed by the addition of fresh BME containing no serum and PPMO at specified concentrations. After 3 days, the cultures were frozen and thawed to make lysates. The lysates were used to infect fresh cell monolayers in culture media containing the specified concentrations of the appropriate PPMOs. These cultures were then frozen and thawed after 3 days, and putative drug resistant viruses were isolated after five cycles of such virus amplification in the presence of PPMO.
Western blot analysis.
BHK-21 cell monolayers in six-well plates were incubated for 3 h with 2.5 μM of specific PPMOs, followed by a 1-h infection period with FMDV A24Cruz at a MOI of 5 PFU/cell, as described above. Five hours p.i., the cells were washed with PBS and lysed with lysis buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 15 mM MgCl2, 1% NP-40, 1% sodium deoxycholate, and 1 mM phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. After clarification, the samples were divided for protein or RNA quantification. The total protein concentrations in the cell extracts were determined by the Coomassie method (Pierce, Rockford, IL) according to the manufacturer's recommendations. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using a 10% NuPAGE precast gel system (Invitrogen, Carlsbad, CA). Ten micrograms of the total proteins from each sample was loaded per lane, followed by transfer of the gel to Immobilon-P (polyvinylidene difluoride) transfer membranes (Millipore, Bedford MA). Viral structural protein VP3 was detected with FMDV-specific polyclonal rabbit sera (generously provided by Marvin Grubman) and secondary goat anti-rabbit horseradish peroxidase conjugate (Pierce). Cellular tubulin, employed as an internal control protein, was detected with a monoclonal antibody (tubulin-alpha AB-2; Lab Vision, Freemont, CA). Binding of the horseradish peroxidase conjugate was detected with a chemiluminescence reagent system (ECL; Amersham, Piscataway, NJ) used according to the manufacturer's instructions and was visualized on X-ray film (X-OMAT; Kodak, NY).
FMDV translation reactions in BHK S10 lysates and radioimmunoprecipitation.
A detailed description of S10 BHK-21 extract preparation will be published elsewhere (E. Rieder). Briefly, translation reactions (20 μl) were performed in S10 BHK-21 extracts programmed with A24Cru RNA transcripts prepared as previously described (30) with [35S]-methionine in the absence or presence of PPMO, followed by 1 h of incubation at 32°C. 35S-labeled proteins were then immunoprecipitated with a rabbit Lpro-specific polyclonal antibody (a gift from M. Grubman, USDA) at a 1:40 dilution, and the precipitated proteins products were separated by SDS-12% PAGE and visualized by autoradiography.
RNA isolation, real-time RT-PCR, and sequencing.
Total RNA was extracted from the supernatants of infected cell cultures, using an RNeasy mini kit (QIAGEN, Valencia, CA), followed by cDNA synthesis using a virus-specific oligonucleotide primer (P15, 5′-GGCGGCCGCTTTTTTTTTTTTTTT-3′) and a SuperScript III first-stand synthesis system for reverse transcriptase (RT)-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Real-time PCRs were carried out using AmpliTaq Gold (Applied Biosystems, Foster City, CA) with the following primers: 5′-TCRRNCACTGGTGACAGGCTAAG-3′ and 5′-CCCCTTCTCAGATCCCGAGT-3′ (forward and reverse primers, respectively). For standard PCRs, a Taq DNA polymerase provided with an Advantage 2 PCR enzyme system (Clontech, Palo Alto, CA) was used, and the amplicons were purified by a StrataPrep PCR purification kit (Stratagene). A TaqMan probe (ATGCCCTTCAGGTACC) labeled with fluorescent dye was used to detect the amplicons with an ABI PRISM 7700 detection system. The amplicons were sequenced using gene-specific primers, BigDye terminator cycle sequencing kits (Applied Biosystems, Foster City, CA), and a PRISM 3700 automated sequencer (Applied Biosystems). The primers and probes were designed by using Primer Express software (Applied Biosystems, Foster City, CA). 18S rRNA was used as an internal standard control with primers and probes from TaqMan rRNA control reagent kits (Applied Biosystems, Foster City, CA). Relative RNA quantities were calculated by using the threshold cycle (ΔΔCT) RNA measurement method (ABI user bulletin number 2, Applied Biosystems), normalized against that of 18S rRNA and calibrated against those of the mock-treated uninfected control samples.
Software.
Multiple sequence alignments of FMDV were performed with CLUSTALW 1.7 (39). Analyses of the PPMO sequence design in relation to that of FMDV were done with EditSeq and PrimerSelect, provided by DNASTAR, Inc., Madison, Wisconsin. For statistical analysis, the Student's t test was performed with Microsoft Excel software.
RESULTS
PPMO design.
Six PPMOs were designed to target sequences in the following five regions of positive-strand FMDV genomic RNA (PPMO name[s] follow in parentheses): (i) the first 21 nucleotides of the A24Cru genome (5+), (ii) the cre region in the 5′ UTR (CRE), (iii) the IRES 5D element in the 5′ UTR (5D), (iv) the sequence spanning either of the two AUG start codons of the FMDV polyprotein (AUG1 and AUG2), and (v) the sequence linking the two stem-loop structures found in the 3′-terminal region (3′SL). PPMO 5+, CRE, and 3′SL were designed to duplex with sequences previously described as being involved in RNA synthesis (2, 23). Sequence alignments of viral strains belonging to various FMDV serotypes show that the target sequences of these three PPMOs are highly conserved (Fig. 1B). Of the PPMOs directed against RNA elements involved in viral translation initiation (5D, AUG1, and AUG2), the 5D target sequence is fairly well-conserved, while the AUG1 and AUG2 PPMOs have poorly conserved targets across FMDV serotypes.
PPMO cytotoxicity evaluation.
All PPMOs were first evaluated for their effects on cell viability by MTT assays. BHK-21 cell monolayers were treated with PPMO at incremental concentrations between 1 and 15 μM in the absence of the virus in serum-free media (data not shown) and under conditions similar to those in the antiviral experiments described below. Serum-free media is necessary for efficient uptake of PPMOs containing an R9F2 peptide; therefore, this condition was employed for all PPMO cell culture experiments in this study. Cell viability was over 80% in the presence of PPMO at concentrations of 5 μM or less (see below). As considerable cytotoxicity was observed in this assay when PPMO concentrations of over 5 μM were used, subsequent experiments were carried out at 5 μM or less.
Identification of effective PPMO inhibitors of FMDV replication.
In an initial dose-response evaluation by plaque assay, all of the FMDV-specific PPMOs produced some reduction in A24Cru titers. As shown in Fig. 2A, PPMOs 5+, CRE, and 3′SL showed moderate activity, with a dose-responsive effect of up to a 2 log10 reduction at 5 μM. Remarkably, all three of the PPMOs directed to translation-associated targets, 5D, AUG1, and AUG2, produced potent dose-dependent inhibitions, reaching a maximum titer reduction of greater than 6 log10 at a concentration of 5 μM. The most-potent inhibition was produced by AUG1 and AUG2 PPMOs. Inhibition by all of the above FMDV-targeted PPMOs was apparently sequence-specific, as neither DSscr nor the AUG2scr control PPMOs inhibited viral plaque formation at any concentration tested (Fig. 2A). Further evidence of sequence specificity was obtained by MTT cell-viability assays on uninfected cells treated with the various PPMOs (Fig. 2B) at the same concentrations and under the same experimental conditions as the antiviral experiment of Fig. 2A. None of the PPMOs used in the study reduced cell viability by more than about 20% at 5 μM, and most of the viability readings were well above that (Fig. 2B), indicating that the inhibitory effect of PPMO observed in the antiviral assays was not due to nonspecific effects on cellular health and the ability to produce virus.
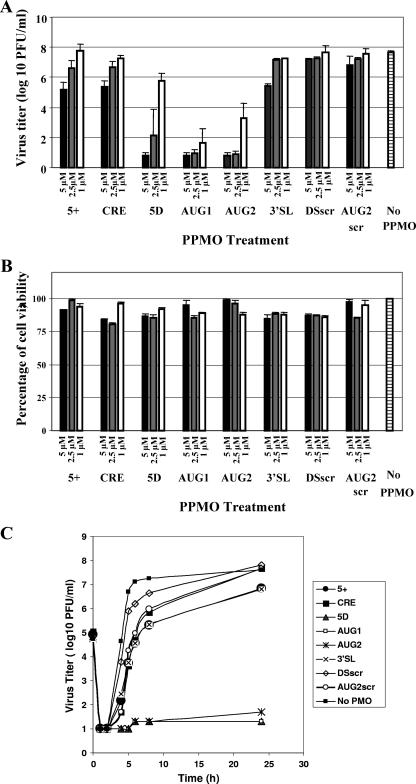
FIG. 2.
Determination of FMDV titers after PPMO treatment. (A) Dose response of viral titers in the presence or absence of PPMO. Cells were treated with PPMO or mock treated for 1 h before inoculation with A24Cru at 0.5 PFU/cell in serum-free media. Following 24 h of incubation under further PPMO treatment, the virus yield was determined by plaque assay and expressed as PFU/ml. Each histogram presents the mean PFU/ml of four samples, and the corresponding standard deviations are shown as error bars. The data from one representative experiment are shown. (B) Uninfected BHK-21 cells were treated with the indicated PPMO at various concentrations under the same experimental conditions as for panel A. Values are presented as percentages of viable cells compared to that of the mock-treated controls, with corresponding standard deviations. (C) Twenty-four hour one-step growth curves of FMDV A24Cru from cells subjected to treatment with a 5 μM concentration of the indicated PPMO. Cell monolayers were mock- or PPMO-treated and infected with A24Cru at a MOI of 5 PFU/cell. The procedures used for PPMO treatment, viral infection, and titration of infectivity are described in Materials and Methods. Each value represents the mean of duplicate assays.
The above highly positive results prompted us to examine the kinetics of PPMO inhibition in a 24-h one-step growth curve, using a high MOI of 5 and PPMO treatment. While the virus titer of the mock-treated cultures reached a plateau at about 6 to 8 h p.i., the DSscr-, AUG2scr-, and CRE-treated cultures displayed delayed virus growth kinetics at early time points but at 24 h p.i. reached about the same titers as that of the mock-treated culture. In agreement with results observed in the antiviral plaque assays described above, cultures treated with 5 μM of AUG1, AUG2, and 5D PPMOs had titers 5 log10 to 6 log10 below that of the controls and exhibited little increase in titers over the 24 h of this experiment (Fig. 2C). The 5+- and 3SL-treated samples showed modest levels of inhibition, with about 1 log reduction in titers compared to that of the controls at 24 h p.i.
Challenge of multiple FMDV serotypes.
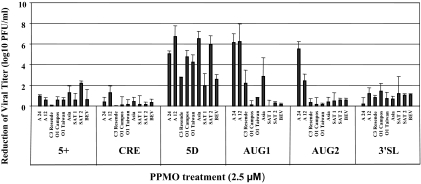
Next, we investigated whether any of the PPMOs could inhibit FMDV serotypes other than type A. We carried out single-dose challenges at 2.5 μM against eight strains of FMDV (Fig. 1B) representing six different serotypes, and the heterologous bovine enterovirus (BEV), and evaluated inhibition levels by plaque assays (Fig. 3). BEV is a picornavirus belonging to the Enterovirus genus (12) and has little sequence similarity with any FMDV strain at the various PPMO target sites (BLAST search analysis, not shown). As shown in Fig. 3, PPMO AUG1 inhibited the growth of three FMDV serotypes: both type A viruses (A12 and A24Cru) by over 6 log10 and type C (C3 Resende) and Asia1 serotype viruses by over 2 log10, compared to that of the AUG1-treated BEV control. However, we note that the pattern of relative inhibition by AUG1 PPMO is not absolutely correlative with sequence agreement. For instance, the sequences of A24Cru and A12 differ considerably at the AUG1 target site (Fig. 1B), yet both strains were strongly inhibited by AUG1 PPMO. The AUG2 compound showed inhibitory activity against only serotype A viruses, an unsurprising result based on the number of sequence mismatches between the AUG2 PPMO and non-serotype A strains at this target site (Fig. 1B). None of the AUG2, AUG1, 5+, CRE, or 3′SL PPMOs significantly reduced BEV titers, providing further indication of the specificity of their inhibitions of FMDV in the various antiviral experiments in this study. Treatment with PPMO 5D resulted in titer reductions from 2 log10 to 7 log10 at 24 h postinfection of the various FMDV serotypes tested, including South African (SAT), Asian, and South American (C3 Resende and O1 Campos) strains (Fig. 3). These results were somewhat surprising, given the number of sequence mismatches (2-4) between the 5D PPMO and its target sites in the various strains (Fig. 1B). Also difficult to explain was that 5D was far more effective against SAT2 than SAT1 despite identical sequences at the 5D target site of these two strains. Further, the 5D PPMO inhibited BEV titers by approximately 2 log10. As sequence agreement between the 5D PPMO and BEV at this target site is not high, this observed partial inhibition is also difficult to reconcile, and additional studies will be required to further characterize this particular PPMO and investigate its specificity. Future efforts to help clarify the above observations could include using a PPMO delivery-peptide composition less prone to nonspecificity than the R9F2 peptide used here, on the same 5D PMO sequence, and/or designing PPMOs that target somewhat different sequences in domain 5 of the FMDV IRES. Despite the peculiarities of some of the data, the results indicate that AUG1 and AUG2 PPMOs work dependably against the type A FMDV strains. The efficacy of PPMO 5D against multiple serotypes indicates that it may have the potential to be broadly applicable; however, further study is required.
FIG. 3.
Effect of PPMO treatment on infectivity of multiple FMDV serotypes and BEV. Inhibition levels are presented relative to that of the mock-treated control for each viral strain and were calculated as 100 × [(virus titer in the presence of PPMO)/(virus titer in the absence of PPMO)]. Results are the mean values from two independent experiments, and the error bars represent the standard deviations.
Effects of PPMO on viral protein expression and RNA synthesis in BHK-21 cells.
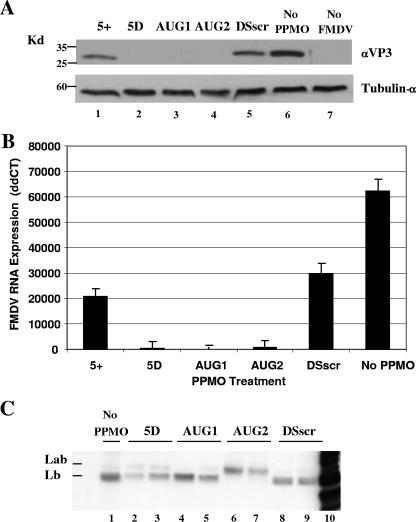
In an attempt to elucidate the mechanism of action of the more-effective PPMOs, three exploratory experiments were performed. First, expression of viral proteins in PPMO- and mock-treated FMDV-infected cells was examined by Western immunoblotting. As shown in Fig. 4A, the 5+ PPMO treatments resulted in only a moderate reduction, whereas the 5D, AUG1, and AUG2 PPMO treatments caused profound reductions in viral protein expression levels compared to those of both the DSscr- and mock-treated controls. The same Western blot membrane was reprobed for cellular tubulin-α (Fig. 4A), showing that similar amounts of cell extract were analyzed for each sample.
FIG. 4.
Effect of PPMO treatment on viral protein and RNA expression. (A) Western blot prepared from cell lysates obtained from BHK-21 cells treated with 2.5 μM of the indicated PPMO. Cell monolayers were PPMO-treated for 3 h and infected with FMDV A24Cru at a MOI of 5 PFU/cell for 1 h, followed by a further 4 h of incubation in a PPMO mixture before cell lysates were prepared (see Materials and Methods). Ten micrograms of total protein was loaded per lane, and the blot was probed with a polyclonal antibody for FMDV VP3 (upper panel) or a monoclonal antibody against tubulin-α (lower panel). (B) Quantitative real-time PCR measurement of FMDV RNA isolated from cell lysates corresponding to those in the above experiment. Values are reported as ΔΔCT, normalized to that of endogenous 18S rRNA and calibrated against mock-treated uninfected control cell lysates. The y axis indicates ΔΔCT (log2 relative units). Each error bar refers to the standard deviation for the respective mean data point (n = 3). Similar results were obtained in at least two independent experiments. (C) FMDV translation products produced in BHK-21 S10 lysates in the presence or absence of PPMO. Translation reactions were programmed with full-length A24Cru RNA in the presence of [35S]-methionine and immunoprecipitated with a polyclonal antibody for Leader protein. PPMOs were included in the translation reactions at concentrations of 0 μM (lane 1), 0.25 μM (lanes 3, 4, 6, and 8), and 2.5 μM (lanes 2, 5, 7, and 9). Lane 10 contains non-PPMO-treated and nonimmunoprecipitated viral RNA translation products. The products were examined by SDS-PAGE on a 12% gel.
To investigate the relationship between PPMO antiviral activity and viral RNA production, quantitative real-time RT-PCR was carried out on samples from the same experiment for viral protein expression by Western blotting described above. A CT measurement method was employed to quantify viral RNA levels. As shown in Fig. 4B, all PPMO-treated samples yielded detectable quantities of FMDV RNA. At 3 h p.i., treatment with 2.5 μM 5D, AUG1, or AUG2 PPMO generated ΔΔCT levels of 564, 91, and 910, indicating a 110-, 686-, or 69-fold reduction, respectively, in the level of viral RNA expression compared to that of the control sample without PPMO (ΔΔCT = 62,432). The 5+- and DSscr-treated samples reduced viral RNA levels three- and twofold (ΔΔCT = 20,882 and 29,532), respectively.
To evaluate the effect of various PPMOs on the translation of the FMDV polyprotein, we performed in vitro translations of A24Cru RNA in BHK-21 S10 extract. Translation reactions in the presence of [35S]-methionine were performed in S10 extracts programmed with full-length A24Cru RNA transcripts with or without PPMO. Radiolabeled proteins were then immunoprecipitated and resolved by SDS-PAGE (Fig. 4C). In the presence of 0.25 or 2.5 μM AUG2 PPMO, a retardation of leader protein migration was observed. The migration of the AUG2 PPMO-treated sample was consistent with that of a protein that had initiated about 80 nt upstream, at the first AUG of Lpro (AUG1, Lab protein). The migrations of translation reactions carried out in the absence of treatment or treatment with AUG1 or 5D PPMO indicate that translation initiated predominantly at AUG2 of the Lpro.
Selection and analysis of PPMO-resistant FMDV.
The ability of viruses in an FMDV population to survive treatment with the three most efficacious PPMOs (5D, AUG1, and AUG2) was investigated by serial passages of A24Cru virus in the presence of gradually increasing concentrations of PPMO (Fig. 5) and quantification by plaque assay. The first two passages employed PPMO at a concentration of 0.5 μM, followed by two passages at 1 μM and a fifth passage (p5) at 2.5 μM. This process was carried out independently with each PPMO twice and with mock-treated cells in triplicate (Fig. 5A). After the fifth passage, plaque size and morphology generated by the 5D- or AUG2-treated (R-5D/p5 and R-AUG2/p5) FMDV-resistant mutant (Rm) viruses were found to be indistinguishable from those of the wild type (WT) virus or A24Cru receiving no PPMO treatment (No-PMO/p5) (data not shown). The effects of various concentrations of PPMO on the growth of R-5D/p5 and R-AUG2/p5 compared to that of WT virus is shown in Fig. 5B. Both R-5D/p5 and R-AUG2/p5 displayed clear advantages in growth over that of the WT virus in the presence of all concentrations of PPMO tested and in a dose-dependent manner (Fig. 5B). To further characterize these Rm variants, sequence analyses were performed on nonpassaged (WT) virus, No-PMO/p5, and the passaged viruses R-5D/p5 and R-AUG2/p5, including plaque-purified viruses (Table 2). After five passages on BHK-21 cells, No-PMO/p5 displayed no nucleotide changes. In contrast, R-AUG2/p5 contained nucleotide changes within the PPMO target site resulting in two nonsynonymous substitutions in Lpro (Table 2). Specifically, a transition at nucleotide 1156 (G to C) caused an amino acid change of glycine to arginine at amino acid residue 27 (G27 to R), whereas a transition at position 1166 (A to C) changed a glutamic acid to alanine at residue 30 (E30 to A) (Table 2). Sequence analyses were also performed on 15 and 10 independently plaque-purified viruses derived from R-AUG2/p5 and R-5D/p5, respectively. The results obtained (Table 2) show that quasispecies present in R-AUG2/p5 produced at least six different sequence profiles within the target region for AUG2 PPMO (Table 2, compare R-AUG2/p5 clones to noncloned R-AUG2/p5 and No-PPMO/p5). A similar analysis of R-5D/p5 virus (Table 2) shows variations at nucleotides 1052 (R = A/C) and 1059 (Y = T/C), which fall inside the 5D PPMO target site. We assume that these genetic modifications in the viral population contributed to the PPMO treatment resistance observed in the R-5D/p5 mutants. In 10 plaque purified viruses derived from R-5D/P5, five different mutant profiles were found (Table 2).
FIG. 5.
Passage history, derivation, and characterization of Rm viruses isolated in this study. (A) Virus passages in the presence or absence of PPMO are indicated by vertical lines; “P” denotes the passage number, and “bp” denotes the blind-passage number. For each passage, 2 × 106 cells were infected at a MOI of 0.5 PFU/cell. A24Cru plaque formation was drastically reduced with AUG1 PPMO treatment, and undiluted cell lysates of passage 3 caused no CPE when applied to fresh cells. After five passages in the presence or absence of PPMO, cell lysates corresponding to the R-AUG2/p5, R-5D/p5, and No-PPMO viruses were analyzed by plaque assay and RT-PCR (see explanation of panel C below). Analyses of two independent experiments with PPMO- and No-PPMO-treated cells produced consistent results. (B) Effects of subsequent PPMO treatment on WT and PPMO-treated passage 5 R-AUG2/p5 and R- 5D/p5 viruses. The inhibition value in the presence of PPMO is relative to that of the mock-treated control in each case and was calculated as described in the legend to Fig. 4. The results are the means from two experiments, and the error bars represent the standard deviation. (C) PCR gel from RT-PCRs of PPMO-treated or untreated viruses. Cells were infected with A24Cru at a MOI of 0.5 PFU/cell in the presence of increasing amounts (0.5 to 1 μM) of PPMO AUG1. PCR products were amplified from cDNA synthesized from WT unpassaged virus, first and third passage virus produced under selection with AUG1 PPMO, and cells of each of three consecutive blind passages of lysates from passage 3 AUG1 PPMO-treated cells. The results show one representative experiment. Amplified products were visualized by ethidium bromide staining on 2% agarose gels. WT A24Cru virus yielded a single band migrating at 1 kb (lane A) amplified with sense (5′-[C]17TAAGTTCT-3′) and antisense (5′-TGCCGTCCACCATGCACACCTCGCT-3′) primers. Lanes: B, A24Cru passaged five times in the absence of PPMO; C and D, first and third passages of A24Cru under PPMO AUG1treatment; E, F, and G, cell lysates from cells subjected to three rounds of treatment with AUG1/p3 cell lysate; first and last lanes, 1-kb DNA ladder (Invitrogen); H, reverse transcript control provided by the Superscript III first-strand synthesis system for RT-PCR (Invitrogen, Carlsbad, CA); I, PCR control provided by the Advantage 2 PCR enzyme system (Clontech, Palo Alto, CA); J, a no-template PCR control reaction. Neg, negative. (D) Hairpin loop structure (ΔG at −12.2) corresponding to FMDV IRES domain 5, generated by mfold (43), with observed mutations indicated by arrows.
TABLE 2.
Comparison of PPMO-resistant A24Cruz virus recovered from PPMO-treated BHK-21 cells
| Virus source | Passage history (clone no.)a | Locations of mutant nucleotidesb | Nucleotide sequenceb,c | Frequencyd |
|---|---|---|---|---|
| Parental A24virus (WT) | P0 | GCCACAGGAAGGATGGAATTCAe | ||
| R-AUG2/p5 | P5 | 1127, 1137 | ------C---------C----- | |
| R-AUG21-7 | P5 (clones 1-7) | 1122, 1126-1128, 1136-1138 | -A---GCU-------CCC---- | 7/15 |
| R-AUG28 | P5 (clone 8) | 1123, 1131, 1137 | --T-------C-----C----- | 1/15 |
| R-AUG29-10 | P5 (clones 9, 10) | 1123, 1127, 1137 | --T---C---------C----- | 2/15 |
| R-AUG211 | P5 (clone 11) | 1123, 11310, 1137 | --T-------A-----C----- | 1/15 |
| R-AUG212-13-14 | P5 (clones 12-14) | 1126, 1137 | -----C----------C----- | 3/15 |
| R-AUG215 | P5 (clone 15) | 1123, 1127 | --T---C--------------- | 1/15 |
| No-PPMO1-3 | P5 (clones 1-3) | --------------------- | 3/3 | |
| Parental A24 virus (WT) | P0 | AGGCCGGCACCTTTCTTTTAAf | ||
| R-5D/p5 | P5/5D | 1023, 1030 | --------R------Y----- | |
| R-5D1-5 | P5/clones 1-5 | 1023, 1030 | --------G------C----- | 5/9 |
| R-5D6 | P5/clone 6 | 1032 | -----------------C--- | 1/9 |
| R-5D7 | P5/clone 7 | 1017 | --C------------------ | 1/9 |
| R-5D8 | P5/clone 8 | 1017, 1022 | --C----T------------- | 1/9 |
| R-5D9 | P5/clone 9 | 1018 | ---A----------------- | 1/9 |
| No-PPMO1-3 | P5/clones 1-3 | --------------------- | 3/3 |
FMDV A24Cru WT viruses were passaged and selected in the presence of the indicated PPMO, as described in Materials and Methods and Results. Infections were repeated twice with each PPMO treatment and in triplicate for the No-PPMO controls. P0, unpassaged clone; P5, clone passaged five times in the presence of the indicated PPMO; clone no., plaque-purified clone producing a particular mutation profile. Unpurified virus stocks are shown in bold.
Numbering is based on the complete FMDV A24Cru sequence (GenBank accession number AY593768).
Sequence data corresponding to cDNAs of individual plaque-purified viruses derived from either R-AUG2/p5, R-5D/p5, or No-PPMO/p5. R, A or G; Y, C or T.
Frequency of each particular resistant mutant virus per total number of mutant viruses from indicated PPMO-treated cultures.
Nucleotides 1121 to 1142.
Nucleotides 1015 to 1035.
An interesting pattern emerged from repeated exposure of A24Cru virus to AUG1 PPMO. Two virus passages with AUG1 PPMO at 0.5 μM followed by one passage at 1 μM resulted in no observable cytopathology, indicating that no viable viruses were present at passage 3 (Fig. 5A). This experiment was repeated three times with identical results (Fig. 5A). Consistent with the lack of a viral-induced cytopathic effect was the complete absence of plaque formation observed after passage 3 with AUG1 PPMO (Fig. 5A), suggesting that the virus population had been extinguished. To further investigate whether the above treatment had indeed caused viral extinction, two more strategies to determine if any virus had survived treatment were employed. First, 1/3 of the cell culture supernatant harvested after the third passage under AUG1 treatment was used to infect fresh BHK-21 monolayers for 72 h at 37°C, and two additional blind passages carried out in the same manner in the absence of PPMO were then performed. In these experiments, we allowed infections to proceed for 72 h prior to determining the titers by plaque assay. As shown in Fig. 5A, three blind passages of AUG1/p3, performed in triplicate, produced AUG1/p3/bp3[a,b,c] with no detectable viral infectivity. Second, RT-PCR was used to assay for the presence of FMDV RNA in passage 1 and passage 3 cells treated with AUG1 PPMO, as well as in each of the three blind-passage derivatives (Fig. 5C). RNA extracted from BHK-21 cell lysates corresponding to AUG1/p1 produced an amplicon visible by agarose gel electrophoresis, although the intensity of the band was far weaker than that of the untreated and No-PMO/p5 A24 controls (Fig. 5C, compare lane C to lanes A and B). Remarkably, no PCR product was detected from either AUG1/p3 or the three blind passages (Fig. 5C, compare lanes D, E, F, and G to lanes A and B). Together, these results show that, under the conditions of our experiment, treatment of cells with 1 μM or less of AUG1 PPMO during three consecutive passages eliminated any detectable FMDV.
DISCUSSION
In this study, we investigated the effects of PPMO on replication of FMDV in cell cultures. We used PPMO designed to duplex with genetic elements in the 5′ and 3′ ends of the FMDV genome thought to be essential for viral RNA synthesis or translation.
Two PPMO compounds complementary to the first and second functional AUG initiation codons of the FMDV polyprotein, AUG1 and AUG2, generated a 6-log10 specific decrease in viral titer and provided protection from two different strains within FMDV serotype A (A24 and A12) (Fig. 3). A low level of sequence agreement between serotypes in the region containing the first and second AUG codons precluded efficacy of these two PPMO against other serotypes.
PPMO 5D was also potent, generating a 5-log10 specific reduction of a serotype A strain (Fig. 2A). 5D was also able to generate 2 log10 to 4 log10 titer reductions of strains from four other FMDV serotypes (Fig. 3), compared to its effect on the heterologous BEV. The level of inhibition generated by each of the most active PPMOs (5D>AUG1>AUG2) against multiple FMDV serotypes reflected the degree of agreement in the respective PPMO target site sequences between the six serotypes challenged (Fig. 1B). PPMO 5D did cause some nonspecific inhibition of BEV-1 growth for reasons that remain unclear. 5D PPMO did not appear inordinately cytotoxic as determined by MTT assay (Fig. 2B and data not shown). Examination of BEV RNA by BLAST search did not reveal any of the 21 nucleotide regions with high levels of agreement with PPMO 5D. It is most likely that the cytotoxicity generated by all of the PPMOs in this study at concentrations over 5 μM was due to the peptide component of the PPMO. We note the recent development of an alternate arginine-rich peptide (often named P7) (5, 9, 42) shown to be equivalent to the R9F2 peptide used in this study in its ability to deliver PMO into cells yet less toxic (1), more stable (27), and less affected by serum (9). Clearly, improvements of this nature will contribute to the utility of PPMO technology.
PPMO inhibition of FMDV titers was, largely at least, sequence specific, dose-responsive, and effective at both low (0.5) and high (5 to 10) MOIs (Fig. 2 and 3). Overall, the three methods employed for evaluating FMDV inhibition by PPMO, plaque formation, VP3 protein expression, and genomic RNA synthesis, generated results that were consistent with one another (Fig. 2 and 4).
We found that the PPMO-targeting regions of the FMDV genome involved in translation initiation (5D, AUG1, and AUG2) were highly efficacious, whereas those targeting the more highly conserved sequences associated with regulation of RNA synthesis (5+, CRE, and 3′SL) were markedly less so (Fig. 2A). These different efficacies may reflect the levels of accessibility of the various target locations to PPMO hybridization and/or the ability of PPMO, once hybridized, to interfere in events of the viral life cycle associated with the particular RNA target locations. In vitro translation experiments showing that when AUG2 PPMO is present, FMDV polyprotein synthesis is shifted and starts at the AUG1 codon (Fig. 4C) clearly demonstrate that PPMO can act by affecting translation. This result is consistent with studies showing that FMDV preferentially selects the AUG2 over the AUG1 start site (6). Other reports have also noted that 5′ UTR- and AUG region-targeted PPMOs generally have a higher likelihood of efficacy than PPMOs targeted against other locations in the genomes of positive-strand RNA viruses (29, 40, 42) and have implicated inhibition of translation as a likely mechanism of action. Yuan et al. (42) documented that a PPMO targeted to the AUG region of coxsackievirus B3 was ineffective, whereas PPMOs targeted to the stem-loop V region were highly effective. By comparison, in our study PPMOs targeted to the FMDV AUG regions were highly effective. The different results suggest agreement with the current belief that during translation initiation, the 40S ribosomal subunit joins viral RNA in the vicinity of stem-loop V in the picornaviral type I IRES, but at the AUG region in type II IRES (35).
Our results agree with those of Gutíerrez et al. (18), who investigated antisense oligonucleotides directed against each of the two functional FMDV AUGs and other targets implicated in viral translation regulation. Their study showed that microinjection of antisense DNA or RNA targeting the AUG2 region into the cytoplasms of BHK-21 cells caused transient inhibition of FMDV infections. The magnitude of viral inhibition produced by FMDV-specific PPMO compares quite favorably with results from studies with other nucleic-acid-based agents evaluated against FMDV, such as small interfering RNA (8, 10, 19, 21, 25), antisense DNA (18), and antisense RNA (3, 17).
Viral populations partially resistant to inhibition by AUG2 or 5D PPMO were produced by serial passaging of FMDV A24Cru in cell cultures under the pressures of PPMO treatment. Mutations present in the PPMO target regions (Table 2), and the reduced virus sensitivity to the relevant PPMO (Fig. 5B), strongly suggesting that PPMO treatment contributed to amplification of the FMDV mutants that were isolated. The lack of any such mutations in the WT or No-PPMO/p5 viruses sequenced also contributes to such a conclusion. As the AUG2 and 5D PPMO target sites are located in regions of secondary structure considered important to viral functions, we presume that mutants that escaped from these PPMOs were able to maintain typical RNA structural conformation while limiting the ability of PPMO to hybridize efficiently. Note that an Asia1 (GenBank accession number A4593797) strain and O1 strain HLJOC12/03 (GenBank accession number DQ119643) contain the two predominant mutations (AGGCCGGCGCCTTTCCTTTAA; changes are indicated in bold) found in R/A24-5D variants. Figure 5D shows an mfold prediction of the RNA secondary structure of the IRES domain 5 hairpin where these two mutations are found and provides an indication that neither mutation would disrupt the overall configuration of this region. Similarly, the R27 residue found in R-AUG2/p5 Lpro is also present in SAT1 (GenBank accession number AY593839), SAT2 (GenBank accession number NC003992), and SAT3 (GenBank accession number AY593850) viruses. Alteration of residue 30, as in our E30A R-AUG2/p5 mutant, although rarely found among FMDV isolates (7), has been previously reported. Guarne et al. (16) reported a K residue instead of the typical E residue in Lpro following the second AUG in a SAT2 sequence. Our ability to generate numerous escape mutants in PPMO-treated cell cultures in this study is consistent with the high adaptability of FMDV to selective pressure previously observed in vitro (30, 33) and in vivo (11, 22) and provides further evidence that FMDV possesses a remarkable potential for genetic variation, likely through positive selection and random drift acting on heterogeneous viral populations (11).
AUG1 was the most efficacious PPMO in this study, generating a 6-log10 reduction in viral titers when present at 1 μM (Fig. 2A). Furthermore, when present at 1 μM or less for only three passages, it was capable of reducing FMDV infectivity to below detectable levels. This finding suggests that mutations caused by AUG1 PPMO result in nonviable progeny and that this PPMO represents a strong candidate for further development as a potential drug against the A serotypes.
This study, at the very least, provides targeting guidance for PPMO against FMDV. Considering the agricultural and socioeconomic importance of FMD and the current need for early disease interventions, the results herein suggest that PPMO could be considered a candidate technology for therapeutic development against FMD. Further modifications of the composition of the peptide component of the PPMO seem advisable, to attempt to reduce the potential toxicity. Once the toxicity profile is acceptable, PPMO warrants consideration for evaluation in an FMDV animal model.
Acknowledgments
We thank Lisa Aschenbrenner and Robert Blouch for excellent technical assistance and the chemistry group at AVI BioPharma for the expert production of all PPMOs used in this study.
Ariel Vagnozzi was supported by the Plum Island Animal Disease Center Research Participation Program administrated by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and U.S. Department of Agriculture. This work was supported by the U.S. Department of Agriculture, Agricultural Research Service, through CRIS project no. 1940-32000-035-00D.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Abes, S., H. M. Moulton, P. Clair, P. Prevot, D. S. Youngblood, R. P. Wu, P. L. Iversen, and B. Lebleu. 2006. Vectorization of morpholino oligomers by the (R-Ahx-R)4 peptide allows efficient splicing correction in the absence of endosomolytic agents. J. Control Release 116:304-313. [DOI] [PubMed] [Google Scholar]
- 2.Agol, V. I., A. V. Paul, and E. Wimmer. 1999. Paradoxes of the replication of picornaviral genomes. Virus Res. 62:129-147. [DOI] [PubMed] [Google Scholar]
- 3.Bigeriego, P., M. F. Rosas, E. Zamora, E. Martinez-Salas, and F. Sobrino. 1999. Heterotypic inhibition of foot-and-mouth disease virus infection by combinations of RNA transcripts corresponding to the 5′ and 3′ regions. Antivir. Res. 44:133-141. [DOI] [PubMed] [Google Scholar]
- 4.Brown, F. 2003. The history of research in foot-and-mouth disease. Virus Res. 91:3-7. [DOI] [PubMed] [Google Scholar]
- 5.Burrer, R., B. W. Neuman, J. P. Ting, D. A. Stein, H. M. Moulton, P. L. Iversen, P. Kuhn, and M. J. Buchmeier. 2007. Antiviral effects of antisense morpholino oligomers in murine coronavirus infection models. J. Virol. 81:5637-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, X., I. E. Bergmann, R. Fullkrug, and E. Beck. 1995. Functional analysis of the two alternative translation initiation sites of foot-and-mouth disease virus. J. Virol. 69:560-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrillo, C., E. R. Tulman, G. Delhon, Z. Lu, A. Carreno, A. Vagnozzi, G. F. Kutish, and D. L. Rock. 2005. Comparative genomics of foot-and-mouth disease virus. J. Virol. 79:6487-6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, W., W. Yan, Q. Du, L. Fei, M. Liu, Z. Ni, Z. Sheng, and Z. Zheng. 2004. RNA interference targeting VP1 inhibits foot-and-mouth disease virus replication in BHK-21 cells and suckling mice. J. Virol. 78:6900-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deas, T. S., I. Binduga-Gajewska, M. Tilgner, P. Ren, D. A. Stein, H. M. Moulton, P. L. Iversen, E. B. Kauffman, L. D. Kramer, and P. Y. Shi. 2005. Inhibition of flavivirus infections by antisense oligomers specifically suppressing viral translation and RNA replication. J. Virol. 79:4599-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de los Santos, T., Q. Wu, S. de Avila Botton, and M. J. Grubman. 2005. Short hairpin RNA targeted to the highly conserved 2B nonstructural protein coding region inhibits replication of multiple serotypes of foot-and-mouth disease virus. Virology 335:222-231. [DOI] [PubMed] [Google Scholar]
- 11.Domingo, E., C. Escarmis, E. Baranowski, C. M. Ruiz-Jarabo, E. Carrillo, J. I. Nunez, and F. Sobrino. 2003. Evolution of foot-and-mouth disease virus. Virus Res. 91:47-63. [DOI] [PubMed] [Google Scholar]
- 12.Earle, J. A., R. A. Skuce, C. S. Fleming, E. M. Hoey, and S. J. Martin. 1988. The complete nucleotide sequence of a bovine enterovirus. J. Gen. Virol. 69:253-263. [DOI] [PubMed] [Google Scholar]
- 13.Enterlein, S., K. L. Warfield, D. L. Swenson, D. A. Stein, J. L. Smith, C. S. Gamble, A. D. Kroeker, P. L. Iversen, S. Bavari, and E. Muhlberger. 2006. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob. Agents Chemother. 50:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge, Q., M. Pastey, D. Kobasa, P. Puthavathana, C. Lupfer, R. K. Bestwick, P. L. Iversen, J. Chen, and D. A. Stein. 2006. Inhibition of multiple subtypes of influenza A virus in cell cultures with morpholino oligomers. Antimicrob. Agents Chemother. 50:3724-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubman, M. J. 2005. Development of novel strategies to control foot-and-mouth disease: marker vaccines and antivirals. Biologicals 33:227-234. [DOI] [PubMed] [Google Scholar]
- 16.Guarné, A., J. Tormo, R. Kirchweger, D. Pfistermueller, I. Fita, and T. Skern. 1998. Structure of the foot-and-mouth disease virus leader protease: a papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 17:7469-7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutiérrez, A., E. Martínez-Salas, B. Pintado, and F. Sobrino. 1994. Specific inhibition of aphthovirus infection by RNAs transcribed from both the 5′ and the 3′ noncoding regions. J. Virol. 68:7426-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutiérrez, A., A. Rodríguez, B. Pintado, and F. Sobrino. 1993. Transient inhibition of foot-and-mouth disease virus infection of BHK-21 cells by antisense oligonucleotides directed against the second functional initiator AUG. Antivir. Res. 22:1-13. [DOI] [PubMed] [Google Scholar]
- 19.Kahana, R., L. Kuznetzova, A. Rogel, M. Shemesh, D. Hai, H. Yadin, and Y. Stram. 2004. Inhibition of foot-and-mouth disease virus replication by small interfering RNA. J. Gen. Virol. 85:3213-3217. [DOI] [PubMed] [Google Scholar]
- 20.Kinney, R. M., C. Y. Huang, B. C. Rose, A. D. Kroeker, T. W. Dreher, P. L. Iversen, and D. A. Stein. 2005. Inhibition of dengue virus serotypes 1 to 4 in Vero cell cultures with morpholino oligomers. J. Virol. 79:5116-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, M., W. Chen, Z. Ni, W. Yan, L. Fei, Y. Jiao, J. Zhang, Q. Du, X. Wei, J. Chen, Y. Liu, and Z. Zheng. 2005. Cross-inhibition to heterologous foot-and-mouth disease virus infection induced by RNA interference targeting the conserved regions of viral genome. Virology 336:51-59. [DOI] [PubMed] [Google Scholar]
- 22.Martínez, M. A., J. Dopazo, J. Hernández, M. G. Mateu, F. Sobrino, E. Domingo, and N. J. Knowles. 1992. Evolution of the capsid protein genes of foot-and-mouth disease virus: antigenic variation without accumulation of amino acid substitutions over six decades. J. Virol. 66:3557-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason, P. W., M. J. Grubman, and B. Baxt. 2003. Molecular basis of pathogenesis of FMDV. Virus Res. 91:9-32. [DOI] [PubMed] [Google Scholar]
- 24.McClorey, G., H. M. Moulton, P. L. Iversen, S. Fletcher, and S. D. Wilton. 2006. Antisense oligonucleotide-induced exon skipping restores dystrophin expression in vitro in a canine model of DMD. Gene Ther. 13:1373-1381. [DOI] [PubMed] [Google Scholar]
- 25.Mohapatra, J. K., A. Sanyal, D. Hemadri, C. Tosh, R. M. Kumar, and S. K. Bandyopadhyay. 2005. Evaluation of in vitro inhibitory potential of small interfering RNAs directed against various regions of foot-and-mouth disease virus genome. Biochem. Biophys. Res. Commun. 329:1133-1138. [DOI] [PubMed] [Google Scholar]
- 26.Moulton, H. M., M. H. Nelson, S. A. Hatlevig, M. T. Reddy, and P. L. Iversen. 2004. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjug. Chem. 15:290-299. [DOI] [PubMed] [Google Scholar]
- 27.Nelson, M. H., D. A. Stein, A. D. Kroeker, S. A. Hatlevig, P. L. Iversen, and H. M. Moulton. 2005. Arginine-rich peptide conjugation to morpholino oligomers: effects on antisense activity and specificity. Bioconjug. Chem. 16:959-966. [DOI] [PubMed] [Google Scholar]
- 28.Neuman, B. W., D. A. Stein, A. D. Kroeker, M. J. Churchill, A. M. Kim, P. Kuhn, P. Dawson, H. M. Moulton, R. K. Bestwick, P. L. Iversen, and M. J. Buchmeier. 2005. Inhibition, escape, and attenuated growth of severe acute respiratory syndrome coronavirus treated with antisense morpholino oligomers. J. Virol. 79:9665-9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman, B. W., D. A. Stein, A. D. Kroeker, H. M. Moulton, R. K. Bestwick, P. L. Iversen, and M. J. Buchmeier. 2006. Inhibition and escape of SARS-CoV treated with antisense morpholino oligomers. Adv. Exp. Med. Biol. 581:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieder, E., B. Baxt, and P. W. Mason. 1994. Animal-derived antigenic variants of foot-and-mouth disease virus type A12 have low affinity for cells in culture. J. Virol. 68:5296-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rieder, E., T. Bunch, F. Brown, and P. W. Mason. 1993. Genetically engineered foot-and-mouth disease viruses with poly(C) tracts of two nucleotides are virulent in mice. J. Virol. 67:5139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieder, E., T. Henry, H. Duque, and B. Baxt. 2005. Analysis of a foot-and-mouth disease virus type A24 isolate containing an SGD receptor recognition site in vitro and its pathogenesis in cattle. J. Virol. 79:12989-12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas, E. R., E. Carrillo, M. Schiappacassi, and R. Campos. 1992. Modification of foot-and-mouth disease virus O1 Caseros after serial passages in the presence of antiviral polyclonal sera. J. Virol. 66:3368-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosas, M. F., E. Martínez-Salas, and F. Sobrino. 2003. Stable expression of antisense RNAs targeted to the 5′ non-coding region confers heterotypic inhibition to foot-and-mouth disease virus infection. J. Gen. Virol. 84:393-402. [DOI] [PubMed] [Google Scholar]
- 35.Rueckert, R. R. 1996. Picornaviridae: the viruses and their replication, p. 609-654. In B. N. Fields, D. M. Knipe, and P. H. Howley (ed.), Field's virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 36.Stein, D., E. Foster, S. B. Huang, D. Weller, and J. Summerton. 1997. A specificity comparison of four antisense types: morpholino, 2′-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 7:151-157. [DOI] [PubMed] [Google Scholar]
- 37.Summerton, J., and D. Weller. 1997. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 7:187-195. [DOI] [PubMed] [Google Scholar]
- 38.Sutmoller, P., and R. Casas Olascoaga. 2003. The risks posed by the importation of animals vaccinated against foot and mouth disease and products derived from vaccinated animals: a review. Rev. Sci. Tech. 22:823-835. [DOI] [PubMed] [Google Scholar]
- 39.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van den Born, E., D. A. Stein, P. L. Iversen, and E. J. Snijder. 2005. Antiviral activity of morpholino oligomers designed to block various aspects of Equine arteritis virus amplification in cell culture. J. Gen. Virol. 86:3081-3090. [DOI] [PubMed] [Google Scholar]
- 41.Van Rensburg, H. G., and P. W. Mason. 2002. Construction and evaluation of a recombinant foot-and-mouth disease virus: implications for inactivated vaccine production. Ann. N. Y. Acad. Sci. 969:83-87. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, J., D. A. Stein, T. Lim, D. Qiu, S. Coughlin, Z. Liu, Y. Wang, R. Blouch, H. M. Moulton, P. L. Iversen, and D. Yang. 2006. Inhibition of coxsackievirus B3 in cell cultures and in mice by peptide-conjugated morpholino oligomers targeting the internal ribosome entry site. J. Virol. 80:11510-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]