Abstract
The DNA genomes of geminiviruses have a limited coding capacity that is compensated for by the production of small multifunctional proteins. The AL2 protein encoded by members of the genus Begomovirus (e.g., Tomato golden mosaic virus) is a transcriptional activator, a silencing suppressor, and a suppressor of a basal defense. The related L2 protein of Beet curly top virus (genus Curtovirus) shares the pathogenicity functions of AL2 but lacks transcriptional activation activity. It is known that AL2 and L2 can suppress local silencing by interacting with adenosine kinase (ADK) and can suppress basal defense by interacting with SNF1 kinase. However, how the activities of these viral proteins are regulated remains an unanswered question. Here, we provide some answers by demonstrating that AL2, but not L2, interacts with itself. The zinc finger-like motif (CCHC) is required but is not sufficient for AL2 self-interaction. Alanine substitutions for the invariant cysteine residues that comprise the motif abolish self-interaction or cause aberrant subnuclear localization but do not abolish interaction with ADK and SNF1. Using bimolecular fluorescence complementation, we show that AL2:AL2 complexes accumulate primarily in the nucleus, whereas AL2:ADK and L2:ADK complexes accumulate mainly in the cytoplasm. Further, the cysteine residue mutations impair the ability of AL2 to activate the coat protein promoter but do not affect local silencing suppression. Thus, AL2 self-interaction correlates with nuclear localization and efficient activation of transcription, whereas AL2 and L2 monomers can suppress local silencing by interacting with ADK in the cytoplasm.
The geminiviruses are a diverse family of plant pathogens that replicate their single-stranded DNA genomes in the host cell nucleus through double-stranded DNA (dsDNA) replicative forms that associate with histone proteins in minichromosomes. They have a limited coding capacity that varies from four to seven genes, depending on the virus, and do not encode polymerases. Instead, host machinery is recruited to express viral mRNAs and amplify viral genomes. These properties distinguish geminiviruses as paragons of genetic efficiency and useful models for fundamental cellular processes such as DNA replication, transcription, and epigenetic regulation of gene activity (12).
Geminivirus replication requires only the AL1 gene product (also known as AC1, C1, or Rep, for replication initiator protein) (8). AL1 binds sequences within the viral origin and introduces a site- and strand-specific nick in the dsDNA replicative form that generates a template primer for DNA synthesis (11, 20). In viruses belonging to the genus Begomovirus, the AL2 protein (also known as AC2, C2, or TrAP, for transcriptional activator protein) acts as a transcription factor required for the expression of viral genes needed at late times in infection (31, 32). Thus, the core functions of AL1 and AL2, which are expressed early in infection, involve redirecting the cellular replication and transcription machinery to initiate on viral templates. However, more recent work has established that these proteins also have secondary functions that allow them to interface with multiple cellular pathways to create an environment favorable to virus replication (3, 12, 35). These host control activities, which include altering the cell cycle, affecting cellular metabolism, and suppressing antiviral defenses, can be mechanistically unrelated to the core activities but are nevertheless critical to viral pathogenesis (19, 26, 36, 39). Thus, in order to understand how geminiviruses are able to successfully counter multiple defense systems, commandeer biosynthetic machinery, and reprogram host cells, it is necessary to ask how functional modulation of viral proteins is achieved. Answers to this question will no doubt provide a greater appreciation of viral genetic economy and new insight into mechanisms by which multifunctional proteins can be regulated.
The AL2 protein of Tomato golden mosaic virus (TGMV) and related begomoviruses is a model multifunctional protein that alters the activities of a number of cellular proteins or protein complexes and is itself influenced by these interactions. AL2 regulates the viral gene expression program by enabling expression of the coat protein (CP) and nuclear shuttle protein genes. AL2-mediated stimulation of these promoters is complex and involves both activation and derepression mechanisms (29, 30, 32). The 15-kDa AL2 resembles a typical transcription factor in several respects: it has a nuclear localization signal (NLS), a zinc finger-like domain composed of cysteine and histidine residues, and an acidic activation domain (6, 15, 27, 37). However, dsDNA-binding activity is weak and not sequence specific, and AL2 likely is targeted to responsive promoters by interactions with cellular proteins. The identities of these proteins, and of those contacted by the activation domain, are not yet known. As expected, AL2 accumulates in the nucleus, although it also is present in the cytoplasm of infected cells. Following expression in insect cells from a baculovirus vector, phosphorylated AL2 accumulates predominately in the nucleus, while nonphosphorylated forms are found in both the nucleus and the cytoplasm, suggesting that subcellular localization is influenced by as yet unidentified cellular kinases (41).
In addition to its core role in viral transcription, AL2 also is a pathogenicity factor that suppresses more than one host defense pathway. Constitutive expression of truncated AL2 (lacking the activation domain) or the related L2 protein from Beet curly top virus (BCTV; genus Curtovirus) in transgenic Nicotiana benthamiana plants conditions a novel enhanced susceptibility phenotype characterized by a reduction in mean latent period (time to first appearance of symptoms) and by a decrease in the inoculum concentration required to elicit infection without a significant increase in disease symptoms or virus replication (33). Enhanced susceptibility correlates with the ability of AL2 and L2 to interact with and inactivate SNF1 kinase, a global regulator of metabolism that responds to the cellular energy charge (13). However, the exact nature of the defense mediated by SNF1, which appears to influence the property of viral infectivity rather than virulence, remains uncharacterized.
Begomovirus AL2 proteins and BCTV L2 protein also are suppressors of RNA silencing, an antiviral defense first observed in plants (2, 5, 23, 38). AL2 can reverse previously established silencing when expressed from an RNA virus vector such as Potato virus X (PVX) and also can inhibit silencing when expressed from plasmids delivered by particle bombardment or by agroinfiltration of leaves (34, 36, 39, 40). The available evidence suggests that AL2 suppresses silencing by both transcription-dependent and transcription-independent mechanisms (3). The transcription-dependent mechanism is believed to involve the activation of host genes (e.g., WEL-1) that may act as endogenous, negative regulators of RNA silencing (34). Not surprisingly, this mechanism requires an intact NLS and activation domain, as well as the zinc- and DNA-binding activities (6, 34, 37). The second mechanism does not require the activation domain and correlates with the ability of AL2 (and BCTV L2) to interact with and inactivate adenosine kinase (ADK), an enzyme primarily localized in the cytoplasm (40, 41). The BCTV L2 protein, which, unlike AL2, is not required for late viral gene expression, is probably restricted to this mechanism (16, 28). Because ADK is required for efficient production of the methyltransferase cofactor S-adenosylmethionine, ADK deficiency reduces cellular transmethylation activity (21). Methylation of DNA sequences complementary to target RNA or target gene promoters is associated with posttranscriptional gene silencing and transcriptional gene silencing, respectively. Thus, it is possible that AL2 acts in a transcription-independent manner to interfere with RNA-directed methylation of the viral genome. Methylation has been shown to markedly reduce the replication of geminivirus DNA in transfected protoplasts and to inhibit the activity of geminivirus promoters when they are used in transgenes (4, 25).
While it is evident that AL2 interacts with a number of cellular proteins (both known and unknown) to execute its several roles in the geminivirus replication cycle, it is unclear how the activities of this viral protein are regulated. Here, we begin to address this issue by showing that AL2, but not L2, can interact with itself to form dimers and higher-order multimers. Complexes containing monomeric and multimeric forms of AL2 are differentially localized and have different functional capabilities. Self-interaction correlates with nuclear localization of AL2 complexes and the ability to efficiently activate transcription, whereas monomeric AL2 and L2 interact with ADK mainly in the cytoplasm.
MATERIALS AND METHODS
Yeast two-hybrid analysis.
The yeast two-hybrid system was used to identify interactions between geminivirus AL2 and L2 proteins (7, 14). Positive interaction between bait and prey was indicated by growth of Y190 cells on medium lacking histidine and containing 50 mM 3-aminotriazole. Media also lacked leucine and tryptophan to ensure maintenance of expression plasmids. Additional confirmation of interactions was obtained by assessing β-galactosidase activity using a filter-lift assay. Bait and prey constructs containing AL2, AL21-114, AL21-83, AL21-83, Δ33-43, L2, SNF1, and ADK have been previously described (13, 41). AL2 substitution mutants C33A, C35A, H40A, and C43A were generated by site-directed mutagenesis using the primers 5′-GACCTGAACGCTGGCTGTTCC (C33A), 5′-GAACTGTGGCGCTTCCATATAC (C35A), 5′-CCATATACATTGCCATCGACTGC (H40A), and 5′-CACATCGACGCCAGAAACAATGG (C43A) (alanine codons are underlined).
Protein expression and gel blot assays.
Construction of expression plasmids and conditions for the expression, partial purification, and analysis of six-histidine-tagged AL2 (His-AL2) and AL2 fused to glutathione S-transferase (GST-AL2) from Escherichia coli have been described previously (15). Standard protein gel blots (Western blots) were performed using anti-His tag or anti-GST antibody, as appropriate.
For protein-protein gel blot analysis (far Western blots), samples of partially purified GST and GST-AL2 were fractionated pairwise on a 12% polyacrylamide gel containing sodium dodecyl sulfate (SDS) and were separated after transfer to a membrane to create three identical blots. One blot was probed with anti-GST antibody to verify the presence of GST and GST-AL2. The other two blots were incubated with soluble protein extracts from E. coli cells expressing either His-AL2 or His-CAT (primary probes) at room temperature for 2 h. After being blocked with PBST buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 0.1% Tween 20, 5% nonfat milk) (1), both membranes were probed with anti-His tag antibody.
Glycerol gradient centrifugation.
Sedimentation analysis was carried out essentially as described by Gallo et al. (10). Partially purified His-AL2 protein (10 μg) was loaded on 5 to 20% or 10 to 30% glycerol gradients in buffer containing 150 mM NaCl and general protease inhibitor cocktail (Sigma), with or without 1 mM dithiothreitol (DTT). For each experiment, parallel gradients were loaded with 200 μg each of lysozyme (14 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), and β-galactosidase (120 kDa) as molecular mass standards. Gradients were centrifuged in a Beckman SW41 rotor at 38,000 rpm for 51 h at 4°C. Twenty-nine fractions of 390 μl each were collected from the top of the gradients using an Auto Densi-flow IIC fractionator (Buchler Instruments). Sedimentation of molecular mass standards was monitored by measuring the absorbance of each fraction at 280 nm. For gradients containing His-AL2, a 30-μl aliquot of each fraction was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by immunoblotting with anti-His tag antibody.
BiFC analysis of interactions.
The bimolecular fluorescence complementation (BiFC) protocol used was based on the method of Hu et al. (17). The BiFC expression vectors p2YN and p2YC were constructed for fusing codons 1 to 158 or 159 to 238, respectively, of the yellow fluorescent protein (YFP) to the 3′ end of a gene of interest. A mutant version of the enhanced YFP gene (pEYFP; Clontech) in which codon 69 was modified from glutamine to methionine (YFP Q69M) was used as the source of the YFP sequence.
Vectors p2YN and p2YC were constructed using standard PCR-based mutagenesis and cloning procedures. The relevant features of these plasmids are as follows: (i) plasmid backbones are from the binary Ti plasmid pBC301 (42); (ii) both plasmids have the enhanced Cauliflower mosaic virus (CaMV) 35S promoter, the Tobacco etch virus 5′ untranslated leader sequence, and the CaMV 3′ terminator sequence (22) in the transfer DNA region; (iii) both have PacI and SpeI restriction endonuclease sites for insertion of DNA immediately downstream of the Tobacco etch virus leader; and (iv) immediately upstream of the YFP sequences in each vector is the coding sequence for a linker and hemagglutinin epitope of the following amino acid sequence: TSGGGGSGGGGSGYPYDVPDYAGAAA (the hemagglutinin epitope is underlined).
The wild-type or mutant versions of the TGMV AL2 gene were amplified by PCR with forward primer 5′-CCCTTAATTAAC.ATG.CGA.AAT.TCG.TCT.TCC.TCA and reverse primer 5′-GGGACTAGT.TTT.AAA.TAA.GTT.CTC.CCA.GAA. The L2 gene of BCTV was amplified with forward primer 5′-CCCTTAATTAACATG.GAA.CCA.CGT.TCA.T and reverse primer 5′-GGGACTAGT.TCC.AAG.TAT.ATC.TCT.AGT.CTC. The ADK2 gene from Arabidopsis thaliana, in the plasmid pRSET-ADK (40), was PCR amplified with forward primer 5′-CCCTTAATTAACATG.GCT.TCT.TCT.TCT.AAC.TAC and reverse primer 5′-GGGACTAGT.GTT.AAA.GTC.GGG.TTT.CTC.AGG. In all primer sequences provided, periods separate codons of the amplified genes, and the recognition sequences for the restriction endonucleases PacI or SpeI are underlined. The resulting PCR products were digested with PacI and SpeI and ligated into PacI-SpeI-digested p2YN and p2YC.
For BiFC experiments, plasmids were transformed into Agrobacterium tumefaciens GV311, and cultures were used to infiltrate N. benthamiana leaves as described previously (40). Cultures containing p2YN- and p2YC-based plasmids were mixed 1:1 immediately prior to infiltration. Leaf tissue was analyzed by microscopy approximately 36 h postinfiltration using a Leica DM IRB epifluorescent microscope equipped with an Optronics digital camera (Magnafire model S99802). Leaf tissue was photographed under either white light or UV light. To record YFP fluorescence, a Chroma filter (I3) with a 450- to 490-nm excitation wavelength and 515-nm emission wavelength was used.
Transcriptional studies.
Analysis of transcription activation was carried out with N. benthamiana suspension culture protoplasts isolated and transfected as described previously (29). Constructs capable of generating a replicating TGMV DNA A with the β-glucuronidase (GUS) reporter in place of the CP coding region and a null mutation of the AL2 gene (TGMV-GUS/AL2−) (pTGA55) and an expression plasmid containing the TGMV AL2 gene (35S-AL2) also have been described previously (30-32).
Expression of AL2 proteins in insect cells.
Wild-type and CCHC mutant AL2 proteins were expressed in insect cells from a baculovirus vector, and nuclear fractions were purified as described previously (41). The presence of wild-type or CCHC mutant AL2 proteins was detected by Western blotting using a primary antibody directed against the full-length AL2 protein as described previously (41).
Silencing suppression studies.
Local silencing suppression assays based on A. tumefaciens-mediated transient expression in N. benthamiana or N. benthamiana line 16c containing a GFP transgene have been described previously (18, 24). Expression plasmids containing GFP, GUS, p19, or AL2 and conditions for agroinfiltration, photographic analysis, and GFP mRNA analysis also have been previously described (40).
RESULTS
Conserved residues needed for AL2 self-interaction in yeast cells are not required for interaction with SNF1 or ADK.
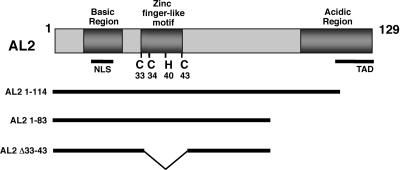
AL2 protein has several distinct features, including a basic region near the N terminus that contains an NLS, a central region that contains a zinc finger-like motif (C-X1-C-X4-H-X2-C), and an acidic C-terminal region that contains the minimal transcription activation domain (6, 15, 27, 37) (Fig. 1). The central region is required for DNA and zinc binding. This region contains the invariant cysteines as well as the conserved histidine residue that is present in most, but not all, begomovirus AL2 proteins. In contrast, BCTV L2 protein lacks recognizable basic and acidic regions but shares the central zinc finger-like motif with AL2.
FIG. 1.
Diagram of wild-type AL2 protein and deletion mutants. Illustrated is the 129-amino-acid TGMV AL2 protein showing the relative locations of the basic domain that contains an NLS, the C-terminal acidic region that contains the minimal transcriptional activation domain (TAD), and the amino acid coordinates of invariant cysteines and conserved histidine residues in the zinc finger-like motif. Truncated AL2 proteins lacking the TAD (AL21-114 and AL21-83) or lacking the TAD and the zinc finger-like motif (AL21-83, Δ33-43) were employed in yeast two-hybrid experiments.
Following expression of TGMV AL2 in E. coli, we often observe a minor fraction of recombinant TGMV AL2 that migrates at a rate consistent with that of a dimer in SDS-PAGE (data not shown). To confirm that AL2 is capable of self-interaction and to identify sequences that might be required for this property, truncated forms of the protein as well as mutant versions containing individual amino acid substitutions were examined in the yeast two-hybrid system (7, 9). Various bait proteins consisting of the GAL4 DNA-binding domain fused to AL2 lacking the activation domain were coexpressed with prey proteins consisting of similarly truncated AL2 fused to the GAL4 activation domain (Fig. 1). The yeast Y190 reporter strain contains lacZ and HIS3 genes under control of the GAL1 promoter, which contains GAL4-binding sites. Interaction between bait and prey proteins, resulting in reconstitution of the GAL4 transcriptional activator in the yeast nucleus, is indicated by growth of Y190 cells on synthetic medium lacking histidine and containing 50 mM 3-aminotriazole. Confirmation and approximate quantitation of interaction strength can be obtained by assessing β-galactosidase activity.
While AL2 does not interact with a standard panel of negative control proteins, including lamin, CDK2, p53, and chloramphenicol acetyltransferase (CAT) (13, 41), self-interaction was clearly evident between bait and prey proteins consisting of either AL21-114 or AL21-83 (Table 1). Neither AL21-114 nor AL21-83 interacted with TGMV AL1 (Rep) protein, which served as a negative control in these experiments. However, additional deletion of the zinc finger-like motif (Δ33-43) in AL21-83 abolished self-interaction. Further analysis naturally focused on the conserved cysteine and histidine residues contained in this region. As summarized in Table 1, individual substitution mutations to alanine, including C33A, C35A, H40A, and C43A in the AL21-83 background, also abolished apparent interaction in this system.
TABLE 1.
Conserved cysteine and histidine residues are required for AL2 self-interaction in the yeast two-hybrid systema
| Bait | Prey | Interactionb | Activationd |
|---|---|---|---|
| AL21-114 | AL1 (Rep) | − | |
| AL21-83 | AL1 (Rep) | − | |
| AL21-114 | AL21-114 | ++ | |
| AL21-83 | AL21-83 | ++ | |
| AL21-83, Δ33-43 | AL21-83 | − | |
| L2 | L2 | − | |
| AL2 C33Ac | AL21-83 | − | |
| AL2 C35A | AL21-83 | − | |
| AL2 H40A | AL21-83 | − | |
| AL2 C43A | AL21-83 | − | |
| AL21-129 | Vector | ++ | |
| L2 | Vector | − |
The indicated bait proteins were expressed as GAL4 DNA-binding domain fusions, and prey proteins were expressed as GAL4 activation domain fusions in yeast Y190 cells. TGMV AL2 constructs used for interaction testing lacked the activation domain located within the C-terminal residues 115 to 129.
Interaction was indicated by the ability of cells to grow on medium lacking His and containing 50 mM 3-aminotriazole. As an additional indicator of interaction, colonies were monitored for LacZ activity (blue color) using a filter-lift assay. Interaction symbols are as follows: −, no interaction, no evidence of blue color after overnight incubation; +, interaction, blue color developed within 2 to 6 h; ++, strong interaction, blue color developed within minutes to 2 h.
AL2 substitution mutants were expressed in an AL21-83 background.
Ability of full-length AL2 and L2 bait proteins to activate transcription in the absence of prey protein was assessed as described in footnote b for interactions.
AL2 and L2 previously were shown to interact with the cellular kinases SNF1 and ADK (13, 41). We therefore asked if these interactions also are sensitive to the same substitutions of cysteine and histidine residues. The data summarized in Table 2 clearly show that they are not. In the AL21-83 background, the C33A, C35A, H40A, and C43A mutant bait proteins all maintained their ability to interact with SNF1 and ADK. Based on the β-galactosidase assay, interactions between the kinases and mutant AL2 proteins actually appeared somewhat stronger than that with the comparable nonsubstituted AL21-83. Neither SNF1 nor ADK interacted with p53 or CAT, which were used as negative controls in these experiments.
TABLE 2.
Cysteine and histidine mutant AL2 proteins interact with SNF1 and ADKa
| Bait | Prey | Interaction |
|---|---|---|
| CAT | SNF1 | − |
| p53 | SNF1 | − |
| AL21-114 | SNF1 | ++ |
| AL21-83 | SNF1 | + |
| AL2 C33Ab | SNF1 | ++ |
| AL2 C35A | SNF1 | ++ |
| AL2 H40A | SNF1 | ++ |
| AL2 C43A | SNF1 | ++ |
| L2 | SNF1 | ++ |
| CAT | ADK | − |
| p53 | ADK | − |
| AL21-114 | ADK | ++ |
| AL21-83 | ADK | + |
| AL2 C33Ab | ADK | ++ |
| AL2 C35A | ADK | ++ |
| AL2 H40A | ADK | ++ |
| AL2 C43A | ADK | ++ |
| L2 | ADK | ++ |
Yeast two-hybrid experimental design and symbols are the same as those described in the footnotes to Table 1.
AL2 substitution mutants were expressed in an AL21-83 background.
These studies allowed us to conclude that AL2 can interact with itself and that residues essential for this interaction in yeast cells lie between amino acids 1 and 83. In addition, the conserved cysteine and histidine residues of the zinc finger-like motif are necessary for AL2 self-interaction in this system but are not required for interaction with SNF1 and ADK.
BCTV L2 does not self-interact in the yeast two-hybrid system.
AL2 pathogenicity functions are shared by the related curtovirus L2 protein, and both proteins have similar zinc finger-like motifs (13, 33, 40). It was therefore logical to ask if self-interaction is common to AL2 and L2. Before doing this, however, it was necessary to investigate whether BCTV L2 can independently activate transcription in yeast when expressed as a GAL4 DNA-binding domain fusion (bait) independently of a prey protein.
As shown previously, full-length AL2 with an intact activation domain is a strong transcriptional activator in yeast cells (Table 1). In contrast, a full-length L2 bait protein did not cause significant transcriptional activation, consistent with the observation that it is not required for expression of late viral genes (16, 28). However, the same L2 bait construct was able to interact with ADK and SNF1 (Table 2). Also unlike AL2, BCTV L2 protein did not exhibit self-interaction when coexpressed as bait and prey (Table 1), suggesting that the zinc finger-like motif alone is not sufficient for this property and that additional contacts absent in L2 also are involved.
Biochemical confirmation of AL2 self-interaction.
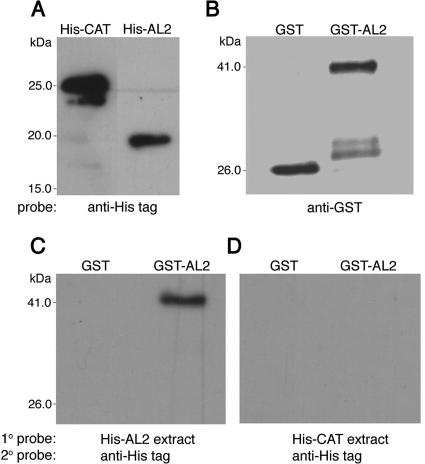
For reasons that are not clear, it proved difficult to reproducibly detect TGMV AL2 self-interaction in standard pull-down experiments. However, self-interaction was easily observed by protein-protein gel blot (far Western) analysis. These studies employed protein extracts from E. coli cells expressing full-length AL2 (129 amino acids) as a GST-tagged (GST-AL2) or a His-tagged (His-AL2) fusion protein. Extracts from cells expressing GST- or His-tagged CAT (His-CAT) were included as negative controls.
As shown in Fig. 2A, standard protein gel blot analysis (Western blotting) using an anti-His tag antibody probe revealed the presence of His-AL2 and His-CAT in total protein extracts from E. coli cells transformed with plasmids designed to express these fusion proteins. Similar gel blot analysis using an anti-GST probe indicated the presence of GST and GST-AL2 in samples purified from extracts of E. coli transformed with the cognate expression plasmids (Fig. 2B). When identical protein blots of GST and GST-AL2 were first incubated with extracts containing His-AL2 (primary probe) followed by an anti-His tag antibody (secondary probe), no signal was observed in the GST sample, but a signal corresponding to His-AL2 was clearly visible at the position of the substantially larger GST-AL2 fusion protein, indicating GST-AL2-His-AL2 interaction (Fig. 2C, far Western blot). However, when His-CAT-containing extract was used as the primary probe with the same anti-His tag antibody as the secondary probe, no signal was observed in either the GST or GST-AL2 sample (Fig. 2D). This analysis provided verification of AL2 self-interaction by a separate methodology and demonstrated that interaction is independent of the GST tag, His tag, GAL4 DNA-binding domain, or GAL4 activation domain moieties that were fused to AL2 in these or the yeast two-hybrid experiments.
FIG. 2.
Protein-protein blot analysis of AL2 self-interaction. (A) Immunoblot of total protein extracts from E. coli cells expressing either His-AL2 or His-CAT, probed with anti-His tag antibody, is shown. For experiments depicted in the remaining panels, three identical blots of partially purified GST and GST-AL2 were used: a blot probed with anti-GST antibody (B); a blot incubated with His-AL2-containing extract (primary probe) followed by being immunoblotted with anti-His tag antibody (secondary probe) (C); and a blot incubated with His-CAT-containing extract (primary probe, negative control) followed by being immunoblotted with anti-His tag antibody (secondary probe) (D). The positions of molecular mass standards are shown to the left of panel A. The positions of GST (∼26 kDa) and GST-AL2 (∼41 kDa) are indicated to the left of panels B and C. His-AL2 (∼16 kDa) consistently migrates at a slightly lower-than-expected rate in polyacrylamide gels. Additional faint bands observed in the GST-AL2 sample (B) are breakdown products.
AL2 forms dimers and higher-order multimers in solution.
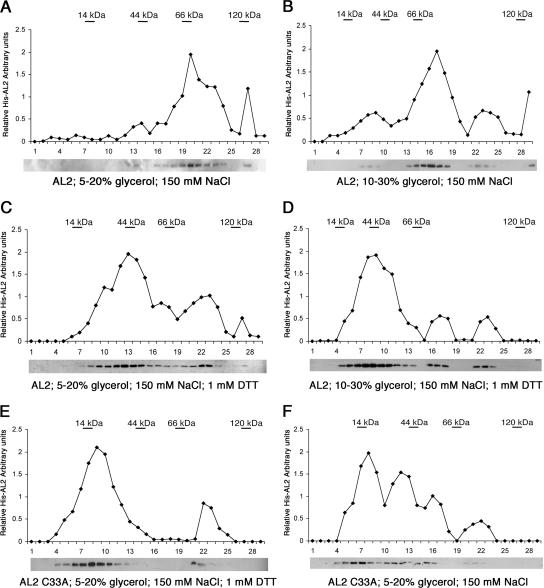
Glycerol density gradient centrifugation was chosen as an additional method of biochemical analysis, because it permits the multimerization state of a protein to be examined in solution. These studies used full-length His-AL2 proteins expressed in E. coli and purified from extracts by nickel nitrilotriacetic acid-agarose affinity chromatography. The identity of the protein was confirmed by separate protein gel blot analyses using anti-His tag and anti-AL2 antiserum (data not shown). Following centrifugation through 5 to 20% or 10 to 30% glycerol gradients, aliquots of gradient fractions were subjected to SDS-PAGE, blotted, and probed with anti-His tag antibody to determine the position of His-AL2 in the gradients. Molecular mass standards were analyzed in parallel gradients, and the optical density (at 280 nm) of gradient fractions was determined to monitor the sedimentation of the marker proteins lysozyme (14 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), and β-galactosidase (120 kDa).
Surprisingly, centrifugation through 5 to 20% gradients containing 150 mM NaCl revealed that most AL2 sedimented at a rate consistent with that of a tetramer (ca. 64 kDa), although a considerable amount of larger material also was present. Essentially no monomeric AL2 (ca. 16 kDa) was evident, and only a relatively small amount of it appeared to be in dimeric form (ca. 32 kDa) (Fig. 3A). Subsequent analysis on 10 to 30% gradients permitted resolution of the larger material into forms consistent with tetramers (most abundant) and hexamers (ca. 96 kDa). Again, only relatively small amounts of dimeric AL2 were evident (Fig. 3B). However, when reducing conditions were maintained by the inclusion of DTT in the gradients (1 mM DTT), dimeric AL2 became the most abundant form, although apparent tetramers and hexamers still remained (Fig. 3C and D). In contrast, when mutant AL2 protein containing an alanine substitution for cysteine 33 (His-AL2 C33A) was subjected to glycerol gradient centrifugation under the same reducing conditions, the protein was mostly monomeric, confirming the importance of this invariant cysteine for self-interaction (Fig. 3E). Monomeric AL2 C33A protein also was the most abundant form in the absence of DTT, although in this case a substantial amount of larger material also was present, suggesting that reducing conditions must be maintained in order to prevent the stabilization of aggregates by oxidation (Fig. 3F).
FIG. 3.
Glycerol gradient analysis of AL2 complexes. His-AL2 protein was sedimented on 5 to 20% or 10 to 30% glycerol gradients in 150 mM NaCl in the presence or absence of 1 mM DTT, as indicated. Gradient fractions were subjected to immunoblot analysis using an anti-His tag antibody as shown below each graph, which illustrates the relative amount of AL2 in each fraction. Molecular mass standards included lysozyme (14 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), and β-galactosidase (120 kDa), which were sedimented in parallel gradients and monitored by absorbance at 280 nm. The positions of peak fractions corresponding to each marker protein are indicated in the graphs. (A to D) Sedimentation of wild-type AL2; (E and F) sedimentation of AL2 C33A.
Inclusion of 1,10-phenanthrolene (10 mM) or EDTA (10 mM), both capable of chelating zinc, had little or no effect on the sedimentation profile of wild-type AL2 under reducing conditions in 5 to 20% glycerol gradients (data not shown). However, because it can be difficult to remove zinc from proteins, these results do not provide definitive information about the role of zinc ions in self-interaction.
We concluded that AL2 protein has a strong tendency to form dimers in solutions of moderate salt in the presence of 1 mM DTT, and its stability in these reducing conditions indicates that dimerization does not depend on disulfide bridges. However, the dimers have a propensity to aggregate into higher-order multimers, particularly if reducing conditions are not maintained. The biological significance of the larger, apparently tetrameric and hexameric complexes is unclear, as their existence could be due to spurious aggregation stabilized by oxidation. The formation of similar aggregates by the non-self-interacting C33A mutant protein under nonreducing conditions supports this interpretation.
Analysis of self-interaction and ADK interaction in planta.
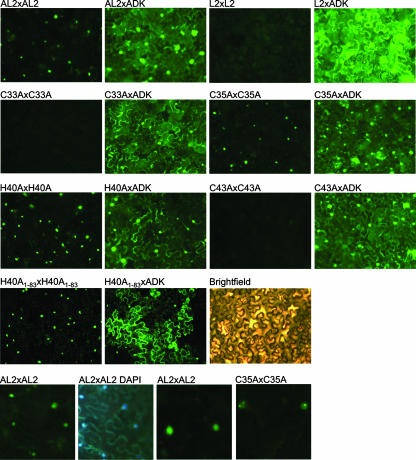
BiFC was employed as a means of verifying AL2 self-interaction in vivo (17). With this method, potentially interacting proteins are fused to the N- or C-terminal portions of YFP and are introduced into living cells. Association of interacting partners reconstitutes YFP, resulting in fluorescence, which indicates interaction and reveals where the interacting proteins accumulate in the cell. We adopted a transient system in which N. benthamiana leaves are coinfiltrated with A. tumefaciens cultures containing disarmed Ti plasmids designed to express the fusion proteins to be tested. Cells were viewed by fluorescence microscopy 36 h postinfiltration. The test proteins consisted of the N-terminal fragment of the YFP open reading frame (YN, encoding amino acids 1 to 158) or the C-terminal YFP fragment (YC, encoding amino acids 159 to 238) fused to either wild-type TGMV AL2, AL2 proteins containing alanine substitutions for the invariant cysteine and conserved histidine residues (CCHC mutants), BCTV L2, or Arabidopsis ADK. All viral proteins used in these studies were full length. Both N- and C-terminal fusion proteins were constructed, and these were tested in all pairwise combinations. Interactions were observed regardless of whether the YN and YC fusions were N- or C-terminal or, in the case of ADK interactions, which partner was fused to YN or YC (data not shown). Thus, all YN and YC fusion proteins were expressed and were stable, although some combinations produced stronger signals than others.
Following agroinfiltration, strong self-interaction of TGMV AL2 was observed with most of the AL2:AL2 complexes accumulating in the nuclei of N. benthamiana epidermal cells (Fig. 4). Nuclear localization was confirmed by 4′,6′-diamidino-2-phenylindole (DAPI) staining. In contrast, no significant self-interaction of BCTV L2, or of the AL2 C33A or AL2 C43A mutant proteins, was evident. Surprisingly, however, both the C35A and H40A mutant proteins were observed to self-interact and accumulate in the nucleus, despite the fact that these mutations abolished self-interaction in the yeast two-hybrid system (Fig. 4). This discrepancy is not due to the use of truncated proteins (amino acids 1 to 83) in yeast and full-length proteins here, because an abbreviated mutant protein (AL2 H40A1-83) was capable of self-interaction in N. benthamiana cells (Fig. 4). It is possible that AL2:AL2 interaction is stabilized by posttranslational modification or by cellular proteins in the plant cell environment. However, while wild-type AL2 and AL2 H40A accumulated in a more or less uniform manner in the nucleoplasm, AL2 C35A displayed a unique, punctate localization pattern (Fig. 4). Of 35 nuclei examined, punctate nuclear localization of AL2 C35A complexes was observed in all 35, whereas this pattern was not seen with AL2 complexes in over 50 nuclei examined.
FIG. 4.
BiFC analysis of AL2:AL2, AL2:ADK, and L2:ADK complexes in N. benthamiana epidermal cells. Constructs expressing wild-type and mutant AL2, L2, or ADK fused to the N- or C-terminal portion of YFP (YN or YC) were delivered by agroinfiltration to N. benthamiana leaves. In the experiments shown, YFP was fused to the C terminus of the test proteins, and ADK fusion proteins included the C-terminal portion of YFP. Cells were photographed 36 h postinfiltration under UV light. The exposure time was 4 s, except for those of AL2 H40A:ADK, AL2 H40A1-83:ADK, AL2 H40A1-83:AL2 H40A1-83, and L2:ADK (2 s) as well as L2:L2 (8 s). Protein combinations are indicated above each photograph. The UV photographs in the bottom row show higher-magnification views (×10 instead of ×5 objective) of nuclear AL2:AL2 complexes, DAPI-stained nuclei, and AL2 C35A:AL2 C35A complexes exhibiting aberrant subnuclear localization.
The wild-type AL2 and L2 proteins and all of the AL2 CCHC mutant proteins were capable of interacting with ADK in N. benthamiana cells (Fig. 4). Consistent with the predominantly cytoplasmic nature of ADK, AL2:ADK complexes mostly accumulated in the cytoplasm, although some nuclear accumulation also was observed, suggesting that AL2 and the AL2 CCHC mutant proteins are capable of relocalizing a fraction of ADK to the nucleus. In contrast, nuclear accumulation of L2:ADK complexes was rarely if ever seen. These observations suggest that the AL2 NLS is not masked by ADK interaction and that AL2 self-interaction is not required for nuclear entry.
The results of BiFC studies allow us to conclude that AL2 self-interaction occurs in vivo and that AL2:AL2 complexes preferentially accumulate in the nucleus. They also are consistent with our previous reports of AL2:ADK and L2:ADK interaction and the observation that a significant fraction of AL2 accumulates in the cytoplasm of TGMV-infected cells, in this case as a consequence of interaction with ADK.
Mutations that disrupt AL2 self-interaction in vivo impair transcription activation activity.
To examine the functional consequences of self-interaction, we investigated the ability of wild-type AL2 and AL2 CCHC mutant proteins to activate transcription from the TGMV CP promoter. N. benthamiana protoplasts were transfected with a construct containing a TGMV DNA A dimer that generates a replicating monomeric A genome component in plant cells. The DNA A released contains a null mutation of the AL2 gene and the GUS reporter in place of the CP coding region (TGMV A-GUS/AL2−) (pTGA55). Cotransfections included a second plasmid that expresses wild-type AL2 or a mutant AL2 protein (AL2 C33A, C35A, H40A, or C43A) from the CaMV 35S promoter. Extracts were prepared 3 days posttransfection, and fluorometric GUS assays were performed as previously described (29, 31).
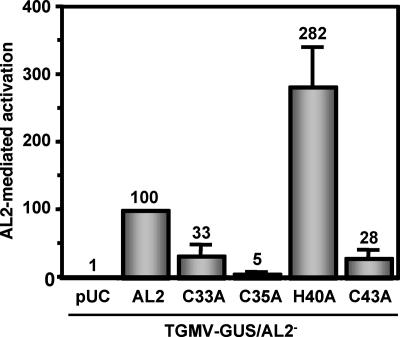
Stimulation of the CP promoter by AL2 protein typically ranges from 5- to 10-fold relative to basal expression levels. The activity of the AL2 mutant proteins relative to that of wild-type AL2 is illustrated in Fig. 5. Mutation of residues required for self-interaction or correct subnuclear localization in planta (C33A, C35A, and C43A) resulted in a significant reduction of CP promoter activation (33% or less of the level of wild-type activity). Surprisingly, the H40A substitution mutant, which displayed wild-type self-interaction and nuclear localization in vivo, consistently exhibited increased activity (ca. 2.5-fold) relative to that of wild-type AL2. Similar results were obtained in an experiment in which protoplasts were cotransfected with the AL2 expression plasmids and a nonreplicating plasmid containing the CP promoter (−657 to +1) fused to the GUS reporter (data not shown).
FIG. 5.
Activation of the CP promoter by wild-type and mutant AL2 proteins. The ability of wild-type and mutant AL2 proteins to activate the CP promoter was tested by cotransfecting N. benthamiana protoplasts with plasmids expressing the proteins from the 35S promoter (or empty vector, pUC, as a negative control) and TGMV-GUS/AL2−. In plant cells, TGMV-GUS/AL2− generates a monomeric, replicating TGMV DNA A containing a GUS replacement of the CP coding region and a null mutation in the AL2 gene. Wild-type AL2-mediated activation was assigned a value of 100%. Values shown are the averages from three experiments. Error bars represent standard errors of the means.
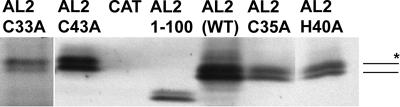
Because transcription activation activity depends on nuclear localization, the ability of AL2 CCHC mutant proteins to enter the nucleus, under conditions in which detection does not demand self-interaction, was tested. This was done by expressing each of them in insect cells from a baculovirus vector, followed by cell fractionation and protein gel blot analysis. Consistent with previous results, wild-type AL2 was observed in both the nuclear and cytoplasmic fractions, with phosphorylated forms accumulating preferentially in the nucleus (41). As shown in Fig. 6, none of the mutations prevented AL2 from entering the nucleus, confirming that self-interaction is not a prerequisite for nuclear entry. Thus, we concluded that the invariant cysteine residues, but not the less conserved histidine residue, are necessary for maximal transcription activation activity.
FIG. 6.
CCHC mutant AL2 proteins localize to the nuclei of insect cells. Fractionated protein extracts from Sf9 cells infected with recombinant baculoviruses expressing either native TGMV AL2, CCHC mutant AL2, AL21-100, or CAT (a negative control protein) are shown. Nuclear fractions were prepared 48 h postinoculation, and equivalent amounts (representing 5 × 105 cells) were electrophoresed on 4 to 20% polyacrylamide-SDS gels and subjected to Western blot analysis using an AL2-specific antibody. The positions of nonphosphorylated and phosphorylated (asterisk) AL2 are indicated to the right. WT, wild type.
The AL2 C33A substitution does not abolish local silencing suppression activity.
In previous work, we demonstrated that AL2, L2, and AL21-100 can suppress local silencing in transient infiltration assays and that suppression strongly correlated with the ability of these proteins to interact with ADK and to inhibit its activity (40). Because we showed that AL2 C33A interacts with ADK but is compromised with respect to its ability to interact with itself in vivo and in vitro, we chose this mutant protein to test whether self-interaction was necessary for suppression of local silencing.
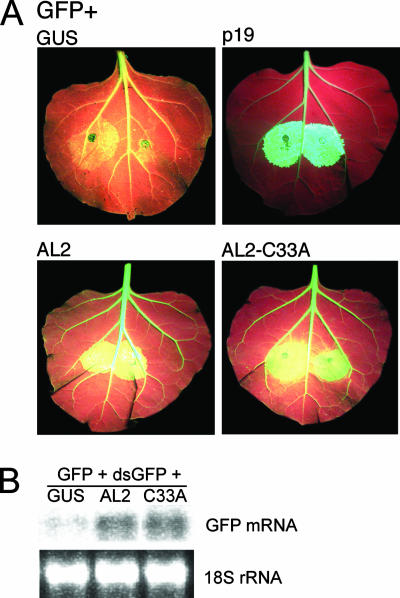
Suppression activities of wild-type AL2 and AL2 C33A were compared in a two-component system that utilizes N. benthamiana line 16c, which expresses a GFP transgene (24). Leaves were infiltrated with A. tumefaciens cultures containing Ti plasmids capable of expressing GFP (the silencing trigger) and a test or control. A construct expressing p19 protein, an efficient silencing suppressor from the RNA-containing Cymbidium ringspot virus, was used as a positive control, and a construct expressing GUS was used as a negative control. Enhanced GFP expression due to silencing suppression in infiltration zones was visualized under UV light as yellow-green fluorescence against a light-red background caused by a combination of red chlorophyll autofluorescence and normal GFP expression from the 16c transgene. Suppression activity also was examined in a three-component system in which wild-type N. benthamiana plants were coinfiltrated with constructs expressing GFP, double-stranded GFP RNA (the silencing trigger), and AL2 or AL2 C33A.
Results from representative experiments are presented in Fig. 7. Wild-type AL2 displayed characteristically weak silencing suppression activity compared to that of the p19-positive control, as indicated by a light yellow-green fluorescence that nevertheless was greater than that of the GUS-negative control, and by greater accumulation of GFP mRNA relative to that of the GUS control. By both of these measures, the activity of AL2 C33A appeared comparable to that of wild-type AL2 protein, indicating that self-interaction is not required for local silencing suppression.
FIG. 7.
Local silencing suppression by AL2 and AL2 C33A. (A) Leaves of N. benthamiana line 16c, expressing a 35S-GFP transgene, were coinfiltrated with Agrobacterium cultures delivering Ti plasmids expressing GFP (silencing trigger) and AL2, AL2 C33A, GUS (negative control), or p19 (positive control). Representative leaves were photographed under UV light 6 days postinfiltration. Under these conditions, high-level GFP expression in the infiltration zone (relative to the level of GFP expression from the transgene) appears yellow-green. (B) Leaves of wild-type N. benthamiana plants were coinfiltrated with constructs expressing GFP, double-stranded GFP RNA (dsGFP) (silencing trigger), and AL2, AL2 C33A, or GUS (negative control). Total RNA was isolated from infiltration zones and was subjected to RNA gel blot hybridization using a 32P-labeled GFP-specific probe. 18S rRNAs are shown as loading controls.
DISCUSSION
The studies presented here represent initial steps toward understanding the regulation of the multifunctional geminivirus AL2 protein. We demonstrate for the first time that begomovirus AL2 protein, but not its curtovirus counterpart L2 from BCTV, is capable of self-interaction and that AL2 complexes accumulate primarily in the nucleus. Mutations in the zinc finger-like CCHC motif that disrupt self-interaction in plant cells (C33A and C43A) or cause aberrant subnuclear localization of AL2 complexes (C35A) impair transcriptional activation activity. Thus, efficient transcription activation correlates with formation and correct subnuclear localization of AL2:AL2 complexes. The exception is residue H40, which is not required for self-interaction or wild-type nuclear localization in N. benthamiana cells and exhibits greater transcriptional activation than wild-type AL2. Why the H40A substitution confers greater activity is not clear, but this mutant protein stands in sharp contrast to the cysteine substitution mutants and underscores the correlation between self-association and efficient transactivation of the CP promoter. On the other hand, AL2, the AL2 CCHC mutants, and L2 all efficiently interact with ADK in the cytoplasm. The viral proteins also suppress local silencing in a manner that correlates with ADK interaction, as demonstrated previously for AL2 and L2 and here for AL2 C33A (40). Thus, monomeric AL2 and L2 participate in the formation of cytoplasmic AL2:ADK and L2:ADK complexes, which are correlated with transcription-independent, local silencing suppression. Taken together, the findings reported here are consistent with AL2 and L2 sharing pathogenicity functions, including the ability to suppress silencing through ADK inactivation, while only AL2 is a transcriptional activator.
It is curious that all of the AL2 CCHC mutants were unable to self-interact in yeast, yet two of them (AL2 C35A and AL2 H40A) were found to self-interact in plant cells by BiFC analysis. While we cannot explain this difference, we speculate that AL2:AL2 interaction could be stabilized by posttranslational modification (e.g., phosphorylation) that might occur correctly in plant but not yeast cells or that the complexes are stabilized by plant cellular proteins. In any case, it appears that contacts lying outside of the zinc finger-like domain contribute to self-interaction, since L2 is unable to self-interact in yeast or in planta despite the presence of an intact CCHC motif.
Evidence from BiFC experiments indicated that in all cases in which self-interaction was detected, AL2 complexes accumulated in the nucleus. However, self-interaction is not a requirement for nuclear import, since all of the AL2 CCHC mutant proteins were detected in the nucleus following expression in insect cells. This is further supported by the accumulation of a portion of AL2:ADK and CCHC mutant AL2:ADK complexes in the nucleus. However, the C35 residue may play a role in subnuclear localization, as AL2 C35A complexes exhibited a punctate distribution not seen with wild-type AL2. Aberrant subnuclear localization may explain why this mutant protein dimerizes but does not show wild-type levels of transcription activation. At this time, we cannot say whether C33 and C43 also are required for correct subnuclear accumulation, because the method we used to analyze localization (BiFC) requires self-interaction. It should be noted that our results are consistent with those of a previous analysis of similar cysteine mutant AL2 (C2) proteins from Tomato yellow leaf curl virus China (TYLCV-C). Analysis of GFP fusion proteins showed that the invariant cysteine residues were not required for nuclear localization, but the mutant proteins displayed aberrant subnuclear localization (35).
BiFC analysis showed that AL2:ADK and L2:ADK complexes accumulate predominantly in the cytoplasm, providing a rationale for the presence of AL2 in both the nucleus and the cytoplasm of TGMV-infected cells (41). As noted above, detection of signal attributable to AL2:ADK and CCHC mutant AL2:ADK complexes in the nucleus indicates that the ADK interaction, and the CCHC mutations, do not obscure or inactivate the AL2 NLS. The biological significance of ADK relocalization, if any, is unclear. Because both AL2 and L2 can inactivate ADK in vitro and in vivo and L2:ADK complexes appear entirely cytoplasmic, it seems unlikely that relocalization is a major mechanism of ADK inactivation.
One of the most prominent features of AL2 is the zinc finger-like CCHC motif that it shares with the related L2 protein (15). It has been demonstrated for the TYLCV-C AL2 homologue that the invariant cysteine residues of this motif are involved in binding zinc, single-stranded DNA, and dsDNA (37). The role of the conserved histidine residue in these activities was not investigated, as it is not present in the TYLCV-C protein. Here, we show that all of the residues comprising the CCHC motif in the TGMV AL2 protein are essential for self-interaction in yeast cells, while two of them (C33 and C43) also are essential for self-interaction in plant cells. Taken together, these observations identify the zinc finger-like motif as an interaction platform that alone or together with other domains mediates interactions with molecular partners, linking and facilitating regulation of the several biochemical and functional activities of AL2. More specifically, we speculate that the zinc finger-like domain acts as a rheostat to modulate the relative propensity of AL2 to interact with itself, with cellular proteins, and with DNA, favoring different interactions under different conditions. This in turn could influence the localization and/or alter the functionality of the viral protein. The molecular details of such a mechanism are not yet clear. Further analysis of the role of the CCHC motif in the AL2:SNF1 interaction should provide some insight into this question, and these studies are in progress.
Acknowledgments
We thank Deborah Parris for helpful discussions and advice on glycerol gradient protocols. We also thank Janet Sunter for maintenance of the N. benthamiana suspension cell line and assistance with protoplast transfection experiments and Shannon Woody for technical assistance with BiFC studies.
This work was supported by grants from the National Research Initiative of the USDA Cooperative State Research and Extension Service (number 04-35301-14508; D.M.B.), a National Institutes of Health MBRS/SCORE Grant (GM-08194; G.S.), and the OARDC SEEDS Program (J.A.L.).
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Siedman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley and Sons, New York, NY.
- 2.Baulcombe, D. C. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 3.Bisaro, D. M. 2006. Silencing suppression by geminivirus proteins. Virology 344:158-168. [DOI] [PubMed] [Google Scholar]
- 4.Brough, C. L., W. E. Gardiner, N. Inamdar, X. Y. Zhang, M. Ehrlich, and D. M. Bisaro. 1992. DNA methylation inhibits propagation of tomato golden mosaic virus DNA in transfected protoplasts. Plant Mol. Biol. 18:703-712. [DOI] [PubMed] [Google Scholar]
- 5.Ding, S.-W., H. Li, R. Lu, F. Li, and W.-X. Li. 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102:109-115. [DOI] [PubMed] [Google Scholar]
- 6.Dong, X., R. van Wezel, J. Stanley, and Y. Hong. 2003. Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77:7026-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durfee, T., K. Becherer, P.-L. Chen, S.-H. Yeh, Y. Yang, A. E. Kilburn, W.-H. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 8.Elmer, J. S., L. Brand, G. Sunter, W. E. Gardiner, D. M. Bisaro, and S. G. Rogers. 1988. Genetic analysis of tomato golden mosaic virus. II. The product of the AL1 coding sequence is required for replication. Nucleic Acids Res. 16:7043-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 10.Gallo, M. L., D. H. Jackwood, M. Murphy, H. S. Marsden, and D. S. Parris. 1988. Purification of the HSV-1 65 kilodalton DNA binding protein: properties of the protein and evidence of its association with the viral encoded DNA polymerase. J. Virol. 62:2874-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gladfelter, H. J., P. A. Eagle, E. P. B. Fontes, L. Batts, and L. Hanley-Bowdoin. 1997. Two domains of the AL1 protein mediate geminivirus origin recognition. Virology 239:186-197. [DOI] [PubMed] [Google Scholar]
- 12.Hanley-Bowdoin, L., S. Settlage, and D. Robertson. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus replication. Mol. Plant Pathol. 5:149-156. [DOI] [PubMed] [Google Scholar]
- 13.Hao, L., H. Wang, G. Sunter, and D. M. Bisaro. 2003. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinase. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 15.Hartitz, M. D., G. Sunter, and D. M. Bisaro. 1999. The geminivirus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1-14. [DOI] [PubMed] [Google Scholar]
- 16.Hormuzdi, S. G., and D. M. Bisaro. 1995. Genetic analysis of beet curly top virus: examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206:1044-1054. [DOI] [PubMed] [Google Scholar]
- 17.Hu, C.-D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 18.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot: induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and ligation at the viral origin of replication by the replication protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffatt, B. A., Y. Y. Stevens, M. S. Allen, J. D. Snider, L. A. Periera, M. I. Todorova, P. S. Summers, E. A. Weretilnyk, L. Martin-Caffrey, and C. Wagner. 2002. Adenosine kinase deficiency is associated with developmental abnormalities and reduced transmethylation. Plant Physiol. 128:812-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Restrepo, M. A., D. D. Freed, and J. C. Carrington. 1990. Nuclear transport of plant potyviral proteins. Plant Cell 2:987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 24.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemanpillai, M., I. Dry, J. Randles, and A. Rezaian. 2003. Transcriptional silencing of geminiviral promoter-driven transgenes following homologous virus infection. Mol. Plant-Microbe Interact. 16:429-438. [DOI] [PubMed] [Google Scholar]
- 26.Shen, W., and L. Hanley-Bowdoin. 2006. Geminivirus infection up-regulates the expression of two Arabidopsis protein kinases related to yeast SNF1- and mammalian AMPK-activating kinases. Plant Physiol. 142:1642-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shivaprasad, P. V., R. Akbergenov, D. Trinks, R. Rajeswaran, K. Veluthambi, T. Hohn, and M. M. Pooggin. 2005. Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J. Virol. 79:8149-8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanley, J., J. Latham, M. S. Pinner, I. Bedford, and P. G. Markham. 1992. Mutational analysis of the monopartite geminivirus beet curly top virus. Virology 191:396-405. [DOI] [PubMed] [Google Scholar]
- 29.Sunter, G., and D. M. Bisaro. 2003. Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305:452-462. [DOI] [PubMed] [Google Scholar]
- 30.Sunter, G., and D. M. Bisaro. 1997. Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): evidence for activation and derepression mechanisms. Virology 232:269-280. [DOI] [PubMed] [Google Scholar]
- 31.Sunter, G., and D. M. Bisaro. 1991. Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416-419. [DOI] [PubMed] [Google Scholar]
- 32.Sunter, G., and D. M. Bisaro. 1992. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunter, G., J. Sunter, and D. M. Bisaro. 2001. Plants expressing tomato golden mosaic virus AL2 or beet curly top virus L2 transgenes show enhanced susceptibility to infection by DNA and RNA viruses. Virology 285:59-70. [DOI] [PubMed] [Google Scholar]
- 34.Trinks, D., R. Rajeswaran, P. V. Shivaprasad, R. Akbergenov, E. J. Oakeley, K. Veluthambi, T. Hohn, and M. Pooggin. 2005. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 79:2517-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanitharani, R., P. Chellappan, and C. M. Fauquet. 2005. Geminiviruses and RNA silencing. Trends Plant Sci. 10:144-151. [DOI] [PubMed] [Google Scholar]
- 36.Vanitharani, R., P. Chellappan, J. S. Pita, and C. Fauquet. 2004. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 78:9487-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Wezel, R., H. Liu, Z. Wu, J. Stanley, and Y. Hong. 2003. Contribution of the zinc finger to zinc and DNA binding by a suppressor of posttranscriptional gene silencing. J. Virol. 77:696-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 39.Voinnet, O., Y. M. Pinto, and D. C. Baulcombe. 1999. Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96:14147-14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, H., K. J. Buckley, X. Yang, R. C. Buchmann, and D. M. Bisaro. 2005. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 79:7410-7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H., L. Hao, C.-Y. Shung, G. Sunter, and D. M. Bisaro. 2003. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang, C., P. Han, I. Lutziger, K. Wang, and D. J. Oliver. 1999. A mini binary vector series for plant transformation. Plant Mol. Biol. 40:711-717. [DOI] [PubMed] [Google Scholar]