Abstract
Although lineage I West Nile virus (WNV) strain Eg101 induced beta interferon (IFN-β) production as early as 12 h after infection in primary mouse embryo fibroblasts and did not inhibit the JAK-STAT signaling pathway, it was still able to replicate efficiently. To gain insights about possible viral countermeasures used by this virus to suppress the host response, the cell transcriptional profile and the kinetics of IFN regulatory factor (IRF) expression and activation were examined at early times after infection. By 12 h after WNV infection, the majority of the up-regulated genes were ones involved in IFN pathways. However, comparison of IFN-stimulated gene (ISG) expression levels in mock-infected, IFN-treated, and virus-infected cells indicated that WNV infection suppressed the up-regulation of a subset of ISGs, including genes involved in transcriptional regulation, apoptosis, and stress responses, prior to 24 h after infection. Analysis of mRNA and protein levels for representative genes indicated that suppression was at the transcriptional and posttranscriptional levels. Translocation of IRF-3 to the nucleus was observed beginning at 8 h, IRF-7 expression was detected by 12 h, but IRF-1 expression was not detected until 24 h after infection. Virus-induced gene suppression was sufficient to overcome the effect of exogenous IFN pretreatment for 1 h but not for 4 h prior to infection. These data indicate that WNV can selectively counteract the host response at early times after infection by previously unreported mechanisms.
West Nile virus (WNV) is a single-stranded, positive-sense RNA virus that is maintained in nature via a mosquito-bird-mosquito transmission cycle. A member of the family Flaviviridae, genus Flavivirus, WNV is closely related to other human pathogens, such as yellow fever virus, dengue (DEN) virus, tick-borne encephalitis virus, Japanese encephalitis virus (JEV), and Murray Valley encephalitis virus. Flaviviruses can replicate in a wide variety of cultured cells of vertebrate and arthropod origin. In mammalian and avian cells, the replication kinetics of flaviviruses are significantly slower than those of many other RNA viruses, such as encephalomyocarditis virus, Sendai virus, and Newcastle disease virus, that are typically utilized in studies of the activation of interferon (IFN) pathways in cells (37). WN progeny virions begin to be released from infected cells between 10 and 12 h after infection, and peak titers of extracellular virus are observed by 24 to 36 h after infection (46).
In response to virus infection, cells produce cytokines, such as type I IFNs, as a result of activation/up-regulation of various transcription factors including members of the IFN regulatory factor (IRF) family (2, 34, 43, 44). IRFs constitute a family of transcriptional activators and repressors that regulate the innate host response, and cell growth. Nine members of the mammalian IRF family (IRFs 1 to 9) are known, all of which contain a conserved N-terminal DNA binding domain that interacts with the 5′-AANNGAAA-3′ promoter element that is similar to the IFN-stimulated response element (ISRE). The carboxy-terminal regions of IRFs, with the exception of IRF-1 and IRF-2, contain a domain that mediates interactions with other family members as well as other transcription factors. The IRFs are activated by phosphorylation in their C-terminal regions that results in their translocation from the cytoplasm to the nucleus and transcriptional activation of ISRE-containing genes. The various IRFs differ in their cellular location, structural properties, the stimuli that activate them, and the functions they evoke. During a viral infection, IRF-3 has an early role in inducing the transcription of IFN-β, which in turn induces the expression of IRF-7. Newly synthesized IRF-7 promotes the transcription of members of the IFN-α gene family, thereby creating a positive-feedback loop that augments the host antiviral response. IRF-1, IRF-3, and IRF-7 together with histone transacetylases form the active IFN-α enhanceosome (1).
The binding of IFNs to cell surface IFN receptors results in activation of JAK, which in turn phosphorylates STAT1 and STAT2 (41). Phosphorylation of Tyr on these STATs leads to the formation of two transcription activator complexes, the IFN-α-activated factor (AAF) complex, which consists of a homodimer of pSTAT1, and the IFN-stimulated gene factor 3 (ISGF3) complex, which is composed of heterotrimers of STAT1, STAT2, and IRF-9. Once formed, AAF and ISGF3 complexes translocate to the nucleus, where they bind to promoters that contain the IFN-γ-activated sequence (GAS) and ISRE, respectively. Both IRF-5 and IRF-7 have an ISRE in their promoters, while the IRF-1, IRF-2, IRF-8, and IRF-9 genes have a GAS element in their promoters (42). IFNs induce the formation of a number of additional STAT-containing complexes, including STAT3 and STAT5 homodimers, as well as STAT2-STAT1 and STAT5-CrkL (v-Crk sarcoma virus CT10 oncogene homologue avian-like) heterodimers, that are involved in mediating gene transcription (6). IFN-mediated activation of ISRE and GAS elements results in the transcriptional activation of a large number of target genes involved in a variety of biological activities.
Members of different families of RNA viruses have been shown to directly induce ISGs prior to IFN production by the activation of several transcription factors, including NF-κB, AP-1 (ATF-2/c-Jun), and IRF-3 (5, 26, 38). A recent study showed that infection of cells with the flaviviruses JEV or DEN type 2 (DEN2) virus, induced IFN-β gene expression by RIG-I-dependent IRF-3 and phosphatidylinositol 3-kinase-dependent NF-κB activation pathways (7).
Previously reported microarray studies of mosquito-borne flavivirus infections were done on transformed cell cultures at relatively late stages of infection. The cellular response to WNV infection was analyzed at 24, 36, and 48 h after infection in human embryonic kidney 293 cells (14) and at 24 h in human glioblastoma cells (24). In 293 cells, the expression of IFN-β and several ISGs was detected at 24 and 36 h after WNV infection (14). The response to DEN2 virus infection in cultures of human umbilical vein endothelial cells was analyzed at 48 h (51) and 120 h (25) after infection. Multiple functional cell pathways, including stress, antiviral defense, immune, cell adhesion, wound, and inflammatory pathways were activated by these times. In the single published in vivo study, a number of host genes were reported to be differentially expressed in the brains, livers, and spleens of mice 5 days after a subcutaneous WNV inoculation (50). Neuroinvasive WNV strains induced higher levels of expression from these genes than did attenuated virus strains. Up-regulated genes included those associated with IFN, T-cell response, inflammation, and apoptosis pathways.
Viruses have evolved various means of counteracting the antiviral action of IFN, including inhibiting IFN induction and/or production, inhibiting JAK-STAT signaling, and altering ISG expression (15, 37). Results from several recent studies indicate that flaviviruses regulate JAK-STAT signaling in infected cells. Studies with WNV and Kunjin virus provided evidence that viral protein(s) can block JAK-STAT signaling (17, 23, 29), and various flavivirus nonstructural proteins were subsequently implicated as possible regulators of JAK-STAT signaling (4, 28, 32).
In a previous study, we showed that even though infection of primary mouse embryo fibroblasts (MEFs) with WNV strain Eg101 induced IFN-β production as early as 12 h after infection and did not interfere with the STAT signaling pathway, virus replicated efficiently (39). To investigate whether a WNV infection might be able to counteract host innate responses at early times after infection, the kinetics of activation/expression of IRFs and the expression of ISGs in primary MEFs were examined beginning at 2 h after infection. IRF-3 activation/up-regulation and IRF-7 expression were detected before 12 h after infection, while the induction of IRF-1 expression was delayed until 24 h. Although induction of a number of ISGs was observed, WNV infection suppressed the up-regulation of a subset of ISGs prior to 24 h after infection.
MATERIALS AND METHODS
Cells and virus.
Primary C3H/He (He) MEFs were prepared from 14- to 18-day-old embryos and frozen after one passage in culture. Only cells from passages 2 or 3 were used for experiments. MEFs were grown in minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum and 10 μg/ml gentamicin. Baby hamster kidney-21/WI2 cells (hereafter referred to as BHK cells) (49) were maintained in minimal essential medium supplemented with 5% heat-inactivated fetal calf serum and 10 μg/ml gentamicin. All cells were grown at 37°C in a 4.5% CO2 atmosphere. A stock of WNV lineage I strain Eg101 was prepared by infecting BHK cells at a multiplicity of infection (MOI) of 0.1 and harvesting culture fluid 32 h after infection, a time when the cell monolayer was still intact. To remove IFN produced by BHK cells, viral particles were pelletted by ultracentrifugation, resuspended in medium with 0.5% serum (4 × 107 PFU/ml), aliquoted, and stored at −80°C.
Sample preparation and microarray hybridization.
Replicate confluent primary MEF monolayers in T25 flasks were either mock infected or infected with WNV Eg101 strain at an MOI of 10 for 2, 6, or 12 h (samples M, W2, W6, and W12). Other sets of cultures were treated with 1,000 U/ml of universal type I IFN (PBL Biomedical Laboratories, Piscataway, NJ) for 1 h and then either incubated with fresh medium for 2 h before cell RNA isolation (I samples) or infected with WNV at an MOI of 10 for 1 h and then incubated with medium for 1 h (I/W2 samples). Cells in a replicate flask were counted prior to infection in each experiment to precisely calculate the amount of virus needed to achieve the desired MOI. WNV was adsorbed for 1 h at room temperature, and the monolayers were washed with 5 ml of medium three times to remove unbound virus before fresh medium was added. Samples of culture fluid from infected replicate flasks were removed at various times and stored at −80°C until titration by plaque assay as described previously (39).
Total cellular RNA was extracted by lysing cells with TriReagent (Molecular Research Center, Cincinnati, OH). RNA purification, quality control, labeling, GeneChip hybridization, data acquisition, and preliminary data analysis were performed by Expression Analysis (Durham, NC) according to standard protocols available from Affymetrix (Santa Clara, CA). Briefly, RNA quality was assessed with an Agilent 2100 Bioanalyzer (Palo Alto, CA). Biotin-labeled cRNAs were quantified with a spectrophotometer, their integrity was assessed on test chips (Test3 array; Affymetrix), and they were then loaded onto mouse genomic 430A chips (Affymetrix). After hybridization, the chips were washed, stained using a GeneChip Fluidics Station 400 (Affymetrix), and then scanned with a GeneChip Scanner 3000 (Affymetrix). The 430A chip contains 22,690 transcripts (RefSeq database sequences and sequences related to the U74Av2 array). Data were collected for each sample from two independent experiments (biological replicates).
Microarray data analysis.
GeneChip Operating System software (version 1.1.1; Affymetrix) was used to analyze the data. For comparisons across different arrays, the data from each array were first normalized by a global scaling strategy, using a scaling target intensity of 500. By using the Affymetrix-defined comparison mathematical algorithms, the relative change in expression between each of the infected samples in comparison to a mock-infected control of the same cell type was calculated, log 2 transformed, and further classified as not changed, increased (signal log ratio change P value of <0.005), decreased (signal log ratio change P value of >0.995), or marginally increased or decreased. To classify a gene as significantly up-regulated or down-regulated after infection or IFN treatment, two additional criteria were used: (i) the relative change had to be greater than or equal to 2 (signal log ratio of 1 if up-regulated or −1 if down-regulated) to be classified as increased or decreased, and (ii) genes that were classified as up-regulated had to be flagged as present in the infected/treated samples, while genes that were classified as down-regulated had to be flagged as present in the mock-infected control sample.
Microarray data sets (CEL files) were further analyzed at the Emory Biomolecular and Computing Resource Center using the R-Bioconducting package (http://www.bioconductor.org) for Affymetrix array analysis. Briefly, data sets were loaded into the R-Bioconductor package and RMA (robust multichip analysis) values were generated using the following parameters: RMA background correction method, quantile normalization method, perfect match only values, and the median polish summary method for signal calculation (20). RMA values were imported into GeneSpring, version 6.2 (Silicon Genetics, Redwood City, CA), and expression values for each gene were normalized across chips to the median value of each gene. In each experiment, relative change values (experimental versus control) were determined, and a cutoff value of 2.0 was arbitrarily applied to ascertain genes that were differentially expressed across experiments and between the conditions within each experiment.
Real-time qRT-PCR.
The reaction mixture contained 500 ng of cellular RNA, the primer pair (1 μM), and the probe (0.2 μM) in a total volume of 50 μl. Real-time quantitative reverse transcription-PCR (qRT-PCR) analysis of mouse genes was performed with Assays-on-Demand 20× primer and fluorogenic TaqMan FAM/TAMRA (6-carboxyfluorescein/6-carboxytetramethylrhodamine)-labeled hybridization probe mixes from Applied Biosystems (Foster City, CA), The catalog identification numbers of assays used for the genes indicated in parentheses were Mm00437762_m1 (B2m), Mm00492089_m1 (Daxx), Mm00516788_m1 (Irf7), Mm004912265_m1 (Rsad2 or Vig1), Mm00726868_m1 (Oas1g), Mm00455081_m1 (Oasl1), Mm00469582_m1 (Tyki), Mm00432307_m1 (Casp11), Mm00515191_m1 (Irf1), Mm00782550_s1 (SOCS1), Mm00447364_m1 (Trim21), and Mm00556509_ m1 (Zc3hdc1). The RNAs were quantified using an Applied Biosystems 7500 sequence detection system. The mRNA of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used as an endogenous control and was detected using TaqMan mouse GAPDH Control Reagents primers and probe (Applied Biosystems). One-step RT-PCR was performed for each target gene and for the endogenous control in a singleplex format using 200 ng of RNA and the TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). The cycling parameters were as follows: reverse transcription at 48°C for 30 min, AmpliTaq activation at 95°C for 10 min, denaturation at 95°C for 15 s, and annealing/extension at 60°C for 1 min (cycle repeated 40 times). Triplicate cycle threshold CT values were analyzed with Microsoft Excel using the comparative CT (ΔΔCT) method of the SDS software (Applied Biosystems). Statistical analysis of the data was done using the TINV test in Microsoft Excel. The values were first normalized to the endogenous reference gene (GAPDH) and then presented as relative change in comparison to the uninfected calibrator sample in relative quantification (RQ) units. Error bars indicate the calculated maximum (RQMax) and minimum (RQMin) expression levels and represent the standard error (SE) of the mean of the expression levels. The error bars are based on an RQMin/Max of the 95% confidence level. If the error bars for any two samples do not overlap, the expression levels of these samples are significantly different (P value of <0.05).
Confocal microscopy.
He MEFs grown to 50% confluence on 15-mm glass coverslips in wells of a 24-well plate were infected with WNV at an MOI of 5. The cells were fixed by incubation with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min and then permeabilized by ice-cold methanol for 10 min. Coverslips were washed with PBS and then blocked overnight with 5% horse serum (Invitrogen, Carlsbad, CA) in PBS. Coverslips were incubated with rabbit anti-IRF-3 (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:50 in PBS containing 5% horse serum for 1 h at 37°C and then washed three times with PBS. Coverslips were next incubated with chicken anti-rabbit immunoglobulin G-fluorescein isothiocyanate (Santa Cruz Biotechnology) diluted 1:300 in PBS containing 5% horse serum. After a washing step with PBS, the coverslips were mounted on glass slides with Prolong mounting medium (Invitrogen) and visualized with a 100× oil immersion objective on an LSM 510 laser confocal microscope using LSM 5 (version 3.2) software (Carl Zeiss Inc., Thornwood, NY). The images compared were obtained using the same instrument settings.
Western blotting.
Replicate cell monolayers were washed with ice-cold PBS and then scraped into radioimmunoprecipitation assay buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]), containing Complete, mini, EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN) and phosphatase Cocktail Inhibitor II (Sigma-Aldrich, St. Louis, MO). Following separation by 7.5% SDS-polyacrylamide gel electrophoresis (PAGE), the proteins were electrophoretically transferred to a nitrocellulose membrane. The membrane was blocked with 1× Tris-buffered saline containing 5% dry milk (or bovine serum albumin when phosphorylated proteins were to be detected) and 0.1% Tween-20 for 1 h at 22°C and then incubated with a polyclonal primary antibody specific for IRF-1, IRF-3, IRF-7, actin (Santa Cruz Biotechnology), caspase 11 (Abcam, Cambridge, MA), phospho-IRF-3 (Ser390), phospho-STAT5 (Tyr694), phospho-CrkL (Tyr207), or eIF4E (Cell Signaling, Beverly, MA) overnight at 4°C in the presence of blocking buffer. The blots were then incubated with the secondary antibody (anti-rabbit horseradish peroxidase; Santa Cruz Biotechnology) for 1 h at 22°C and processed for enhanced chemiluminescence using a Super-Signal West Pico detection kit (Pierce, Rockford, IL) according to the manufacturer's instructions.
RESULTS
Comparison of gene expression in WNV-infected MEFs and IFN-treated MEFs.
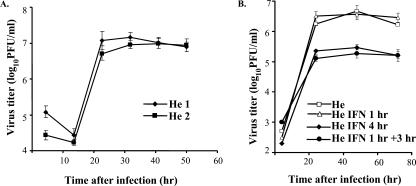
Our previous study showed that incubation of primary He MEFs with 1,000 IU/ml of universal type I IFN for 4 h prior to infection with WNV Eg101 at an MOI of 1 decreased the virus yield by about 10-fold (39). When cells were infected 3 h after pretreatment with 1,000 IU/ml of universal type I IFN for 1 h, a similar decrease in the viral yield was observed (Fig. 1B). However, when cells were pretreated for only 1 h prior to infection with the same concentration of IFN, no effect on virus yield was observed (Fig. 1B), suggesting that the viral infection could overcome the establishment of an antiviral state induced by exogenous IFN under these conditions. To obtain a comprehensive view of the changes in cell gene expression occurring at early times after WNV infection, microarray assays were utilized. Previous microarray analyses reported for flavivirus-infected cells were done at later times after infection and in transformed cell cultures. Many transformed cell lines have mutations in IFN pathway genes (8). Third-passage primary He MEFs were mock infected or infected with WNV (MOI of 10) in two independent experiments. These biological replicates were designated He1 and He2. In both experiments, a duplicate flask was used to generate a viral growth curve (Fig. 1A). Also, duplicate flasks of primary MEFs were incubated with universal type I IFN (1,000 IU/ml) or medium for 1 h and then incubated with fresh medium for two more hours. Total cell RNA was harvested from infected cells at 2, 6, and 12 h after infection and from IFN-treated cells 3 h after initiation of IFN treatment and hybridized to mouse expression 430A chips. Data were collected and analyzed as described in Materials and Methods. In IFN-treated MEFs, 98 probe sets (representing 84 genes) showed up-regulation by more than twofold. The majority of the up-regulated genes were known ISGs (9). Data obtained from multiple probe sets for a single gene were consistent. For example, three probe sets for Stat1 showed induction levels of 3.0-, 2.6-, and 2.3-fold. These data provided an internal control for the hybridization conditions as well as for chip integrity. Several of the genes up-regulated in He MEFs after IFN treatment were not previously reported to be activated by IFN. These included chloride channel calcium-activated genes 1 and 2 (Clca1 and 2) and the plectin 1 (Plec1), nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta (Nfkbiz), and caspase 11 (Casp11, mouse homolog of human CASP4) genes.
FIG. 1.
Kinetics of WNV replication in primary He MEFs with or without IFN pretreatment. (A) Confluent monolayers of He MEFs were infected with WNV at an MOI of 10. (B) Confluent monolayers of He MEFs were treated as follows and then infected with WNV at an MOI of 10: untreated, incubated with 1,000 IU/ml of universal type I IFN for 1 or 4 h prior to infection, or treated with IFN for 1 h and then incubated with medium without IFN for 3 h prior to infection. Culture fluid samples harvested at the indicated times after infection were titrated for infectivity by plaque assay. Each data point is the average of duplicate titrations from two experiments. Error bars indicate standard deviations.
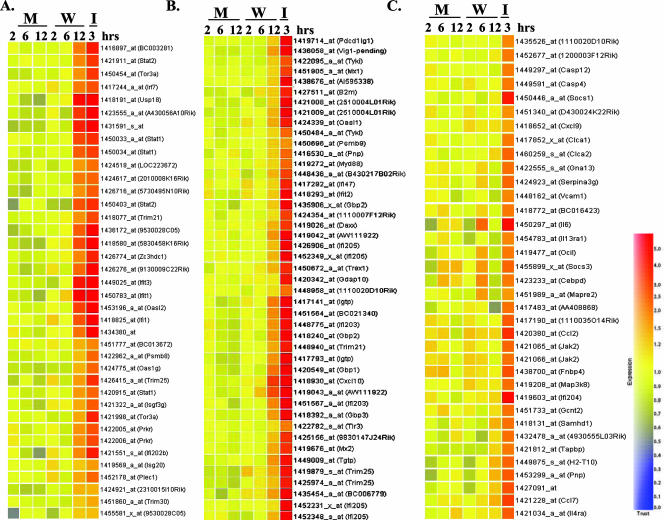
The basic script of GeneSpring software was used to compare the expression data from WNV-infected and IFN-treated cells. A gene tree, an experimental tree, a k-means classification, and a self-organizing map were generated using genes that were twofold overexpressed or twofold underexpressed under at least one condition in the RMA. The data from biological replicates (He1 and He2) were averaged by the software for each of the 256 genes presented in the tree. Hierarchical clustering was performed for those genes with changes in their expression levels of about or more than twofold in IFN-treated (I) or WNV-infected (W) cells compared to the control cells. Three clusters of ISGs were revealed (Fig. 2). The first cluster included probe sets that showed a high level of induction in both WNV-infected and IFN-treated cells. These genes are listed in Table 1 in the same order as in Fig. 2. IFN-induced genes, such as the Oas family genes and the Irf7, Irf9, Prkr, ISG20, Stat1, and Stat2 genes, were present in this cluster, further confirming that the IFN pathway was efficiently activated by 12 h after infection with WNV. The expression of three genes, the G1p2 (human homolog, ISG15), Ifit1 (ISG56 or p56), and Ifit3 (ISG49) genes, which are known targets for IRF-3 (16), was induced to high levels by 12 h after WNV infection (Fig. 2 and Table 1). Human ISG54 (mouse homolog, Ifit2), ISG56, and ISG15 genes were previously shown to be Jak1 (hence IFN) independent but IRF-3 dependent (10). Although Ifit1 was induced to high levels by WNV infection, Ifit2 (ISG54) was not induced by 12 h, suggesting that the expression of various members of the p56 family of ISGs is differentially regulated at early stages of WNV infection in MEFs. A recent study reported that vesicular stomatitis virus infection up-regulated the expression of the ISG56 gene but not the ISG54 gene in the livers of infected mice (45). This study also showed that the regulation of these two genes varied in different mouse organs and with different stimuli.
FIG. 2.
Hierarchical clustering of genes showing altered expression levels after WNV infection or IFN treatment in primary He MEFs. Total cellular RNA from mock-infected (M), WNV-infected (W), or IFN-treated (I) He MEFs was extracted and analyzed on 430A microarray chips as described in Materials and Methods. Only genes with about or more than a twofold increase in their expression levels in WNV-infected or IFN-treated cells compared to mock-infected MEFs were included in the hierarchical clustering shown. The color scale on the right indicates normalized expression values from the highest (5.0) to the lowest (0.0) levels.
TABLE 1.
Genes up-regulated to a higher level in WNV-infected MEFs than in IFN-treated cells
| Gene symbola | Gene name (accession no.)b | UniGene no. | Avg change in expression (n-fold)c
|
|
|---|---|---|---|---|
| I cells | W12 cells | |||
| Parp9 | Poly(ADP-ribose) polymerase family, member 9 (BC003281) | Mm.49074 | 3.8 | 2.7 |
| Stat2 | Signal transducer and activator of transcription 2 | Mm.293120 | 5.1 | 3.4 |
| Tor3a | Torsin family 3, member A | Mm.206737 | 3.3 | 2.5 |
| Irf7 | IFN regulatory factor 7 | Mm.3233 | 2.2 | 2.2 |
| Usp18 | Ubiquitin specific protease 18 | Mm.326911 | 18.0 | 12.9 |
| Ifi44 | IFN-induced protein 44 (A430056A10) | Mm.30756 | 8.4 | 6.6 |
| G1p2 | IFN-α-inducible protein (AK018325) | Mm.4950 | 8.6 | 5.9 |
| Stat1 | Signal transducer and activator of transcription 1 | Mm.277406 | 3.0 | 3.9 |
| ApoL | Apoliprotein L (LOC223672) | Mm.243758 | 1.9 | 1.9 |
| Ifi35 | IFN-induced protein 35 (2010008K16Rik) | Mm.45558 | 2.2 | 2.1 |
| Tdrd7 | Tudor domain containing 7 (5730495N10Rik) | Mm.275413 | 1.5 | 2.1 |
| Trim21 | Tripartite motif protein 21 | Mm.321227 | 3.9 | 2.1 |
| 9530028C05 | Hypothetical protein; similar to major histocompatibility complex class II E beta | Mm.258507 | 10.8 | 3.3 |
| Ifrg28 | 28-kDa IFN-α responsive; Rtp4 (5830458K16Rik) | Mm.180157 | 9.9 | 6.1 |
| Zc3hdc1 | Zinc finger CCCH-type domain-containing 1 | Mm.268462 | 4.3 | 3.6 |
| Ifih1 (MDA5) | IFN induced with helicase C domain 1 (9130009C22Ric) | Mm.136224 | 8.3 | 3.8 |
| Ifit3 | IFN-induced protein with tcp repeats 3 | Mm.271850 | 22.6 | 9.2 |
| Ifit1 | IFN-induced protein with tcp repeats 1 | Mm.6718 | 59.4 | 14.7 |
| Oasl2 | 2′-5′ Oligoadenylate synthetase-like 2 | Mm.228363 | 7.2 | 4.8 |
| Ifi1 | IFN-inducible protein 1 | Mm.29938 | 11.4 | 3.2 |
| 9830147J24Rik | Similar to guanylate nucleotide binding protein | Mm.45740 | 7.4 | 2.7 |
| BC013672 | cDNA sequence BC013672; DEAD/DEAH box helicase | Mm.275233 | 2.1 | 1.4 |
| Psmb8 | Proteosome subunit, beta type 8 | Mm.180191 | 2.6 | 1.7 |
| Oas1g | 2′-5′ Oligoadenylate synthetase 1G | Mm.14301 | 2.2 | 2.0 |
| Trim25 | Tripartite motif protein 25 | Mm.248445 | 5.3 | 2.2 |
| Isgf3g | IFN-dependent positive acting transcription factor 3 gamma | Mm.2032 | 2.7 | 2.1 |
| Prkr | Protein kinase, IFN-inducible double-stranded RNA dependent | Mm.311918 | 2.7 | 2.0 |
| Ifi202b | IFN-activated gene 202B | Mm.218770 | 3.7 | 3.5 |
| Isg20 | IFN-stimulated protein | Mm.322843 | 1.8 | NC |
| Bst2 | Bone marrow stromal cell antigen 2 | Mm.260325 | 1.8 | 2.2 |
| Plec1 | Plectin 1 | Mm.307601 | 2.1 | 1.9 |
| Trim30 | Tripartite motif protein 30 | Mm.295578 | 1.9 | 1.9 |
Genes are ordered by hierarchical clustering as in Fig. 2A. Underlined genes were detected as up-regulated 8 h after infection.
GenBank accession number refers to the sequence used to design a gene's probe for the Affymetrix 430A chips.
Cells were treated with 1,000 IU/ml of universal type I IFN (I) or infected with WNV for 12 h (W12). Average relative change was calculated by comparison to values obtained in the mock-infected cells. NC, not changed.
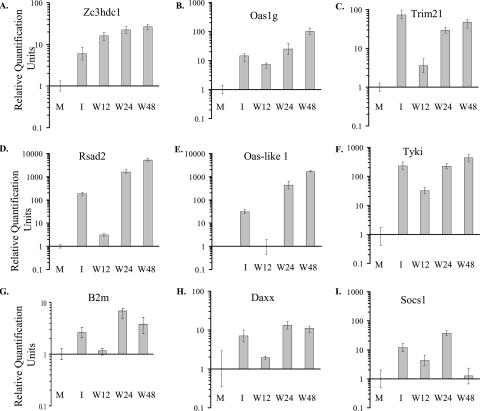
The time course of expression of three genes from cluster 1, zinc finger CCCH-type domain-containing 1 (Zc3hdc1), tripartite motif protein 21 (Trim21), and 2′-5′ oligoadenylate synthetase 1G (Oas1g), was analyzed by real-time qRT-PCR. Zc3hdc1 and Oas1g mRNA levels were increased by 12 h after infection and by 3 h after initiation of IFN treatment. The expression of these genes continued to increase through 48 h after infection (Fig. 3A and B). At 12 h after infection, Trim21 expression was significantly lower than that observed 3 h after IFN treatment, but the level of expression in infected cells increased to that in IFN-treated cells by 24 h (Fig. 3C). The real-time qRT-PCR data for each of the genes tested corroborated the microarray data, and consistent results were obtained for RNA samples from independent biological replicates. However, the absolute values obtained from the microarray and RT-PCR assays were not identical due to differences intrinsic to these two techniques.
FIG. 3.
Analysis of changes in the expression levels of selected genes after IFN treatment or WNV infection by real-time qRT-PCR. Primary He MEFs were mock infected (M), infected with WNV at an MOI of 10, or incubated with 1,000 IU/ml of universal type I IFN for 1 h followed by a 2-h incubation in medium without IFN (I). Changes in intracellular mRNA levels of the Zc3hdc1, Oas1g, Trim21, Rsad2, Oas-like 1, Tyki, B2m, Daxx, and Socs1 genes are shown. Infected cell samples were harvested 12, 24, or 48 h after infection (W12, W24, or W48, respectively). The mRNA level for each gene is expressed in RQ units as the log10 relative change in expression compared to the level of the same mRNA present in mock-infected He cells or, in the case of the Oas-like 1 gene, in 12-h WNV-infected cells. Error bars represent the SE (n = 3) and are based on an RQMin/Max of the 95% confidence level. If the error bars for any two samples do not overlap, the expression levels of these samples are significantly different (P value of <0.05).
The second cluster (Table 2) included genes for which expression levels increased by only about twofold in 12-h WNV-infected cells (Fig. 2B) but for which strong up-regulation by IFN treatment was observed. The level of expression of the Rsad2 gene (also called Vig1, Cig5, and viperin) was confirmed by qRT-PCR. At 12 h after WNV infection, Rsad2 expression was more than 10-fold lower than that in IFN-treated cells, but by 24 and 48 h after infection, the expression of this gene was induced to much higher levels (Fig. 3D). Real time qRT-PCR analysis of the mRNAs levels of two other genes from the second cluster, Oas-like 1 and the thymidylate kinase family lipopolysaccharide-inducible member (Tyki), confirmed a reduced level of expression of these genes at 12 h after WNV infection compared to cells treated with IFN (Fig. 3E and F). Three of the five p65 guanylate binding protein (Gbp) gene family members (Gbp1 to Gbp3) were also in the second cluster. These genes are classical secondary response genes that require de novo synthesis of transcription factors activated by a JAK2-dependent pathway for their up-regulation (30). The detected upregulation of these genes indicated that activation of JAK2 had occurred in WNV-infected MEFs by 12 h. Genes associated with apoptosis such as proapoptotic transcription factor Irf1, programmed cell death 1 ligand 1 (Pdcd1lg1), Fas death domain-associated protein (Daxx), and poly(ADP-ribose) polymerase family member 14 (Parp14) as well as major histocompatibility complex class I antigen presentation pathway genes, such as beta-2 microglobulin (B2m) and proteasome (prosome and macropain) subunit beta 9 (Psmb9) were also in the second cluster (Table 2). The lower expression levels of the B2m and Daxx genes at 12 h after WNV infection were confirmed by real-time qRT-PCR (Fig. 3G and H). However, the maximum induction of both of these genes after either IFN treatment or WNV infection was relatively low (about 10-fold) even at later times after infection. These data showed that efficient activation of the genes in cluster 2 was delayed compared to that of genes in cluster 1 in WNV-infected cells.
TABLE 2.
Genes up-regulated to a lower level in WNV-infected cells than in IFN-treated cells
| Gene symbola | Gene name (accession no.)b | UniGene no. | Avg change in expression (n-fold)c
|
|
|---|---|---|---|---|
| I | W12 | |||
| Pdcd1lg1 | Programmed cell death 1 ligand 1 | Mm.245363 | 5.4 | NC |
| Rsad2, Vig1 | Radical S-adenosyl domain methionine containing 2 | Mm.24045 | 10.6 | 2.0 |
| Tyki | Thymidylate kinase family LPS-inducible member | Mm.27183 | 2.8 | NC |
| Mx1 | Myxovirus (influenza virus) resistance 1 | Mm.33996 | 3.2 | NC |
| AI595338 | Expressed sequence AI595338 (GBP, Mpa2l) | Mm.275893 | 8.1 | NC |
| B2m | Beta-2 microglobulin | Mm.163 | 4.4 | NC |
| Oasl1 | 2′-5′ Oligoadenylate synthetase-like 1 | Mm.95479 | 4.7 | 1.7 |
| Psmb9 | Proteasome (prosome, macropain) subunit, beta 9 | Mm.787 | 2.5 | NC |
| Pnp | Purine-nucleoside phosphorylase | Mm.17932 | 1.9 | NC |
| Myd88 | Myeloid differentiation primary response gene 88 product | Mm.213003 | 2.3 | NC |
| Irf1 | IFN regulatory factor 1 (B430217B02Rik) | Mm.105218 | 1.9 | NC |
| Ifi47 | IFN-γ- inducible protein 47 | Mm.24769 | 3.4 | NC |
| Ifit2 | IFN-induced protein with tcp repeats 2 | Mm.2036 | 5.1 | NC |
| Gbp2 | Guanylate nucleotide binding protein 2 | Mm.24038 | 10.0 | 2.6 |
| 1110007F12Rik | RIKEN cDNA 1110007F12 gene product | Mm.248440 | 3.3 | NC |
| Daxx | Fas death domain-associated protein | Mm.271809 | 3.4 | NC |
| Iigp1 | IFN-inducible GTPase 1(AW111922) | Mm.261140 | 37.7 | 5.8 |
| Ifi205 | IFN-activated gene 205 product | Mm.255414 | 7.0 | 1.5 |
| Trex1 | Three prime repair exonuclease 1 | Mm.347553 | 4.2 | NC |
| Gdap10 | Ganglioside-induced differentiation-assoc-protein 10 | Mm.358597 | 3.8 | NC |
| Ifrg15 | IFN-α-responsive gene product | Mm.351270 | 1.9 | NC |
| Igtp | IFN-γ-induced GTPase product | Mm.33902 | 11.7 | 3.4 |
| Parp14 | Poly(ADP-ribose) polymerase family, member 14 (BC021340) | Mm.89210 | 7.7 | 2.9 |
| Ifi203 | IFN-activated gene 203 product | Mm.261270 | 11.2 | 3.1 |
| Gbp1 | Guanylate nucleotide binding protein 1 | Mm.250 | 11.9 | 3.9 |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | Mm.877 | 25.4 | 5.8 |
| Gbp3 | Guanylate nucleotide binding protein 3 | Mm.1909 | 4.8 | 1.8 |
| Tlr3 | Toll-like receptor 3 | Mm.33874 | 1.6 | 1.4 |
| 9830147J24Rik | RIKEN cDNA 9830147J24 gene (Cd274, Pdcd1l1) | Mm.45740 | 2.5 | NC |
| Mx2 | Myxovirus (influenza virus) resistance 2 | Mm.14157 | 4.1 | 1.7 |
| Tgtp | T-cell specific GTPase | Mm.338891 | 4.9 | 2.0 |
| BC006779 | PPAR-α-interacting complex protein 285 | Mm.315872 | 6.4 | 1.7 |
Genes are ordered by hierarchical clustering as in Fig. 2B.
GenBank accession number refers to the sequence used to design a gene's probe for the Affymetrix 430A chips.
Average relative change was calculated by comparison to the value obtained in mock-infected cells. I, IFN-treated cells; W12, 12-h WNV-infected cells; NC, not changed.
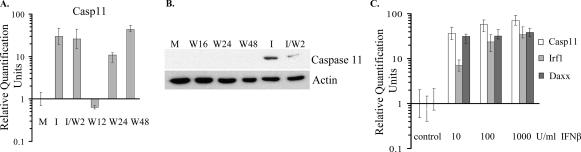
Genes in the third cluster were efficiently induced by IFN treatment but were activated to less than twofold in WNV-infected cells by 12 h (Table 3 and Fig. 2C). Included in this cluster were genes encoding stress-related serine proteinase inhibitors (Serpina3g and Serping1); genes from calcium-regulated pathways, such as the Clca1 and Clca2 genes; and genes involved in apoptosis pathways, such as the Casp11 (mouse homolog of human CASP4) and Casp12 genes. Genes involved in transcriptional regulation, such as the Nfkbiz and CCAAT/enhancer binding protein (C/EBP) delta (Cebpd) genes; several genes encoding cytokines, such as the chemokine (C-X-C motif) ligand 9 (Cxcl9), chemokine (C-C motif) ligand 2 (Ccl2), chemokine (C-C motif) ligand 7 (Ccl7), and interleukin 6 (Il-6) genes; and genes regulating the cytokine response, such as the suppressor of cytokine signaling 1 and 3 (Socs1 and Socs3) genes, were in this cluster. Socs1 and Socs3 gene expression was induced by IFN treatment but not at 12 h after WNV infection (Fig. 2C and Table 3). Socs1 is an important in vivo inhibitor of type I IFN signaling and contributes to balancing the beneficial antiviral versus detrimental proinflammatory effects of the innate immune responses (11). Real-time RT-PCR data showed that treatment of cells with IFN for 3 h resulted in more than a 10-fold increase in Socs1 mRNA expression compared to control cells, while the amount of Socs1 mRNA detected in 12-h WNV-infected cells was threefold lower than in IFN-treated cells (Fig. 3I). At 24 h after infection, the level of Socs1 expression was significantly higher than that detected in IFN-treated cells, but the expression of Socs1 mRNA was suppressed by 48 h after infection to the level observed in control uninfected cells. The timing of this suppression suggested that it might be modulated by the accumulation of viral protein(s) between 24 and 48 h in infected cells. The kinetics of induction of another gene from cluster 3, the Casp11 gene, was also analyzed by real-time qRT-PCR (Fig. 4A). At 12 h after infection, the expression of Casp11 mRNA was suppressed below the level observed in mock-infected cells, while IFN treatment induced the expression of this gene by more than fivefold. By 24 after infection, Casp11 mRNA levels had increased by 10-fold, and by 48 h the levels were higher than those in the IFN-treated cells (Fig. 4A). A strong Casp11 protein band was detected in the IFN-treated sample by Western blotting (Fig. 4B). However, no protein was detected at 16, 24, or 48 h after WNV infection (Fig. 4B), indicating that viral infection suppressed the expression of protein levels even after the mRNA levels were up-regulated.
TABLE 3.
Genes up-regulated in IFN-treated but not WNV-infected MEFsa
| Gene symbolb | Gene name (accession no.c or alternative name) | UniGene no. | Avg increase in expression (n-fold)d |
|---|---|---|---|
| 1110020D10Rik | Torsin A interacting protein 2 (lumenal domain like) | Mm.253335 | 1.7 |
| Pnpt1 | Polyribonucleotide nucleotidyltransferase 1 | Mm.211131 | 2.1 |
| Casp12 | Caspase 12 | Mm.42163 | 2.0 |
| Casp11 | Caspase 11, apoptosis-related cysteine protease | Mm.1569 | 2.0 |
| Socs1 | Suppressor of cytokine signaling 1 | Mm.130 | 3.7 |
| Arid5a | AT-rich interactive domain 5A (Mrf1 like) (D430024K22Rik) | Mm.34316 | 1.9 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9 | Mm.766 | 1.7 |
| Clca1 | Chloride channel calcium activated 1 | Mm.275745 | 2.3 |
| Gna13 | Guanine nucleotide binding protein, alpha 13 | Mm.193925 | 2 |
| Serpina3g | Serine (or cysteine) proteinase inhibitor, clade A, 3G | Mm.264709 | 2 |
| Vcam1 | Vascular cell adhesion molecule 1 | Mm.76649 | 2 |
| BC016423 | cDNA sequence BC016423 (peripherial benzodiazepine receptor associated protein) | Mm.221609 | 2 |
| Il-6 | Interleukin 6 | Mm.1019 | 7.8 |
| Il-13ra1 | Interleukin 13 receptor, alpha 1 | Mm.24208 | 2 |
| Ocil | Osteoclast inhibitory lectin | Mm.197536 | 3.5 |
| Socs3 | Suppressor of cytokine signaling 3 | Mm.3468 | 3.2 |
| Cebpd | CCAAT/enhancer binding protein (C/EBP), delta | Mm.347407 | 2.6 |
| Mapre2 | Microtubule-associated protein, RP/EB family, member 2 | Mm.132237 | 1.3 |
| Nfkbiz | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta (AA408868) | Mm.247272 | 2 |
| Pbef1 | Pre-B-cell colony-enhancing factor 1 (nicotinamide phosphoribosyltransferase) | Mm.202727 | 2 |
| Ccl2 | Chemokine (C-C motif) ligand 2 | Mm.290320 | 2.9 |
| Jak2 | Janus kinase 2 | Mm.275839 | 2 |
| Fnbp4 | Formin binding protein 4 | Mm.314887 | 3.5 |
| Map3k8 | Mitogen-activated protein kinase kinase kinase 8 | Mm.3275 | 2 |
| Ifi204 (Mnda, Ifi16) | Myeloid cell nuclear differentiation antigen (IFN-γ-inducible protein 16) | Mm.212870 | 4 |
| Samhd1 | SAM domain and HD domain, 1 | Mm.248478 | 2 |
| Ibrdc3 | IBR domain containing 3 | Mm.259672 | 2.3 |
| Tapbp | TAP binding protein | Mm.154457 | 2.3 |
| H2-T10 | histocompatibility 2, T region locus 10 | Mm.87776 | 2.8 |
| Ccl7 | Chemokine (C-C motif) ligand 7 | Mm.341574 | 3.3 |
| Nmi | N-myc (and STAT) interactor | Mm.7491 | 2.1 |
| Lgals3bp | Lectin, galactoside-binding, soluble, 3 binding protein | Mm.3152 | 1.9 |
| Il-4ra | Interleukin 4 receptor, alpha | Mm.233802 | 2.8 |
Genes were up-regulated in cells treated with 1,000 IU/ml of IFN type I but not in cells infected with WNV for 12 h.
GenBank accession number refers to the sequence used to design a gene's probe for the Affymetrix 430A chips.
Genes are ordered by hierarchical clustering as in Fig. 2C.
Average relative change was calculated by comparison to the value obtained in the mock-treated cells.
FIG. 4.
Real-time qRT-PCR and Western blot analysis of Casp11 expression in response to IFN treatment and/or WNV infection. Primary He MEFs were mock infected for 1 h (M), infected with WNV at an MOI of 10 (W), incubated with 1,000 IU/ml of universal type I IFN for 1 h and then either incubated with medium without IFN for 2 h (I) or infected with WNV for 1 h and incubated with medium for 1 h (I/W2). Replicate infected cultures were harvested at the indicated times after infection. (A) Relative quantification of intracellular Casp11 mRNA by real-time qRT-PCR. RNA levels are expressed in RQ units as the log10 relative change in expression compared to the level of this mRNA in mock-infected cells. Each mRNA was normalized to GAPDH mRNA. Error bars represent the SE (n = 3). (B) Caspase 11 proteins in cell lysates were detected by Western blotting as described in Materials and Methods. The blots shown are representative of three independent experiments. (C) Primary He MEFs were treated with 10, 100, or 1,000 U/ml of mouse IFN-β, and intracellular mRNA levels expressed from the Casp11, Irf1, and Daxx genes were quantified by qRT-PCR.
Analysis of gene expression in IFN-pretreated WNV-infected cells.
To investigate whether pretreatment with IFN could abrogate the observed suppression of the expression of a subset of cell genes by WNV infection, gene expression was analyzed in cells pretreated with IFN for 1 h and then infected with WNV, a condition shown not to affect viral yield (Fig. 1B). MEFs were incubated with universal type I IFN or medium for 1 h and then either incubated with fresh medium for two more hours (I) or infected with WNV (MOI 10) for 1 h and then incubated with medium for an additional hour (I/WZ) before cell RNA was harvested. The list of genes obtained with the I/W2 samples showing differential expression compared to untreated cells was very similar to the list of genes obtained with samples from cells treated only with IFN (I samples) (data not shown), suggesting that viral infection did not interfere with the up-regulation of the majority of the IFN-regulated genes. Most of these transcripts showed either the same or a slightly higher level of induction in I/W2 samples compared to I samples. However, a few genes, including the Irf1, B2m, and Socs3 genes, showed a lower relative change in I/W2 samples than in I samples. A pairwise comparison using I samples as the baseline and I/W2 samples as the experiment revealed a cluster of genes induced to a lower level in the presence of WNV infection (data not shown). This cluster included the Irf1 and B2m genes, the myeloid differentiation primary response gene 88 (Myd88) (Table 2 genes), and the Cebpd, Socs1 and Socs3 genes (Table 3 genes). Each of these genes was about 1.5- to 2-fold down-regulated in I/W2 samples compared to I samples. A strong Casp11 band was detected in the I sample by Western blotting, but only a faint band was detected in the I/W2 sample (Fig. 4B, compare lanes I and I/W2), indicating that WNV infection could still efficiently suppress Casp11 protein levels after a 1-h IFN pretreatment of cells prior to infection.
To rule out the possibility that the activation of genes in the I and I/W2 cells was due to nonphysiologic effects caused by the large amount of nonhomologous universal type I IFN used, cells were treated with 10, 100, or 1,000 IU/ml of mouse IFN-β, which is the first IFN produced in response to a viral infection. Samples were harvested after 3 h of IFN treatment, and the levels of Irf1, Casp11, and Daxx mRNA were analyzed by real-time qRT-PCR (Fig. 4C). For each of these genes, similar levels of induction were observed with each of the different concentrations of mouse IFN-β used, with the exception of 10 IU/ml of IFN-β, for which a lower level of Irf1 mRNA induction was observed. The mRNA levels induced for each of these genes were higher than those seen after incubation of cells with 1,000 IU/ml of universal type I IFN (Fig. 4C).
Analysis of IRF expression patterns in WNV-infected cells.
The microarray analysis showed that Irf1 mRNA was not up-regulated at 12 h after WNV Eg101 infection in primary MEFs even though IRF-1 was previously reported to be rapidly up-regulated by other virus infections and IFN (43). In virus-infected cells, IRF-3 typically is rapidly activated and induces the transcription of IFN-β and ISGs. Both IRF-3 and IRF-7 are direct transducers of virus-mediated signaling that results in the induction of a number of other ISGs (19, 22). The activation and expression kinetics of IRF-3 and the expression kinetics of IRF-1 and IRF-7 were next analyzed in WNV-infected MEFs.
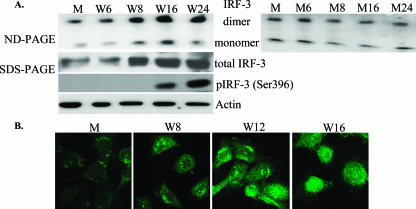
IRF-3 activation was first analyzed in WNV-infected (MOI of 10) He MEFs by detection of dimer formation. Dimerized IRF-3 has a significantly retarded mobility on nondenaturing gels compared to the monomeric form (21). Low levels of both IRF-3 monomer and dimer bands were consistently detected in uninfected and mock-infected cells. This was expected since previous studies reported constitutive expression of IFN-α/β, albeit at very low levels, in the absence of virus or other IFN inducers in normal primary fibroblasts, splenocytes, and bone marrow cells obtained from mouse embryos (42). Consistent with this, some phosphorylated STAT1 protein was also previously detected in mock-infected primary MEFs (39). The amounts of IRF-3 monomer and dimer increased between 6 and 8 h after WNV infection and remained elevated through 24 h (Fig. 5A). In contrast, in mock-infected cells, the levels of IRF-3 monomers and dimers were similar at each of the times analyzed. The increase in the level of IRF-3 dimers observed by 8 h after infection was consistent with our previous data, showing up-regulation of IFN-β mRNA between 8 and 12 h and increased phosphorylation of STAT1 by 16 h after WNV infection (39). IRF-3 is activated by phosphorylation at multiple sites, including Ser396, within the carboxy-terminal portion of the protein (31, 40). The phosphorylation of IRF-3 Ser396 (phosphoSer396 IRF-3) in primary He MEFs was examined by immunoblotting using an antibody that recognizes IRF-3 phosphorylated at this position. Lysates were prepared from cells infected with WNV for 6, 8, 16, and 24 h and from mock-treated cells. As shown in Fig. 5A, the phosphoSer396 IRF-3 isoform was first detected at 16 h after infection. Activated IRF-3 is known to translocate to the nucleus (26). The kinetics of IRF-3 nuclear translocation in WNV-infected He MEFs were evaluated by confocal microscopy. As expected, IRF-3 was detected only in the cytoplasm of mock-infected cells (Fig. 5B). Focal nuclear localization of IRF-3 was observed in WNV-infected cells at 8 and 12 h after WNV infection. By 16 h, IRF-3 was predominantly located in the nucleus and was diffusely distributed. These results indicate that WNV Eg101 infection rapidly activates IRF-3.
FIG. 5.
Analysis of expression and activation of IRF-3 in WNV-infected He MEFs. (A) Primary He MEFs were mock infected for 1 h (M) or infected with WNV at an MOI of 10 (W). Replicate infected cultures were harvested at the indicated times after mock or WNV infection. IRF-3 dimers were detected by Western blotting after separation by electrophoresis on 7.5% nondenaturing (ND) polyacrylamide gels (left and right upper panels). Total IRF-3 and phosphoSer396 IRF-3 were detected by Western blotting after separation in 10% SDS-PAGE (middle panels). Actin was used as a cell protein control. (B) He MEFs were mock infected or infected with WNV for 8, 12, or 16 h. The cells were fixed, permeabilized, and incubated with anti-IRF-3 antibody at a dilution of 1:50, followed by incubation with secondary fluorescein isothiocyanate-conjugated antibody and visualized by confocal microscopy. Representative images are shown for each time point.
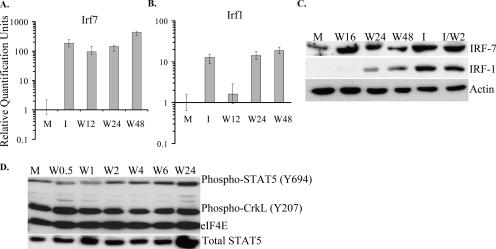
The expression patterns of Irf1 and Irf7 in WNV-infected MEFs were analyzed by real-time qRT-PCR using total RNA from mock- or WNV-infected He MEFs extracted at 12, 24, or 48 h after infection. Irf7 mRNA levels increased by more than 100-fold in samples from 12-h infected and universal type I IFN-treated (1,000 IU/ml) cells (Fig. 6A). Pretreatment of cells with 10 or 100 U/ml of mouse IFN-β induced Ifr7 mRNA to similar levels (data not shown). In contrast, Irf1 mRNA was induced by less than twofold by 12 h after WNV infection (Fig. 6B) but by more than 10-fold after IFN treatment. By 24 h after infection, Irf1 mRNA expression levels had increased by 10-fold and remained at this level through 48 h. A low level of IRF-7 was detected in mock-infected cells by Western analysis. By 16 h after infection, the level of IRF-7 had increased significantly and was similar to the level detected in the IFN-treated sample (Fig. 6C). IRF-7 protein expression remained elevated through 48 h after infection. In contrast, IRF-1 was not detected in mock-infected cells, and a very weak IRF-1 band was detected at 16 h. Thereafter, although IRF-1 protein levels increased, they did not reach the levels seen in IFN-treated cells at either 24 or 48 h (Fig. 6C, exposure time of 15 min), even though the levels of IRF-1 mRNA at both these times were similar to those in IFN-treated cells (Fig. 6B). When IRF-1 levels were analyzed at earlier times after infection, a very faint band was detected at 1 h after infection, but no bands were detected at 2, 4, 6, or 8 h after infection even after an exposure time of 1 h (data not shown).
FIG. 6.
Analysis of IRF-1, IRF-7, and STAT5 expression and STAT5 and CrkL phosphorylation in WNV-infected He MEFs. Primary He MEFs were mock infected for 1 h (M), infected with WNV at an MOI of 10 (W), incubated with 1,000 IU/ml of universal type I IFN for 1 h and then incubated with medium without IFN for 2 h (I), or incubated with 1,000 IU/ml of universal type I IFN for 1 h and then infected with WNV (MOI of 10) for 2 h (I/W2). (A) Relative quantification of intracellular Irf7 mRNA by real-time qRT-PCR. RNA levels are expressed in RQ units as the log10 relative change in expression compared to the level of this mRNA in mock-infected cells. Error bars represent the SE (n = 3). (B) Relative quantification of intracellular Irf7 mRNA by real-time qRT-PCR. Each mRNA was normalized to GAPDH mRNA. (C) IRF-1 and IRF-7 expression detected by Western blotting after separation by 10% SDS-PAGE. (D) Phosphorylated forms of STAT5 and CrkL as well as total STAT5 were detected by Western blotting after separation by 10% SDS-PAGE. Actin or eIF4E were used as cell protein controls. The blots shown are representative of three independent experiments.
STAT5 plays an important role in IFN signaling and participates in the induction of type I IFN-dependent responses (47). Because the STAT5-CrkL heterodimer regulates GAS-mediating gene transcription (12) and IRF-1 is known to have a GAS element in its promoter (36, 48), the time courses of STAT5 and CrkL phosphorylation were next examined in WNV-infected cells. STAT5 is associated with Tyk2, whereas CrkL associates with the guanine-nucleotide exchange factor C3G. Following type I IFN stimulation, CrkL is recruited to Tyk2, which phosphorylates both STAT5 and CrkL, causing them to form heterodimers that are translocated to the nucleus, where they activate GAS-mediated gene induction (12). The level of phosphorylated STAT5 was reduced shortly after adsorption of virus to cells (0.5- and 1-h time points) compared to mock-infected cells, but by 2 h (1 h after addition of fresh medium), the levels had increased and were similar to those seen in mock-infected cells. By 6 h after infection, the levels had increased slightly but by 24 h had increased significantly (Fig. 6D). A similar amount of total STAT5 was observed in mock- and WNV-infected cells at 0.5, 1, 2, 4, and 6 h after infection and increased notably by 24 h after infection (Fig. 6D). CrkL phosphorylation levels were high in all samples tested (Fig. 6D). The data suggest that formation of the STAT5-CrkL heterodimer is not blocked by WNV Eg101 infection.
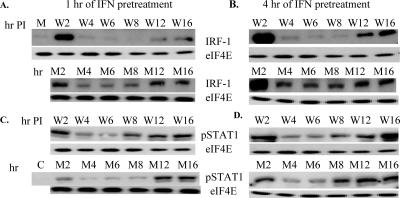
The data indicate that WNV infection delayed induction of Irf1 mRNA prior to 24 h after infection (Fig. 6B) and significantly reduced the level of IRF-1 protein through 48 h (Fig. 6C). To test whether virus infection could suppress IRF-1 expression in IFN-pretreated cells, the time course of IRF-1 expression was examined in cells pretreated with universal type I IFN (1,000 IU/ml) for 1 h (Fig. 7A, upper panel) or 4 h (Fig. 7B, upper panel) before infection with WNV at an MOI of 10. Samples from cells treated only with IFN were analyzed at the same time points (Fig. 7A and B, bottom panels). A very faint band of IRF-1 was detected in extracts from mock-infected cells. The same dilution of IRF-1 antibodies (1:500) was used as in the experiment shown in Fig. 5, where no band in the mock-infected cells was detected. A faint band was detected in these experiments due to an increase in the exposure time from 10 to 30 min (Fig. 7A, lane M). Increased IRF-1 expression was observed in cells treated with IFN for 1 h and then incubated with medium for 2 h (Fig. 7A, lower panel). A much higher amount of IRF-1 was detected in extracts from cells pretreated with IFN for 4 h and then with medium for 2 h (Fig. 7B, lower panel). By 4 h, after either 1 or 4 h of IFN pretreatment, the levels of IRF-1 had decreased significantly (Fig. 7A and B, lower panels). The short half-life of the IRF-1 protein (30 min) (43) ensures that IFN gene induction is transient. In 1-h IFN-pretreated cells, the levels of IRF-1 were slightly increased at 8 h of incubation and further increased at 12 and 16 h, probably in response to newly produced IFN-β (Fig. 7A, lower panel). In the 4-h IFN-pretreated cells, IRF-1 levels were similar from 4 through 8 h of incubation and then increased at 12 and 16 h (Fig. 7B, lower panel). At 2 h after the removal of IFN, IRF-1 levels in extracts from uninfected cells and cells infected with WNV were similar (Fig. 7A and B, upper panels; compare W2 and M2). At 4, 6, and 8 h, the levels of IRF-1 were lower in the infected cells than those in uninfected cells (Fig. 7A and B, upper panels). Although the levels of IRF-1 increased in infected cells at 12 and 16 h, the levels remained much lower than those seen at 2 h. This secondary up-regulation of IRF-1 in response to infection was stronger in cells pretreated with IFN for 4 h than in cells pretreated with IFN for 1 h (Fig. 7A and B, compare lanes W12 and W16). These results indicate that the expression of IRF-1 was also suppressed in WNV-infected cells pretreated with IFN but to a lesser extent and for a shorter time than in cells that were not IFN pretreated. AAF (STAT1 homodimer) is the major transcription complex that activates Irf1 transcription, and STAT1 phosphorylation is a prerequisite for homodimer formation. To determine whether IRF-1 suppression was due to inhibition of STAT1 phosphorylation, the levels of STAT1 phosphorylation were analyzed by Western blotting using the same set of samples. The levels of STAT1 phosphorylation were increased at 2 h of incubation for all of the extracts compared to the levels in the control cells (Fig. 7C and D). In contrast to what was observed with the IRF-1 protein, the amount of STAT1 phosphorylation observed in the infected samples was similar to or increased compared to that in uninfected cells pretreated for either 1 or 4 h with IFN (Fig. 7C and D). Also, no differences in the levels of STAT5 and CrkL phosphorylation were observed between the 1- or 4-h IFN-pretreated, uninfected or infected samples (data not shown). These results indicate that WNV infection suppresses IRF-1 protein expression without inhibiting STAT1 activation.
FIG. 7.
Effect of different lengths of IFN pretreatment on IRF-1 expression and STAT1 phosphorylation after WNV infection. Primary He MEFs were mock infected for 1 h (M) or untreated (C) or incubated with 1,000 IU/ml of universal type I IFN for 1 h or 4 h and then either incubated with medium without IFN (M2 to M16) or infected with WNV (W2 to W16) for the indicated times. IRF-1 (A and B) or phospho-STAT1 (C and D) protein expression was detected by Western blotting after protein separation by 10% SDS-PAGE. eIF4E was used as a protein control. The blots shown are representative of three independent experiments.
DISCUSSION
The kinetics of activation of IRF-3, induction of IRF-1, and expression of IRF-7 and induction of the IFN-induced antiviral response in MEFs infected with the lineage I WNV strain Eg101 were investigated in the present study. WNV Eg101 and NY99 are closely related lineage I strains (differing by 21 amino acid residues). Although Eg101 is less neuroinvasive than NY99 because it lacks an E protein glycosylation site, both viruses show similar levels of neurovirulence (3). All strains of WNV replicate efficiently in a wide variety of cultured cells of vertebrate and arthropod origin. How WNV overcomes innate antiviral defenses in so many different host cells is currently being investigated. Results from several recent studies indicate that flaviviruses, including some WNV strains, can evoke processes that negatively regulate JAK-STAT signaling and IFN-induced responses in various infected cell lines (4, 17, 23, 29, 32). These events occur with the accumulation of viral proteins, which begins between 12 and 24 h after infection. The various levels of resistance to the antiviral actions of IFN observed for different strains of WNV and other flaviviruses have been mainly attributed to the efficiency with which particular viral nonstructural proteins, such as NS4B of DEN virus (32), NS5 of tick-borne encephalitis virus (4), or NS2A of WNV (Kunjin) (28), control the host cell JAK-STAT signaling pathway (23).
Early events that occur prior to significant amplification of WNV Eg101 viral RNA or proteins were analyzed in the present study. The data obtained suggest that WNV Eg101 employs other countermeasures than the previously reported inhibition of JAK-STAT signaling by viral nonstructural proteins to attenuate the host antiviral response. Phosphorylation of both STAT1 isoforms was observed by 16 h and increased to peak levels by 40 h after WNV Eg101 infection in MEFs (39), indicating activation rather than blockage of the IFN-induced JAK-STAT pathway. Even though an antiviral state was induced, WNV Eg101 established and maintained efficient levels of replication.
Functional classification of all of the genes detected as up-regulated by 12 h after WNV infection by microarray analysis showed that many were known ISGs. Comparison of ISG expression levels in mock-infected cells, cells treated with IFN, and cells infected with WNV for 12 h revealed three clusters of genes. The first cluster included genes showing a high level of induction in both 12-h WNV-infected and 3-h IFN-treated cells. The target genes for IFN-β, components of the ISGF3 transcription activator complex, such as the Stat1, Stat2, and Irf9 genes, as well as ISGF3 target genes, such as the Oas, Prkr, Mx2, and Irf7 genes (33), were present in this cluster. We previously reported up-regulation of the IFN-β gene in WNV Eg101-infected He MEFs by 12 h after infection (39). In the present study, the kinetics of activation of IRF-3, the transcription factor essential for the induction of type I IFN and other ISGs, were directly examined by analysis of IRF-3 dimer formation, IRF-3 phosphorylation at Ser396, and IRF-3 nuclear translocation. IRF-3 dimer levels increased between 6 and 8 h after WNV infection and remained elevated through 24 h (Fig. 5A). IRF-3 translocation to the nucleus was detected by 8 h after infection, and by 16 h the majority of IRF-3 was located in the nucleus. Consistent with this time course of IRF-3 activation, up-regulation of known IRF-3-targeted genes, such as mouse homologs of human ISG56 (Ifit1), ISG49 (Ifit3), and ISG15 (G1p2) (16), was detected by 8 h after infection with WNV (data not shown). IRF-3 has been reported to be activated by phosphorylation of Ser/Thr residues located between residues 382 to 408 (GGASSLENTVDLHISNSHPLSLTSDQY) (40). Phosphorylation of both IRF-3 Ser396 and Ser398 was observed following Sendai virus infection or double-stranded RNA treatment, but neither of these serines was phosphorylated after lipopolysaccharide treatment even though IRF-3 was activated and translocated to the nucleus (40). Another study reported that only Ser386 was phosphorylated after infection with Newcastle disease virus or treatment with either double-stranded RNA or lipopolysaccharide and that phosphorylated Ser386 was detected only in dimers, suggesting that this phosphorylation site is the critical determinant of IRF-3 activation (31). In the present study, phosphorylation of Ser396 was not detected until 16 h after WNV infection. However, IRF-3 dimerization and nuclear localization were observed starting at 8 h after infection. These results suggest that phosphorylation of other IRF-3 Ser/Thr residues, such as Ser386, may be responsible for the initial activation of IRF-3 in WNV Eg101-infected MEFs. A previous study with WNV NY99 also reported the inability to detect phosphorylation of IRF-3 at Ser396 prior to 36 or 20 h after infection in two human cell lines, 293 (embryonic kidney) and A549 (lung carcinoma), respectively (13, 14).
The second cluster of genes revealed by comparison of ISG expression levels in mock-infected cells, cells treated with IFN, and cells infected with WNV for 12 h included ones that were induced to only about twofold in infected cells but were strongly up-regulated by IFN treatment. Genes in the third cluster were induced by IFN treatment but not up-regulated prior to 24 h in WNV-infected cells. Genes in these clusters included ones encoding stress-related serine proteinase inhibitors and genes in calcium-regulated and apoptosis pathways. A number of genes involved in transcriptional regulation such as the Irf1, Nfkbiz (NF-κB inhibitor zeta [IκBζ]), and Cebpd genes were also in these clusters. Suppression of these three transcription factors by WNV Eg101 infection would be expected to inhibit the expression of the genes they regulate. Studies with IκBζ knockout mice showed that IκBζ is essential for the expression of a number of genes, including lipopolysaccharide-inducible genes such as the Il-6, Casp11, Ccl7, and Cebpd genes (52). Several genes encoding cytokines (Cxcl9, Ccl2, Ccl7, and Il-6 genes) were not induced in WNV Eg101-infected MEFs. The decreased expression of several ISGs associated with apoptosis, such as proapoptotic transcription factor Irf1, Pdcd1lg, Daxx, and Parp14, in WNV-infected MEFs would be expected to sustain cell viability and enhance virus production. Only two caspases, the Casp11 and Casp12 genes, were induced by type I IFN treatment in MEFs (Fig. 2C), and the expression of both of these genes was suppressed by WNV Eg101 infection. The human homolog of mouse Casp11, CASP4, has been shown to mediate endoplasmic reticulum stress-induced apoptosis (18), and suppression of its expression would be expected to prolong the survival of WNV-infected cells.
It was previously reported that DEN2 virus infection induced both IRF-1 and IRF-7 expression in human A549 cells, while JEV infection did not induce these IRFs even though IFN-β was induced in both JEV and DEN2 virus infections before 24 h (7). JEV, but not DEN2 virus infection, efficiently blocked the IFN-induced JAK-STAT signaling cascade by preventing Tyk2 and STAT phosphorylation (27). Chang et al. (7) proposed that in DEN2 virus-infected cells, AAF (STAT1 homodimer) and ISGF3 (STAT1/STAT2/IRF-9 complex) complexes formed and induced Irf1 and Irf7 transcription, respectively, while in JEV-infected cells, blockade of Tyk2 and STAT activation prevented formation of these transcription complexes (7). WNV Eg101 infection of MEFs also did not block STAT activation (39). In DEN2 virus-infected cells, IRF-1 protein expression was observed by 12 h after infection, while IRF-7 expression was not detected until 36 h (7). In WNV Eg101-infected cells, IRF-7 was detected by 16 h after infection, indicating rapid formation of ISGF3 complexes. However, IRF-1 expression was not detected until 24 h. Because STAT1 and STAT5 were phosphorylated in WNV-infected cells and because these are components of the AAV and STAT5-CrkL complexes, respectively, that are required for transcriptional activation of genes with GAS elements in their promoters (such as Irf1), the observed delay in IRF-1 expression was likely not due to an inability of AAV and STAT5-CrkL complexes to form. Crucial steps downstream of STAT activation could be the targets of the WNV Eg101-induced blockage. It was previously reported that histone deacetylase 1 activity is required for full transcriptional activation of the Irf1 gene (53).
For the Casp11 and Irf1 genes, an additional level of regulation was observed in WNV-infected cells. For both of these genes, protein levels detected in cells infected for 16, 24, or 48 h with WNV were low or undetectable even though the mRNA levels detected for these genes at these times were as high or higher than those in IFN-treated cells, which expressed high levels of these proteins. Decreased IRF-1 protein levels were also evident in cells treated with IFN for 1 or 4 h and then infected with WNV (Fig. 7A and Fig. 7B). Four-hour IFN-pretreated, WNV-infected MEFs produced virus yields that were 10-fold lower than control cells, and up-regulation of IRF-1 by viral infection was detected by 12 h after infection. In contrast, in 1-h IFN-pretreated and untreated infected cells, up-regulation of IRF-1 expression was not detected until 16 to 24 h after infection and was lower at these times than in cells treated only with IFN (Fig. 6C and 7A). Virus titers produced by the 1-h IFN-pretreated cells were similar to those produced by control infected cells, supporting an inverse correlation between the levels of IRF-1 expression and virus yields. Whether suppression of the expression of a subset of ISGs is mediated directly or indirectly by WNV Eg101 infection is currently under investigation. Known mechanisms of differential regulation of cellular protein levels include phosphorylation/dephosphorylation of translation factors by protein kinases, regulation of mRNA stability, and accelerated protein degradation (35).
Acknowledgments
This work was supported by Public Health Service research grant AI045135 to M.A.B. from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
We thank Ali Pirani and Kim Gernert, BimCore Facility, Emory University School of Medicine, for assistance with microarray data analysis; Mohamed Emara for assistance with confocal microscopy; Dmitriy Scherbik for assistance with graphics; and Gertrude Radu for proofreading.
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Au, W. C., and P. M. Pitha. 2001. Recruitment of multiple interferon regulatory factors and histone acetyltransferase to the transcriptionally active interferon a promoters. J. Biol. Chem. 276:41629-41637. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, B., B. Lubyova, and P. M. Pitha. 2002. On the role of IRF in host defense. J. Interferon Cytokine Res. 22:59-71. [DOI] [PubMed] [Google Scholar]
- 3.Beasley, D. W., L. Li, M. T. Suderman, and A. D. Barrett. 2002. Mouse neuroinvasive phenotype of West Nile virus strains varies depending upon virus genotype. Virology 296:17-23. [DOI] [PubMed] [Google Scholar]
- 4.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biron, A. B., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-351. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 6.Brierley, M. M., and E. N. Fish. 2005. Stats: multifaceted regulators of transcription. J. Interferon Cytokine Res. 25:733-744. [DOI] [PubMed] [Google Scholar]
- 7.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8:157-171. [DOI] [PubMed] [Google Scholar]
- 8.Collins, S. E., R. S. Noyce, and K. L. Mossman. 2004. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J. Virol. 78:1706-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elco, C. P., J. M. Guenther, B. R. Williams, and G. C. Sen. 2005. Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-κB, and interferon but not Toll-like receptor 3. J. Virol. 79:3920-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenner, J. E., R. Starr, A. L. Cornish, J. G. Zhang, D. Metcalf, R. D. Schreiber, K. Sheehan, D. J. Hilton, W. S. Alexander, and P. J. Hertzog. 2006. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 7:33-39. [DOI] [PubMed] [Google Scholar]
- 12.Fish, E. N., S. Uddin, M. Korkmaz, B. Majchrzak, B. J. Druker, and L. C. Platanias. 1999. Activation of a CrkL-Stat5 signaling complex by type I interferons. J. Biol. Chem. 274:571-573. [DOI] [PubMed] [Google Scholar]
- 13.Fredericksen, B. L., and M. Gale, Jr. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 80:2913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 16.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hitomi, J., T. Katayama, Y. Eguchi, T. Kudo, M. Taniguchi, Y. Koyama, T. Manabe, S. Yamagishi, Y. Bando, K. Imaizumi, Y. Tsujimoto, and M. Tohyama. 2004. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and Aβ-induced cell death. J. Cell Biol. 165:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 20.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 21.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 22.Juang, Y. T., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller, B. C., B. L. Fredericksen, M. A. Samuel, R. E. Mock, P. W. Mason, M. S. Diamond, and M. Gale, Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J. Virol. 80:9424-9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh, W. L., and M. L. Ng. 2005. Molecular mechanisms of West Nile virus pathogenesis in brain cell. Emerg. Infect. Dis. 11:629-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liew, K. J., and V. T. Chow. 2006. Microarray and real-time RT-PCR analyses of a novel set of differentially expressed human genes in ECV304 endothelial-like cells infected with dengue virus type 2. J. Virol. Methods 131:47-57. [DOI] [PubMed] [Google Scholar]
- 26.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, W. J., X. J. Wang, D. C. Clark, M. Lobigs, R. A. Hall, and A. A. Khromykh. 2006. A single amino acid substitution in the West Nile virus nonstructural protein NS2A disables its ability to inhibit alpha/beta interferon induction and attenuates virus virulence in mice. J. Virol. 80:2396-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacMicking, J. D. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25:601-609. [DOI] [PubMed] [Google Scholar]
- 31.Mori, M., M. Yoneyama, T. Ito, K. Takahashi, F. Inagaki, and T. Fujita. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279:9698-9702. [DOI] [PubMed] [Google Scholar]
- 32.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 35.Proud, C. G. 2007. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J. 403:217-234. [DOI] [PubMed] [Google Scholar]
- 36.Rein, T., M. Muller, and H. Zorbas. 1994. In vivo footprinting of the IRF-1 promoter: inducible occupation of a GAS element next to a persistent structural alteration of the DNA. Nucleic Acids Res. 22:3033-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarkar, S. N., and G. C. Sen. 2004. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol. Ther. 103:245-259. [DOI] [PubMed] [Google Scholar]
- 39.Scherbik, S. V., J. M. Paranjape, B. M. Stockman, R. H. Silverman, and M. A. Brinton. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Servant, M. J., N. Grandvaux, B. R. tenOever, D. Duguay, R. Lin, and J. Hiscott. 2003. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J. Biol. Chem. 278:9441-9447. [DOI] [PubMed] [Google Scholar]
- 41.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 42.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 43.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 44.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 45.Terenzi, F., C. White, S. Pal, B. R. Williams, and G. C. Sen. 2007. Tissue-specific and inducer-specific differential induction of ISG56 and ISG54 in mice. J. Virol. 81:8656-8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trent, D. W., and C. W. Naeve. 1980. Biochemistry and replication, p.159-199. In T. P. Monath (ed.), St. Louis encephalitis. American Public Health Association, Washington, DC.
- 47.Uddin, S., F. Lekmine, A. Sassano, H. Rui, E. N. Fish, and L. C. Platanias. 2003. Role of Stat5 in type I interferon-signaling and transcriptional regulation. Biochem. Biophys. Res. Commun. 308:325-330. [DOI] [PubMed] [Google Scholar]
- 48.Upreti, M., and P. C. Rath. 2005. Expression and DNA binding activity of the recombinant interferon regulatory factor-1 (IRF-1) of mouse. Mol. Biol. Rep. 32:103-116. [DOI] [PubMed] [Google Scholar]
- 49.Vaheri, A., W. D. Sedwick, S. A. Plotkin, and R. Maes. 1965. Cytopathic effect of rubella virus in RHK21 cells and growth to high titers in suspension culture. Virology 27:239-241. [DOI] [PubMed] [Google Scholar]
- 50.Venter, M., T. G. Myers, M. A. Wilson, T. J. Kindt, J. T. Paweska, F. J. Burt, P. A. Leman, and R. Swanepoel. 2005. Gene expression in mice infected with West Nile virus strains of different neurovirulence. Virology 342:119-140. [DOI] [PubMed] [Google Scholar]
- 51.Warke, R. V., K. Xhaja, K. J. Martin, M. F. Fournier, S. K. Shaw, N. Brizuela, N. de Bosch, D. Lapointe, F. A. Ennis, A. L. Rothman, and I. Bosch. 2003. Dengue virus induces novel changes in gene expression of human umbilical vein endothelial cells. J. Virol. 77:11822-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamamoto, M., S. Yamazaki, S. Uematsu, S. Sato, H. Hemmi, K. Hoshino, T. Kaisho, H. Kuwata, O. Takeuchi, K. Takeshige, T. Saitoh, S. Yamaoka, N. Yamamoto, S. Yamamoto, T. Muta, K. Takeda, and S. Akira. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430:218-222. [DOI] [PubMed] [Google Scholar]
- 53.Zupkovitz, G., J. Tischler, M. Posch, I. Sadzak, K. Ramsauer, G. Egger, R. Grausenburger, N. Schweifer, S. Chiocca, T. Decker, and C. Seiser. 2006. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 26:7913-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]