Abstract
Human cytomegalovirus (HCMV) induces profound changes in infected cell morphology, including a large cytoplasmic inclusion that corresponds to the virion assembly complex (AC). In electron micrographs, the AC is a highly vacuolated part of the cytoplasm. Markers of cellular secretory organelles have been visualized at the outer edge of the AC, and we recently showed that a marker for early endosomes (i.e., early endosome antigen 1) localizes to the center of the AC. Here, we examined the relationship between the AC and components of the secretory apparatus, studied temporal aspects of the dramatic infection-induced cytoplasmic remodeling, examined the three-dimensional structure of the AC, and considered the implications of our observations for models of HCMV virion maturation and egress. We made three major observations. First, in addition to being relocated, the expression levels of some organelle markers change markedly during the period while the AC is developing. Second, based on three-dimensional reconstructions from z-series confocal microscopic images, the observed concentric rings of vesicles derived from the several compartments (Golgi bodies, the trans-Golgi network [TGN], and early endosomes) are arranged as nested cylinders of organelle-specific vesicles. Third, the membrane protein biosynthetic and exocytic pathways from the endoplasmic reticulum to the Golgi bodies, TGN, and early endosomes are in an unusual arrangement that nonetheless allows for a conventional order of biosynthesis and transport. Our model of AC structure suggests a mechanism by which the virus can regulate the order of tegument assembly.
As suggested by its name, human cytomegalovirus (HCMV) induces profound changes in infected cell morphology. The readily visible cytoplasmic and nuclear inclusions in HCMV-infected cells in human tissues (15, 21, 27, 44, 55) are part of the overall effect infection has on the cell. HCMV biology includes a protracted lytic replication cycle. After a 2- to 3-day eclipse phase, infected cells produce infectious progeny virus at a rate of approximately one infectious unit per hour per cell for a week or more (46). The large cytoplasmic inclusions correspond to a structure that is the site of final virion assembly and maturation, the virion assembly complex (AC; also known as the “viral field”) (20, 45, 47, 50). ACs are large circular structures that are generally present one per infected cell, even in syncytia containing up to 10 or so nuclei (9, 10, 20). Similar structures have been observed in infected cultured primary fibroblasts, endothelial cells, and smooth muscle cells (21, 24). One side of the structure has a perinuclear localization, with the nucleus taking on a kidney-like shape as it bends partially around the AC (20, 25, 34, 59). Electron microscopic evidence indicates that the AC is in a highly vacuolated part of the cytoplasm and is the site of final tegumentation and envelope acquisition (20, 50, 51, 54, 57).
The substantial literature based on morphological and chemical identification of intracellular structures by light and electron microscopy is being joined with observations made using well-defined molecular markers. Thus, viral tegument, envelope, and some nonstructural proteins have been localized to the AC, and cytoskeletal filaments radiate from a microtubule organizing center in the AC (9, 23, 24, 40, 47). The AC is relatively devoid of endoplasmic reticulum (ER) markers (9, 24, 47). Markers of the ER-to-Golgi intermediate compartment and the Golgi apparatus appear to be radially displaced away from the AC, and markers of the trans-Golgi network (TGN) are present at the outer edge of the AC (9, 24, 47, 49). It had been thought that viral proteins occlude cellular proteins from the AC (47, 49), but recent evidence indicates otherwise. Gaspar and Shenk found that a cellular cell cycle control protein that is normally resident in nuclei relocates to the AC during infection (19). We found that a marker of early endosomes (early endosome antigen 1 [EEA1]) concentrates at the center of the AC (9).
The observation that early endosomes, components of the secretory apparatus normally considered to be intermediaries between the TGN and the cell surface, are concentrated at the center of a cytoplasmic structure (the AC) that is ringed by the ER-to-Golgi intermediate compartment, Golgi bodies, and TGN was striking and unexpected. In the present study, we examined the relationship between the AC and these components of the secretory apparatus, studied the temporal aspects of the dramatic infection-induced cytoplasmic remodeling, determined the three-dimensional structure of the AC, and considered the implications of our observations for models of HCMV virion maturation and egress.
MATERIALS AND METHODS
Virus and cells.
HCMV strain AD169 (American Type Culture Collection, Manassas, VA) was propagated, and titers were determined in low-passage primary human lung fibroblasts (HLF; obtained from the Centers for Disease Control and Prevention) or human diploid fibroblasts (MRC-5; American Type Culture Collection, Manassas, VA). The experiments reported here were done in MRC-5 cells that were propagated in Earle's modified Eagle medium containing 10% fetal bovine serum (HyClone, Logan, UT), 2 mM l-glutamine, 50 U of penicillin/ml, and 50 μg of streptomycin/ml. Infections were done in the same medium with 5% serum and no antibiotics.
Confocal immunofluorescence analysis.
Confocal immunofluorescence microscopy was done as previously described (9, 10). Briefly, cell growth and infections were done in eight-well Labtek chamber slides (Nunc, Naperville, IL). Infections were performed at a multiplicity of infection (MOI) of 0.01. Prior to staining, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), incubated in 50 mM ammonium chloride to quench autofluorescence, permeabilized in PBS containing 0.2% Triton X-100 and normal goat serum for 15 min, and then incubated for 1 h in blocking buffer (10% normal goat serum and 5% glycine in PBS). Primary antibodies were diluted in blocking buffer. Staining was for 1 h at room temperature, followed by three washes with PBS and then reaction with the secondary antibody for 1 h. After being washed, cells were mounted using Vectashield with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA).
Primary antibodies included anti-peptide rabbit polyclonal antibodies directed at the C-terminal domain of HCMV pUS17 and the N-terminal domain of pUS18 (9, 10), rabbit polyclonal antibodies against the Golgi protein mannosidase II (Abcam, Inc., Cambridge, MA) and rab5 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), and a mouse monoclonal antibody (MAb) to HCMV IE2/pUL122 (MAb810; Chemicon, Inc., Temecula, CA). Additional MAbs were against an ER-to-Golgi chaperone (BiP/GRP78), a Golgi protein (GM130), a TGN protein (p230), and an early endosome protein (EEA1) (all from BD Biosciences, San Jose, CA). Secondary antibodies included polyclonal goat anti-rabbit (Alexa Fluor 488 conjugated) and goat anti-mouse (Alexa Fluor 568 conjugated) (Molecular Probes, Eugene, OR).
Images were captured with a Leica TCS-SP digital confocal microscope. Quantitative image analysis was done using ImagePro version 6.1 (Media Cybernetics, Inc., Silver Springs, MD), and the digital images were processed by using Adobe Photoshop CS. Three-dimensional reconstructions and a movie (see Video S1 in the supplemental material) were made from the z-series confocal pictures by using Velocity software (64-bit version; Improvision, Inc., Lexington, MA).
Immunoblots.
HLF cells infected with HCMV(AD169) at 0.5 PFU per cell were harvested in a buffer containing 10 mM Tris-Cl (pH 7.5), 25% glycerol, 1.5 mM MgCl2, 800 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 1% NP-40, and a protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). Clarified extract was mixed with sodium dodecyl sulfate (SDS) sample buffer (6×) containing Tris-HCl (pH 6.8), 0.2% 2-mercaptoethanol, 30% glycerol, and 10% SDS and then boiled for 5 min. Proteins were separated by SDS-polyacrylamide gel electrophoresis in a 10% gel, followed by electrical transfer of the proteins to a nitrocellulose membrane (0.2-μm pore size; Schleicher & Schuell, Keene, NH). After blocking for 1 h at room temperature in buffer containing of 5% fat-free milk powder and 0.05% Tween 20 in PBS, the blot was reacted overnight with primary MAbs in a buffer containing 0.05% Tween 20 and 1% normal goat serum (Caltag, Burlingame, CA) in PBS. After three washes in PBS with 0.05% Tween 20, the blot was incubated with horseradish peroxidase-conjugated goat anti-mouse antibody (Pierce Biotechnology) for 1 h, followed by three washes in PBS with 0.05% Tween 20. Detection was by chemiluminescence (Pierce Biotechnology) and exposure to film. The relative band densities were measured by using Image Pro 6.1 software.
RESULTS
Time course of changes in cytoplasmic abundance and distribution of markers of components of the secretory apparatus.
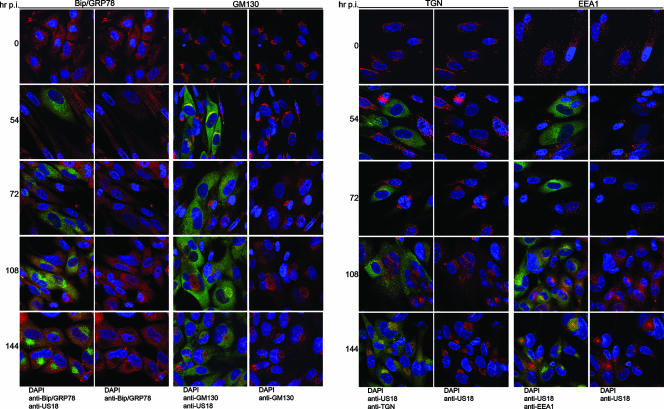
In the first series of experiments, we studied changes in the arrangement and abundance of components of the secretory apparatus during the course of infection at relatively low MOIs. Although this results in a degree of asynchronicity of infection, it offers on-slide direct comparisons of fluorescence intensities between infected and uninfected cells. For identification of infected cells, we did not use a mouse MAb against a viral protein such as IE-2 because of the need to use mouse MAb against cellular markers. HCMV recombinants that express green fluorescent protein were tested, but the level of green fluorescent protein expression was high enough that spectral overlap interfered with the visualization of other colors. We chose to mark infected cells with previously described rabbit polyclonal antibodies to pUS17 and pUS18 (9, 10). The results from the pUS18 experiments are shown in Fig. 1; similar results were obtained in parallel experiments done using the US17 antibody (data not shown). Note that pUS18 is expressed as a viral late gene and becomes detectable by immunofluorescence assay approximately 36 h after infection (10, 22). Although infected cell nuclei enlarge early during infection, it is possible that some unstained cells with normal-sized nuclei are in fact at early stages of infection. Importantly, the conclusions from the lower-MOI experiments shown in Fig. 1 and 2 are consistent with the results obtained in the higher-MOI experiments shown in Fig. 3.
FIG. 1.
Time course of cytoplasmic remodeling. HLF cells were mock infected or infected for 144 h at an MOI of 0.01. The indicated organelle markers (ER, Bip/GRP78; Golgi, GM130; TGN, p230; and early endosomes, EEA1) were detected by confocal microscopy using MAb (red stain), and infected cells were detected with a rabbit polyclonal antibody to US18 (green). DNA was stained with DAPI. For each organelle and time point, the left panel is a merge of all three colors, and the right panel is a merge of just the green and blue channels.
FIG. 2.
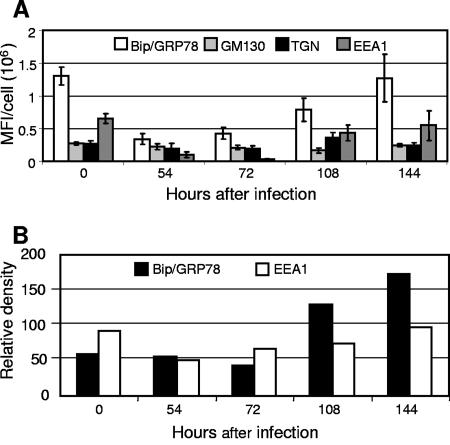
Time course of expression of components of the secretory apparatus in HCMV-infected cells. (A) Images collected in the course of experiments similar to those shown in Fig. 1 were analyzed to quantitate the mean fluorescence intensity (MFI) in a single confocal cross-section per cell for each marker. Ten infected cells were analyzed for each marker. (B) Relative immune blot reactivity of Bip/GRP78 and EEA1 as a function of time after infection, as measured by densitometry of chemiluminescence images. Organelle markers: ER, Bip/GRP78; Golgi, GM130; TGN, p230; and early endosomes, EEA1.
FIG. 3.
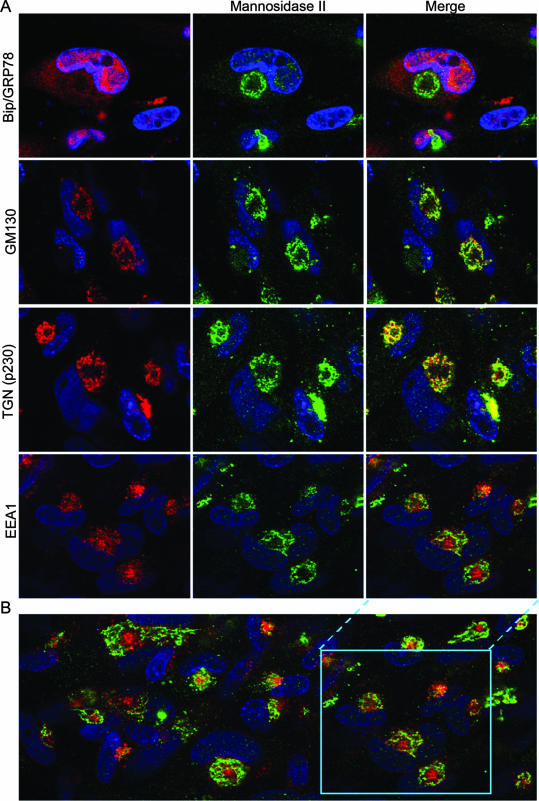
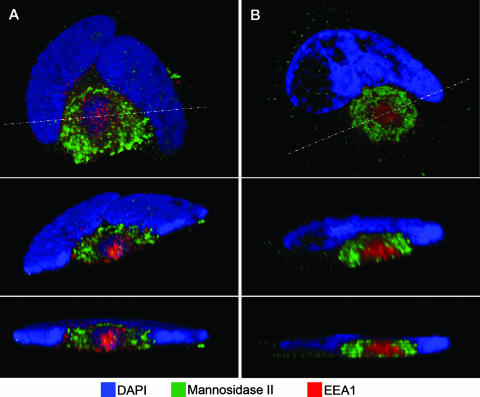
Localization of secretory apparatus components relative to a Golgi marker. (A) HLF cells mock infected or infected with HCMV(AD169) for 144 h were fixed and then stained with a rabbit polyclonal antibody against a Golgi marker (mannosidase II) (green), MAb against the indicated organelle markers (red), and DAPI prior to confocal microscopy. The organelle markers detected with the MAb were Bip/GRP78 (ER), GM130 (Golgi), p230 (TGN), and EEA1 (early endosomes). (B) A larger field from the early endosome/Golgi staining is shown to demonstrate that the patterns shown are representative.
Figure 1 shows confocal microscopic images of the time course of an infection during which cells were fixed and then stained with antibodies to pUS18 and markers of the ER (Bip), Golgi apparatus (GM130), the TGN (p230), and early endosomes (EEA1). Because of the imperfect synchronicity of the infections, images were chosen from those that represented the most advanced stage of infection at each time point. The mean fluorescence intensities (per cell per confocal cross-section) of the cellular markers in infected cells at each time point are plotted in Fig. 2A, and the results of densitometric analysis of immunoblots of proteins extracted from similar time points of other experiments are shown in Fig. 2B.
Two major points can be made from these experiments. (i) Based on visual inspection of the confocal images (Fig. 1), the fluorescence intensity for all of the cellular markers in infected cells appears to be reduced from 54 to 74 h postinfection (hpi). As can be seen in the graphs of fluorescence intensities (Fig. 2A), for some proteins, this is likely the result of dilution or diffuse dispersal of a nearly fixed amount of protein throughout the generally enlarged cytoplasm of infected cells (GM130 and p230); for others (Bip and EEA1), it is due to reductions in the total amount of the protein. The immunoblot results (Fig. 2B) for Bip/GRP78 and EEA1 are consistent with the quantitative fluorescence data.
(ii) ACs are evident in some infected cells at 108 hpi and are present in most infected cells by 144 hpi. At these times, the intracellular distributions of the cellular markers are markedly different from those in uninfected cells. The ER-to-Golgi chaperone, BiP/GRP78, is present in some infected cell nuclei, which can occur when the N-terminal hydrophobic sequence of BiP/GRP78 is cleaved (33). In addition, BiP/GRP78 is more diffusely distributed in the cytoplasm of infected cells than in uninfected cells and is almost completely occluded from the AC (illuminated by the US18 antibody). BiP/GRP78 fluorescence per confocal cross-section drops to about a third of its level in uninfected cells from 54 to 72 hpi and returns to its uninfected cell level by 144 hpi, a finding consistent with the increase in overall levels seen in the immune blot (Fig. 2B).
As previously observed (9, 24, 47, 49), the Golgi (GM130) and TGN (p230) markers stain in a ring-like pattern around the AC periphery starting at about 4 dpi, with this becoming a predominant feature by 144 hpi. The mean fluorescence intensities for these proteins are relatively constant throughout the time course examined. At 54 hpi, GM130 is concentrated directly adjacent to one side of the nucleus, with the distribution becoming more diffuse at 72 hpi, before formation of the ring around the AC. p230 appears to progress toward a perinuclear focus that then develops into the ring around the AC. The early endosome marker, EEA1, underwent dramatic changes in abundance and localization over the time course studied. From a scattered and often punctate “starry night” distribution in uninfected cells (readily visible in Fig. 1 and 4), EEA1 staining became diffuse, and the protein almost disappeared from cells at 54 and 72 hpi (Fig. 1 and 2). As its abundance increased at the 108- and 144-hpi time points, it concentrated at the center of the AC. Live-cell microscopy of fluorescently tagged organelle markers and pulse-chase experiments will be required for definitive assessment of the paths to final localization.
FIG. 4.
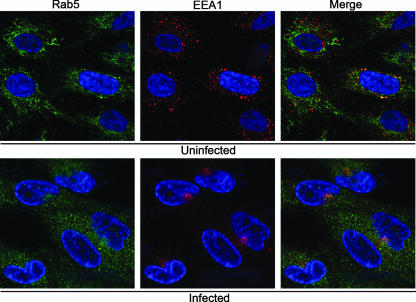
Relative localization of the early endosome-associated proteins, EEA1 and rab5. HLF cells were infected with HCMV(AD169) for 144 h before being fixed and stained with DAPI (blue), a mouse MAb against the early endosomal marker EEA1 (red), and a rabbit polyclonal antibody the rab5 (green). The images were obtained by confocal microscopy.
Spatial distribution of components of the secretory apparatus relative to a Golgi marker.
To assess the relative intracellular localizations of the organelle markers, infected cells were stained at 144 hpi with a rabbit polyclonal antibody against a Golgi marker (mannosidase II) (green) and MAb against individual organelle markers (red). In the confocal micrographs shown in Fig. 3, the rabbit antibody against mannosidase II stains infected cells with a characteristic ring at the outer edge of the AC, as seen previously (9, 24, 47, 49). With the mannosidase II staining as a point of reference, the concentric arrangement of components of the secretory apparatus is readily apparent. The ER is diffusely distributed in the cytoplasm away from the AC and interspersed with the Golgi stain in the ring at the AC periphery. Staining of the two independent Golgi markers is very highly coincident in the dotted ring-like structure bounding the AC. The TGN marker has substantial overlap with the Golgi marker but is partially offset toward the AC interior, suggesting a closely intertwined or interlaced juxtaposition of these compartments. The presence of the early endosomal marker to the interior of the AC is clearly illustrated here, with the generality of the observation being demonstrated in the lower-power field shown in panel B. These results indicate that during the remodeling of the cytoplasm in HCMV-infected cells, early endosomes (an organelle that is normally found near the cell periphery as a near-terminal component of the exocytic and endocytic pathways) have been relocated to the center of a concentric structure in which the ER is the outermost component, followed by the Golgi bodies, TGN, and then the early endosomal compartment at the center (Fig. 3 and 6).
FIG. 6.
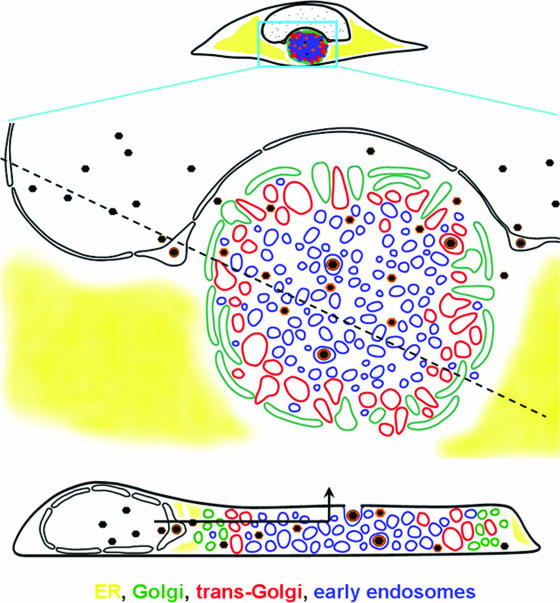
Schematic representation of AC structure and the maturational path of nascent virions. The diagram is based on data such as shown in Fig. 1, 3, and 5, coupled with prior ultrastructural analyses (see, for example, reference 50) and other information cited in the text about herpesvirus maturation. In this representation, the AC is the large circular structure that is bounded by Golgi vesicles. The concentric arrangement of the Golgi, TGN, and early endosomal compartments is shown. The cross-sectional representation through the cell at the bottom of the figure shows the nested cylindrical arrangement of the vesicular compartments (sectioning along the dotted line in the upper panel). In this model, nascent capsids acquire a subset of tegument proteins prior to nuclear egress. In the cytoplasm, tegumentation occurs during migration from the AC periphery to the exit vesicle, which is transported vertically to the cell surface without needing to traverse the secretory pathway in reverse. The path of egress is indicated in the lower panel by the arrow.
Absence of rab5 staining in the AC.
The small G protein, rab5, is often used as a marker of early endosomes. In HCMV-infected cells, rab5 had a modest association with virus-containing vacuoles and dense bodies but was not observed to associate with the AC in the manner we observed for EEA1 (24). To examine this directly, we stained uninfected and infected cells with a rabbit polyclonal antibody to rab5 and a MAb to EEA1 (Fig. 4). Although there was some colocalization between the markers, it varied from cell to cell, and in most cells the staining for each marker was largely independent. Thus, in infected cells, the EEA1 antibody strongly stains the AC, while rab5 has limited AC localization and is predominantly present in the cytoplasm as diffusely scattered small granular punctae.
Three-dimensional structure of the AC.
Figure 5 shows sections through two three-dimensional images reconstructed from z-series confocal images; a movie showing the structure from panel A being rotated in space is available (see Video S1 in the supplemental material). These images make it clear that the AC is not composed of nested spheres of different types of vesicles, as might have been imagined based on single cross-sectional images. Instead, the AC is better described as a set of concentric nested cylinders, with each cylinder being composed of a particular type of vesicle. The schematic diagram shown in Fig. 6 illustrates the relationship of maturing virions to the AC-associated vesicular structures. The conundrum of how vesicles containing mature virions might make their way from the center of this compartment to the cell surface is easily explained when the structure of the AC is considered in three dimensions (bottom panel of Fig. 6).
FIG. 5.
Three-dimensional reconstructions of HCMV-infected cell structure. HLF cells were infected with HCMV(AD169) for 108 h (A) or 144 h (B and C) before being fixed and stained with DAPI (blue), a mouse MAb against the early endosomal marker EEA1 (red), and a rabbit polyclonal antibody the Golgi marker mannosidase II (green). A z-series of confocal microscopic images was obtained at 1-μm increments. In each panel, the top image is a projection of the complete z-series. The middle images are tilted cross-sections taken along the dotted line in panel A. The dotted boxes outline the cut face. The bottom images in each panel are edge-on views of the same section (perpendicular to the z-axis). Note that the green Golgi marker does not extend across the top of the cut face, which is evidence for a nested cylindrical, rather than nested spherical, arrangement of organelle-specific vesicles.
In addition to the vesicle-associated proteins, we note the presence in some AC of diffuse DAPI staining (readily visible in Fig. 5A but not in Fig. 5B), which indicates the presence of DNA. Pulse-chase bromodeoxyuridine staining results in staining of the AC (1), suggesting that at least some of this is viral DNA. We do not know whether it is encapsidated; it does not have a punctate appearance in our images.
DISCUSSION
We made three major observations relating to the structure and biogenesis of the HCMV AC. First, in addition to being dramatically relocated, the expression of some organelle markers changes markedly during the period while the AC is developing. Second, based on three-dimensional reconstructions from z-series confocal microscopic images, it is clear that the observed concentric rings of vesicles from the several compartments (Golgi, TGN, and early endosomes) are arranged as nested cylinders of organelle-specific vesicles. Third, the membrane protein biosynthetic and exocytic pathways from the ER to the Golgi, TGN, and early endosomes are in an unusual spatial arrangement that nonetheless allows for a conventional order of biosynthesis and transport. These observations and some of their connections to known properties of virion maturation and egress are summarized in Fig. 6.
AC development.
As illuminated by phase-contrast time-lapse cinematography, the remodeling of infected cells during formation of the AC is a dynamic process (59). Several requirements for AC formation have been identified. The failure to form AC in the absence of viral DNA synthesis (40; our unpublished observations) indicates that its formation is dependent on expression of one or more viral late genes. Treatment of cells with nocodazole, which depolymerizes microtubules, leads to AC disruption (47); this is possibly a direct effect and/or an effect relating to the disrupted transport of AC components or regulators. The kinase activity of pUL97 is required for formation of perinuclear complexes that correspond to AC (2, 40). Although alternative paths for virion maturation exist, viral yields are reduced by >100-fold in the absence of UL97 (2, 41). ACs form in cells infected with HCMV strains AD169 and Towne, indicating that genes absent in these highly passaged laboratory strains (6, 14, 37, 42) are not required for the process. In the absence of the abundant tegument protein ppUL32 (pp150), viral genes from all kinetic classes are expressed, and apparently encapsidated viral DNA is transported to the cytoplasm, but infectious virions are either not produced or not released (1). pp28 (pUL99) is a myristoylated AC-associated protein that is essential for the production of infectious virions (52). Although not examined in detail, images in these studies are consistent with AC-like structures forming in the absence of ppUL32 and pUL99. Nothing is known of the cellular effectors of AC development.
We studied some of the kinetic aspects of AC development. We found different patterns of expression of markers for cytoplasmic organelles, such that some have relatively constant levels during development of the AC, whereas others undergo substantial decreases in the amount detected 2 to 3 days after infection, with the net amount increasing sharply thereafter. The specific regulatory mechanisms responsible for this remain to be identified. The results shown here, plus our previous description of nuclear relocalization of a normally cytoplasmic protein (Golgin-97) that is involved in regulation of TGN structure and early endosome-to-TGN retrograde transport (3, 4, 29, 32, 36, 60), suggest that a diverse set of regulatory mechanisms are used during AC development.
AC structure.
A product of the extensive cytoplasmic remodeling that occurs during the first 3 to 4 days after HCMV infection is a dramatic reorientation of the cellular endocytic and exocytic machinery. This is manifest in the structure we have determined for the AC. In this structure, the ER is distributed throughout much of the cytoplasm and in a ring at the AC periphery. Golgi and TGN vesicles form tightly nested rings around the AC; the AC interior contains vesicles that consistently stain with the early endosomal marker, EEA1. Interestingly, a lysosomal marker did not concentrate in the AC (47). In the standard lysosomal degradation pathway, cargos are transported in vesicles from the cell surface or other organelles to early endosomes and then to late endosomes and finally to lysosomes. The spatial reconfiguration in HCMV-infected cells suggests that at least some aspects of the lysosomal degradation pathway are altered and possibly uncoupled. In addition, consistent with a prior report (24), little AC staining was seen for rab5, a small G protein EEA1 effector that is often found in association with EEA1 on early endosomes. Early endosomes are not monolithic entities; rather, they come in tubular and vesicular forms and vary in the complement of associated proteins. In some cells, EEA1 is a more specific marker for early endosomes than is rab5 (17), but EEA1-negative/rab5-positive early endosomes have been identified (7). The structure of the cytoplasmic inclusion in naturally infected renal tubule cells (15) is at least superficially similar to its structure in infected cultured fibroblasts. In subsequent studies, it will be important to use organelle-specific markers to learn the details of AC structure in the three-dimensional matrix of human tissue.
AC and virion maturation and egress.
Exactly how virions mature in the AC has remained a mystery. Early ultrastructural analyses indicated that nucleocapsids could acquire an envelope at the inner nuclear membrane and at cytoplasmic vesicles, but the relationship between the processes was not understood (25, 28, 34, 45, 54). Severi and coworkers (39, 50, 51) were the first to propose that HCMV nucleocapsids acquire an envelope during the process of budding from the inner nuclear membrane to the lumen of the nuclear membrane and then lose that envelope upon “infecting” the cytoplasm. In their model, the partially tegumented nucleocapsid acquires its full coat of tegument as it migrates through a field of circularly arrayed vesicles that were thought to be derived from the Golgi apparatus, followed by acquisition of its final envelope by budding into a cytoplasmic vesicle near the center of this complex for ultimate transport to the cell surface for release. The path of egress for the exit vesicle from the center of the complex was not identified. The envelopment/de-envelopment/re-envelopment process is similar to a model originally proposed by Stackpole for a herpesvirus of frogs (56), has been confirmed by other investigators for HCMV (20), and has been extended to other herpesviruses (reviewed in reference 35).
Although it is clear that final maturation of HCMV takes place in vesicles of the later components of the vesicular transport system, whether the site of final envelopment is the TGN or early endosomes remains an item of discussion (8, 18, 24, 26, 31, 58). Virion glycoproteins such as gB accumulate in the TGN (8, 26, 58), but this need not be the exit vesicle, because glycoproteins might be transported from a TGN staging area to an early endosomal exit vesicle. Tooze et al. (57) treated cells with soluble horseradish peroxidase, which is taken up by recycling early endosomes, and localized the endocytosed enzyme at the ultrastructural level on the basis of the electron-dense product it produces in reactions with diaminobenzidine. The reaction product concentrated in a cytoplasmic region corresponding to the AC, in vesicles that contained what appeared to be fully tegumented and enveloped HCMV virions, thus providing evidence that at least some exit vesicles are derived from early endosomes. In pulse-chase experiments, virions and the enzyme were cleared from vacuoles within 2 h, indicating that these are indeed actively trafficked exit vesicles. This, coupled with our observation of EEA1 staining in the same part of the cell, constitutes strong evidence that early endosomes can serve as HCMV exit vesicles but do not eliminate TGN vesicles from also contributing.
As illustrated schematically in Fig. 6, a possible path of virion maturation and egress through the AC would involve nucleocapsid migration from the periphery toward the center of the AC. The migration would occur in the cytoplasmic space and not inside of membrane-bounded vesicles. It has been calculated a herpesvirus capsid would take 23 years to move 1 mm through material of cytoplasmic complexity and structure in a purely diffusion-driven process (13). Thus, like herpes simplex virus nucleocapsids (5, 11-13, 16, 30, 43, 48, 53), and for transport of HCMV nucleocapsids to the nucelus during virion entry (38), intracellular motility of HCMV nucleocapsids during virion maturation is likely to be mediated via attachment to microtubule-associated motors. The model suggests that the order of addition of tegument proteins might be regulated in part by reversible specific interactions of tegument proteins with proteins that are themselves specifically attached to one or another of the various organelles that form the highly ordered AC.
Supplementary Material
Acknowledgments
We thank Judy Drazba and the staff of the LRI Imaging Core for help with image analysis; Fu-Zhang Wang, Pamela Berk, and Rachel Leahy for their contributions to this study; and Cynthia Hilston and James Finke for sharing the rab5 antibody.
Footnotes
Published ahead of print on 22 August 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.AuCoin, D. P., G. B. Smith, C. D. Meiering, and E. S. Mocarski. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol. 80:8199-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azzeh, M., A. Honigman, A. Taraboulos, A. Rouvinski, and D. G. Wolf. 2006. Structural changes in human cytomegalovirus cytoplasmic assembly sites in the absence of UL97 kinase activity. Virology 354:69-79. [DOI] [PubMed] [Google Scholar]
- 3.Barr, F. A. 1999. A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol. 9:381-384. [DOI] [PubMed] [Google Scholar]
- 4.Barr, F. A., and B. Short. 2003. Golgins in the structure and dynamics of the Golgi apparatus. Curr. Opin. Cell Biol. 15:405-413. [DOI] [PubMed] [Google Scholar]
- 5.Benboudjema, L., M. Mulvey, Y. Gao, S. W. Pimplikar, and I. Mohr. 2003. Association of the herpes simplex virus type 1 Us11 gene product with the cellular kinesin light-chain-related protein PAT1 results in the redistribution of both polypeptides. J. Virol. 77:9192-9203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coumailleau, F., V. Das, A. Alcover, G. Raposo, S. Vandormael-Pournin, S. Le Bras, P. Baldacci, A. Dautry-Varsat, C. Babinet, and M. Cohen-Tannoudji. 2004. Over-expression of Rififylin, a new RING finger and FYVE-like domain-containing protein, inhibits recycling from the endocytic recycling compartment. Mol. Biol. Cell 15:4444-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crump, C. M., C. H. Hung, L. Thomas, L. Wan, and G. Thomas. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105-11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, S., and P. E. Pellett. 2007. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology 361:263-273. [DOI] [PubMed] [Google Scholar]
- 10.Das, S., Y. Skomorovska-Prokvolit, F. Z. Wang, and P. E. Pellett. 2006. Infection-dependent nuclear localization of US17, a member of the US12 family of human cytomegalovirus-encoded seven-transmembrane proteins. J. Virol. 80:1191-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diefenbach, R. J., M. Miranda-Saksena, E. Diefenbach, D. J. Holland, R. A. Boadle, P. J. Armati, and A. L. Cunningham. 2002. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 76:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohner, K., K. Radtke, S. Schmidt, and B. Sodeik. 2006. Eclipse phase of herpes simplex virus type 1 infection: efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 80:8211-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dohner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolan, A., C. Cunningham, R. D. Hector, A. F. Hassan-Walker, L. Lee, C. Addison, D. J. Dargan, D. J. McGeoch, D. Gatherer, V. C. Emery, P. D. Griffiths, C. Sinzger, B. P. McSharry, G. W. Wilkinson, and A. J. Davison. 2004. Genetic content of wild-type human cytomegalovirus. J. Gen. Virol. 85:1301-1312. [DOI] [PubMed] [Google Scholar]
- 15.Donnellan, W. L., S. Chantra-Umporn, and J. M. Kidd. 1966. The cytomegalic inclusion cell: an electron microscopic study. Arch. Pathol. 82:336-348. [PubMed] [Google Scholar]
- 16.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1998. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 17.Foster, L. J., C. L. de Hoog, Y. Zhang, Y. Zhang, X. Xie, V. K. Mootha, and M. Mann. 2006. A mammalian organelle map by protein correlation profiling. Cell 125:187-199. [DOI] [PubMed] [Google Scholar]
- 18.Fraile-Ramos, A., A. Pelchen-Matthews, T. N. Kledal, H. Browne, T. W. Schwartz, and M. Marsh. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218-232. [DOI] [PubMed] [Google Scholar]
- 19.Gaspar, M., and T. Shenk. 2006. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc. Natl. Acad. Sci. USA 103:2821-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilloteaux, J., and M. R. Nassiri. 2000. Human bone marrow fibroblasts infected by cytomegalovirus: ultrastructural observations. J. Submicrosc. Cytol. Pathol. 32:17-45. [PubMed] [Google Scholar]
- 21.Grefte, A., N. Blom, M. van der Giessen, W. van Son, and T. H. The. 1993. Ultrastructural analysis of circulating cytomegalic cells in patients with active cytomegalovirus infection: evidence for virus production and endothelial origin. J. Infect. Dis. 168:1110-1118. [DOI] [PubMed] [Google Scholar]
- 22.Guo, Y. W., and E. S. Huang. 1993. Characterization of a structurally tricistronic gene of human cytomegalovirus composed of U(s)18, U(s)19, and U(s)20. J. Virol. 67:2043-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hertel, L., and E. S. Mocarski. 2004. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of pseudomitosis independent of US28 function. J. Virol. 78:11988-12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homman-Loudiyi, M., K. Hultenby, W. Britt, and C. Soderberg-Naucler. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasaki, Y., T. Furukawa, S. Plotkin, and H. Koprowski. 1973. Ultrastructural study on the sequence of human cytomegalovirus infection in human diploid cells. Arch. Gesamte Virusforsch. 40:311-324. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis, M. A., K. N. Fish, C. Soderberg-Naucler, D. N. Streblow, H. L. Meyers, G. Thomas, and J. A. Nelson. 2002. Retrieval of human cytomegalovirus glycoprotein B from cell surface is not required for virus envelopment in astrocytoma cells. J. Virol. 76:5147-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jesionek and Kiolemenoglou. 1904. Über einen befund von protozoenartigen gebilden in den organen eines feten. Munch. Med. Wochenschr. 51:1905-1907. [Google Scholar]
- 28.Kimura, A., T. Tamura, and T. Nakao. 1976. Intracytoplasmic uncoated capsids of human cytomegalovirus. Tohoku J. Exp. Med. 119:223-236. [DOI] [PubMed] [Google Scholar]
- 29.Kjer-Nielsen, L., R. D. Teasdale, C. van Vliet, and P. A. Gleeson. 1999. A novel Golgi-localization domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 9:385-388. [DOI] [PubMed] [Google Scholar]
- 30.Koshizuka, T., Y. Kawaguchi, and Y. Nishiyama. 2005. Herpes simplex virus type 2 membrane protein UL56 associates with the kinesin motor protein KIF1A. J. Gen. Virol. 86:527-533. [DOI] [PubMed] [Google Scholar]
- 31.Lee, G. E., J. W. Murray, A. W. Wolkoff, and D. W. Wilson. 2006. Reconstitution of herpes simplex virus microtubule-dependent trafficking in vitro. J. Virol. 80:4264-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, L., G. Tai, and W. Hong. 2004. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-Golgi network. Mol. Biol. Cell 15:4426-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto, A., and P. C. Hanawalt. 2000. Histone H3 and heat shock protein GRP78 are selectively cross-linked to DNA by photoactivated gilvocarcin V in human fibroblasts. Cancer Res. 60:3921-3926. [PubMed] [Google Scholar]
- 34.McGavran, M. H., and M. G. Smith. 1965. Ultrastructural, cytochemical, and microchemical observations on cytomegalovirus (salivary gland virus) infection of human cells in tissue culture. Exp. Mol. Pathol. 76:1-10. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 2005. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 36.Munro, S., and B. J. Nichols. 1999. The GRIP domain: a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9:377-380. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogawa-Goto, K., K. Tanaka, W. Gibson, E. Moriishi, Y. Miura, T. Kurata, S. Irie, and T. Sata. 2003. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J. Virol. 77:8541-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pignatelli, S., P. Dal Monte, M. P. Landini, B. Severi, R. Nassiri, J. Gilloteaux, J. M. Papadimitriou, G. R. Shellam, T. Mertens, C. Buser, D. Michel, and P. Walther. 2007. Cytomegalovirus primary envelopment at large nuclear membrane infoldings: what's new? J. Virol. 81:7320-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prichard, M. N., W. J. Britt, S. L. Daily, C. B. Hartline, and E. R. Kern. 2005. Human cytomegalovirus UL97 kinase is required for the normal intranuclear distribution of pp65 and virion morphogenesis. J. Virol. 79:15494-15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prichard, M. N., N. Gao, S. Jairath, G. Mulamba, P. Krosky, D. M. Coen, B. O. Parker, and G. S. Pari. 1999. A recombinant human cytomegalovirus with a large deletion in UL97 has a severe replication deficiency. J. Virol. 73:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191-200. [DOI] [PubMed] [Google Scholar]
- 43.Radtke, K., K. Dohner, and B. Sodeik. 2006. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell. Cell Microbiol. 8:387-400. [DOI] [PubMed] [Google Scholar]
- 44.Resnik, K. S., M. DiLeonardo, and M. Maillet. 2000. Histopathologic findings in cutaneous cytomegalovirus infection. Am. J. Dermatopathol. 22:397-407. [DOI] [PubMed] [Google Scholar]
- 45.Ruebner, B. H., T. Hirano, R. J. Slusser, and D. N. Medearis, Jr. 1965. Human cytomegalovirus infection. electron microscopic and histochemical changes in cultures of human fibroblasts. Am. J. Pathol. 46:477-496. [PMC free article] [PubMed] [Google Scholar]
- 46.Sampaio, K. L., Y. Cavignac, Y. D. Stierhof, and C. Sinzger. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol. 79:2754-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satpute-Krishnan, P., J. A. DeGiorgis, and E. L. Bearer. 2003. Fast anterograde transport of herpes simplex virus: role for the amyloid precursor protein of Alzheimer's disease. Aging Cell 2:305-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seo, J. Y., and W. J. Britt. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 51.Severi, B., M. P. Landini, M. Musiani, and M. Zerbini. 1979. A study of the passage of human cytomegalovirus from the nucleus to the cytoplasm. Microbiologica 2:265-273. [Google Scholar]
- 52.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, G. A., S. P. Gross, and L. W. Enquist. 2001. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc. Natl. Acad. Sci. USA 98:3466-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith, J. D., and E. De Harven. 1973. Herpes simplex virus and human cytomegalovirus replication in WI-38 cells I. Sequence of viral replication. J. Virol. 12:919-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, M. G. 1959. The salivary gland viruses of man and animals (cytomegalic inclusion disease). Prog. Med. Virol. 2:171-202. [Google Scholar]
- 56.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 58.Turcotte, S., J. Letellier, and R. Lippe. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847-8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wright, H. T., Jr., F. H. Kasten, and R. M. McAllister. 1968. Human cytomegalovirus, observations of intracellular lesion development as revealed by phase contrast, time-lapse cinematography. Proc. Soc. Exp. Biol. Med. 127:1032-1036. [DOI] [PubMed] [Google Scholar]
- 60.Yoshino, A., B. M. Bieler, D. C. Harper, D. A. Cowan, S. Sutterwala, D. M. Gay, N. B. Cole, J. M. McCaffery, and M. S. Marks. 2003. A role for GRIP domain proteins and/or their ligands in structure and function of the trans-Golgi network. J. Cell Sci. 116:4441-4454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.