Abstract
West Nile virus (WNV) is a human pathogen that can cause symptomatic infections associated with meningitis and encephalitis. Previously, we demonstrated that replication of WNV inhibits the interferon (IFN) signal transduction pathway by preventing the accumulation of phosphorylated Janus kinase 1 (JAK1) and tyrosine kinase 2 (Tyk2) (J. T. Guo et al., J. Virol. 79:1343-1350, 2005). Through a genetic analysis, we have now identified a determinant on the nonstructural protein 4B (NS4B) that controls IFN resistance in HeLa cells expressing subgenomic WNV replicons lacking the structural genes. However, in the context of infectious genomes, the same determinant did not influence IFN signaling. Thus, our results indicate that NS4B may be sufficient to inhibit the IFN response in replicon cells and suggest a role for structural genes, or as yet unknown interactions, in the inhibition of the IFN signaling pathway during WNV infections.
West Nile virus (WNV) is a member of the flavivirus family, which includes hepatitis C virus, Japanese encephalitis virus (JEV), and dengue virus (30). After binding to cellular receptors, these viruses enter the cell via active endocytosis and release their genomes into the cytoplasm through a pH-dependent mechanism, allowing translation to occur immediately. Genome replication occurs at the membrane of the endoplasmic reticulum (ER) within a multiprotein complex (14, 34, 57). The genome is packaged into a virion core composed of the structural proteins in the ER. Immature virions move through the trans-Golgi network to the plasma membrane. Infectious virus is released through exocytosis at early times postinfection and by virus-induced cell death later (14, 18).
Virions are small icosahedral particles that contain a single positive-stranded RNA molecule of approximately 11 kilobases. The viral genome encodes a single polyprotein, which is then proteolytically processed into 10 individual proteins. The cleavage of the precursor protein is facilitated by cellular peptidase and furin (or furin-like enzyme) and the viral protease NS2B/3 (13, 54, 55). The proteins include three structural proteins (the capsid C, membrane M, and envelope E) and seven nonstructural (NS) proteins (glycoprotein NS1, NS2A, protease cofactor NS2B, protease and helicase NS3, NS4A, NS4B, and the polymerase NS5). Although incompletely characterized, the NS proteins all seem important to replication. Thus, only a few deletion and complementation studies have been possible as most mutations totally disable virus replication (21-23, 32). Enzymatic properties have been assigned to only two NS proteins. NS5 is a large protein containing a domain coding the RNA-dependent RNA polymerase responsible for genome replication as well as a methyltransferase required for the formation of the cap structure at the 5′ end of the viral genome (10, 13, 17).
WNV has emerged as a major cause of viral encephalitis. Since its introduction into North America in 1999, WNV has rapidly spread across the continental United States and has recently been identified in Mexico and South America (26). WNV isolates are generally separated into two lineages (I and II) based on phylogenetic analysis of their envelope sequences (2). To date, only lineage I viruses have been shown to cause neurologic disease (26). The virus is generally maintained in an enzootic cycle between mosquitoes and birds (4). Mosquitoes act as the primary vectors of virus transfer. Mammals are incidental hosts that cannot act as reservoirs due to low virus levels in the blood. Upon infection, humans first develop a febrile illness that is generally resolved quickly (5). In approximately 10% of symptomatic cases, more serious sequelae occur, including meningoencephalitis. The immunocompromised, elderly, and children are at highest risk for symptomatic infections progressing to more severe central nervous system disease and even death. Antiviral therapies or vaccines are not yet available to treat or prevent WNV infections. In contrast to hepatitis C virus infections, interferon (IFN)-based therapies appear to be ineffective against WNV infections (6, 20, 36).
IFNs act as the earliest immune mediators against virus infections. The alpha IFN (IFN-α) receptor (IFNAR) has two major subunits, IFNAR1 and IFNAR2c, which dimerize upon IFN-α binding (51). Associated with the cytoplasmic domains of the subunits are members of the Janus kinase (JAK) family. In the absence of ligand, tyrosine kinase 2 (Tyk2) is associated with IFNAR1 while JAK1, STAT1, and STAT2 are associated with IFNAR2 (27). Dimerization of the receptor components leads to the activation of JAK1, which leads to a cross-phosphorylation cascade with Tyk2 (7, 8, 12). Activated JAK1 and Tyk2 tyrosine phosphorylate the STAT proteins STAT1 and STAT2, which then interact with IFN regulatory factor 9 to form the heterotrimeric transcription factor IFN-stimulated gene factor 3 (ISGF-3). ISGF-3 binds to specific DNA sequences known as IFN-specific response elements (ISRE) that are present in the promoters of select genes (19).
Recently, we along with others discovered that WNV and various members of the genus Flavivirus in the Flaviviridae family of viruses, including dengue virus and JEV, block the IFN signal transduction pathway (3, 15, 29, 38). Replication of these viruses inhibits the phosphorylation and activation of the IFN receptor-associated kinases, JAK1 and Tyk2, thus blocking phosphorylation of the STAT proteins and the activation of IFN-induced genes. These findings raised questions about the nature of the viral determinants that play a role in the observed inhibition of the IFN response. Although several reports addressing this question have been published, they have not yet yielded firm conclusions. For example, Munoz-Jordan and colleagues reported that, as with dengue virus, NS4B of WNV is the major determinant for inhibition (37, 38). In contrast, Liu and colleagues reported that all NS proteins, save for NS1 and NS5, could inhibit the IFN response (33). Alternatively, Lin et al. concluded from their investigations that JEV, the closest relative of WNV (>80% amino acid identity in NS5), inhibits the IFN response through NS5 (28). The latter study was based on more detailed work from Best and colleagues demonstrating that NS5 of Langat virus, a tick-borne encephalitis virus, could antagonize the IFN response through direct binding to the IFN-α and IFN-γ receptor complexes (3).
While previous investigations relied on transient expression assays, we used an alternative strategy based primarily on replication-competent subgenomic replicons and infectious WNV. Our results revealed that the mechanism by which WNV inhibits the IFN response depends on multiple factors and is much more complex than anticipated from previous studies.
MATERIALS AND METHODS
Plasmid construction and mutagenesis.
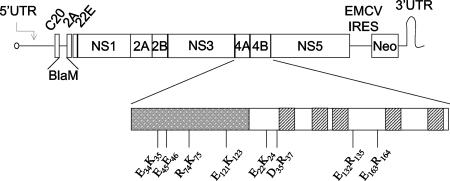
For mutagenesis experiments, we used plasmid C20DX-BlaM. This plasmid was derived from plasmid C20Dxrep/neo expressing a subgenomic WNV lineage I ([WNI] Kunjin) replicon and neomycin phosphotransferase (24). The BLaM (beta-lactamase) gene including the autoprotease site 2A from foot and mouth disease virus was incorporated into the 5′ end of the C20DXrep/neo replicon as previously described (52, 53). The final construct is described in Fig. 1.
FIG. 1.
Schematic of C20DX-BLaM. WNV subgenomic replicon containing the beta-lactamase (BLaM) transgene. The coding regions for NS4A (gray box) and NS4B are magnified. The transmembrane domains of NS4B are represented by dashed boxes. The double-alanine site-directed substitutions are highlighted. UTR, untranslated region; IRES, internal ribosome entry cite; EMCV, encephalomyocarditis virus.
To create NS4B-E22K24, we mutated NS4B residues E22K24 in C20DX-BLaM to alanines using a QuickChange mutagenesis kit (Stratagene), according to the manufacturer's instruction. The primers used for mutagenesis are available from the authors on request. The mutations were verified by nucleotide sequence analysis. To minimize the chances for the presence of unwanted mutations, a 1,216-nucleotide fragment was excised with BsiWI and AgeI and back-cloned into naïve C20DX-BlaM. The NS4B-E22K24 mutation was introduced into C20DXrep/neo without the beta-lactamase gene; the BsiWI-AgeI fragment was excised from the C20DX-BLaM plasmid and cloned into the corresponding region of C20DXrep/neo to produce C20DX-EK (where EK indicates the mutation of the E22K24 residues to alanines).
To produce WNII-ER, NS4B residues E22R24 were replaced with alanines in the WNV lineage II (WNII) infectious clone pSP6WN/Xba. To create pWNII-KUN4B and pWNII-KUN4B-EK infectious clones, we replaced nucleotides 6931 to 7000 of pSP6WN/Xba, encoding NS4B, with the corresponding nucleotides from Kunjin replicon C20DXrep/neo and C20DX-BLaM-EK, respectively, by a two-step PCR mutagenesis protocol (41). The sequences were verified by restriction endonuclease digestion and DNA sequencing. The primers used for plasmid construction are available on request from the authors.
To create pUNO-NS4B constructs, NS4B coding regions were cloned from C20DX-BlaM and NS4B-E22K24 replicons. Both constructs contained the 2-kDa segment with the forward primer (restriction sites are highlighted in boldface) 5′-GCACCGGTCATCATGCAACGTTCGCAGACAGACAAC-3′. The reverse primer was 5′-TACCTCTAGATCATCTTTTTAGTCCTGGTTTTTCC-3′. After PCR amplification, PCR products were digested with AgeI and XbaI. These fragments were cloned into pUNO using AgeI and NheI sites.
Cell culture and virus infections.
HeLa, Vero, and BHK-21 cells were maintained at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco's modified Eagle medium (DMEM) (Gibco) supplemented with 10% fetal calf serum and 100 μg/ml penicillin, 100 μg/ml streptomycin, 1% nonessential amino acids (Gibco), and 50 μg/ml glutamine. Mouse embryo fibroblasts (MEFs) were maintained under similar conditions as above except in DMEM supplemented with 10% heat-inactivated fetal calf serum and 100 μg/ml glutamine, as described previously (45, 47).
Prior to infection, HeLa or Vero cells were seeded at a density of 1 × 106 cells in 100-mm plates. Cells were infected with virus in phosphate-buffered saline containing 2% fetal bovine serum for 1 h. Unadsorbed virus was removed by washing, and DMEM supplemented with 2% fetal bovine serum was added to the cells. After 24 h, cells were treated with IFN-α2a (Schering-Plough) or left untreated, as described below. For growth curves, Vero cells or MEFs were seeded at 5 × 104 cells/well in a 24-well plate. After 16 h, cells were infected. Medium was collected at indicated times (see Fig. 7 and 9). Virus titers were determined on BHK-21 cells as previously described (59).
FIG. 7.
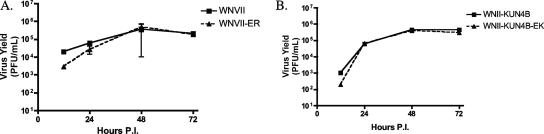
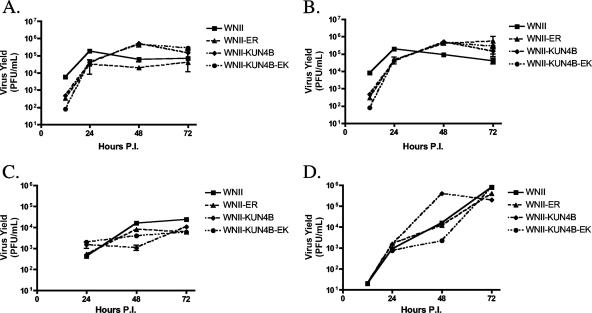
Mutant viruses exhibit growth kinetics similar to wild type. Vero cells were infected at an MOI of 0.5 with WNII, WNII-ER, WNII-KUN4B, or WNII-KUN4B-EK for 12, 24, 48, and 72 h. Virus titers in medium from infected cells were determined by plaque assay on BHK-21 cells.
FIG. 9.
Mutant viruses exhibit growth kinetics comparable to wild type. Wild-type (A and C) or IFN-α/β receptor knockout (B and D) B6 MEFs were infected at an MOI of 0.2 (A and B) or 0.002 (C and D) with WNII, WNII-ER, WNII-KUN4B, or WNII-KUN4B-EK for 12, 24, 48, and 72 h. Virus titers in medium from infected cells were determined by plaque assay on BHK-21 cells. Means are shown with standard error bars.
In vitro transcription and transfection of RNA.
To produce replicon-bearing cells, the C20DX-BLaM plasmid (10 μg) was linearized with XhoI. Linearized plasmid (1 μg) was in vitro transcribed using the SP6 in vitro transcription kit (Promega). HeLa cells were electroporated with 5 μg of C20DX-BLaM RNA. Forty-eight hours posttransfection, cells were selected with 500 μg/ml G418. After 3 weeks of selection, colonies were harvested and expanded as pools. The same procedure was followed to produce replicon-bearing cells without the beta-lactamase transgene.
To produce infectious virus, WNII was produced from the plasmid construct pSP6WN/Xba or mutants as previously described (59). BHK-21 cells were electroporated with 2 μg of in vitro transcribed RNA. Medium was collected 3 days posttransfection, cleared of cell debris, and stored at −70°C. Titers were determined by plaque assay on BHK-21 cells as previously described (59).
Beta-lactamase assay.
HeLa or replicon-bearing cells were seeded into 24-well tissue culture dishes at 5 × 104 cells/well for 24 h. The cells were then assayed for beta-lactamase activity using a beta-lactamase loading solutions kit (Invitrogen) according to the manufacturer's instructions. Briefly, the cells were washed two times in Opti-Mem (Gibco), incubated for 1.5 h at room temperature in Opti-MEM containing the fluorescent substrate CCF2A, and then viewed on a Nikon fluorescence microscope.
Antibodies.
Rabbit polyclonal antibodies to WNV NS protein NS4B were produced by Pacific Immunology (San Diego, CA) against a peptide spanning positions 2 to 12 (EMGWLDKTKSD) of WNV. Goat polyclonal antibodies against actin (sc1616; Santa Cruz Biotechnology), mouse monoclonal antibodies against STAT1α p91 (sc417; Santa Cruz Biotechnology), and rabbit polyclonal antibodies against phosphotyrosine 701-STAT1 (9171; Cell Signaling Technologies) were used in Western blotting experiments.
VSV cytopathic effect (CPE) assay.
Parental HeLa and replicon cells were seeded in 24-well tissue culture dishes at 5 × 104 cells/well. After 24 h, the cells were treated with indicated concentration of IFN for 24 h and then infected with vesicular stomatitis virus ([VSV] Indiana strain) at a multiplicity of infection (MOI) of 1. Twenty-four hours postinfection, the cells were washed with phosphate-buffered saline, fixed with 10% formaldehyde for 30 min, and stained with 2% crystal violet for 20 min. Excess stain was removed with multiple rounds of washes with water.
Western blotting.
Cells were lysed in 1% Triton lysis buffer (1% Triton X-100, 150 mM sodium chloride, 50 mM Tris, pH 8.0), and protein amounts were quantified by Bradford assay (Bio-Rad). Proteins (15 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P (Millipore) membranes. Membranes were cut according to molecular weight markers, and membrane strips were probed with the indicated antibodies at the following dilutions: actin, 1:600; STAT1α p91, 1:500; P-STAT1, 1:750. Horseradish peroxidase-conjugated secondary antibodies against goat, mouse, and rabbit immunoglobulin G (Amersham) were used at a dilution of 1:2,500. The bands were visualized using SuperSignal West Pico solutions (Pierce).
RNA analysis.
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For Northern blotting, 5 μg of total RNA was fractionated on a 1% agarose gel containing 2.2 M formaldehyde and transferred to nitrocellulose. Membranes were hybridized with a riboprobe specific for the neomycin phosphotransferase II gene. For PCR, total RNA (1 μg) was used for cDNA synthesis with random primers. Real-time PCR was performed in triplicate using 200 pg of amplified cDNA and primer-probe sets specific to β-actin, IFIT1, IFI27, and MxA (ABI Biosystems) on an ABI7000 sequence detection system. Results were normalized to β-actin prior to determination of relative induction.
Reporter gene assays.
HeLa cells were transfected using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. As a positive control, pEF-HA-HPIV2 expressing the V protein of human parainfluenza virus type 2 (HPIV2) was used (16, 39). Each transfection of 7.5 × 104 cells contained 0.8 μg of the plasmid of interest (HPIV2 V protein or pUNO-NS4B constructs), 0.2 μg of pISRE-Luc (Clontech), and 0.02 μg of pCMV-RL (Promega). Transfections were performed in triplicate. At 24 h posttransfection, cells were mock treated or treated with 100 U of IFN-α2a. Cells were maintained for 24 h and then harvested and lysed. Luciferase assays were performed using a dual luciferase assay system (Promega). Firefly luciferase expression was normalized to Renilla luciferase.
Nucleotide sequence accession number.
The nucleotide sequence of the C20DX-BlaM construct has been deposited in the GenBank database under accession number EF536932.
RESULTS
Mutations in the NS4 region.
To analyze the role of WNV NS proteins in the inhibition of IFN signaling, a panel of mutants was created in C20DX-BLaM, a WNV subgenomic replicon expressing the beta-lactamase transgene. We targeted the NS4 coding regions to more clearly define their functions in viral replication and IFN signaling. Unlike other NS proteins, the NS4 proteins do not encode specific enzymatic activities. Because both NS4A and NS4B are required in cis for viral replication, we created mutants with only two amino acid changes to maximize the chances for obtaining mutants sensitive to IFN but competent for viral replication. We selected regions containing clusters of five charged amino acids and mutated two charged residues to alanine (1) (Fig. 1). Because clusters of charged residues are generally located on the surface of proteins, altering them increases the chances of disrupting protein-protein interactions or functions while leaving the overall structures intact. In vitro transcribed RNA derived from the wild-type and the mutants was electroporated into HeLa cells. Twenty-four hours after electroporation, more than 25% of the cells expressed beta-lactamase, indicating that all the genomes used in these experiments were translated with similar efficiency (data not shown).
To identify genomes that were competent for RNA replication, we maintained the transfected HeLa cells in medium containing G418. Only one of the eight mutants produced colonies after 3 weeks of selection. This replicon had a mutation in amino acids E22K24 in the NS4B protein (NS4B-E22K24). The colonies were pooled and expanded for all subsequent analyses. Analyzing the cells for beta-lactamase activity revealed that >95% of the cells were positive and, hence, replicated the mutant genomes (Fig. 2B and C). Northern blot analysis confirmed this result and demonstrated that the mutant replicated in HeLa cells with an efficiency that was comparable to wild-type replicons (Fig. 2D). Similarly, Western blot analysis revealed that cells expressing wild-type and mutant replicons expressed equivalent levels of NS4B proteins, indicating proper genome replication and translation (see Fig. 4). Finally, transfection of BHK-21 cells with wild-type and NS4B-E22K24 replicons produced stable colonies, as observed with HeLa cells, demonstrating that the results were not influenced by cell-type-specific factors (data not shown).
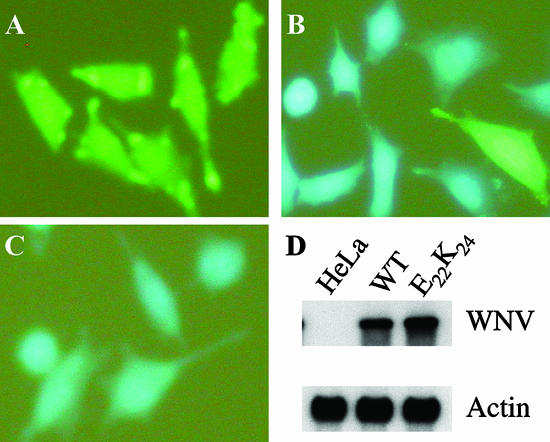
FIG. 2.
C20DX-BLaM replicons are stable in HeLa cells. HeLa cells were electroporated with wild type or mutant RNA. The transfected cells were selected with G418. Colonies were pooled and passed as cell lines. Parental HeLa (A), C20DX-BLaM (B), or NS4B-E22K24 (C) cells were analyzed for beta-lactamase activity. (D) Northern blot analysis of cell lines for the amount of WNV RNA present. Actin served as the loading control. WT, wild type; E22K24, NS4B-E22K24.
FIG. 4.
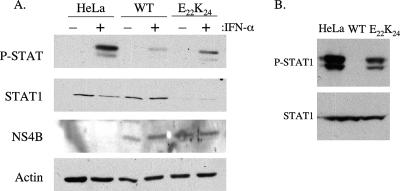
The NS4B-E22K24 mutation cannot inhibit IFN-induced STAT phosphorylation in replicon cells. (A) Parental HeLa, C20DX-BLaM (wild type [WT]) and NS4B-E22K24 (E22K24) cells were left untreated or treated with 1,000 IU of IFN-α for 20 min. (B) A second pool of replicon-bearing HeLa cells was generated with the C20DXrep/neo replicon harboring the NS4B-E22K24 mutation without the beta-lactamase transgene. Cells were treated with IFN-α. Equal amounts of cell lysates were separated by SDS-PAGE and transferred to Immobilon-P membranes. The expression of STAT1, phosphorylated STAT (P-STAT), and NS4B proteins was detected by Western blotting with the corresponding specific antibodies. Actin served as the lane loading control. One representative experiment of three is shown.
Mutation in NS4B relieves inhibition of IFN signaling.
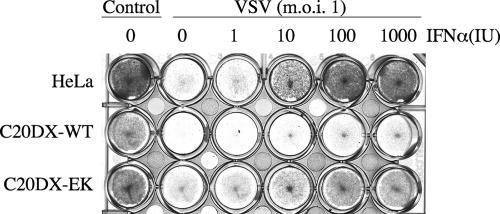
In the next step, we investigated whether expression of the NS4B-E22K24 mutant in HeLa cells inhibited the IFN response, as observed in cells expressing the wild-type replicon. For this purpose, we took advantage of the known antiviral activity of IFN against VSV. We incubated cells with IFN prior to VSV infection and determined the protective effects by observing virus-induced cell death (Fig. 3). The results showed that IFN protected HeLa cells from VSV-induced CPE at 10 U; in contrast, wild-type replicon cells were not protected at any concentration. The NS4B-E22K24 mutant replicon-containing cells were protected against lysis by 100 U of IFN, a slightly higher amount than needed to prevent lysis in parental HeLa cells. The results indicated that the IFN signaling pathway remained functional in the presence of the NS4B-E22K24 replicon and was able to induce downstream effects resulting in protection from VSV lysis.
FIG. 3.
The NS4B-E22K24 mutation relieves WNV repression of IFN antiviral response. Parental HeLa, C20DX-BLaM, or NS4B-E22K24 (C20DX-EK) cells were incubated for 24 h with the indicated concentrations of IFN-α and then infected with VSV at an MOI of 1. Twenty-four hours postinfection, cells were stained with crystal violet to determine CPE. The cells in the control column were not infected or treated with IFN. WT, wild type.
To validate the results obtained with VSV infection and correlate these findings with the molecular components of the IFN activation cascade, we incubated cells with IFN and determined the levels of phosphorylated STAT1. As previously seen with KUNCD20 cells, wild-type C20DX-BLaM replicon cells significantly inhibited the levels of phosphorylated STAT1 (Fig. 4) (15). In contrast, HeLa cells harboring the NS4B-E22K24 replicon still exhibited stimulation of the IFN pathway, as seen by induced phosphorylation of the STAT1 protein (Fig. 4). The level of STAT1 phosphorylation was increased in comparison to cells with the wild-type replicon but slightly reduced in relation to the parental HeLa cells (Fig. 4). Surprisingly, the overall level of STAT1 protein was decreased in the mutant replicon cells. Although the basis for reduced STAT1 expression in cells harboring the NS4B-E22K24 replicon is currently unclear, the ratio of phosphorylated to total protein is comparable to that found in parental HeLa cells.
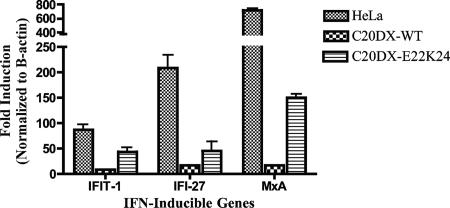
To better quantitate the effects of wild-type and mutant replicons on IFN signaling, we examined the induction of ISGs in parental HeLa cells and replicon cells treated with IFN. Using TaqMan quantitative PCR, we analyzed the expression of three genes known to be up-regulated by IFN treatment, the IFIT1, IFI27, and MxA genes (Fig. 5) (9). The wild-type replicon inhibited ISG induction to almost the background level. Induction of ISGs in the NS4B-E22K24 replicon cells was less than in HeLa cells but much greater than in wild-type replicon cells. For instance, the MxA gene, which has been shown to play an important role in antiviral activities against a number of viruses, is induced fivefold less in NS4B-E22K24 replicon cells than in parental HeLa cells, but expression is eightfold higher than in wild-type replicon cells. These results agree with those above demonstrating that the mutation of two amino acids in the NS4B coding region of the WNV replicon does not suppress genome replication but relieves the inhibition of IFN signaling. To validate our results, we produced a second pool of HeLa cells expressing the NS4B-E22K24 mutant in the context of a C20DXrep/neo vector lacking the beta-lactamase transgene (Fig. 1). As before, the cells expressing the mutant were sensitive to IFN, demonstrating that the outcomes of our experiments were not influenced by the selection of the vector. Moreover, these cells expressed normal levels of STAT1 (Fig. 4B).
FIG. 5.
IFN-stimulated gene expression in replicon cells. Parental HeLa, C20DX-BLaM (C20DX-WT), and NS4B-E22K24 (C20DX-E22K24) cells were left untreated or treated with 100 IU of IFN-α for 6 h. Total RNA was isolated by TRIzol reagent extraction. Equivalent amounts of total RNA were reverse transcribed with gene-specific primers to create cDNA pools. Real-time PCR with ISG-specific TaqMan primer-probe sets was performed. Results shown are the average of two independent experiments.
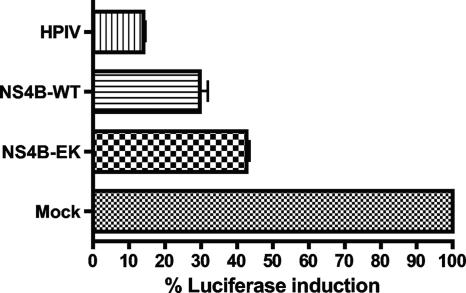
To determine the effect individually expressed NS4B proteins had on IFN signaling, wild-type and mutant NS4B-E22K24 proteins were exogenously expressed in HeLa cells treated with IFN. As a reporter for the activation of the IFN signal transduction pathway, we used the luciferase gene expressed from the IFN-responsive ISG54 promoter. Cells transfected with a plasmid expressing the V protein of HPIV, leading to the degradation of STAT2, served as a positive control for the inhibition of IFN signaling (39). As reported previously, NS4B inhibited activation of the IFN-responsive promoter, but the inhibition was weaker than observed with the V protein (Fig. 6) (37, 38). Consistent with the results described above, wild-type NS4B blocked IFN signaling more efficiently than the mutant (Fig. 6).
FIG. 6.
Individually expressed NS4B proteins reflect activity in replicons. HeLa cells were transfected with each of indicated plasmids. At 24 h posttransfection, cells were treated with 100 U of IFN-α. The firefly luciferase activities were normalized to Renilla luciferase to determine ISRE-specific gene induction. Firefly luciferase activities were determined as mean values from three independent experiments performed in triplicate (P values of 0.005 to 0.01). WT, wild type; NS4B-EK, NS4B-E22K24.
Mutations in NS4B do not affect IFN inhibition in infectious virus.
Although the replicon system faithfully mimicked certain aspects of WNV replication, it still had limitations with respect to virus infection. For instance, replicon-bearing cells are selected, and this leads to propagation of variants that are less cytopathic (31, 44). Further, replicons do not express the viral structural proteins, which play an active role in the virus life cycle, thus eliminating their effects as well as possible interactions between NS and structural proteins.
Therefore, to analyze the effects of the NS4B-E22K24 mutation in a more natural context, we introduced the corresponding mutations into a WNII infectious clone. The WNII-derived clone was used because attempts to introduce the mutations into an infectious clone derived from lineage I (NY99) were unsuccessful due to instability of the plasmid during mutagenesis (48). Although WNII and Kunjin virus (lineage I) exhibit very high overall amino acid homology (∼90%), they differ between residues 11 and 32 in NS4B. Moreover, NS4B in WNII carries an arginine (R) residue in lieu of lysine (K) at position 24. To minimize the chance for any lineage- or context-specific effects, we also produced chimeric WNII infectious clones with Kunjin sequences spanning amino acids 10 to 33 in NS4B and introduced the E22K24 mutations, yielding mutant WNII-KUN4B-EK. The resulting RNAs were transfected into BHK-21 cells, and virus was collected 3 days later. The presence of the mutations was verified by sequence analysis (data not shown). The titer of the WNII-ER mutant virus was approximately fivefold less than the wild type. The wild-type and mutant chimeric viruses (WNII-KUN4B and WNII-KUN4B-EK) replicated to levels equivalent to parent WNII. Further, in single-step growth curves in Vero cells, both mutants replicated to identical levels compared to their respective wild types (Fig. 7).
HeLa cells were incubated with wild-type or mutant virus at an MOI of 1 to ensure that a majority of cells were infected, and then cells were treated with IFN to analyze signaling. Western blotting for phosphorylated STAT1 showed that wild-type and mutant viruses all completely inhibited IFN stimulation (Fig. 8). The duration of infection did not influence these results. Moreover, the viruses behaved identically in Vero cells, suggesting that the effects were not cell type specific (data not shown). Further, the chimeric viruses WNII-KUN4B and WNII-KUN4B-EK were analyzed for their ability to block IFN signaling. Both chimeric viruses inhibited STAT1 phosphorylation in both HeLa and Vero cells (data not shown). These observations were surprising since they did not reflect the results seen with the replicon-bearing cells.
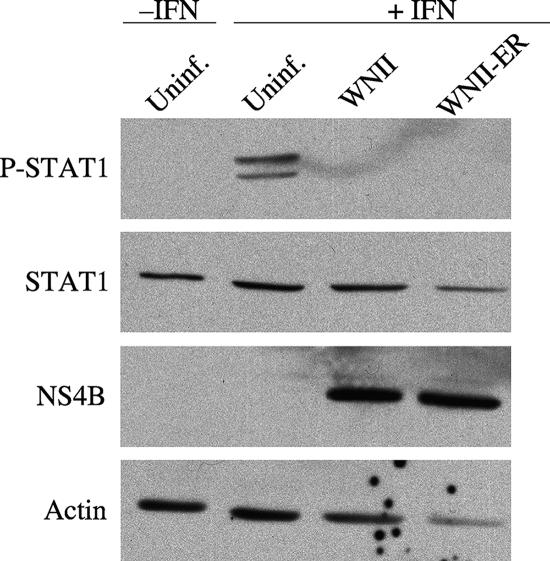
FIG. 8.
Mutant viruses inhibit STAT1 phosphorylation. HeLa cells were infected at an MOI of 1 with WNII or WNII-ER for 24 h. The infected (lanes 3 and 4) or uninfected (Uninf; lane 2) cells were treated with 1,000 U of IFN-α (+IFN) for 20 min. As a control, cells were not infected (Uninf; lane 1) and left untreated (−IFN). Equal amounts (15 μg) of cell lysates were separated by SDS-PAGE and transferred to Immobilon-P membranes. The expression of phosphorylated STAT1 (P-STAT1), STAT1, and NS4B proteins was detected by Western blotting with the corresponding specific antibodies. Actin served as the lane loading control. One representative experiment of three is shown.
The infection experiments in HeLa cells could have masked a subtle effect of the NS4B mutations, since they were performed with a relatively high infectious dose. To further characterize the NS4B virus mutant, we analyzed the impact of IFN production and response on mutant virus replication and spread. We infected MEFs derived from wild-type B6 mice, which are competent for IFN production and activation. To control for any effects not related to the IFN response, we infected MEFs from congenic IFN-α/β receptor knockout animals. Low-passage MEFs were infected with WNII and mutant viruses at low MOIs of 0.2 and 0.002 and assayed for virus production. The mutant viruses exhibited slightly lower virus titers at early times (12 h postinfection) but produced levels of virus equivalent to wild-type later (24 to 72 h postinfection). Since the mutants exhibited a similar delay in MEFs lacking the IFN-α/β receptor, this effect was not caused by activity of the IFN system but most likely reflected reduced replication following the infection. Although the starting inocula did not appear to make a significant difference in the outcome of the experiments, IFN-α/β receptor knockout MEFs produced higher amounts of virus than wild-type cells. This observation is consistent with the proposed activation of IFN-β following infection of cells with WNV (Fig. 9), (11). Interestingly, the WNII-ER mutant viruses caused less cell death than observed with the wild type, suggesting that the mutations in NS4B reduced the cytopathic potential of this virus (data not shown).
DISCUSSION
In this report, we established a biologically relevant system in which to examine the effects of WNV NS proteins on the IFN signaling pathway. Our findings demonstrate that alanine substitution mutations at residues E22K24 in the NS4B coding region resulted in a WNV replicon that was replication competent yet unable to completely inhibit the IFN phosphorylation cascade. Further, homologous mutations in infectious WNV do not mirror these results, suggesting a complex interplay between the structural and NS proteins. Although it has been hypothesized that NS4B is the protein responsible for WNV-mediated IFN inhibition, these data are the first to define the importance of NS4B for the inhibition of IFN signaling in the biological context of replicating genomes. In fact, our results are surprising in view of previous studies with ectopically expressed proteins of WNV and other flaviviruses that suggest roles for essentially all other NS proteins in the inhibition of the IFN response (3, 28, 33). One major difference between our experimental approach and that of other investigators was that we relied primarily on cells replicating viral genomes rather than transient transfection assays. Nevertheless, we also observed that wild-type NS4B blocks IFN signaling and that the NS4B-E22K24 mutant partially relieves the inhibition. Our results show, similar to others, that wild-type NS4B can block the majority of IFN signaling, whereas the NS4B-E22K24 mutant partially relieves the inhibition. Although our in vitro reporter assays confirm previously published data concerning the role of NS4B in the inhibition of IFN signaling as well as support our own work with replicon cells, we must interpret these overexpression studies with caution. We cannot exclude the possibility that transfection and overexpression disrupt other mechanisms that lead to alteration of the cell. Moreover, as our studies with full-length infectious virus show, there may be more than one player important in blocking IFN signaling pathways.
NS4B is a small, predominantly helical, membrane-associated protein. Hydrophobicity plots based on primary amino acid sequence indicate that the protein consists of four or five transmembrane domains (42, 58). Recently, Miller and colleagues showed that the NS4B protein of the closely related dengue virus has three transmembrane domains (35). Although neither the crystal structure nor topology of WNV NS4B has been elucidated, the targets we selected for mutagenesis are located in predicted exposed regions of the protein, based on the above criteria (Fig. 1). Further, deletion mutagenesis of overexpressed dengue virus NS4B indicated that amino acids 1 to 77 failed to mediate the IFN signaling block (37). Our results demonstrated that amino acids in the N-terminal domain are required for this inhibition in WNV. It is important that both regions are predicted to reside in the lumen of the ER (35). The WNV NS4B-E22K24 mutation may prevent important charge-charge interactions necessary for protein binding. Further, the mutation might alter the conformation of a critical domain for function. This possibility is intriguing due to the finding that dengue NS4B can have multiple conformations, which are dictated by the N terminus (35).
Our results demonstrate that IFN inhibition by WNV is more complex than previously envisioned. Although NS4B is capable of blocking IFN signaling in transient expression and replicon systems, the exact mechanism is unclear. There are a number of possibilities, both direct and indirect, through which NS4B can block IFN signaling. First, reducing the amount of IFN receptor would cause an inhibition. However, we have found that expression of IFNAR is unaffected by the presence of the replicon (15). Second, NS4B could bind to the receptor or the JAK proteins to physically block activation. Third, NS4B could activate a negative regulatory protein such as a phosphatase or the suppressor of cytokine signaling proteins to act at the level of the receptor. Finally, NS4B could cause a generalized cellular stress that leads to down-regulation of a number of cell signaling pathways. The mechanism of NS4B-induced inhibition may be determined by dissecting the biochemical composition of the IFN receptor in the presence of virus.
Although mutations in NS4B in subgenomic replicons allowed for IFN signaling, infections with full-length viruses containing corresponding mutations abrogated IFN pathway activation. As mentioned above, the replicon cells were pools, and the effects seen were global; thus, it is unlikely that other WNV proteins were mutated in the replicon. It is more plausible that a structural protein not present in the replicon cells is involved in blocking STAT phosphorylation. However, apart from their role in virus entry and egress, not much has been discovered to date about the structural proteins during virus replication. Even though they are not necessary to maintain functionally replicating replicons, they may play an as yet unknown role in virus pathogenesis (25). For example, it has been reported that capsid proteins could cause apoptosis and inflammation under certain experimental conditions (60). Also, Munoz-Jordan and colleagues demonstrated that the inhibition of IFN signaling by NS4B could be augmented by the presence of NS2A and NS4A (38). In the case of infectious virus, it is possible that NS2A or NS4A can compensate for NS4B during the viral life cycle. In other words, protein interactions may dictate the degree of IFN response inhibition. Further, the level of replication may affect the IFN signaling pathway. However, we think that this possibility is unlikely, since Northern blot analysis showed that levels of WNV genomic RNA were similar in infected and replicon cells (data not shown).
It is important to note that the replicon is maintained in cells without the CPE that is a hallmark of WNV infection (40, 43, 46, 49). Interestingly, the mutant WNII viruses were less cytopathic than the wild type, yet virus titers were not reduced. These data would directly link NS4B as a mediator of cell death separate from other functions. NS4B is known to expand and modify the ER, which facilitates virus replication (56). It may be that NS4B alters the ER such that cells incur death from an unfolded protein response or ER stress (50, 61). Whatever the mechanism might be, our data suggest that inhibition of IFN innate immunity during infection is more complex than previously suggested. Future studies will elucidate the mechanism(s) through which WNV can evade and influence the immune system.
Acknowledgments
We thank Kerry Campbell and William Mason for their helpful comments and critical reading of the manuscript. We acknowledge the services provided by the Fox Chase Cancer Center tissue culture and DNA sequencing facilities. We thank Alexander Khromykh (Brisbane, Australia) for the C20DXrep/neo plasmid, Vladimir Yamshchikov (University of Kansas) for the pSP6WN/Xba plasmid, Curt Horvath (Northwestern University) for the pEF-HA-HPIV2 plasmid, and Michael Diamond (Washington University, St. Louis, MO) for the wild-type and IFN-α/β receptor knockout MEFs essential to conduct this study. We acknowledge J. David Stiffler for producing replicon mutants and Paul Gramlich for screening NS antibodies.
This work was supported by grants from the NIH and an appropriation from the Commonwealth of Pennsylvania. J.D.E. was supported by grant T32 CA-09035-30.
Footnotes
Published ahead of print on 22 August 2007.
REFERENCES
- 1.Babb, R., C. C. Huang, D. J. Aufiero, and W. Herr. 2001. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA-binding structure. Mol. Cell. Biol. 21:4700-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berthet, F. X., H. G. Zeller, M. T. Drouet, J. Rauzier, J. P. Digoutte, and V. Deubel. 1997. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 78:2293-2297. [DOI] [PubMed] [Google Scholar]
- 3.Best, S. M., K. L. Morris, J. G. Shannon, S. J. Robertson, D. N. Mitzel, G. S. Park, E. Boer, J. B. Wolfinbarger, and M. E. Bloom. 2005. Inhibition of interferon-stimulated JAK-STAT signaling by a tick-borne flavivirus and identification of NS5 as an interferon antagonist. J. Virol. 79:12828-12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinton, M. A. 2002. The molecular biology of West Nile Virus: a new invader of the western hemisphere. Annu. Rev. Microbiol. 56:371-402. [DOI] [PubMed] [Google Scholar]
- 5.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1125. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 6.Chan-Tack, K. M., and G. Forrest. 2005. Failure of interferon alpha-2b in a patient with West Nile virus meningoencephalitis and acute flaccid paralysis. Scand. J. Infect. Dis. 37:944-946. [DOI] [PubMed] [Google Scholar]
- 7.Colamonici, O., H. Yan, P. Domanski, R. Handa, D. Smalley, J. Mullersman, M. Witte, K. Krishnan, and J. Krolewski. 1994. Direct binding to and tyrosine phosphorylation of the alpha subunit of the type I interferon receptor by p135tyk2 tyrosine kinase. Mol. Cell. Biol. 14:8133-8142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colamonici, O. R., H. Uyttendaele, P. Domanski, H. Yan, and J. J. Krolewski. 1994. p135tyk2, an interferon-alpha-activated tyrosine kinase, is physically associated with an interferon-alpha receptor. J. Biol. Chem. 269:3518-3522. [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredericksen, B. L., M. Smith, M. G. Katze, P. Y. Shi, and M. Gale, Jr. 2004. The host response to West Nile virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78:7737-7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauzzi, M. C., L. Velazquez, R. McKendry, K. E. Mogensen, M. Fellous, and S. Pellegrini. 1996. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J. Biol. Chem. 271:20494-20500. [DOI] [PubMed] [Google Scholar]
- 13.Grun, J. B., and M. A. Brinton. 1986. Characterization of West Nile virus RNA-dependent RNA polymerase and cellular terminal adenylyl and uridylyl transferases in cell-free extracts. J. Virol. 60:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grun, J. B., and M. A. Brinton. 1988. Separation of functional West Nile virus replication complexes from intracellular membrane fragments. J. Gen. Virol. 69:3121-3127. [DOI] [PubMed] [Google Scholar]
- 15.Guo, J. T., J. Hayashi, and C. Seeger. 2005. West Nile virus inhibits the signal transduction pathway of alpha interferon. J. Virol. 79:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, J. T., Q. Zhu, and C. Seeger. 2003. Cytopathic and noncytopathic interferon responses in cells expressing hepatitis C virus subgenomic replicons. J. Virol. 77:10769-10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 18.Hanna, S. L., T. C. Pierson, M. D. Sanchez, A. A. Ahmed, M. M. Murtadha, and R. W. Doms. 2005. N-linked glycosylation of West Nile virus envelope proteins influences particle assembly and infectivity. J. Virol. 79:13262-13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalil, A. C., M. P. Devetten, S. Singh, B. Lesiak, D. P. Poage, K. Bargenquast, P. Fayad, and A. G. Freifeld. 2005. Use of interferon-alpha in patients with West Nile encephalitis: report of 2 cases. Clin. Infect. Dis. 40:764-766. [DOI] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 2000. cis- and trans-acting elements in flavivirus RNA replication. J. Virol. 74:3253-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khromykh, A. A., P. L. Sedlak, and E. G. Westaway. 1999. trans-complementation analysis of the flavivirus Kunjin ns5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J. Virol. 73:9247-9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 27.Li, X., S. Leung, I. M. Kerr, and G. R. Stark. 1997. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol. Cell. Biol. 17:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, R. J., B. L. Chang, H. P. Yu, C. L. Liao, and Y. L. Lin. 2006. Blocking of interferon-induced Jak-Stat signaling by Japanese encephalitis virus NS5 through a protein tyrosine phosphatase-mediated mechanism. J. Virol. 80:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced Jak-Stat signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., and C. M Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, PA.
- 31.Liu, W. J., H. B. Chen, and A. A. Khromykh. 2003. Molecular and functional analyses of Kunjin virus infectious cDNA clones demonstrate the essential roles for NS2A in virus assembly and for a nonconservative residue in NS3 in RNA replication. J. Virol. 77:7804-7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, W. J., X. J. Wang, V. V. Mokhonov, P. Y. Shi, R. Randall, and A. A. Khromykh. 2005. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J. Virol. 79:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 281:8854-8863. [DOI] [PubMed] [Google Scholar]
- 36.Morrey, J. D., C. W. Day, J. G. Julander, L. M. Blatt, D. F. Smee, and R. W. Sidwell. 2004. Effect of interferon-alpha and interferon-inducers on West Nile virus in mouse and hamster animal models. Antivir. Chem. Chemother. 15:101-109. [DOI] [PubMed] [Google Scholar]
- 37.Munoz-Jordan, J. L., M. Laurent-Rolle, J. Ashour, L. Martinez-Sobrido, M. Ashok, W. I. Lipkin, and A. Garcia-Sastre. 2005. Inhibition of alpha/beta interferon signaling by the NS4B protein of flaviviruses. J. Virol. 79:8004-8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 40.Parquet, M. C., A. Kumatori, F. Hasebe, K. Morita, and A. Igarashi. 2001. West Nile virus-induced Bax-dependent apoptosis. FEBS Lett. 500:17-24. [DOI] [PubMed] [Google Scholar]
- 41.Pont-Kingdon, G. 1994. Construction of chimeric molecules by a two-step recombinant PCR method. BioTechniques 16:1010-1011. [PubMed] [Google Scholar]
- 42.Puig-Basagoiti, F., M. Tilgner, C. J. Bennett, Y. Zhou, J. L. Munoz-Jordan, A. Garcia-Sastre, K. A. Bernard, and P. Y. Shi. 2007. A mouse cell-adapted NS4B mutation attenuates West Nile virus RNA synthesis. Virology 361:229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramanathan, M. P., J. A. Chambers, P. Pankhong, M. Chattergoon, W. Attatippaholkun, K. Dang, N. Shah, and D. B. Weiner. 2006. Host cell killing by the West Nile virus NS2B-NS3 proteolytic complex: NS3 alone is sufficient to recruit caspase-8-based apoptotic pathway. Virology 345:56-72. [DOI] [PubMed] [Google Scholar]
- 44.Rossi, S. L., Q. Zhao, V. K. O'Donnell, and P. W. Mason. 2005. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology 331:457-470. [DOI] [PubMed] [Google Scholar]
- 45.Samuel, M. A., and M. S. Diamond. 2005. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 79:13350-13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samuel, M. A., J. D. Morrey, and M. S. Diamond. 2007. Caspase 3-dependent cell death of neurons contributes to the pathogenesis of West Nile virus encephalitis. J. Virol. 81:2614-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samuel, M. A., K. Whitby, B. C. Keller, A. Marri, W. Barchet, B. R. Williams, R. H. Silverman, M. Gale, Jr., and M. S. Diamond. 2006. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shrestha, B., D. Gottlieb, and M. S. Diamond. 2003. Infection and injury of neurons by West Nile encephalitis virus. J. Virol. 77:13203-13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Su, H. L., C. L. Liao, and Y. L. Lin. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 76:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell Microbiol. 8:907-922. [DOI] [PubMed] [Google Scholar]
- 52.Varnavski, A. N., and A. A. Khromykh. 1999. Noncytopathic flavivirus replicon RNA-based system for expression and delivery of heterologous genes. Virology 255:366-375. [DOI] [PubMed] [Google Scholar]
- 53.Varnavski, A. N., P. R. Young, and A. A. Khromykh. 2000. Stable high-level expression of heterologous genes in vitro and in vivo by noncytopathic DNA-based Kunjin virus replicon vectors. J. Virol. 74:4394-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wengler, G. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 55.Wengler, G., G. Czaya, P. M. Farber, and J. H. Hegemann. 1991. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J. Gen. Virol. 72:851-858. [DOI] [PubMed] [Google Scholar]
- 56.Westaway, E. G., A. A. Khromykh, M. T. Kenney, J. M. Mackenzie, and M. K. Jones. 1997. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology 234:31-41. [DOI] [PubMed] [Google Scholar]
- 57.Westaway, E. G., J. M. Mackenzie, M. T. Kenney, M. K. Jones, and A. A. Khromykh. 1997. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 71:6650-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wicker, J. A., M. C. Whiteman, D. W. Beasley, C. T. Davis, S. Zhang, B. S. Schneider, S. Higgs, R. M. Kinney, and A. D. Barrett. 2006. A single amino acid substitution in the central portion of the West Nile virus NS4B protein confers a highly attenuated phenotype in mice. Virology 349:245-253. [DOI] [PubMed] [Google Scholar]
- 59.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 60.Yang, J. S., M. P. Ramanathan, K. Muthumani, A. Y. Choo, S. H. Jin, Q. C. Yu, D. S. Hwang, D. K. Choo, M. D. Lee, K. Dang, W. Tang, J. J. Kim, and D. B. Weiner. 2002. Induction of inflammation by West Nile virus capsid through the caspase-9 apoptotic pathway. Emerg. Infect. Dis. 8:1379-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, C. Y., Y. W. Hsu, C. L. Liao, and Y. L. Lin. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. J. Virol. 80:11868-11880. [DOI] [PMC free article] [PubMed] [Google Scholar]