Abstract
The broad spectrum of antiviral activity of ribavirin (RBV) lies in its ability to inhibit IMP dehydrogenase, which lowers cellular GTP. However, RBV can act as a potent mutagen for some RNA viruses. Previously we have shown a lack of correlation between antiviral activity and GTP repression for Hantaan virus (HTNV) and evidence for RBV's ability to promote error-prone replication. To further explore the mechanism of RBV, GTP levels, specific infectivity, and/or mutation frequency was measured in the presence of RBV, mycophenolic acid (MPA), selenazofurin, or tiazofurin. While all four drugs resulted in a decrease in the GTP levels and infectious virus, only RBV increased the mutation frequency of viral RNA (vRNA). MPA, however, could enhance RBV's mutagenic effect, which suggests distinct mechanisms of action for each. Therefore, a simple drop in GTP levels does not drive the observed error-prone replication. To further explore RBV's mechanism of action, we made a comprehensive analysis of the mutation frequency over several RBV concentrations. Of importance, we observed that the viral population reached a threshold after which mutation frequency did not correlate with a dose-dependent decrease in the level of vRNA, PFU, or [RTP]/[GTP] (where RTP is ribavirin-5′-triphosphate) over these same concentrations of RBV. Modeling of the relationship of mutation frequency and drug concentration showed an asymptotic relationship at this point. After this threshold, approximately 57% of the viral cDNA population was identical to the wild type. These studies revealed a lethal threshold, after which we did not observe a complete loss of the quasispecies structure of the wild-type genome, although we observed extinction of HTNV.
Hantaviruses represent an important and growing source of disease emergence in both established and developing countries (27). Hantaviruses are enveloped, negative-stranded RNA viruses with a tripartite, segmented genome (28). The genome consists of S (small), M (medium), and L (large) segments of RNA, which encode the nucleocapsid protein (N), glycoproteins (Gn and Gc), and L protein (RNA-dependent RNA polymerase [RdRp]), respectively. When virus is transmitted to humans through inhalation of aerosols of virus shed in rodent excreta, hantaviruses may cause either of two serious illnesses: hemorrhagic fever renal syndrome (HFRS) (21) or hantavirus pulmonary syndrome (HPS) (18, 26). Hantaan virus (HTNV), an Old World hantavirus, is the primary agent of HFRS, while Sin Nombre and Andes viruses are the primary agents in New World cases of HPS in North and South America, respectively.
At present, there are no vaccines or antivirals approved by the U.S. Food and Drug Administration for treatment of any of the hemorrhagic fever viruses or HPS. A limited number of antivirals have been tested, and few have been effective against viruses within the family Bunyaviridae. Ribavirin (1-β-d-ribofuranosyl-1,2,4-triazole-3-carboamide) (RBV), a broad-spectrum antiviral agent, and related C-nucleoside analogues ribamidine, selenazofurin (SEL), and tiazofurin (TIA) show potent antiviral activity in vitro against HTNV (17, 19). RBV was proven effective in the treatment of lethal encephalitic suckling mice infected with HTNV (16). Moreover, studies performed in China with HFRS patients suggest that RBV provides an improved prognosis when given early in the course of disease (15). As no other antiviral drugs with this level of activity for the hantaviruses have been reported, RBV remains an important lead for the development of new and better drugs for HTNV and perhaps other members of the Bunyaviridae. However, RBV's mechanism of action has remained elusive; therefore, efforts toward understanding its mechanism should bring new insights into the design of new drugs. For example, one could use the mechanistic information to explore chemical modifications that would increase its selectivity toward RdRps and decrease selectivity for IMP dehydrogenase (IMPDH).
A cursory review of the literature may suggest that RBV's mechanism of action is understood, but an intensive review of the literature reveals a complex profile of activity and perhaps a pleiotropic mechanism for this compound. These seemingly diverse mechanisms of action may derive from RBV's unique interaction with the polymerase machinery for each virus family and the dependence or requirements of that virus family on the nucleotide pool levels, specifically those of GTP. RBV decreases cellular GTP pools by competitive inhibition with IMP for the active site of IMPDH (2, 35). This has been reported to be the mechanism of action for RBV for several viruses (22, 24). However, viral enzymatic activities are also targeted, as suggested by several reports that show RBV's effect on capping (11), translation efficiency of viral mRNA (41), and viral polymerase activity (9). These activities may be due to direct incorporation of RBV 5′-monophosphate into mRNA or viral genomic RNAs since it resembles GMP, although it can pseudo-base pair with C and U. Recently, error-prone replication mediated by RBV has been reported for several RNA viruses (reviewed in reference 42), which would also require direct incorporation of RBV. We have also reported that RBV induced error-prone replication in HTNV (31). RBV can act as a potent RNA virus mutagen in poliovirus-infected cells, and it has been proposed that its incorporation into viral RNAs (vRNAs) causes the virus to enter error catastrophe (5). It is apparent that RBV acts as a mutagen for several viruses, including hantaviruses, but from the studies reported to date, it is not clear for most viruses how or if GTP repression via IMPDH contributes to the observed antiviral effect. Recently we have reported that, at least for HTNV, RBV's activity did not correlate with inhibition of IMPDH but rather with production of RBV triphosphate (RTP) (38). This suggests the interaction of RTP with the hantaviral RdRp, and while consistent with the observed increase in mutation frequency reported earlier for HTNV (31), it does not provide additional insight into the operational mechanism of RBV.
Recently, alternative theoretical models to error catastrophe have been developed for drug-induced mutation frequency in viruses (3, 37). One of the most recent theoretical models, lethal mutagenesis, differs from error catastrophe conceptually. In essence, the theory of lethal mutagenesis is based on a demographic process reflected by an apparent decline in the absolute abundance of the quasispecies population rather than an evolutionary shift in genotype space (3). Furthermore, lethal mutagenesis mandates the extinction of a population, while error catastrophe theory can shift the population toward higher fitness as well as driving what is often referred to as the mutational meltdown of the genome. Clearly, the design of effective antivirals and treatment strategies should be guided by these theoretical considerations. With that in mind, we have designed experiments to reveal which model, if any, was operational for RBV's inhibitory activity against HTNV. We undertook a comprehensive analysis of the mutation frequency over a wide range of RBV concentrations. We also examined the antiviral effects of several additional IMPDH inhibitors, including mycophenolic acid (MPA), selenazofurin (SEL), and tiazofurin (TIA), of which SEL and TIA are C-nucleoside analogues of RBV. This enhanced mutation frequency was observed only with RBV and not with other IMPDH inhibitors. Importantly, we observed that a relatively narrow lethal threshold was sufficient for extinction of HTNV. In essence, this threshold reflects a point in the drug concentration at which additional mutations are not tolerated and hence the virus is unable to replicate. However, this point did not lead to a complete loss of the genomic information, as predicted by the alternative hypothesis of error catastrophe.
MATERIALS AND METHODS
Cells, virus, and compound treatments.
Vero E6 cells (CRL-1586; ATCC) were propagated and used as described previously (29). HTNV infections were performed by the adsorption of diluted stock virus (76-118 strain) at a multiplicity of infection of 0.1 for 1 h on 3-day-old, confluent Vero E6 monolayers. Treatment of Vero E6 cells with guanosine (Guo), RBV, or MPA was carried out by the replacement of medium with the appropriate concentration of the compound or compound mixtures in Dulbecco's modified Eagle's medium with 10% fetal bovine serum after infection. Virus and RNA were harvested at 72 h after infection. PFU were measured by agarose overlay as described previously (39). Total RNA was isolated with Trizol (Invitrogen) as suggested by the manufacturer.
Quantitative real-time RT PCR.
Quantification of HTNV S-segment vRNA within virus-infected cells was carried out with a real-time reverse transcription (RT)-PCR assay using the comparative cycle threshold method 2−ΔΔCT as described elsewhere (38). Real-time RT-PCRs were performed in triplicate for each sample and prepared with TaqMan universal PCR master mix (Applied Biosystems).
Nucleotide sequencing.
cDNA prepared as described above was amplified by PCR with Phusion high-fidelity DNA polymerase (Finnzyme Oy, Finland) according to the manufacturer's protocols. HS24 (forward, 5′-TACTAGAACAACGATGGCAACTATG-3′) and HS1336 (reverse, 5′-GTGCAAATATGATTGATAATGATTCAGTAG-3′) primers were used to amplify the open reading frame of N within the S-segment gene. The amplified product was cloned into plasmid pCR-4 (TOPO cloning kits for sequencing; Invitrogen) after A tailing by using Taq polymerase (Promega). Ninety-six colonies per group were picked, and each colony was subjected to colony PCR by using M13 forward and M13 reverse primers. PCR products were used as the template for BigDye 3.1 automated cycle sequencing (Applied Biosystems) with either M13 forward or M13 reverse primers. The background mutation frequency was measured using the same enzymes with plasmid DNA which encodes the HTNV S-segment cDNA. Sequence analysis included positions 91 to 1329 of the HTNV S-segment cDNA and was done with the Seqscape program (Applied Biosystems) by comparison with the published sequence (GenBank accession number M14626) (30).
vRNA size profiling.
HTNV S-segment cDNAs were synthesized with one of the four different primers which were complementary to the regions 441 to 461, 963 to 985, 1441 to 1465, and 1673 to 1696 in viral sense RNA. The copy number of the transcribed cDNA was measured by use of a SYBR green real-time PCR method (DyNAmo HS SYBR green quantitative PCR kit; NEB) employing a 160-bp amplicon covering a 302- to 461-bp region in the viral sense RNA. The copy number was calculated from a regression of standard DNA samples.
Extraction and analysis of drug metabolism.
Vero cells were incubated at 37°C with RBV or MPA for the indicated times. Cells were washed twice with sterile phosphate-buffered saline and extracted with perchloric acid as described previously (23). The samples were centrifuged at 12,000 × g for 10 min, and the supernatant was neutralized with 4 M of KOH buffered with 1 M of potassium phosphate (pH 7.4). KClO4 was removed by centrifugation. Separation and detection of nucleotides were performed using a high-pressure liquid chromatograph equipped with a Partisil-10 strong anion-exchange column (10 μM, 250 by 4.6 mm; Keystone Scientific, Inc., Bellefonte, PA). Elution of the nucleotides was accomplished with a 50-min linear gradient from 5 mM of NH4H2PO4 (pH 2.8) to 750 mM of H4H2PO4 (pH 3.7) buffer, with a flow rate of 2 ml/min. Purine standards were detected by their absorbance at 260 nm as they eluted from the column.
RESULTS
RBV, but not SEL, TIA, or MPA, showed a dose-dependent response with viral PFU that did not correlate with GTP levels.
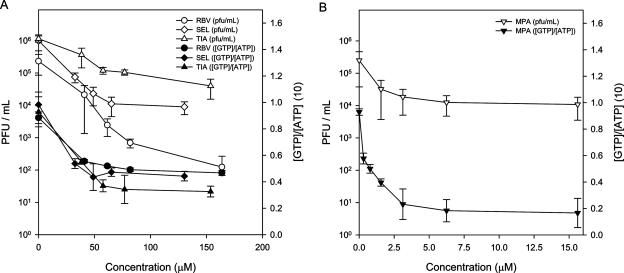
RBV, SEL, TIA, and MPA (Fig. 1) are known inhibitors of IMPDH. To define how the observed antiviral activity of RBV correlates with the inhibition of IMPDH, we measured the intracellular levels of GTP and the relative levels of HTNV PFU from infected Vero E6 cells for several concentrations of RBV, SEL, TIA, and MPA (Fig. 2A and B). GTP levels were reduced by 30% to 60% in Vero E6 cells over the concentrations examined (Fig. 2A). The levels of GTP were reduced by 80% in the presence of 3.1 μM MPA (Fig. 2B). In the SEL, TIA, and MPA drug treatments, the levels of HTNV PFU correlated closely with the levels of GTP (Fig. 2A and B). In contrast, increasing RBV reduced HTNV PFU by approximately 100-fold after the GTP levels reached their lowest; hence, RBV did not show any clear correlation with GTP levels. Drug cytotoxicity was not observed over the range of drug concentrations employed (data not shown). This indicates that RBV's inhibitory effect on HTNV differed from that of the other IMPDH inhibitors and further corroborates our earlier study (38).
FIG. 1.
Structures of RBV, SEL, TIA, and MPA.
FIG. 2.
Dose-response curves showing PFU and relative [GTP]/[ATP] following treatment of HTNV-infected cells. Following infection with HTNV, Vero E6 cells were treated with RBV, SEL, or TIA (A) and MPA (B) at several concentrations over a period of 3 days. Vero cells and supernatants were measured for PFU and GTP/ATP content, respectively.
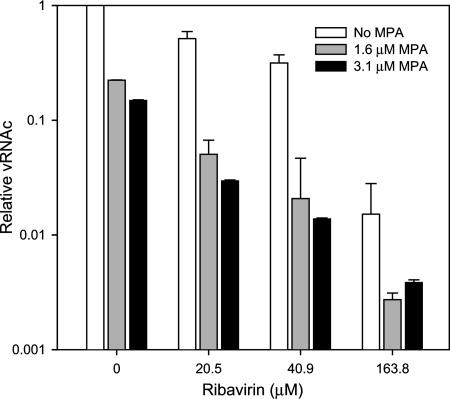
MPA's antiviral activity was additive when used in combination with RBV, decreasing HTNV vRNA and increasing mutation frequency.
In the study described above, MPA's antiviral activity, but not RBV's, correlated with GTP levels. MPA's, but not RBV's, antiviral activity can be completely reversed with exogenous guanosine (38). These results indicated that the antiviral activity of RBV was not due to the inhibition of IMPDH but was likely due to its conversion to RTP and its subsequent effect on vRNA polymerase. Since the antiviral activity of these two agents was a result of interaction with two different targets, we hypothesized that we would observe an increased antiviral effect upon the addition of both drugs to HTNV-infected cells. As shown in Fig. 3, the combination of RBV with MPA showed an increased antiviral effect. For example, 20 μM (5 μg/ml) of RBV with 1.6 μM (0.5 μg/ml) of MPA showed better antiviral activity than 3.1 μM (1 μg/ml) of MPA alone. We have shown previously that GTP levels in Vero E6 cells treated with RBV and MPA were similar to GTP levels in cells treated with MPA alone (38).
FIG. 3.
Antiviral effects of RBV and MPA on cellular concentrations of HTNV vRNA (vRNAc). HTNV vRNA levels were measured at various combinations of RBV in the absence or presence of MPA.
To explore whether the mutation frequency would stay the same for RBV/MPA mixtures versus RBV, we measured the mutation frequency by cloning and sequencing the S segment from HTNV-infected Vero E6 cells treated with RBV at various concentrations or treated with a combination of 41 μM (10 μg/ml) RBV and 3.1 μM (1 μg/ml) MPA (Table 1). We developed a high throughput sequencing system that allowed us to obtain much greater accuracy in measurements of the mutation frequency (see Materials and Methods). This system allowed rapid sequencing of over 100,000 nucleotides (nt) per experimental group to obtain the average mutation frequency. The wild-type virus showed an average mutation frequency of 1 per 10,000 nt (Table 1). We noted no increase in background mutation frequency from PCR with Phusion high-fidelity DNA polymerase (data not shown). The mutation frequency of MPA was similar to that of the untreated or wild-type HTNV. However, we observed an increase in the mutation frequency with the combination treatment of 41 μM of RBV with 3.1 μM MPA. In light of the previous result which showed that these two drugs act through different mechanisms (Fig. 2), this result suggests that MPA potentiated the activity of RBV.
TABLE 1.
Summary of mutation frequencies of HTNV vRNA in the absence or presence of various chemical treatments
| Treatment of HTNV-infected Vero E6 cells | No. of cDNAs examined | No. of nt analyzed | % of cDNAs with mutations | % of C→U or G→A mutations of total | Mutation frequencyb
|
|||
|---|---|---|---|---|---|---|---|---|
| C→U, G→A | U→C, A→G | Othersc | Total | |||||
| No treatment | 85 | 105,315 | 11.8 | 36.4 | 0.4 | 0.2 | 0.5 | 1.0 |
| RBV | ||||||||
| 41 μM | 87 | 107,793 | 23.0 | 77.3 | 1.6¶ | 0.2 | 0.3 | 2.0 |
| 82 μM | 88 | 109,032 | 36.4 | 53.8 | 1.9§ | 0.8 | 0.8 | 3.6§ |
| 123 μM | 170 | 210,630 | 34.7 | 73.4 | 2.8§ | 0.4 | 0.6 | 3.8§ |
| 164 μM | 91 | 112,749 | 41.8 | 75.0 | 3.5§ | 0.6 | 0.5 | 4.6§ |
| 246 μM | 182 | 225,498 | 44.0 | 66.0 | 3.0§ | 0.7 | 0.9 | 4.6§ |
| MPA, 15.6 μM | 95 | 117,705 | 15.8 | 26.3 | 0.4 | 0.7 | 0.5 | 1.6 |
| RBV + MPAa | 92 | 113,988 | 42.4 | 77.0 | 4.1§ | 0.7 | 0.5 | 5.4§ |
| TIA, 154 μM | 90 | 111,510 | 18.9 | 50.0 | 0.8 | 0.4 | 0.8 | 1.6 |
| SEL, 130 μM | 93 | 115,227 | 18.3 | 35.0 | 0.6 | 0.3 | 1.1 | 1.7 |
Cotreatment of 41 μM RBV with 3.1 μM MPA.
Mutations per 10,000 nt sequenced. §, P < 0.001; ¶, P < 0.01 (Student's t test).
Any mutations other than transition mutations, including frameshift.
Mutation frequency of RBV reaches a threshold, while PFU and vRNA levels show a dose-dependent response.
Previously, we have shown that RBV increased the mutation frequency of HTNV RNAs (31), and this suggested that error catastrophe may be involved. However, since our previous report, other models for these observations have been reported (3, 37). To further probe our earlier observations and those described above, we set out to determine the mutation frequency by cloning and sequencing the S segment from HTNV-infected Vero E6 cells treated with RBV at various concentrations.
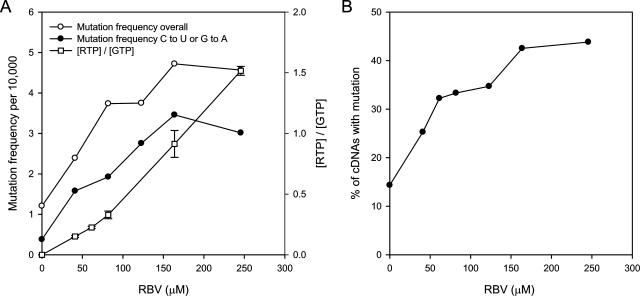
In up to five separate experiments, we cloned and analyzed cDNA sequences derived from approximately 90 independent clones per group depending on the treatment (Table 1 and data not shown). For each treatment group, we made a detailed mutational analysis, and we show these data for one representative experiment (Table 1 and data not shown). The overall mutation frequency and the C→U and G→A mutation frequency showed a linear increase to 82 μM (20 μg/ml), after which neither the overall relative mutation frequency nor the C→U and G→A mutation frequency showed a linear dose response (Fig. 4A). Rather, in this representative experiment and others, we observed three distinct slopes, which suggest a zone corresponding to a threshold in treatment groups from 82 to 246 μM (20 to 60 μg/ml). This is further demonstrated when one plots the percentages of cDNAs with mutation increases (with respect to the mutation frequency of the wild type) over the concentrations of RBV (Fig. 4B).
FIG. 4.
Analyses of mutation spectrum as a function of [RBV]. (A) Mutation frequency was plotted as a function of [RBV] for one representative experiment. [RTP]/[GTP] was assessed and is also presented. Standard deviations are shown for [RTP]/[GTP]. (B) The percentage of cDNAs as a function of the number of mutations relative to that for the wild-type virus is plotted against [RBV].
An additional breakdown of the cDNA population spectrum is presented in Table 2. These data show the cumulative population structures from four independent experiments. We examined the data for the ability to fit a Poisson distribution. Hence, this analysis allowed us to address the question as to whether sequencing of this region of the genome would reflect the population overall. Briefly, we used the average mutation frequency (Table 1) to calculate the theoretical Poisson distribution for each concentration of drug we examined. For example, for 0 and 41 μM RBV, the average values were 0.15 and 0.296 mutations for this length of cDNA, respectively. The values given in Table 1 reflect the average numbers of mutations per 10,000 nt sequenced. The Poisson analysis then provides the theoretical proportion of mutations predicted within the population. This output was then compared to the actual data (Table 2), which were derived from two to four separate experiments. These two outputs were compared by criteria for assessing the goodness of fit using SAS. This value/degrees of freedom was 0.6241, which supports the fit of the model to the data. We also examined the data with SAS GENMOD to do a generalized estimating equations analysis of the repeated experiments. The output Pr> Z was 0.2831, which supports the overall fit of the model for each experiment. These results support a random distribution with respect to the incorporation of RBV throughout the genome.
TABLE 2.
Average distribution of mutations in cDNAs as a function of drug treatment
| Treatment | No. of analyzed cDNAs | % of cDNAsa with:
|
||||
|---|---|---|---|---|---|---|
| No mutations | One mutation | Two mutations | Three mutations | Four mutations | ||
| No treatment | 460 | 85.7 | 12.4 | 1.5 | 0.4 | 0.0 |
| RBV | ||||||
| 41 μM | 360 | 74.7 | 20.8 | 4.4 | 0.0 | 0.0 |
| 61 μM | 180 | 67.8 | 27.2 | 3.9 | 1.1 | 0.0 |
| 82 μM | 276 | 66.7 | 27.5 | 4.3 | 1.1 | 0.4 |
| 123 μM | 170 | 65.3 | 24.1 | 9.4 | 1.2 | 0.0 |
| 164 μM | 348 | 57.5 | 29.3 | 10.6 | 1.7 | 0.9 |
| 246 μM | 162 | 56.2 | 34.6 | 4.9 | 4.3 | 0.0 |
| MPA, 15.6 μM | 176 | 77.8 | 18.8 | 3.4 | 0.0 | 0.0 |
| RBV + MPAb | 185 | 62.7 | 26.5 | 8.6 | 2.2 | 0.0 |
| TIA, 154 μM | 90 | 81.1 | 17.8 | 1.1 | 0.0 | 0.0 |
| SEL, 130 μM | 93 | 81.7 | 16.1 | 1.1 | 1.2 | 0.0 |
Values represent averages from up to four independent experiments; each experiment contained a minimum of 90 independent cDNAs.
Cotreatment of 41 μM RBV with 3.1 μM MPA.
To further explore possible reasons for the observed threshold, we examined the kinetics of [RTP]/[GTP] at different RBV concentrations and the spectrum of vRNA products being synthesized (i.e., full-length versus truncations due to incomplete synthesis). As shown in Fig. 4A, the ratio of [RTP] and [GTP] showed a dose response and hence ruled out the possibility of [RTP]/[GTP] affecting mutation frequency.
We made a statistical evaluation of total mutation (TM) frequency as a function of RBV concentration presented in Fig. 4. The mutation frequency per 10,000 nt was regressed for all RBV concentrations via simple linear, bivariate, and quadratic regressions. The R2 value for the simple regression relationship of TM versus concentration was 0.82. For the bivariate analyses, concentrations were analyzed by the two apparent groups, 0 to 41 μM and 82 to 246 μM. The R2 value for the simple regression relationship of TM versus concentration was 0.98 for 0 to 41 μM or 0.76 for 82 to 246 μM. The R2 value for the quadratic regression relationship of TM versus concentration was 0.98. These statistical analyses support a significant change in mutation response as the RBV concentration increased past a lethal threshold. Finally, we subjected the data to a second-degree polynomial, which confirms that a linear fit of the data is not appropriate (data not shown). The polynomial fit confirmed the suggestion that the relationship of mutation frequency and drug concentration was asymptotic at this point.
Second, our studies shown in Table 1 relied on the cloning of the entire S-segment open reading frame. Hence, it is possible that our data reflect our sampling strategy in that we were not accounting for truncated vRNA products. To address this, we designed primer sets that measured the numbers of copies of four vRNA lengths (Fig. 5). We found that the copy number ratios of cDNAs primed from the position of nt 446 and from the very end position of nt 1,670 remained constant regardless of RBV treatments (13-fold difference at 0 μM and 16-fold difference at 82 μM) (Fig. 5). This suggests that our sampling method accurately reflected the population spectrum within the cell. This strongly supports that the vRNA populations within these thresholds were undergoing extinction; however, these experiments do not address how.
FIG. 5.
HTNV S vRNA length profile in the absence or presence of various concentrations of RBV. The copy numbers of cDNA primed with the primers described in the text were measured by using a SYBR green quantitative real-time PCR method.
DISCUSSION
Since RBV's synthesis in 1972 by Sidwell et al. (32), numerous studies have shown RBV to have a remarkably broad spectrum of antiviral activity. It has become increasingly clear that its mechanisms of action may differ, however, for different virus families. Considerable evidence has shown that cellular GTP levels are reduced by treatment with RBV for several viruses (22, 36). However, there has been no obvious correlation between GTP levels and RBV's antiviral effect across RNA viruses. For example, in the case of Flaviviridae and Paramyxoviridae, GTP reduction is suggested as the primary mechanism since exogenous guanosine reverses the antiviral effect proportionally (22). Similarly, GB virus shows reduced specific infectivity when grown in the presence of RBV; however, activity is completely reversed by the addition of guanosine (20). In these examples, the antiviral activity parallels MPA, which inhibits IMPDH (22, 24, 33). In striking contrast to results with these viruses, Wray et al. found no relationship among GTP reduction, RTP, and influenza virus, and their work suggested a direct effect of RTP on viral polymerase (43). Recently, we have reported a similar finding for HTNV (38). The complexity of the mechanism of RBV is further demonstrated in the report that RBV is not effective against members of the Filoviridae (14). Finally, RBV has been suggested to cause error catastrophe for several viruses (for examples, see recent reviews in references 1 and 42). Our studies sought to clarify its mechanism for HTNV, with the recognition that this mechanism may be specific for this family of viruses.
Two independent mechanisms of action for RBV's antiviral activity are suggested for HTNV.
Three lines of evidence support two independent targets of RBV's anti-HTNV activity: IMPDH and the hantaviral polymerase. First, MPA, SEL, and TIA levels correlated with GTP repression and antiviral activity, measured as infectious HTNV particles released by plaque assay. In contrast, RBV did not. Importantly, RBV treatments of HTNV-infected cells caused an additional 100-fold decrease in vRNA levels at a time when levels of GTP had already reached a plateau. Second, Guo rescue experiments are consistent with two targets for RBV (38). Unlike MPA, the antiviral effect of RBV was not fully recovered by the addition of exogenous Guo. Finally, we observed an increase in antiviral activity and mutation frequency when both RBV and MPA were added to HTNV-infected Vero E6 cells. It is plausible to suggest that RTP is incorporated more efficiently when GTP levels are further reduced by MPA. This suggests that while RBV's overall mechanism is targeted at RdRp, an additional drop in GTP levels could enhance its mutagenic potential. Future exploration of drug combinations in HTNV animal models would be very insightful and perhaps suggest alternative approaches to treatment of HFRS.
RBV increased the mutation frequency of HTNV and showed a threshold response.
For several viruses, including HTNV, RBV can act as an RNA mutagen, which would be expected to reduce the specific infectivity of HTNV. We have explored the kinetics of the loss of specific infectivity of HTNV with an increase in RBV concentration. Our results showed that RBV reduced specific infectivity in a dose-response manner, which did not correlate with the decrease in GTP levels promoted by RBV. To further explore the mechanism for the loss of specific infectivity for HTNV, we exhaustively analyzed the mutation frequency and type as a function of RBV concentration. The mutation frequency of the wild-type population of HTNV was 1.0 mutation per 10,000, which is comparable to that of poliovirus or lymphocytic choriomeningitis virus (4, 13). RBV at 41 to 61 μM increased the overall mutation frequency twofold (1.0 versus 2.0 mutations per 10,000) (Table 1). A detailed sequence analysis showed that frequencies of G→A and C→U mutations, which are predicted to be mediated by RBV incorporation (5), increased similarly. Further analysis revealed that the G→A mutation frequency was much higher than the C→U mutation frequency, as has been reported for West Nile virus (7). The previous study and ours suggest that RBV incorporates more readily into antigenomic than into genomic RNA. Remarkably, the percentage of cDNAs with mutations did not show a dose response after treatment with 82 μM RBV in four separate experiments. The kinetics of the response suggested a threshold had been reached. Our analysis fit the data to a second-degree polynomial, which confirmed that a linear fit of the data is not appropriate. The polynomial fit confirmed the suggestion that the relationship of mutation frequency and drug concentration was asymptotic at this point and confirmed the presence of a threshold.
An obvious question that arises from these studies is why the overall mutation frequency reached a threshold. This value is derived from extraction of total cellular RNA from which a large portion of the S segment is subsequently amplified and cloned. One of the simpler explanations could be that the vRNAs can tolerate only a certain level of mutations before they become unstable, which would result in a conformation that is not recognized by the N protein or the polymerase. Alternatively, the drug could affect the processivity of the L protein, which would result in truncated vRNA products. We observed a dose response of infectious virus produced by the cell and the level of vRNA in the cell. We also showed the absence of truncated vRNA products. A model in which RBV causes extinction of the vRNA is suggested by these data. We can simplify the data as two populations, A1 (wild-type population) and A2 (mutant population). In the absence of RBV, we obtained a ratio of 83:17 for A1:A2 for the wild-type population, which basically means that we observed an increase in mutation frequency in only 20% of the population (5:1) over the baseline mutation frequency established at 80%. Upon the addition of drug, a threshold was reached, in which we observed a ratio of ∼66:∼34 (or 3:1) for A1:A2. This ratio was maintained with increasing concentrations of RBV while viral numbers (PFU and vRNA levels) showed a dose-dependent decrease. Hence, the data argue for the presence of a third population, A3, an inferior genotype, which is unable to replicate. The drug is then promoting the extinction of the virus.
Lethal mutagenesis induced by nucleoside analogs has been suggested as a novel strategy for antiviral drug development for RNA viruses which may exist as a quasispecies (23; see reviews in references 1, 6, 10, and 34). Our data support the hypothesis that lethal mutagenesis of HTNV infection by RBV occurs by an extinction catastrophe rather than an error catastrophe mechanism (3). Challenges facing this approach include the need for intracellular transport of derivatives by active or passive processes, efficient phosphorylation by host or viral enzymes, and high levels of incorporation into the viral genome, thereby increasing error frequency during viral replication and transcription. The combination of antiviral compounds and mutagenic agents to increase the error rate while reducing viral replication provides an additional strategy to promote viral extinction (8). Treatment with 5-fluorouracil and guanidine hydrochloride produces high-frequency extinctions of foot-and-mouth disease virus, but treatment with either drug alone does not (12, 25). Systematic extinction of human immunodeficiency virus type 1 was achieved by combining the reverse transcriptase inhibitor zidovudine with 5-hydroxydeoxycytidine (40). Herein, we show an additive antiviral and mutagenic effect on HTNV with combinations of RBV and MPA. Future efforts in this area call for new molecular probes that will illuminate these mechanisms and provide new insights in the discovery of effective antiviral drugs that can promote lethal mutagenesis.
Acknowledgments
This work was supported by Department of Defense USAMRC grant number W81XWH-04-C-0055 (C. B. Jonsson).
We thank Jim J. Bull, Lauren A. Meyers, Jesse Summers, and Aaron Shatkin for discussions regarding the content of the manuscript and/or comments on the manuscript.
Footnotes
Published ahead of print on 15 August 2007.
REFERENCES
- 1.Anderson, J. P., R. Daifuku, and L. A. Loeb. 2004. Viral error catastrophe by mutagenic nucleosides. Annu. Rev. Microbiol. 58:183-205. [DOI] [PubMed] [Google Scholar]
- 2.Andrei, G., and E. De Clercq. 1993. Molecular approaches for the treatment of hemorrhagic fever virus infections. Antivir. Res. 22:45-75. [DOI] [PubMed] [Google Scholar]
- 3.Bull, J. J., L. A. Meyers, and M. Lachmann. 2005. Quasispecies made simple. PLoS Comput. Biol. 1:e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 6.Daifuku, R. 2003. Stealth nucleosides: mode of action and potential use in the treatment of viral diseases. BioDrugs 17:169-177. [DOI] [PubMed] [Google Scholar]
- 7.Day, C. W., D. F. Smee, J. G. Julander, V. F. Yamshchikov, R. W. Sidwell, and J. D. Morrey. 2005. Error-prone replication of West Nile virus caused by ribavirin. Antivir. Res. 67:38-45. [DOI] [PubMed] [Google Scholar]
- 8.Domingo, E., C. Escarmis, E. Lazaro, and S. C. Manrubia. 2005. Quasispecies dynamics and RNA virus extinction. Virus Res. 107:129-139. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson, B., E. Helgstrand, N. G. Johansson, A. Larsson, A. Misiorny, J. O. Noren, L. Philipson, K. Stenberg, G. Stening, S. Stridh, and B. Oberg. 1977. Inhibition of influenza virus ribonucleic acid polymerase by ribavirin triphosphate. Antimicrob. Agents Chemother. 11:946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freistadt, M. S., G. D. Meades, and C. E. Cameron. 2004. Lethal mutagens: broad-spectrum antivirals with limited potential for development of resistance? Drug Resist. Update 7:19-24. [DOI] [PubMed] [Google Scholar]
- 11.Goswami, B. B., R. Crea, J. H. Van Boom, and O. K. Sharma. 1982. 2′-5′-Linked oligo(adenylic acid) and its analogs. A new class of inhibitors of mRNA methylation. J. Biol. Chem. 257:6867-6870. [PubMed] [Google Scholar]
- 12.Grande-Perez, A., E. Lazaro, P. Lowenstein, E. Domingo, and S. C. Manrubia. 2005. Suppression of viral infectivity through lethal defection. Proc. Natl. Acad. Sci. USA 102:4448-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grande-Perez, A., S. Sierra, M. G. Castro, E. Domingo, and P. R. Lowenstein. 2002. Molecular indetermination in the transition to error catastrophe: systematic elimination of lymphocytic choriomeningitis virus through mutagenesis does not correlate linearly with large increases in mutant spectrum complexity. Proc. Natl. Acad. Sci. USA 99:12938-12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huggins, J. W. 1989. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Rev. Infect. Dis. 11(Suppl. 4):S750-S761. [DOI] [PubMed] [Google Scholar]
- 15.Huggins, J. W., C. M. Hsiang, T. M. Cosgriff, M. Y. Guang, J. I. Smith, Z. O. Wu, J. W. LeDuc, Z. M. Zheng, J. M. Meegan, Q. N. Wang, et al. 1991. Prospective, double-blind, concurrent, placebo-controlled clinical trial of intravenous ribavirin therapy of hemorrhagic fever with renal syndrome. J. Infect. Dis. 164:1119-1127. [DOI] [PubMed] [Google Scholar]
- 16.Huggins, J. W., G. R. Kim, O. M. Brand, and K. T. McKee, Jr. 1986. Ribavirin therapy for Hantaan virus infection in suckling mice. J. Infect. Dis. 153:489-497. [DOI] [PubMed] [Google Scholar]
- 17.Huggins, J. W., R. K. Robins, and P. G. Canonico. 1984. Synergistic antiviral effects of ribavirin and the C-nucleoside analogs tiazofurin and selenazofurin against togaviruses, bunyaviruses, and arenaviruses. Antimicrob. Agents Chemother. 26:476-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan, A., and A. S. Khan. 2003. Hantaviruses: a tale of two hemispheres. Panminerva Med. 45:43-51. [PubMed] [Google Scholar]
- 19.Kirsi, J. J., J. A. North, P. A. McKernan, B. K. Murray, P. G. Canonico, J. W. Huggins, P. C. Srivastava, and R. K. Robins. 1983. Broad-spectrum antiviral activity of 2-beta-d-ribofuranosylselenazole-4-carboxamide, a new antiviral agent. Antimicrob. Agents Chemother. 24:353-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford, R. E., D. Chavez, B. Guerra, J. Y. Lau, Z. Hong, K. M. Brasky, and B. Beames. 2001. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 75:8074-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, H. W., and G. van der Groen. 1989. Hemorrhagic fever with renal syndrome. Prog. Med. Virol. 36:62-102. [PubMed] [Google Scholar]
- 22.Leyssen, P., J. Balzarini, E. De Clercq, and J. Neyts. 2005. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 79:1943-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loeb, L. A., J. M. Essigmann, F. Kazazi, J. Zhang, K. D. Rose, and J. I. Mullins. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. USA 96:1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinoski, F., and V. Stollar. 1981. Inhibitors of IMP dehydrogenase prevent Sindbis virus replication and reduce GTP levels in Aedes albopictus cells. Virology 110:281-289. [DOI] [PubMed] [Google Scholar]
- 25.Pariente, N., S. Sierra, P. R. Lowenstein, and E. Domingo. 2001. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 75:9723-9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters, C. J., and A. S. Khan. 2002. Hantavirus pulmonary syndrome: the new American hemorrhagic fever. Clin. Infect. Dis. 34:1224-1231. [DOI] [PubMed] [Google Scholar]
- 27.Schmaljohn, C., and B. Hjelle. 1997. Hantaviruses: a global disease problem. Emerg. Infect. Dis. 3:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1633. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 29.Schmaljohn, C. S., S. E. Hasty, S. A. Harrison, and J. M. Dalrymple. 1983. Characterization of Hantaan virions, the prototype virus of hemorrhagic fever with renal syndrome. J. Infect. Dis. 148:1005-1012. [DOI] [PubMed] [Google Scholar]
- 30.Schmaljohn, C. S., G. B. Jennings, J. Hay, and J. M. Dalrymple. 1986. Coding strategy of the S genome segment of Hantaan virus. Virology 155:633-643. [DOI] [PubMed] [Google Scholar]
- 31.Severson, W. E., C. S. Schmaljohn, A. Javadian, and C. B. Jonsson. 2003. Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 77:481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sidwell, R. W., J. H. Huffman, G. P. Khare, L. B. Allen, J. T. Witkowski, and R. K. Robins. 1972. Broad-spectrum antiviral activity of virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177:705-706. [DOI] [PubMed] [Google Scholar]
- 33.Smee, D. F., M. Bray, and J. W. Huggins. 2001. Antiviral activity and mode of action studies of ribavirin and mycophenolic acid against orthopoxviruses in vitro. Antivir. Chem. Chemother. 12:327-335. [DOI] [PubMed] [Google Scholar]
- 34.Smith, R. A., L. A. Loeb, and B. D. Preston. 2005. Lethal mutagenesis of HIV. Virus Res. 107:215-228. [DOI] [PubMed] [Google Scholar]
- 35.Streeter, D. G., J. T. Witkowski, G. P. Khare, R. W. Sidwell, R. J. Bauer, R. K. Robins, and L. N. Simon. 1973. Mechanism of action of 1-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (virazole), a new broad-spectrum antiviral agent. Proc. Natl. Acad. Sci. USA 70:1174-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stridh, S. 1983. Determination of ribonucleoside triphosphate pools in influenza A virus-infected MDCK cells. Arch. Virol. 77:223-229. [DOI] [PubMed] [Google Scholar]
- 37.Summers, J., and S. Litwin. 2006. Examining the theory of error catastrophe. J. Virol. 80:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, Y., D. H. Chung, Y. K. Chu, C. B. Jonsson, and W. B. Parker. 2007. Activity of ribavirin against Hantaan virus correlates with production of ribavirin-5′-triphosphate, not with inhibition of IMP dehydrogenase. Antimicrob. Agents Chemother. 51:84-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takenaka, A., C. J. Gibbs, Jr., and D. C. Gajdusek. 1985. Antiviral neutralizing antibody to Hantaan virus as determined by plaque reduction technique. Arch. Virol. 84:197-206. [DOI] [PubMed] [Google Scholar]
- 40.Tapia, N., G. Fernandez, M. Parera, G. Gomez-Mariano, B. Clotet, M. Quinones-Mateu, E. Domingo, and M. A. Martinez. 2005. Combination of a mutagenic agent with a reverse transcriptase inhibitor results in systematic inhibition of HIV-1 infection. Virology 338:1-8. [DOI] [PubMed] [Google Scholar]
- 41.Toltzis, P., and A. S. Huang. 1986. Effect of ribavirin on macromolecular synthesis in vesicular stomatitis virus-infected cells. Antimicrob. Agents Chemother. 29:1010-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vignuzzi, M., J. K. Stone, and R. Andino. 2005. Ribavirin and lethal mutagenesis of poliovirus: molecular mechanisms, resistance and biological implications. Virus Res. 107:173-181. [DOI] [PubMed] [Google Scholar]
- 43.Wray, S. K., B. E. Gilbert, M. W. Noall, and V. Knight. 1985. Mode of action of ribavirin: effect of nucleotide pool alterations on influenza virus ribonucleoprotein synthesis. Antivir. Res. 5:29-37. [DOI] [PubMed] [Google Scholar]