Abstract
The bioterror threat of a smallpox outbreak in an unvaccinated population has mobilized efforts to develop new antipoxviral agents. By screening a library of known drugs, we identified 13 compounds that inhibited vaccinia virus replication at noncytotoxic doses. The anticancer drug mitoxantrone is unique among the inhibitors identified in that it has no apparent impact on viral gene expression. Rather, it blocks processing of viral structural proteins and assembly of mature progeny virions. The isolation of mitoxantrone-resistant vaccinia strains underscores that a viral protein is the likely target of the drug. Whole-genome sequencing of mitoxantrone-resistant viruses pinpointed missense mutations in the N-terminal domain of vaccinia DNA ligase. Despite its favorable activity in cell culture, mitoxantrone administered intraperitoneally at the maximum tolerated dose failed to protect mice against a lethal intranasal infection with vaccinia virus.
Poxviruses have been at the forefront of advances in medicine for 200 years, from Jenner's paper on the efficacy of vaccination against smallpox in 1798 to the successful use of vaccinia virus to eradicate human smallpox disease, as proclaimed by the World Health Organization in 1977. Throughout the 1990s, the scientific community debated the proposed destruction of the last known stocks of smallpox virus (24, 31). Hanging over this discussion was the fear that undeclared stocks of smallpox could be used as a bioterror weapon against an unvaccinated population. The urgency of this threat was amplified by the terrorist attacks of 2001, followed by the dissemination of anthrax via the postal service. The outbreak of human monkeypox infections in the United States in 2003 further highlighted the risks of reemergence of human poxvirus disease.
Public health and research efforts have been mobilized accordingly to (i) exploit a modified live smallpox vaccine that maintains efficacy while minimizing complications, (ii) pursue alternatives to live vaccination for smallpox prophylaxis, and (iii) discover and bring forward for FDA approval new antipoxviral drugs. Two very different clinical scenarios present different challenges for drug therapy. Prophylaxis of a low-risk population in the event of a threat or actual outbreak mandates an orally available drug with minimal side effects. In contrast, the treatment of confirmed cases of smallpox (which can have a ≥30% fatality rate) need not be hindered by concerns about route of administration and non-life-threatening side effects. The goal is to have at least two approved antipoxviral drugs that act on different molecular targets.
Although many inhibitors of poxvirus replication in culture or animal models have been described previously (50), the initial efforts post-2001 focused on the nucleoside analog cidofovir, an inhibitor of the viral DNA polymerase (28), which was already FDA approved for treatment of cytomegalovirus retinitis. Cidofovir was found to be effective in animal models of orthopoxvirus infection (37, 50, 53). However, because cidofovir is administered intravenously and has significant renal toxicity in humans, emphasis has now shifted to the development of less toxic and orally available derivatives of cidofovir (5). De novo efforts to discover new antipoxviral agents by screening for inhibition of vaccinia replication in culture have yielded an orally available antipoxviral compound, ST-246, that blocks formation of extracellular virus by targeting a protein component of the poxvirus envelope (63). ST-246 performs well in animal models of orthopoxvirus infection (41, 63), is safe in humans, and was recently granted orphan drug designation by the FDA for the prevention and treatment of smallpox.
The complexity of the poxvirus replication cycle and the large number of essential viral proteins present a rich array of additional untapped targets for the discovery of new antipoxviral agents. Novel inhibitors provide leads that could eventuate in a drug, but they are equally valuable as tools for the study of viral replication and host-virus interactions. Accordingly, we have conducted a high-throughput screen of natural-product and synthetic-chemical libraries for antagonists of vaccinia replication. As the first step in this process, we surveyed a collection of 2,880 compounds consisting of known drugs and drug-like molecules, including off-patent substances approved for human, veterinary, cosmetic, or industrial use. This effort yielded 13 confirmed bioactive compounds, including several not known previously to have antipoxviral properties.
We find that the anticancer drug mitoxantrone is a potent inhibitor of vaccinia replication. It acts via a novel mechanism entailing a late-stage block to virus assembly. Isolation of mitoxantrone-resistant vaccinia viruses underscores that a viral protein is the likely target of the drug. We apply whole-genome sequencing (32) to identify the mitoxantrone resistance-conferring mutations.
MATERIALS AND METHODS
Cells and viruses.
BSC40 cells (African green monkey kidney cells) were maintained in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (MEM). Cells were grown at 37°C in a 5% CO2 incubator. The WR strain of vaccinia virus was propagated and titers were determined in BSC40 monolayers at 37°C. A recombinant vaccinia virus, NP-S-EGFP, expressing a nucleus-localized enhanced green fluorescent protein (EGFP) reporter under the control of a vaccinia p7.5 promoter (38), was a gift of Jonathan Yewdell (NIH). The Δtopo virus (10) was a gift of Bernard Moss (NIH).
High-throughput screen.
BSC40 cells (5,000 cells in 30 μl of MEM) were seeded into each well of a 384-well Corning tissue culture microplate. Confluent monolayers were formed after 24 h of incubation at 37°C. Aliquots (5 μl) of 100 μM solutions of test compounds in 100% dimethyl sulfoxide (DMSO) in 384-well plates were mixed with 10 μl of MEM. The test set comprised 2,880 compounds purchased from Microsource and Prestwick. The 15-μl mixtures were transferred to the 384-well plates containing BSC40 monolayers. Aliquots of the vaccinia-EGFP virus (∼32 PFU in 5 μl MEM) were dispensed to each well. The cells were thereby overlaid with a total volume of 50 μl of MEM containing 10% DMSO and 10 μM of the test compound. Each 384-well plate included 16 control wells with no test compound (DMSO only) and 16 wells in which the medium contained 10 μg/ml adenosine N1-oxide (ANO) in 1% DMSO. ANO blocks viral replication and viral protein synthesis without affecting host protein synthesis (26). At 48 h postinfection, the medium was aspirated and the cells washed with phosphate-buffered saline (PBS). The cells were overlaid with 30 μl of 4% paraformaldehyde in PBS for 20 min at room temperature, washed with PBS, and then overlaid with 30 μl of 0.1% Triton X-100 in PBS for 15 min. The solution was aspirated, and the cells were washed again with PBS and then overlaid with 30 μl of 1 μM solution of Hoechst dye in PBS for 30 min in the dark. The dye solution was aspirated, and the cells were washed twice with PBS and overlaid with 50 μl PBS. The plates were sealed and stored at 4°C. The cell layers were scanned for both green fluorescence (488 nM excitation/535 nM emission) and Hoechst fluorescence (364 nM excitation/450 nM emission) with an InCell Analyzer 3000 confocal microscope. Data were captured, images stored, and green fluorescence quantified with a Raven software object intensity module. The DMSO-only controls were set as 100% EGFP expression; the ANO controls were set as 0% EGFP expression.
Secondary screens.
Plaque assays were performed on confluent BSC40 monolayers in six-well plates. The cells were overlaid with 1 ml of medium containing ∼200 PFU of vaccinia WR. The inoculum was removed after 1 h and the cells overlaid with 3 ml of MEM containing serial twofold dilutions of the test compound. Plaque formation was scored at 48 h postinfection by staining with monolayers with crystal violet. The lowest concentrations of the compounds that sufficed to abolish macroscopic plaque formation are reported in Fig. 1.
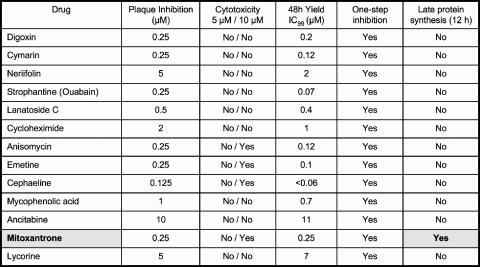
FIG. 1.
Antipoxviral agents identified by high-throughput screening. Thirteen compounds emerging from the screen that passed secondary screens for potency and cytotoxicity are listed in the far-left column. The secondary- and tertiary-assay methods are discussed in the text. Exemplary assays and results are shown for mitoxantrone in Fig. 2 and 4.
Drug-resistant viruses.
BSC40 monolayers (∼2 × 108 cells in 10 150-cm2 culture flasks) were infected with 1 × 108 PFU of vaccinia WR and then overlaid with MEM containing 0.5 μM mitoxantrone. Virus was harvested at 48 h, and 1/20 of the preparation was used to reinfect BSC40 monolayers in the presence of 1 μM mitoxantrone. After a total of six passages in the presence of 1 μM mitoxantrone, the virus stock titers were determined in the absence or presence of 0.5 μM mitoxantrone, which showed that resistant strains had been selected. Clonal isolates were obtained after two rounds of plaque purification from infected cells grown in agarose-containing medium with 0.5 μM mitoxantrone.
Sequencing of viral genomes.
Fifteen 150-cm2 culture flasks of BSC40 cells were infected at a multiplicity of infection (MOI) of 0.1 with either wild-type vaccinia WR or plaque-purified mitoxantrone-resistant strains MX-R1, MX-R2, and MX-R3. Cells were harvested at 48 h postinfection by scraping and then recovered by centrifugation. The cell pellets were resuspended in 10 mM Tris-HCl (pH 8.0) and then lysed mechanically using a Dounce homogenizer. Cell debris and nuclei were removed by low-speed centrifugation. Vaccinia virions were recovered by centrifugation of the postnuclear supernatant through a 36% sucrose cushion. DNA was extracted from the pelleted virions by digesting the material with proteinase K in sodium dodecyl sulfate (SDS), followed by extraction with phenol-chloroform and precipitation of the deproteinized DNA with ethanol. The DNAs were sheared, tagged, amplified, and sequenced with a GS Flex sequencer according to protocols specified by the manufacturer (454 Life Sciences, Branford, CT). The sequences were aligned to the reference genome for vaccinia WR (NCBI accession no. NC_006998) by using software from 454 Life Sciences. Between 49,000 and 95,000 valid viral sequence reads were obtained for each vaccinia genome. The average sequence read was 247 nucleotides. The average depth of coverage per genome varied from a low of 67-fold to a high of 127-fold.
Intranasal infection of mice with vaccinia virus.
Female pathogen-free BALB/c mice (13 to 15 g) were obtained from Charles River Labs, Wilmington, MA. They were quarantined 48 h prior to use and maintained on standard rodent chow and tap water in the AAALAC-accredited laboratory animal research center of Utah State University. Mice were anesthetized with ketamine (100 mg/kg of body weight) by intraperitoneal injection and then infected intranasally with 1 × 105 PFU of vaccinia virus IHD in a 50-μl volume. Animals were treated by intraperitoneal administration of saline solutions of mitoxantrone or cidofovir once a day starting 24 h after virus exposure. Mice were given a total dose of 5 mg/kg mitoxantrone via three schedules: 1.25 mg/kg/day × 4, 2.5 mg/kg/day × 2, and 5 mg/kg/day × 1. Cidofovir was dosed at 100 mg/kg/day × 2. The placebo group received intraperitoneal saline × 4. Ten drug-treated mice for each treatment regimen and 20 placebo control animals were held for death. Five infected mice per treatment group were sacrificed on day 5 to determine lung virus titers (51). Five uninfected mice in each treatment group served as toxicity controls.
RESULTS
High-throughput screen for antipoxviral agents.
Two commercial compound collections (2,000 from Microsource and 880 from Prestwick) were tested for inhibition of replication of vaccinia virus WR in cultured BSC40 cells. Cell monolayers in 384-well plates were infected at an MOI of ∼0.006 with a recombinant virus that expresses karyophilic GFP under the control of a viral early/late promoter (38), and the plates were scanned for green fluorescence at 3 days postinfection. The test compounds were included in the medium at the time of infection at a concentration of 10 μM. ANO (10 μg/ml) was used as a positive control for complete inhibition of viral gene expression (26). The screen was performed twice. A “hit” was defined as any compound that elicited a ≥95% reduction in GFP signal in both iterations of the screen. The initial hit compounds were repurchased individually, retested at 10 μM, and then stratified for potency in the HTS format by testing serial dilutions. Twenty-seven compounds that displayed a dose-dependent inhibition of GFP expression with 90% inhibitory concentration (IC90) values of ≤10 μM were advanced for secondary testing, entailing manual assays of plaque formation by wild-type vaccinia WR on BSC40 monolayers in six-well plates as a function of compound concentration. Cytotoxicity was gauged by morphological changes in the uninfected cells of the monolayer (e.g., rounding and detachment from the plate); any molecules that were cytotoxic at a ≤5 μM concentration were dropped from consideration. Thirteen compounds emerged from the secondary screen, nine of which abolished plaque formation at a concentration of ≤1 μM (Fig. 1).
The 13 verified hits were then subjected to a series of tertiary tests. First, we gauged their effects on the yield of progeny virus at a low MOI (0.01). This assay format will detect either inhibition of virus replication or virus spread. All of the compounds displayed a dose-dependent reduction in virus yield at 48 h postinfection. Potency was quantified as IC99, the concentration at which virus yield was reduced by a factor of 100. Ten of the compounds had IC99 values of ≤1 μM (Fig. 1). The compounds were then tested for effects on virus yield as a function of time during a synchronous infection (MOI of 3). This assay scores specifically for inhibition of virus replication and is insensitive to inhibitors of virus spread. We found that all 13 compounds suppressed formation of infectious progeny by at least 100-fold during one-step growth when tested at or slightly above the IC99 dose (Fig. 1). Finally, the compounds were tested for effects on viral protein synthesis by pulse-labeling the synchronously infected cells with [32S]methionine at 2, 4, 6, 8, and 12 h postinfection and then subjecting whole-cell lysates to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) to gauge the onset of the synthesis of the major late viral structural proteins, which occurs at 6 h in a normal infection. All of the inhibitors except one (mitoxantrone) blocked the progression to late protein synthesis at 12 h postinfection.
Distinct classes of antipoxviral agents.
The 13 poxvirus inhibitors identified in the screen fall into several distinct families. Four of the active compounds are known inhibitors of eukaryal protein synthesis: cycloheximide, anisomycin, emetine, and cephaeline. The fact that these molecules scored in a screen based on suppressing the synthesis of a virus-encoded reporter protein is no surprise and serves as an internal control for the sensitivity of the screen. Nonetheless, it is remarkable that their antipoxviral activities are evident at concentrations well below the cytotoxic dose. Cephaeline (which along with emetine is the active ingredient in ipecac syrup) is extremely potent, with an IC99 of <60 nM. Emetine and anisomycin are also quite potent (IC99s of 100 and 120 nM, respectively). Initial pulse-labeling experiments suggest that viral protein synthesis is more sensitive than cellular protein synthesis to inhibition by the compounds (not shown). These findings raise the prospect that viral mRNAs might be treated differently from bulk cellular mRNAs by the host translation machinery. One striking difference between poxvirus and host mRNAs is that poxvirus late and intermediate transcripts have a poly(A) “head” at their capped 5′ termini (3, 45). Cycloheximide, anisomycin, and emetine were noted previously to elicit selective inhibition of encephalomyocarditis virus protein synthesis versus host translation, while exerting a potent suppression of virus yield (42). Anisomycin was also reported to inhibit the persistence of the double-stranded RNA virus L-A in budding yeast (Saccharomyces cerevisiae) (17). Whereas we do not regard these protein synthesis inhibitors as useful drug leads, given the narrow toxicity window in vivo, they do promise to reveal some interesting aspects of viral translational control upon deeper analysis of their antiviral action.
Five of the inhibitors were cardiac glycosides: digoxin, cymarin, neriifolin, strophantine (ouabain), and lanatoside C. These are steroid glycoside natural products from Digitalis or Strophanthus plant species. Digoxin is used clinically to control ventricular rate and treat congestive heart failure in patients with atrial fibrillation. The mechanism of action of the cardiac glycosides entails inhibition of the Na+-K+ ATPase, and these drugs have been used widely for that purpose in basic cell biology research. Strophantine, cymarin, and digoxin are the most potent of the antipoxviral glycosides identified in the screen (IC99s of 70, 120, and 200 nM, respectively), whereas neriifolin is the least effective (IC99 of 2 μM). Strophantine has been reported to inhibit the growth of Sendai virus (36), and digoxin was recently found to inhibit replication of herpes simplex virus, varicella-zoster virus, cytomegalovirus, and adenovirus (21). Synthetic cardiac glycosides with antiherpes activity have also been reported previously (57, 58). To our knowledge, the antipoxviral activity of cardiac glycosides is a new finding. Pulse-labeling of virus-infected cells with [35S]methionine revealed that several viral early polypeptides were synthesized at 4 to 12 h in the presence of strophantine, digoxin, or cymarin and that little or no synthesis of the major late virion structural proteins had occurred by 12 h (data not shown). Further studies of the role of the Na+-K+ ATPase in poxvirus biology by use of cardiac glycosides should provide new insights into virus-host dynamics, notwithstanding that the narrow therapeutic window of digoxin in humans (limited by life-threatening arrhythmias) precludes their systemic use as antiviral drugs.
Lycorine is a natural product from Amaryllis plant species. It has been reported to have numerous bioactivites, including anticancer, antifungal, antimalarial, and anticholinesterase properties. Several reports highlight an antiviral activity of lycorine against polivirus, SARS coronavirus, and human immunodeficiency virus (30, 54, 62), but we are not aware of any prior studies of antipoxviral activity. Lycorine has reasonable potency (IC99 of 5 μM), and it completely blocked virus replication in a one-step experiment. Metabolic labeling experiments revealed evidence of early protein synthesis at 2 to 8 h postinfection in the presence of lycorine but not progression to late protein synthesis by 12 h (not shown). Lycorine is worthy of further evaluation of its antipoxviral mechanism in cell culture and tests of its efficacy in first-line animal models.
Ancitabine (cyclocytidine) emerged from the screen as a nontoxic antipoxviral agent (IC99 of 11 μM) that completely blocks one-step growth in BSC40 cells. Ancitabine is a derivative of cytosine arabinoside (araC), a widely used anticancer drug. araC inhibits vaccinia virus replication by targeting the viral DNA polymerase (55). Our initial characterization of ancitabine suggests that it acts similarly to araC. The onset of viral early protein synthesis occurs normally (evident at 2 h) in the presence of ancitabine but persists up to 12 h, with no progression to late protein synthesis (data not shown). Dot blot hybridization of DNA from infected cells with virus-specific probes showed that ancitabine blocks viral DNA replication (not shown). Clinical trials of intravenous araC for treatment of smallpox failed to show evidence of efficacy (14, 34). Our finding that vaccinia infection of a murine epidermal dendritic cell line is completely resistant to araC at concentrations that abolish infection of BSC40 cells (13) underscores the caveat that antipoxvirus agents might not be equally effective in all target cells. The existence of an araC-resistant cell population could thereby account for the failure of araC as a single agent for treatment of smallpox.
Mycophenolic acid is an inhibitor of IMP dehydrogenase, a key enzyme in purine biosynthesis. Mycophenolic acid is an orally available immunosuppressant used for prevention of posttransplant organ rejection. Mycophenolic acid has activity against many RNA and DNA viruses (4, 9, 16, 22, 23, 29, 35, 43), including poxviruses (9, 19, 49). Its antiviral activity is linked to inhibition of purine metabolism, insofar as virus replication is rescued by exogenous guanosine. Our identification of mycophenolic acid here attests to the sensitivity of the antipoxviral screen.
Antipoxviral activity of mitoxantrone.
Mitoxantrone is a synthetic anthraquinone derivative used widely, in combination with araC, for treatment of acute myeloid leukemia (27). Mitoxantrone is also FDA approved for treatment of prostate cancer and multiple sclerosis (20). Mitoxantrone is a classical topoisomerase II poison that traps the covalent topoisomerase II-DNA intermediate and thereby triggers lethal double-strand DNA breaks (11). Topoisomerase II is generally accepted to be the target of mitoxantrone's effects on eukaryotic cells and in the clinic. Mitoxantrone has antiprotozoal activity in vitro against Trypanosoma brucei (15), but it has not received much attention as an antiviral. To our knowledge, there are no prior reports of its activity against poxviruses.
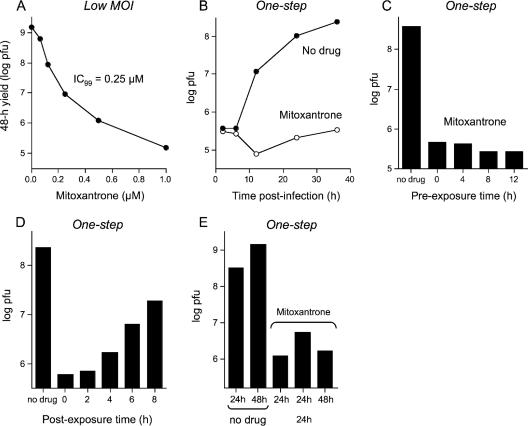
Mitoxantrone blocked plaque formation by vaccinia virus WR at a concentration of ≥0.25 μM (Fig. 1). Virus yield from cell infected at a low MOI was suppressed in a concentration-dependent fashion in the range of 62 nM to 1 μM drug. The yields were reduced by factors of 100, 1,000, and 10,000 at 0.25 μM, 0.5 μM, and 1.0 μM mitoxantrone, respectively (Fig. 2A). The effects of 1 μM mitoxantrone on a synchronous infection of BSC40 cells are shown in Fig. 2B. In this experiment, the cells were preincubated with drug prior to inoculation and maintained in drug thereafter. Mitoxantrone completely blocked the production of infectious progeny virus. The mitoxantrone block to vaccinia replication did not depend on treatment of the cells with mitoxantrone prior to infection (Fig. 2C). Moreover, mitoxantrone prevented virus replication even when added after the infection was initiated (Fig. 2D). Maximal reduction of virus yield was maintained when cells were exposed to the drug 2 h postinfection. The yield increased incrementally as the drug addition time was extended to 4, 6, and 8 h after removal of the inoculum (Fig. 2D). These results suggest that mitoxantrone blocks vaccinia replication at a step subsequent to viral early gene expression (which peaks at 2 to 4 h postinfection).
FIG. 2.
Mitoxantrone inhibits vaccinia virus replication. (A) Virus yield from a low MOI. BSC40 cells were infected with vaccinia WR at an MOI of 0.01 and then overlaid with medium containing mitoxantrone. Virus yield at 48 h (log PFU) is plotted as a function of mitoxantrone concentration. (B) Synchronous infection in the presence of drug. Cells were pretreated for 12 h with medium containing 1 μM mitoxantrone or no drug and then infected at an MOI of 3 in the presence or absence of drug. Virus yield (log PFU) is plotted as a function of time postinfection. (C) Antiviral effect does not require preincubation with mitoxantrone. Cells were pretreated with medium containing 1 μM mitoxantrone for 12, 8, 4, or 0 h prior to infection at an MOI of 3. The infected cells were then overlaid with medium containing 1 μM mitoxantrone. Virus yield at 30 h postinfection (log PFU) is plotted as a function of the time of preincubation and compared to the yield from a control infection in the absence of drug. (D) Cells infected at an MOI of 3 were overlaid with medium containing 1 μM mitoxantrone at 0, 2, 4, 6, or 8 h after removal of the inoculum. Virus yield at 30 h postinfection (log PFU) is plotted as a function of the time of drug addition and compared to the yield from a control infection in the absence of drug. (E) Cells infected at an MOI of 3 were overlaid with drug-free medium or medium containing 1 μM mitoxantrone immediately after removal of the inoculum. Cells were harvested at 24 h or 48 h postinfection as specified. One of the infected monolayers was incubated with mitoxantrone-containing medium for 24 h and then washed three times with drug-free medium and incubated for another 24 h in the absence of drug prior to harvest.
To test whether the mitoxantrone block was reversible, we exposed synchronously infected BSC40 cells to 1 μM mitoxantrone for 24 h (at which point the replication arrest was clearly evident) (Fig. 2E) and then washed the monolayer with drug-free medium and let the infection proceed in the absence of drug for an additional 24 h. The drug-free recovery period resulted in only a modest (threefold) increase in virus yield compared to levels for controls that had been maintained in mitoxantrone continuously for 48 h (Fig. 2E) and controls that were maintained in mitoxantrone for 24 h, washed, and then exposed again to mitoxantrone for an additional 24 h (not shown). Two explanations for the apparent irreversibility of the mitoxantrone block are conceivable: (i) the drug is retained within the cell even after it is withdrawn from the medium or (ii) mitoxantrone elicits a replication catastrophe that cannot be overcome even after the drug is gone.
Mitoxantrone's antipoxviral activity in cell culture is not restricted to the primate-derived BSC40 cell line. The yield of vaccinia progeny 30 h after a synchronous infection of RK13 rabbit kidney cells was reduced 80-fold by 0.063 μM mitoxantrone and 1,000-fold by 0.25 μM mitoxantrone when the drug was applied immediately postinoculation (data not shown). Antipoxviral activity was also not restricted to vaccinia as the infecting agent. Mitoxantrone inhibited the replication of myxoma virus (a member of the Leporipoxvirus genus); 1 μM mitoxantrone reduced the myxoma virus yield from synchronously infected RK13 cells by a factor of 275 (data not shown).
Mitoxantrone does not prevent vaccinia virus protein synthesis.
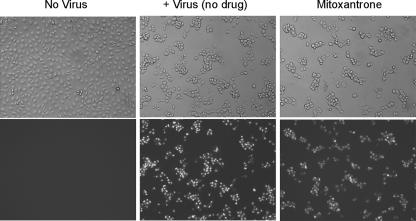
To gauge whether mitoxantrone blocked vaccinia virus gene expression, we performed one-step infection of BSC40 cells with the vaccinia-EGFP virus and examined the cells by light and fluorescent microscopy at 12 h postinfection in the absence or presence of 1 μM mitoxantrone, in parallel with uninfected control cells. Infection of virtually all cells was evident by light microscopy, whereby the cytopathic effect on virus-infected cells (rounding and clumping) contrasted with the confluent flat monolayer in the control sample (Fig. 3, left and middle). All cells displayed bright-green nuclear fluorescence in the infected sample, while none of the uninfected cells were green (Fig. 3). The finding that mitoxantrone-treated virus-infected cells displayed typical cytopathic morphology and were universally green, with intensity similar to that of the untreated control, attested that viral gene expression had occurred.
FIG. 3.
Mitoxantrone does not affect viral gene expression. BSC40 cells were infected with the vaccinia-EGFP virus at an MOI of 10. The cells were pretreated for 12 h with 1 μM mitoxantrone and maintained in mitoxantrone thereafter (mitoxantrone). Control cells were infected in the absence of drug. The cells were examined by bright-field microscopy (top) and fluorescence microscopy (bottom) of the same fields at 12 h postinfection. Mock-infected cells were analyzed in parallel.
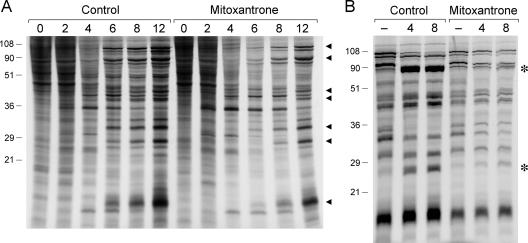
Pulse-labeling synchronously infected BSC40 cells with [35S]methionine at 2, 4, 6, 8, and 12 h postinoculation revealed a typical temporal pattern of vaccinia gene expression in the control and mitoxantrone-treated cells (Fig. 4A), including the appearance of several viral early polypeptides at 2 to 6 h postinfection, a transition to the synthesis of distinctive late proteins by 6 to 12 h, and a decline in the synthesis of host proteins also by 6 to 12 h postinfection. These results show that mitoxantrone does not exert its antiviral activity at the level of viral gene expression. Moreover, because late viral protein synthesis is strictly dependent on viral DNA replication, we surmise that mitoxantrone is not a DNA replication inhibitor. This inference was sustained by dot blot hybridization experiments showing that viral DNA accumulated during synchronous infection in the presence of mitoxantrone (not shown).
FIG. 4.
Mitoxantrone inhibits proteolytic processing of virion structural proteins. (A) BSC40 cells were infected with vaccinia WR at an MOI of 10. The cells were pretreated for 12 h with 1 μM mitoxantrone prior to infection and maintained in mitoxantrone thereafter (mitoxantrone). Control cells were infected in the absence of drug. At the specified times postinfection, medium was removed, and cells were washed with methionine-free MEM and then overlaid with methionine-free MEM containing 30 μCi/ml of [35S]methionine (1,135 Ci/mmol) with or without mitoxantrone. The medium was removed after 30 min and the cells lysed in situ in SDS sample buffer. Aliquots of the lysates were analyzed by SDS-PAGE. An autoradiograph of the gel is shown. The positions and sizes (in kilodaltons) of prestained marker polypeptides are indicated on the left. Synthesis of late viral proteins (indicated by ◂) is evident at 6 to 12 h postinfection in the presence or absence of mitoxantrone. (B) Cells infected in the presence of 1 μM mitoxantrone and control infected cells were pulse-labeled for 30 min with [35S]methionine at 12 h postinfection as described above. The cells were washed twice with MEM after removing the labeling medium and then either lysed immediately (lanes marked −) or overlaid with fresh medium containing unlabeled methionine with or without mitoxantrone. The monolayers were returned to incubate at 37°C and then lysed in situ after 4 or 8 h. Aliquots of the lysates were analyzed by SDS-PAGE. An autoradiograph of the gel is shown. Mitoxantrone blocked the conversion of major structural protein precursors into mature forms migrating at the positions denoted by asterisks.
Mitoxantrone blocks virus assembly.
The major structural proteins of the virion core are synthesized at late times as precursor polypeptides that are processed proteolytically to yield the mature species (56, 60, 61). The largest precursor polypeptides encoded by the vaccinia A10 and A3 genes were identified readily in [35S]methionine pulse-labeling reactions performed at 12 h postinfection (Fig. 4A). These and the other major structure protein precursor (encoded by the L4 gene) were converted to mature forms during an 8-h chase in the presence of unlabeled methionine (Fig. 4B, control). The structural protein precursors were synthesized during virus infection in the presence of mitoxantrone, but there was little or no processing to mature forms, even after 8 h of chase (Fig. 4B, mitoxantrone). Because almost any virus mutation or drug treatment that blocks virus assembly also affects processing of the virion structural precursors, the pulse-chase result provides biochemical evidence that mitoxantrone affects virus morphogenesis. This was evaluated by electron microscopy.
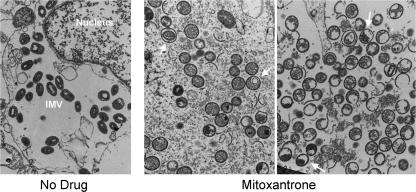
The mature vaccinia virion evolves in stages from microscopically well-characterized immature forms. The earliest of these are crescent-shaped spicule-coated viral membranes, which are observed either free in the cytoplasm or associated with centers of electron-dense viroplasm. Spherical immature viral particles are formed upon closure of the membrane around the granular material. These spherical immature particles develop into brick-shaped intracellular mature virions (IMV), clusters of which were evident in the cytoplasm of cells infected with virus in the absence of drug (Fig. 5, no drug). Whereas IMV were absent in vaccinia-infected cells treated with mitoxantrone, spherical immature virions and membrane crescents were abundant (Fig. 5, mitoxantrone). Thus, mitoxantrone blocked morphogenesis at a step subsequent to formation of spherical immature virions. The drug also promoted accumulation of aberrant forms, including sporadic immature virions containing a smaller enveloped membrane (Fig. 5, middle) and a significant number of “half-moon” particles, in which the diffuse granular material within the sphere had condensed asymmetrically such that one side of the particle appeared empty (Fig. 5, right). Such abnormal half-moon forms were described previously for cells infected at the restrictive temperature with a temperature-sensitive vaccinia virus bearing a mutation in the I7 protein (25), the cysteine protease responsible for processing the major virion structural protein precursors (1, 6, 7, 18, 33).
FIG. 5.
Mitoxantrone blocks formation of IMV. BSC40 cells were infected with the vaccinia WR at an MOI of 10. The cells were pretreated for 12 h with 1 μM mitoxantrone prior to infection and maintained in mitoxantrone thereafter (+ mitoxantrone). Control cells were infected in the absence of drug. At 24 h postinfection, cells were washed with PBS and then overlaid with fixative solution containing glutaraldehyde and paraformaldehyde. The fixed cells were dislodged by scraping and then collected by centrifugation. The specimens were dehydrated and then embedded in epoxy resin. Thin sections were stained with uranyl acetate and lead citrate for visualization by transmission electron microscopy. Middle, arrows indicate sporadic immature virions containing a smaller enveloped membrane; right, arrows indicate half-moon particles.
Selection of drug-resistant viruses implies that mitoxantrone targets a vaccinia protein.
Our findings that mitoxantrone blocks poxvirus assembly at a late stage without affecting viral protein synthesis (and thus without grossly inhibiting viral DNA replication) suggest that the antipoxviral activity of mitoxantrone is distinct from its actions as a topoisomerase II poison. Vaccinia does not encode a type II topoisomerase, and to our knowledge, there is no evidence that the host cell topoisomerase II plays a role in poxvirus infection. Vaccinia does have a type I topoisomerase (48) that is not essential for virus replication or assembly, though its ablation does affect replicative fitness at an early step unrelated to virion morphogenesis (10). Our observation that the vaccinia Δtopo deletion virus was just as sensitive as wild-type vaccinia to inhibition by mitoxantrone (not shown) provides clear evidence that the poxvirus topoisomerase is not the target of this drug.
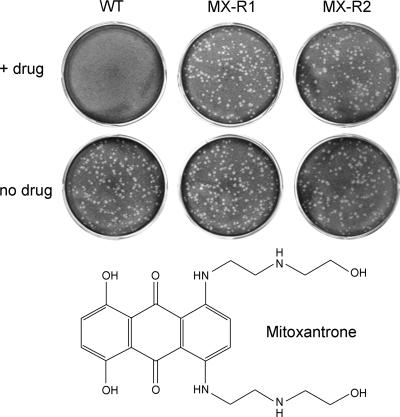
The most straightforward way to determine if a poxvirus protein is the target of mitoxantrone's antiviral activity is to isolate a drug-resistant virus. This was accomplished by six rounds of forced passage of a stock of wild-type vaccinia WR in the presence of 0.5 or 1 μM mitoxantrone. Titration of the virus stock at this stage showed that nearly equal numbers of plaques were formed on BSC40 cells in the presence of mitoxantrone as in the no-drug control, whereas the unselected stock of vaccinia WR did not form plaques in the presence of drug. Viruses isolated from the selected stock by two further rounds of plaque purification in the presence of mitoxantrone were clearly drug resistant, as gauged by plaque formation (Fig. 6) and by one-step growth (not shown). These viruses, designated MX-R strains (mitoxantrone resistant), were just as sensitive as the parental wild-type virus to inhibition by ANO, araC, and rifampin (data not shown).
FIG. 6.
Mitoxantrone-resistant vaccinia viruses. Wild-type vaccinia WR (WT) and two mitoxantrone-resistant variants (MX-R1 and MX-R2) obtained after forced passage in the presence of drug and two rounds of plaque purification were tested for plaque formation on BSC40 cell monolayers. The infected cells were overlaid with medium containing either 0.5 μM mitoxantrone (+ drug) or no additive (no drug). Photographs of the crystal violet-stained cell monolayers are shown. The chemical structure of mitoxantrone is depicted at the bottom.
Mitoxantrone resistance mutations map to a single viral gene.
The morphogenetic block elicited by mitoxantrone resembles that caused by mutations in the I7 cysteine protease. Accordingly, we queried whether I7 might be the immediate target of mitoxantrone by PCR amplifying and sequencing the I7 gene from six plaque-purified MTX-R viruses. No mutations in the I7 polypeptide were found.
We proceeded to identify the mitoxantrone resistance mutations by whole-genome sequencing (32) of our wild-type vaccinia WR strain and three different MX-R strains. Comparison of our wild-type sequence to the genome sequence of vaccinia WR deposited with NCBI (accession no. NC_006998) revealed a single difference at nucleotide 184876 in the VACWR208 gene (G in the reference strain versus T in our wild-type strain). This change in the proline-140 codon of the WR208 polypeptide (from CCC to CCA) is translationally silent. As expected, the same silent change was present in the genomes of the MX-R viruses that were derived from our wild-type strain.
The genome of strain MX-R3 differed from the parental wild-type genome at a single position, entailing a mutation of nucleotide 155989 from G to A. The mutation maps to the gene encoding vaccinia DNA ligase and results in a missense change of cysteine-11 to tyrosine (TGC to TAC). The genome of strain MX-R1 contained a single G-to-A mutation in the DNA ligase gene at nucleotide 156391 that resulted in a missense change of cysteine-145 to tyrosine (TGT to TAT). (MX-R1 also contained an incidental G-to-A mutation at nucleotide 15007 in the VACWR019 gene, causing a change from alanine-21 to valine in the WR019 polypeptide.) The MX-R2 genome contained a 3-nucleotide insertion in the DNA ligase gene that resulted in an extra isoleucine inserted between Ile14 and Tyr15 of an otherwise unperturbed DNA ligase polypeptide.
Structure-activity relationships.
Mitoxantrone consists of an anthracenedione scaffold with 1,4-dihydroxy and 5,8-bis[2-(2-hydroxyethylamino)ethylamino] substituents (Fig. 6). To gain an initial appreciation of the structural features underlying mitoxantrone's antiviral activity, we tested the effects of quinizarin (1,4-dihydroxyanthracenedione), a related compound that lacks the 5,8-bis(alkylamino) groups of mitoxantrone. We found that vaccinia virus plaque size and plaque number were unaffected by up to 20 μM quinizarin (not shown). We conclude that the planar anthraquinone scaffold does not suffice for antipoxviral activity. We also tested solvent Blue 59, an anthracenedione derivative that lacks the dihydroxy groups of mitoxantrone and contains shorter bis(ethylamino) groups in lieu of the bis[2-(2-hydroxyethylamino)ethylamino] moieties of mitoxantrone. Vaccinia plaque size and plaque number were unaffected by up to 20 μM solvent Blue 59 (not shown).
Effect of mitoxantrone in a mouse model of lethal vaccinia virus infection.
The specific and potent effects of mitoxantrone on vaccinia replication in cell culture prompted evaluation of its efficacy in protecting mice against intranasal infection with a lethal dose of vaccinia virus (105 PFU). Mice received daily intraperitoneal doses of mitoxantrone or saline placebo starting 1 day after virus infection. Three mitoxantrone regimens were tested: 1.25 mg/kg/day × 4, 2.5 mg/kg/day × 2, and 5 mg/kg/day × 1. As a positive treatment control, one group of infected animals received intraperitoneal cidofovir (100 mg/kg/day × 2) starting 1 day after infection. Whereas 17/20 mice in the placebo group died (mean time to death was 7.1 ± 2.0 days), 10/10 cidofovir-treated mice survived. By contrast, all 10 mice in each mitoxantrone-treated cohort died (mean times to death were 7.4 ± 0.7 days, 8.2 ± 0.4 days, and 7.0 ± 0.9 days for the four-dose, two-dose, and one-dose mitoxantrone regimens, respectively). Five additional mice in the treated and placebo groups were sacrificed at day 5, and virus titers in the lung were determined (51). Cidofovir-treated mice had 300-fold-lower viral titers per gram of lung tissue than the placebo controls, while titers for the three mitoxantrone-treated groups were nearly identical to that of the placebo group. The same mitoxantrone regimens were given to uninfected mice. Whereas no deaths occurred in the group of five mice receiving 1.25 mg/kg/day × 4, all five mice that received a single dose of 5 mg/kg died, with a mean time to death of 10 ± 0.4 days. We surmise that intraperitoneal treatment with mitoxantrone at and exceeding a maximal tolerated dose had no benefit in the intranasal mouse infection model when therapy was initiated 1 day after inoculation.
DISCUSSION
The threat that smallpox could be used against military and civilian populations has prompted national investment in the development of new antipoxviral therapies. The major pharmaceutical companies are not inclined to initiate searches for new drugs to combat a disease that exits only in theory and for which the best possible outcome would be that the drug is never used. However, the success of SIGA in discovering ST-246, a new antipoxviral agent suitable for smallpox prophylaxis (41, 63), signals that there is an opportunity for niche companies to do good while doing well. Academic and public-sector scientists can also play a role in antipoxviral discovery, especially in the identification and dissemination of information about new inhibitors that could be of use to the research community as probes of poxvirus biology. The route to discovery of novel agents by screening off-patent medicines approved for human use has been advocated as a means to find “new tricks for old drugs” (39) that can bypass the laborious and costly preclinical steps and go straight to clinical trials. This approach is of little interest to the private sector, which is focused on new entities that yield proprietary compositions of matter.
Our screen of 2,880 compounds was “validated” by the output of 13 compounds that inhibited poxvirus replication. The confirmed hits included an agent with known antipoxvirus activity (mycophenolic acid), one related to a known inhibitor (ancitabine), and inhibitors of eukaryal protein synthesis that have a favorable therapeutic index (cycloheximide, anisomycin, emetine, and cephaeline). The antipoxviral activities of cardiac glycosides, lycorine, and mitoxantrone are new findings.
Mitoxantrone is unique among the inhibitors identified here in that it has no major impact on viral gene expression. Viral late protein synthesis proceeds normally in the presence of mitoxantrone, but there is clear biochemical and microscopic evidence of a mitoxantrone-induced block to virion assembly. The antiviral target is likely to be a viral protein, because we were able to isolate mitoxantrone-resistant viruses by passage in the presence of drug. To our knowledge, this is the first instance in which a useful bioactivity of mitoxantrone in eukaryotic cells depends on a target protein other than topoisomerase II.
Our results highlight a “new trick” for mitoxantrone, but it is not the first time that a clinically useful drug with a known mechanism has exerted a highly specific effect on poxvirus replication by a different mechanism. Rifampin, an antibacterial drug used for the treatment of tuberculosis, is an inhibitor of bacterial DNA-dependent RNA polymerase, and yet rifampin's activity against poxviruses entails a block to virion morphogenesis with no effect on viral gene expression (52).
Mitoxantrone now joins several other selective antipoxvirus agents that affect virus assembly: novobiocin, rifampin, TTP-6171, ST-246, and IMCBH. Novobiocin inhibits a very early stage of morphogenesis, so that membrane crescents and spherical immature virions do not accumulate (46). Rifampin causes assembly to arrest after the formation of abnormal membrane-enclosed structures (rifampin bodies), which contain viroplasm but lack the rigid spherical shape of normal immature virus particles (52). Rifampin resistance and hypersensitivity are attributable to point mutations in the D13 gene (2, 59). TTP-6171 targets the I7 protease and causes a morphogenetic arrest similar to that seen in an I7 temperature-sensitive virus at a restrictive temperature (8). IMCBH and ST-246 inhibit the release of infectious progeny virus particles from the cell by preventing the wrapping of IMV by Golgi membranes (40, 63). IMCBH and ST-246 target the F13 gene product, a 37-kDa protein component of the Golgi membrane-derived viral envelope (44, 63).
The results of whole-genome sequencing pinpoint vaccinia DNA ligase as the target of mitoxantrone's antipoxviral activity. Three different MX-R strains have three distinct mutations that result in either amino acid substitutions (C11Y or C145Y) or a single extra amino acid insert within the N-terminal domain of the 552-amino-acid viral ligase polypeptide. This portion of vaccinia DNA ligase is located well upstream of the active site of nucleotidyl transfer and is dispensable for the ligase adenylylation step of the enzyme's nick-sealing reaction (47). Deletion of the N-terminal domain of vaccinia ligase (from amino acids 1 to 200) slows the rate of strand joining and lowers the enzyme's affinity for nicked duplex DNA (47). The Cys11 and Cys145 residues, which are determinants of sensitivity to mitoxantrone, are both strictly conserved in the DNA ligases encoded by nearly all other vertebrate poxviruses, including variola, monkeypox, ectromelia, myxoma, deerpox, and swinepox. The two notable exceptions are the fowlpox and canarypox DNA ligases, in which the equivalent of Cys11 is conserved but the amino acid corresponding to vaccinia Cys145 is replaced by histidine and tyrosine, respectively. Thus, it is possible that canarypox might be naturally impervious to mitoxantrone by virtue of its mimicry of the C145Y ligase mutation that renders vaccinia mitoxantrone resistant.
Although it is not obvious how these DNA ligase mutations could protect vaccinia against mitoxantrone, we are struck by the consilience of our findings with those of DeLange et al. (12), who reported that (i) vaccinia virus replication in BSC40 cells was inhibited by etoposide, another clinically effective anticancer drug that acts as a topoisomerase II poison, (ii) etoposide had no apparent effect on viral gene expression, and (iii) an etoposide-resistant vaccinia virus isolated from a mutagenized stock after six rounds of passage in the presence of etoposide had a resistance-conferring C11Y mutation in the N-terminal domain of vaccinia DNA ligase. We can extrapolate from the published data an IC99 value of ∼100 μg/ml etoposide (diluted from a solution of pure drug in DMSO) for suppression of progeny virus yield in a synchronous infection (12). This dose corresponds to a concentration of ∼130 μM etoposide (molecular weight of 589 g/mol), which explains why our high-throughput screening assays for >90% inhibition of vaccinia replication at 10 μM concentrations of test compounds did not identify etoposide as a hit, even though it was present in both the Prestwick and Microsource compound libraries. Mitoxantrone is apparently more potent than etoposide in blocking vaccinia virus replication.
The C11Y vaccinia ligase mutation that sufficed to confer resistance to etoposide and cross-resistance to amsacrine (another topoisomerase II poison) (12) is identical to the MX-R3 mutation that confers resistance to mitoxantrone. DeLange et al. speculated that the replication block imposed by etoposide might be caused by a defect in forming the hairpin telomeres at the ends of the viral genome (12). However, the observed decrement in the level of monomeric viral genomes in the presence of etoposide seemed insufficient to account for the reduction in the yield of infectious progeny. DeLange et al. did not assess the effects of etoposide on virus morphogenesis or processing of virion structural proteins. In the case of mitoxantrone, the discrete block to virus assembly readily accounts for the reduction in virus yield.
Although mitoxantrone, etoposide, and amsacrine are all topoisomerase II poisons, they have significantly different chemical structures. Neither etoposide nor amsacrine is an anthracenedione (like mitoxantrone). It is not clear whether the antipoxviral activity of these compounds stems from (i) their similar effects on cellular topoisomerase II, which could result in a pretoxic lesion that somehow depends on vaccinia DNA ligase for its antiviral manifestation, or (ii) a common property of these compounds not related directly to topoisomerase II, e.g., their capacity to intercalate into or otherwise bind DNA. At least in the case of etoposide and amsacrine, the antiviral effects cannot be attributed simply to inhibition of vaccinia DNA ligase activity, insofar as deletion of the nonessential vaccinia ligase gene confers resistance to these two drugs, albeit not to the same degree as the C11Y mutation (12). Pulse-field electrophoretic analysis of vaccinia DNA in etoposide-treated infected cells showed the accumulation of subgenomic DNA fragments that were not evident in the absence of the drug (12), consistent with cellular topoisomerase II having access to replicating viral DNA and breaking it in the presence of the drug poison. It is notable that these viral DNA fragments are still observed in etoposide-treated cells infected with the etoposide-resistant C11Y ligase mutant (12), which implies that the drug-induced breakage of viral DNA does not by itself block production of infectious progeny virus. Further studies of the replication of wild-type and mitoxantrone-resistant viruses in the presence and absence of drugs are needed to fully elucidate the basis for the mitoxantrone-induced morphogenetic block.
Finally, this study demonstrates the power of whole-genome sequencing to rapidly locate poxvirus mutations that cause phenotypes of interest. The classical approach to map drug resistance phenotypes entails the use of genomic fragments from the resistant poxvirus strain in a transfection-based assay that scores for marker uptake by the sensitive parental strain to yield drug-resistant progeny (2, 12, 44, 59). This method requires fragment library construction from each virus mutant and is considerably more laborious and time consuming than sequencing and analyzing the genome of a mutant virus, which can be accomplished in several days. The potential drawback of the whole-genome approach is that the analysis could reveal multiple nucleotide changes in different genes that still need to be sorted out by marker rescue before the resistance locus can be assigned. In the present study, we found that the genome sequence of our wild-type vaccinia WR was surprisingly stable compared to that of the reference strain. Moreover, the mitoxantrone-resistant strains accrued amazingly few incidental changes after multiple passages and plaque purification. Thus, we could confidently assign the mitoxantrone resistance phenotype to DNA ligase mutations based on the fact that every mitoxantrone-resistant virus had a mutation in the ligase gene and that two of the resistant strains had no nucleotide changes elsewhere in their genomes.
Acknowledgments
This work was supported by NIH grant U01 AI61456 and contract NO1-AI-15435 from the Virology branch, NIAID. L.D. is the recipient of a physician-scientist career development award from the Dermatology Foundation. S.S. is an American Cancer Society research professor.
Viral genome sequencing was performed in the MSKCC genomics core laboratory, directed by Agnes Viale.
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Ansarah-Sobrinho, C., and B. Moss. 2004. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J. Virol. 78:6335-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldick, C. J., and B. Moss. 1987. Resistance of vaccinia virus to rifampicin conferred by a single nucleotide substitution near the predicted amino terminus of a gene encoding an Mr 62,000 polypeptide. Virology 156:138-145. [DOI] [PubMed] [Google Scholar]
- 3.Baldick, C. J., and B. Moss. 1993. Characterization and temporal regulation of mRNA encoded by vaccinia virus intermediate-stage genes. J. Virol. 67:3515-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borroto-Esoda, K., F. Myrick, J. Feng, J. Jeffrey, and P. Furman. 2004. In vitro combination of amdoxovir and the inosine monophosphate dehydrogenase inhibitors mycophenolic acid and ribavirin demonstrates potent activity against wild-type and drug-resistant variants of human immunodeficiency virus. Antimicrob. Agents Chemother. 48:4387-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buller, R. M., G. Owens, J. Schriewer, L. Melman, J. R. Beadle, and K. Y. Hostetler. 2004. Efficacy of oral active ether lipid analogs of cidofovir in a lethal mousepox infection. Virology 318:474-481. [DOI] [PubMed] [Google Scholar]
- 6.Byrd, C. M., T. C. Bolken, and D. E. Hruby. 2002. The vaccinia virus I7L gene product is the core protein protease. J. Virol. 76:8973-8976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Byrd, C. M., T. C. Bolken, and D. E. Hruby. 2003. Molecular dissection of the vaccinia virus I7L core protein protease. J. Virol. 77:11279-11283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, C. M., T. C. Bolken, A. M. Mjalli, M. N. Arimilli, R. C. Andrews, R. Rothlein, T. Andrea, M. Rao, K. L. Owens, and D. E. Hruby. 2004. New class of orthopoxvirus antiviral drugs that block viral maturation. J. Virol. 78:12147-12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cline, J. C., J. D. Nelson, N. K. Gerzon, R. H. Williams, and D. C. Delong. 1969. In vitro antiviral activity of mycophenolic acid and its reversal by guanine-type compounds. Appl. Microbiol. 18:14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Da Fonseca, F., and B. Moss. 2003. Poxvirus DNA topoisomerase knockout mutant exhibits decreased infectivity associated with reduced early transcription. Proc. Natl. Acad. Sci. USA 100:11291-11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Isabella, P., M. Palumbo, C. Sissi, G. Capranico, N. Carenini, E. Menta, A. Oliva, S. Spinelli, A. P. Krapcho, F. C. Giuliani, and F. Zunino. 1995. Topoisomerase II DNA cleavage stimulation, DNA binding activity, cytotoxicity, and physico-chemical properties of 2-aza- and 2-aza-oxide-anthracenedione derivatives. Mol. Pharmacol. 48:30-38. [PubMed] [Google Scholar]
- 12.DeLange, A. M., M. S. Carpenter, J. Choy, and V. E. Newsway. 1995. An etoposide-induced block in vaccinia virus telomere resolution is dependent on the virus-encoded DNA ligase. J. Virol. 69:2082-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, L., P. Dai, W. Ding, R. D. Granstein, and S. Shuman. 2006. Vaccinia virus infection attenuates innate immune responses and antigen presentation by epidermal dendritic cells. J. Virol. 80:9977-9987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis, D. T., E. B. Doberstyn, S. Awoke, G. L. Royer, and H. E. Renis. 1974. Failure of cytosine arabinoside in treating smallpox: a double-blind study. Lancet ii:377-379. [DOI] [PubMed] [Google Scholar]
- 15.Deterding, A., F. A. Dungey, K. A. Thompson, and D. Steverding. 2005. Antitrypanosomal activities of DNA topoisomerase inhibitors. Acta Trop. 93:311-316. [DOI] [PubMed] [Google Scholar]
- 16.Diamond, M. S., M. Zachariah, and E. Harris. 2002. Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304:211-221. [DOI] [PubMed] [Google Scholar]
- 17.Dinman, J. D., M. J. Ruiz-Echevarria, K. Czaplinski, and S. W. Peltz. 1997. Peptidyl-transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc. Natl. Acad. Sci. USA 94:6606-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ericsson, M., S. Cudmore, S. Shuman, R. Condit, G. Griffiths, and J. K. Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falkner, F. G., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox, E. J. 2004. Mechanism of action of mitoxantrone. Neurology 63:S15-S18. [DOI] [PubMed] [Google Scholar]
- 21.Hartley, C., M. Hartley, I. Pardoe, and A. Knight. 2006. Ionic contra-viral therapy (ICVT): a new approach to the treatment of DNA virus infections. Arch. Virol. 151:2495-2501. [DOI] [PubMed] [Google Scholar]
- 22.Henry, S. D., H. J. Metselaar, R. C. B. Lonsdale, A. Kok, B. L. Haagmans, H. W. Tilanus, and L. J. W. van der Laan. 2006. Mycophenolic acid inhibits hepatitis C virus replication and acts in synergy with cyclosporine A and interferon-alpha. Gastroenterology 131:1452-1462. [DOI] [PubMed] [Google Scholar]
- 23.Hermann, L. L., and K. M. Coombs. 2004. Inhibition of reovirus by mycophenolic acid is associated with the M1 genome segment. J. Virol. 78:6171-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joklik, W. K., B. Moss, B. N. Fields, D. H. Bishop, and L. S. Sandakchiev. 1993. Why the smallpox virus stocks should not be destroyed. Science 262:1225-1226. [DOI] [PubMed] [Google Scholar]
- 25.Kane, E. M., and S. Shuman. 1993. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J. Virol. 67:2689-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kane, E. M., and S. Shuman. 1995. Adenosine N1-oxide inhibits vaccinia virus replication by blocking translation of viral early mRNAs. J. Virol. 69:6352-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koeller, J., and M. Eble. 1988. Mitoxantrone: a novel anthracycline derivative. Clin. Pharmacol. 7:574-581. [PubMed] [Google Scholar]
- 28.Kornbluth, R. S., D. F. Smee, R. W. Sidwell, V. Snarsky, D. H. Evans, and K. Y. Hostetler. 2006. Mutations in the E9L polymerase gene of cidofovir-resistant vaccinia virus strain WR are associated with the drug resistance phenotype. Antimicrob. Agents Chemother. 50:4038-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leyssen, P., J. Balzarini, E. De Clercq, and J. Neyts. 2005. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. J. Virol. 79:1943-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, A., C. Chen, H. Zhang, H. Guo, H. Wang, L. Wang, X. Zhang, S. Hua, J. Yu, P. Xiao, R. Li, and X. Tan. 2005. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 67:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahy, B. W., J. W. Almond, K. I. Berns, R. M. Chanock, D. K. Lvov, R. F. Petersson, H. G. Schatzmayr, and F. Fenner. 1993. The remaining stocks of smallpox virus should be destroyed. Science 262:1223-1224. [DOI] [PubMed] [Google Scholar]
- 32.Margulies, M., M. Egholm, W. E. Altman, A. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. Hem, S. Helgesen, C. H. Ho, G. P. Orzyk, S. C. Jando, M. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moerdyk, M. J., C. M. Byrd, and D. E. Hruby. 2006. Analysis of vaccinia virus temperature-sensitive I7L mutants reveals two potential functional domains. Virol. J. 3:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monsur, K. A., M. S. Hossain, F. Huq, M. M. Rahaman, and M. Q. Haque. 1975. Treatment of variola major with cytosine arabinoside. J. Infect. Dis. 131:40-43. [DOI] [PubMed] [Google Scholar]
- 35.Morrey, J. D., D. F. Smee, R. W. Sidwell, and C. Teng. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antivir. Res. 55:107-116. [DOI] [PubMed] [Google Scholar]
- 36.Nagai, Y., K. Maeno, M. Iinuma, T. Yoshida, and T. Matsumoto. 1972. Inhibition of virus growth by ouabain: effects of ouabain of the growth of HJV in check embryo cells. J. Virol. 9:234-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neyts, J., P. Leyssen, E. Verbeken, and E. De Clercq. 2004. Efficacy of cidofovir in a murine model of disseminated progressive vaccinia. Antimicrob. Agents Chemother. 48:2267-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norbury, C. C., D. Malide, J. S. Gibbs, J. R. Bennink, and J. W. Yewdell. 2002. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 3:265-271. [DOI] [PubMed] [Google Scholar]
- 39.O'Connor, K. A., and B. L. Roth. 2005. Finding new tricks for old drugs: en efficient route for public-sector drug discovery. Nat. Rev. Drug Discov. 4:1005-1014. [DOI] [PubMed] [Google Scholar]
- 40.Payne, L. G., and K. Kristensson. 1979. Mechanism of vaccinia virus release and its specific inhibition by N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine. J. Virol. 32:614-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quenelle, D. C., R. M. L. Buller, S. Parkerm, K. A. Keith, D. E. Hruby, R. Jordan, and E. R. Kern. 2007. Efficacy of delayed treatment with ST-246 given orally against systemic orthopoxvirus infections in mice. Antimicrob. Agents Chemother. 51:689-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramabhadran, T. V., and R. E. Thach. 1980. Specificity of protein synthesis inhibitors in the inhibition of encephalomycocarditis virus replication. J. Virol. 34:293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheidel, L. M., and V. Stollar. 1991. Mutations that confer resistance to mycophenolic acid and ribavirin on Sindbis virus map to the nonstructural protein nsP1. Virology 181:490-499. [DOI] [PubMed] [Google Scholar]
- 44.Schmutz, C., L. G. Payne, J. Gubser, and R. Wittek. 1991. A mutation in the gene encoding the vaccinia virus 37,000 Mr protein confers resistance to an inhibitor of virus envelopment and release. J. Virol. 65:3435-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwer, B., P. Visca, C. Vos, and H. G. Stunnenberg. 1987. Discontinuous transcription or RNA processing of vaccinia virus late messengers results in a 5′ poly(A) leader. Cell 50:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekiguchi, J., and S. Shuman. 1997. Novobiocin inhibits vaccinia replication by blocking virus assembly. Virology 235:129-137. [DOI] [PubMed] [Google Scholar]
- 47.Sekiguchi, J., and S. Shuman. 1997. Domain structure of vaccinia DNA ligase. Nucleic Acids Res. 25:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shuman, S., and B. Moss. 1987. Identification of a vaccinia virus gene encoding a type I DNA topoisomerase. Proc. Natl. Acad. Sci. USA 84:7478-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smee, D. F., M. Bray, and J. W. Huggins. 2001. Anitviral activity and mode of action studies of ribavirin and mcyophenolic acid against orthopoxviruses in vitro. Antivir. Chem. Chemother. 12:327-335. [DOI] [PubMed] [Google Scholar]
- 50.Smee, D. F., and R. W. Sidwell. 2003. A review of compounds exhibiting anti-orthopoxvirus activity in animal models. Antivir. Res. 57:41-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smee, D. F., M.-H. Wong, K. W. Bailey, J. R. Beadle, K. Y. Hostetler, and R. W. Sidwell. 2004. Effects of four antiviral substances on lethal vaccinia virus (IHD strain) respiratory infections in mice. Int. J. Antimicrob. Agents 23:430-437. [DOI] [PubMed] [Google Scholar]
- 52.Sodeik, B., G. Griffiths, M. Ericsson, B. Moss, and R. W. Doms. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 68:1103-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stittelaar, K. J., J. Neyts, L. Naesens, G. van Amerongen, R. F. van Lavieren, A. Holy, E. De Clercq, H. G. M. Niesters, E. Fries, C. Maas, P. G. H. Mulder, B. A. M. van der Zeijst, and A. D. M. E. Osterhaus. 2006. Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature 439:745-748. [DOI] [PubMed] [Google Scholar]
- 54.Szlavik, L., A. Gyuris, J. Minarovits, P. Forgo, J. Molnar, and J. Hohmann. 2004. Alkaloids from Leucojum vernum and antiretroviral activity of Amaryllidaceae alkaloids. Planta Med. 70:871-873. [DOI] [PubMed] [Google Scholar]
- 55.Taddie, J. A., and P. Traktman. 1993. Genetic characterization of the vaccinia virus DNA polymerase: cytosine arabinoside resistance requires a variable lesion conferring phosphonoacetate resistance in conjunction with an invariant mutations localized to the 3′-5′ exonuclease domain. J. Virol. 67:4323-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi, T., M. Oie, and Y. Ichihashi. 1994. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology 202:844-852. [DOI] [PubMed] [Google Scholar]
- 57.Takechi, M., and Y. Tanaka. 1994. Structure-activity relationships of synthetic digitoxigenyl glycosides. Phytochemistry 37:1421-1423. [DOI] [PubMed] [Google Scholar]
- 58.Takechi, M., C. Uno, and Y. Tanaka. 1996. Structure-activity relationships of synthetic cardiac glycosides. Phytochemistry 41:125-127. [DOI] [PubMed] [Google Scholar]
- 59.Tartaglia, J., A. Piccini, and E. Paoletti. 1986. Vaccinia virus rifampicin-resistance locus specifies a late 63,000 Da gene product. Virology 150:45-54. [DOI] [PubMed] [Google Scholar]
- 60.VanSlyke, J. K., C. A. Franke, and D. E. Hruby. 1991. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J. Gen. Virol. 72:411-416. [DOI] [PubMed] [Google Scholar]
- 61.VanSlyke, J. K., S. S. Whitehead, E. M. Wilson, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]
- 62.Vrijsen, R., D. A. Vanden Berghe, A. J. Vlietinck, and A. Boeye. 1986. Lycorine: a eukaryotic termination inhibitor? J. Biol. Chem. 261:505-507. [PubMed] [Google Scholar]
- 63.Yang, G., D. C. Pevear, M. H. Davies, M. S. Collett, T. Bailey, S. Rippen, L. Barone, C. Burns, G. Rhodes, S. Tohan, J. W. Huggins, R. O. Baker, R. L. M. Buller, E. Touchette, K. Waller, J. Schriewer, E. DeClercq, K. Jones, D. Hruby, and R. Jordan. 2005. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J. Virol. 79:13139-13149. [DOI] [PMC free article] [PubMed] [Google Scholar]