Abstract
Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (HHV8) ORF50 encodes a transactivator, K-Rta, which functions as the switch from latent to lytic virus replication. K-bZIP interacts with K-Rta and can repress its transactivation activity for some viral promoters. Both K-Rta and K-bZIP are required for origin-dependent DNA replication. To determine the role of K-bZIP in the context of the viral genome, we generated a recombinant HHV8 bacterial artificial chromosome (BAC) with a deletion in the K-bZIP open reading frame. This BACmid, BAC36ΔK8, displayed an enhanced growth phenotype with respect to virus production and accumulation of virus-encoded mRNAs measured by real-time PCR when K-Rta was used to induce the virus lytic cycle. Conversely, induction of the virus lytic cycle using tetradecanoyl phorbol acetate/n-butyrate resulted in no virus production and an aberrant gene expression pattern from BAC36ΔK8-containing cells compared to wild-type (wt) BAC. This null virus phenotype was efficiently complemented by the expression of K-bZIP in trans, restoring virus production to wt BAC levels. Immunofluorescence staining revealed that subcellular localization of K-Rta was unchanged; however, a disruption of LANA subcellular localization was observed in cells harboring BAC36ΔK8, suggesting that K-bZIP influences LANA localization. Coimmunoprecipitation experiments confirmed that K-bZIP interacts with LANA in BCBL-1 cells and in cotransfection assays. Lastly, the chromatin immunoprecipitation assay revealed that, in an environment where K-Rta is overexpressed and in the absence of K-bZIP, K-Rta binds to CAAT enhancer binding protein α sites within oriLyt, suggesting that it is K-Rta that supplies an essential replication function and that K-bZIP may serve to augment or facilitate the interaction of K-Rta with oriLyt.
Transient assays have shown that Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (HHV8) lytic origin-dependent DNA replication requires the gene products of open reading frames (ORFs) 6, 9, 40/41, 44, 56, and 59 (1). It was also shown that two additional ORFs, 50 and K8, were necessary for efficient replication. The requirement for ORF50 (K-Rta), with respect to oriLyt function, is linked to the presence of an ORF50-responsive promoter present within oriLyt (1, 34). In addition, K-Rta was required in the context of the viral genome for the activation of the entire lytic cycle, as demonstrated by experiments using an HHV8 bacterial artificial chromosome (BAC) recombinant with a deletion of the ORF50 gene (37).
K-bZIP is the proposed homolog of Epstein-Barr virus (EBV) Zta and was shown to interact with oriLyt through a piggyback conformation using the transcription factor CAAT enhancer binding protein α (C/EBPα) (20, 33). In EBV, Zta is required for DNA replication and is the lytic switch protein with transactivation activity (6, 7, 9, 19, 28). However, no associated transactivation activity has been found for K-bZIP. In contrast, transient assays demonstrated that K-bZIP interacts with K-Rta and suppresses the transactivation activity of K-Rta in some viral promoters (10, 11, 18). This repression effect may serve to modulate the activity of K-Rta during a lytic infection. K-bZIP contains a basic amino acid region and a putative leucine zipper that is essential for the associated replication function in transient assays (1). K-bZIP also interacts with several cellular factors, such as transcription factors and proteins that control the cell cycle (12, 13, 18, 26).
Recently, it was shown that K-Rta interacts with oriLyt in a region of HHV8 that contains C/EBPα transcription factor binding sites, presumably through a protein-protein interaction with K-bZIP (35). K-Rta was also shown to interact with members of the core replication complex. These data suggest that K-Rta plays a role in lytic replication in addition to that of a transcriptional activator.
To date, most of the data collected with respect to the activity of K-bZIP have been in the context of transient assays. These assays, for example, the cotransfection replication assay, are invaluable with respect to initial characterization of protein function and interaction with cellular and viral factors. Transient assays, however, are limited in that they cannot address the activities of viral proteins in the context of the viral genome in a dynamic cellular environment. To this end, we generated an HHV8 BAC recombinant that had the entire K-bZIP ORF deleted. This recombinant, BAC36ΔK8, was used to produce stable latently infected Vero cell lines. BAC36ΔK8 and BAC36 (wild type [wt])-containing cell lines treated with tetradecanoyl phorbol acetate (TPA)/n-butyrate to induce viral lytic replication revealed that BAC36ΔK8 was incapable of reactivation and did not produce infectious virus, indicating that K-bZIP is required for virus replication. In addition, although subcellular localization of ORF50 was unchanged, BAC36ΔK8 cell lines displayed a nonpunctate pattern for LANA. Coimmunoprecipitation experiments showed that K-bZIP directly interacted with LANA, suggesting a role for K-bZIP in latency and/or reactivation. trans expression of K-bZIP in BAC36ΔK8-containing cells restored virus production to wt BAC levels and partially redistributed LANA nuclear localization patterns to those observed in wt BAC cell lines, indicating that the null virus phenotype was due to lack of expression of K-bZIP. In contrast, when lytic reactivation was induced using a recombinant adenovirus (Ad) that overexpressed K-Rta, BAC36ΔK8 produced infectious virus at levels many times higher than those observed with BAC36 under the same conditions. Analysis of gene expression revealed an increase in accumulation of mRNA encoding immediate-early, early, and late viral proteins. These data strongly suggest that K-bZIP serves to regulate viral-gene expression in the context of the viral genome and that overexpression of K-Rta in BAC36ΔK8-containing cells can compensate for the replication-associated properties of K-bZIP. A chromatin immunoprecipitation (ChIP) assay performed using BAC36ΔK8-containing cell lines in an environment where K-Rta was overexpressed clearly demonstrated that K-Rta can interact with oriLyt at the C/EBPα sites in the absence of K-bZIP, strongly suggesting that K-Rta, and not K-bZIP, supplies an essential replication function. Hence, K-bZIP may act to amplify or augment the role of K-Rta in initiation of lytic DNA synthesis, but K-bZIP itself is not required for lytic DNA replication under conditions where K-Rta is overexpressed.
MATERIALS AND METHODS
Cells and plasmids.
Vero and Ad293 cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% bovine growth serum. BCBL-1 cells were maintained in RPMI supplemented with 10% fetal bovine serum. BAC36, the wild-type HHV8 BACmid, was provided by S. Gao (University of Texas). Vero cells containing the BAC constructs were maintained in Dulbecco's modified Eagle medium supplemented with 10% bovine growth serum (HyClone, Logan, UT) and 250 μg/ml hygromycin. Plasmid pCMVK8 was generated using the forward primer 5′-cgacttaacagatctcgagctcaagcttcgaattcATGCCCAGAATGAAGGACATACCTACTAAGAGTTC-3′ and the reverse primer 5′-cccgggcccgcggtaccgtcgactgcagaattcTCACTTATCGTCGTCATCCTTGTAATCACATGGTGGGAGTGGCGCGTC-3′ in a PCR using the K-bZIP cDNA clone pcDNA3.1-K-bZIP and the LANA expression plasmid pGTR4-ORF73 LANA, supplied by D. Ganem (University of California—San Francisco).
Construction of a Kaposi's sarcoma-associated herpesvirus K8 (K-bZIP) deletion mutant, BACΔK8.
Mutagenesis of BAC36 was performed using the Red Recombination method as described previously (38). The forward PCR primer, 5′-GTGTAGGCTGGAGCTGCTTCTTCCTCCGTTGTCGACTATAACCTGGCGTGTAAACGTGTAACCCTGCCAAAT-3′, and the reverse primer, 5′-CTATACCTGCTGCAGCTGTCTTGTGTATGCTTTCGATGACACGCGGCCGTATCTGTCAAACATGAGAATTAA-3′, were used to amplify a kanamycin resistance cassette flanked by BAC sequences. This cassette was transformed into BAC36-containing bacteria, and the resulting colonies were plated on kanamycin plates and subsequently screened by Southern blotting. Purified BACΔK8 DNA was transfected into Vero cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Individual cell colonies were selected and expanded, and cells stably harboring the BACΔK8 DNA were selected with 1 mg/ml hygromycin and propagated in 250 μg/ml hygromycin. Once the cells were expanded, the BACmid was isolated from the Vero cells, retransformed into bacteria, and analyzed for the presence of the deletion by sequence analysis and Southern blotting.
High-titer recombinant Ad50 stock preparation.
A recombinant Ad expressing ORF50, Ad type 50 (Ad50), was provided by D. Ganem (University of California—San Francisco) and was expanded in Ad293 cells (Stratagene). The infected Ad293 cells were harvested and centrifuged (5,000 × g) for 10 min to remove the medium. The Ad293 cell pellet was washed with Hanks' balanced salt solution, recentrifuged, and resuspended in 8 ml of the salt solution. The virus was released through four freeze-thaw-vortex cycles. The cell debris was removed by centrifugation at 1,500 × g for 5 min. The recombinant Ad was purified by cesium chloride density gradient centrifugation for 18 h at 100,000 × g. The virus fraction was collected with a syringe and a needle and stored in 2× storage buffer (10 mM Tris, pH 8.0, 100 mM NaCl, 0.1% bovine serum albumin, and 50% glycerol). For control infection experiments, AdTrack (which expresses enhanced green fluorescent protein) viral stocks were prepared as described above.
Construction of a recombinant Ad expressing K-bZIP-FLAG.
The AdEasy Adenoviral Vector System (Stratagene) was used for generation of AdK8. The K-bZIP cDNA ORF was amplified from pTARGET K-bZIP-FLAG and ligated into AdEasy pShuttle vector (Stratagene). Recombinant Ad was generated according to the manufacturer's instructions.
Induction of lytic replication and detection of supernatant virus.
Vero cells stably harboring BAC36 or BAC36ΔK8 were treated with either TPA (25 ng/ml)/n-butyrate (0.3 mM) and high-titer recombinant Ad ORF50 at a multiplicity of infection (MOI) of 4,500, or the cells were treated with the TPA/n-butyrate mixture only. To complement K-bZIP, both BAC36- and BAC36ΔK8-containing mutant cell lines were infected with a recombinant Ad expressing K-bZIP or transfected with a plasmid encoding K-bZIP under the control of the PAN promoter. Medium was harvested and centrifuged twice at 1,000 × g for 10 min. Then, the supernatant was centrifuged for 1 h at 100,000 × g using a SW41Ti rotor. The supernatant was discarded, and the virus pellet was kept at 4°C overnight. The pellet was resuspended in medium, and the virus was used to infect fresh Vero cells. The presence of infectious virus was observed through the expression of green fluorescent protein or quantitated using quantitative PCR (qPCR). For qPCR, pelleted virus was DNase treated (Turbo DNA; Ambion) to remove any contaminating DNA, and then viral DNA was extracted from the purified virus pellet as previously described (38). Briefly, one equal volume of the DNA cell extraction buffer (2% sodium dodecyl sulfate [SDS], 10 mM EDTA, and Tris-HCl, pH 8.0) with proteinase K (150 μg/ml) was added to the virus suspension. DNA was extracted using phenol-chloroform-isoamyl alcohol (24:24:1), followed by ethanol precipitation. An equal volume of DNA was used for qPCR analysis. BAC36 DNA was utilized for a standard curve for the analysis. Each experiment was performed a minimum of three times.
RNA purification and quantitation of the viral transcripts by qPCR.
Vero cells containing BAC36 or BAC36ΔK8 were treated with TPA (25 ng/ml)/n-butyrate (0.3 mM) and high-titer recombinant Ad ORF50 or TPA/n-butyrate alone to induce lytic replication. The cells were harvested at various time points, and total RNA was extracted with the PureLink total RNA purification system kit (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized from 2 μg of total RNA in the presence of random hexamers, deoxynucleoside triphosphates, and superscript III reverse transcriptase (Invitrogen). Data were analyzed as previously described (38).
Generation of a MAb against K-bZIP.
A monoclonal antibody (MAb) for K-bZIP was produced at the University of Nevada Monoclonal Antibody Core Facility. Vero cells infected with AdK8 were prepared using lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% Tween), and recombinant K-bZIP-FLAG protein was affinity purified using FLAG affinity beads. The antibody was an immunoglobulin G1 (IgG1) isotype.
Detection of viral proteins with immunofluorescence assays.
Vero cells containing BAC36 or BAC36ΔK8 were plated on coverslips and reacted with anti-HHV8 specific antibodies as previously described (37). The cells were imaged using an Olympus Fluoview FV1000 microscope with FV10-ASW 1.5 viewer software. The ORF50 antibody used was a gift from D. Ganem. For complementation of LANA subcellular distribution, cells were transfected with pCMVK8 at various concentrations and imaged as described above.
Coimmunoprecipitation assay.
BCBL-1 cells (2 × 108) were treated with TPA (25 ng/ml) and NaB (0.3 mM) for 4 days, and nuclear extract protein was prepared according to the Sigma Cellytic NuCLEAR Extraction Kit protocol without the use of a detergent. K-bZIP, K-Rta, or LANA (ABi catalog no. 13-210-100) specific antibodies (10 μl) were added to 400 μl of BCBL-1 nuclear extract, and the mixture was rotated at 4°C for 2 h, followed by the addition of 50 μl of protein G beads. This mixture was further rotated overnight at 4°C. The beads were washed four times with 1 ml of Tris-buffered saline (Tris-HCl, pH 7.4, 150 mM NaCl) with rotation for 10 min at 4°C after every wash. Thirty microliters of the immunoprecipitated protein was separated through a 10% SDS-polyacrylamide gel electrophoresis (PAGE) gel, which was subsequently transferred to an Immobilion P membrane (Millipore). After an initial blocking step (30 min with Tris-buffered saline plus 5% nonfat milk), the blots were reacted with anti-LANA, anti-K-Rta, or anti-K-bZIP antibody overnight at 4°C, followed by washing and incubation with horseradish peroxidase-conjugated secondary antibody anti-IgG. Protein bands were visualized using a chemiluminescence substrate (Pierce Fempto) and a charge-coupled-device camera.
For cotransfections, 293FT cells (2 × 106/10-cm dish) were transfected with LANA, K-bZIP, and/or K-Rta expression plasmids using TransIT LT1 (Mirrus). Forty-eight hours posttransfection, protein extracts were prepared using lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% NP-40), anti-LANA specific antibody was added, and coimmunoprecipitations were done as described above. For control immunoprecipitation experiments, MAb 84 was used as previously described (4).
ChIP.
Vero cell lines (4 × 107) containing BAC36ΔK8 were infected with Ad50 (MOI = 4,500) for 4 days. Samples were fixed for 10 min with 1% formaldehyde, washed twice with 1× PBS, and lysed for 15 min in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.1% Tween 20, 1 mM EDTA). Samples were sonicated to sheared DNA (∼500 bp), and fragments were diluted sixfold with ChIP buffer (12.5 mM Tris, pH 8, 200 mM NaCl, 1% Triton X-100) and precleared with mouse IgG-agarose conjugate (Santa Cruz Biotechnology) at 4°C for 30 min. For each immunoprecipitation, 2 μg of antibody (anti-ORF50 or anti-K-bZIP) was incubated with the lysate at 4°C overnight. A no-antibody and an antibody isotype control immunoprecipitation were also performed. Protein G-Plus agarose beads (Santa Cruz Biotechnology) were blocked with various amounts of sheared salmon sperm DNA and bovine serum albumin at 4°C overnight and then washed with ChIP buffer. The blocked and washed protein G-Plus beads were incubated with the lysate at 4°C for 1 h. The beads were washed once with low-salt buffer (0.1% SDS, 0.1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 150 mM NaCl), once with high-salt buffer (0.1% SDS, 0.1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 500 mM NaCl), once with LiCl buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris, pH 8), and twice with Tris-EDTA. The beads were resuspended in Tris-EDTA, incubated with RNase A at 37°C for 30 min, and then incubated with proteinase K and 10% SDS at 37°C for 4 h, followed by incubation at 65°C overnight. For input control samples, NaCl was added to the sonicated lysate to a final concentration of 0.3 M, and the mixture was incubated at 65°C overnight. After the antibody-, no-antibody-, and isotype control-immunoprecipitated samples were filtered through a 0.45-μm filter to remove the agarose beads, they were extracted with phenol-chloroform and ethanol precipitated. The primers used for PCR amplification (5′-AATCCCCCATAATCCTCTGC-3′, and reverse, 5′-GGAAAAATCAAAACAAAACTC-3′) corresponded to nucleotides (nt) 23326 to 23572 of HHV8 oriLyt. The control primers, 5′-ACGTCCGGAGAGTTGGAACTGTCA-3′ and 5′-GGGGTCCATGGGATGGGTTAGTCA-3′, were complementrary to the ORF45 region.
RESULTS
Deletion of the K8 (K-bZIP) locus from BAC36.
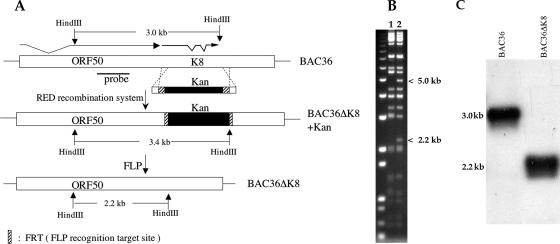
K-bZIP is the product of a spliced transcript that originates just downstream of the ORF50 locus (Fig. 1A). Using the wt BAC36 as a template, we designed PCR primers so that the entire K-bZIP ORF was removed from the genome. This construction did not disrupt the downstream K8.1 ORF or the polyadenylation signal at nt 76713, which is presumably used by both K8.1 and K-Rta (ORF50) (29, 30). The final BAC mutant also left 328 nt upstream of the start of the K8.1 mRNA (22, 31). Figure 1 shows a schematic for the generation of the recombinant, BAC36ΔK8, using PCR primers that integrated the kanamycin resistance gene marker (Kan+) into the HHV8 BAC genome, replacing the entire K-bZIP ORF. Since we designed primers to contain the substrate sequence for FLP recombinase (FRT sites), the Kan+ marker was subsequently removed by expressing FLP recombinase in bacteria (Fig. 1A). This excision of the Kan+ cassette retained only the 15-bp FRT site, which we engineered to contain a HindIII recognition sequence; therefore, the function of any promoter or other cis-acting element upstream or downstream of the K-bZIP ORF was not affected. The resulting BAC recombinants were screened and analyzed by agarose gel electrophoresis and subsequently by Southern blot analysis to show that the K-bZIP ORF was removed from the HHV8 genome (Fig. 1B and C). BACΔK8 HindIII cleavage results in a 2.2-kb band, along with a 5-kb band, which is generated from further downstream, upon removal of the K8 locus (and Kan+ cassette) (Fig. 1B), indicating that the K-bZIP ORF was deleted. A Southern blot showed the presence of a 2.2-kb band in the recombinant BACΔK8 when it was hybridized with a probe from within the ORF50 locus (Fig. 1A and C). Subsequently, the recombinant was sequenced to ensure that the proper insertion/deletion was made.
FIG. 1.
Generation of a recombinant HHV8 BAC with a deletion in the K8 (K-bZIP) ORF. (A) Schematic diagram showing the ORF50 K8 genomic BAC36 locus that was targeted to produce a recombinant with a deletion of the K-bZIP ORF by replacing it with the gene cassette that confers kanamycin resistance flanked by FRT sites. Also shown is the final gene configuration after removal of the Kan cassette using FLP recombinase and the location of the hybridization probe used for Southern blot confirmation. (B) Ethidium bromide-stained gel with BAC36 (lane 1) and the mutated BACmid, BAC36ΔK8 (lane 2), cleaved with HindIII. Shown are the resultant 5.0- and 2.2-kb bands in the BAC36ΔK8 recombinant due to introduction of an additional HindIII site within the FRT recognition sequence. (C) Southern blot of BAC36 and BAC36ΔK8 purified DNA cleaved with HindIII. Shown are the expected bands produced from both BACmid DNAs. The probe used for the experiment is shown in panel A.
BAC36ΔK8-harboring cell lines produce 30-fold more virus than BAC36-containing cells when treated with a K-Rta-expressing Ad.
We used the BAC recombinant, BAC36ΔK8, and BAC36 to generate Vero cell lines that harbored either latent BAC36ΔK8 or BAC36 DNA. Once these cell lines were established, the first step was to induce the lytic cycle in each BACmid-containing cell line and compare virus production and mRNA accumulation levels. We initially chose to induce lytic replication by infecting the BACmid-containing cells with a recombinant Ad (Ad50) expressing the major viral transactivator, the lytic switch protein K-Rta. Previous studies demonstrated that this treatment, using concentrated Ad50, resulted in an extremely efficient reactivation of latent BAC36 from Vero cell lines (8).
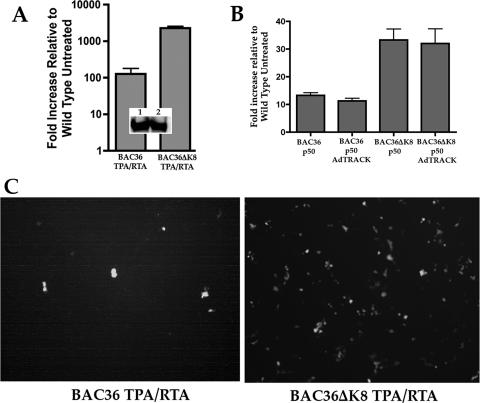
BAC36- or BAC36ΔK8-containing cell lines were infected with Ad50, and supernatant virus was concentrated as previously described (38). The supernatant virus was used for qPCR analysis of viral DNA and for infection of fresh Vero cells to evaluate the number of green cells (infectious virus). qPCR performed on the supernatant virus revealed that an approximately 30-fold increase in supernatant viral DNA was produced from BAC36ΔK8-containing cells treated with Ad50 compared to similarly treated BAC36-containing cells (Fig. 2A). This observation was confirmed when equal volumes of supernatants from each BAC-containing cell line were incubated with fresh Vero cells. We also measured the relative levels of K-Rta protein present in each cell line. Cellular-protein lysates were prepared from each Ad50-infected cell line, and Western blots were performed using an anti-K-Rta specific antibody. The K-Rta protein levels were similar in each cell line, demonstrating that Ad50 efficiently infected both cell lines. In order to rule out possible complementation of BAC36ΔK8 by an Ad protein, we transfected the cell lines with a K-Rta expression plasmid and subsequently infected them with the control Ad AdTRACK (Clontech). The addition of a control Ad had no affect on the level of supernatant virus produced in BAC36- or BAC36ΔK8-containing cell lines, indicating that an Ad-encoded protein did not account for the increase in virus production (Fig. 2B). This control experiment demonstrated that overexpression of K-Rta by plasmid transfection also resulted in an increase in virus production in the BAC36ΔK8-containing cell line; however, the response was not as dramatic as that observed in Ad50-infected samples.
FIG. 2.
BAC36ΔK8 displays an enhanced growth phenotype upon lytic cycle induction with Ad50. Vero cell lines stably carrying BAC36 or BAC36ΔK8 were treated with TPA (25 ng/ml) and Ad50 (MOI = 4,500) at 24 h postplating and incubated for 5 days. Supernatant virus was harvested as described in Materials and Methods, and total cellular RNA was extracted and subjected to qPCR analysis. (A) Quantification of the viral DNA by qPCR. Viral DNA was extracted from supernatant virus, and qPCR was performed using ORF59 primers/probe. The relative increase in viral DNA was determined and expressed as an n-fold increase compared to the untreated BAC36 supernatant virus. (Inset) Western blot reacted with anti-K-Rta antibody from protein lysates from Ad50-infected BAC-containing cell lines. Lanes: 1, protein lysate from a BAC36 cell line infected with Ad50; 2, protein lysate from BAC36ΔK8-containing cells infected with Ad50. The error bars show the standard deviations from at least three separate experiments. (B) Addition of Ad has no effect on HHV8 BAC virus production. Vero cell lines containing either BAC36 or BAC36ΔK8 were transfected with a K-Rta expression plasmid (pTarget K-Rta-FLAG) and subsequently infected with the control Ad AdTRACT. Viral DNA was extracted from the supernatants and analyzed as described for panel A. (C) Ad50-induced BAC36ΔK8 results in an increase in infectious virus. Concentrated supernatant virus from BAC36ΔK8 or BAC36 cell lines was used to infect fresh Vero cells. The cells were imaged 3 to 5 days postinfection for the presence of enhanced green fluorescent protein-positive cells.
In Ad50-treated cell lines, microscopic examination revealed the presence of many more green (virus-infected) cells from BAC36ΔK8 supernatant virus than from cells incubated with BAC36 supernatant virus (Fig. 2C). These results suggested that under conditions where an excess of K-Rta was present, the BACmid that lacked the ability to express K-bZIP produced a much larger amount of infectious virus than a K-bZIP-competent BACmid.
Increased accumulation of virus-specific transcripts from cells containing BAC36ΔK8 treated with Ad50 and TPA/n-butyrate.
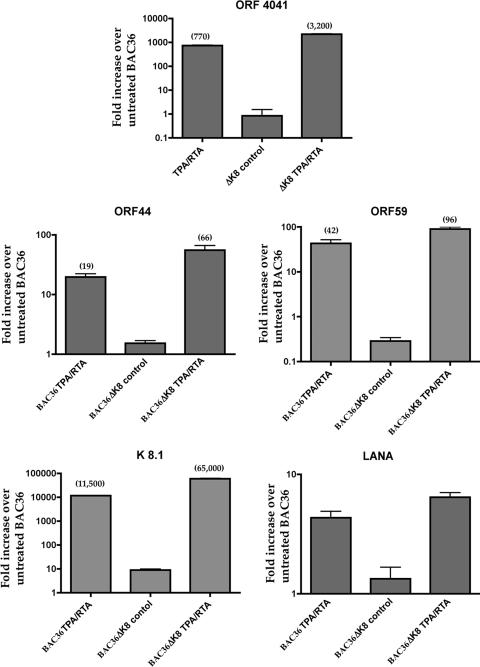
Since we observed an increase in infectious virus and viral DNA in supernatants from cells containing BAC36ΔK8 treated with Ad50 (MOI = 4,500), we investigated the relative accumulation levels of virus-specific mRNAs from both BAC36 and BAC36ΔK8 cell lines. The cell lines were induced with Ad50, total cellular RNA was harvested at 5 days postinduction, and qPCR was performed. We measured mRNA accumulation for transcripts ORF40/41, ORF44, ORF59, K8.1, and LANA. The overall level of mRNA accumulation was about two- to sixfold higher in cells harboring BAC36ΔK8 than in BAC36-containing cells (Fig. 3). The largest difference was observed in the mRNA encoding K8.1, which encodes a late protein that is a structural component of the HHV8 particle (17, 27). This is interesting, since it was suggested that the K8.1 promoter lies within the K8 ORF, but apparently the upstream region in the BAC with K8 deleted was sufficient for K8.1 expression (31). Overall, the transcription data suggest that an increase in accumulation of all kinetic classes of RNA occurred in the cell line containing BAC36ΔK8 compared to BAC36 cell lines treated with K-Rta/TPA.
FIG. 3.
qPCR analysis of several virus-encoded mRNAs from Ad50-induced BAC-containing cell lines. Vero cell lines harboring BAC36ΔK8 or BAC36 were treated with TPA and Ad50 (MOI = 4,500), and total cellular RNA was harvested and, following cDNA synthesis, analyzed by qPCR using primers and probes specific for several viral transcripts. Cyclophilin was used as an internal standard, and data were normalized and analyzed using the comparative threshold cycle method. The data are reported as the relative increase over untreated BAC36. Each analysis was done six times. The error bars show the standard deviations from at least three separate experiments.
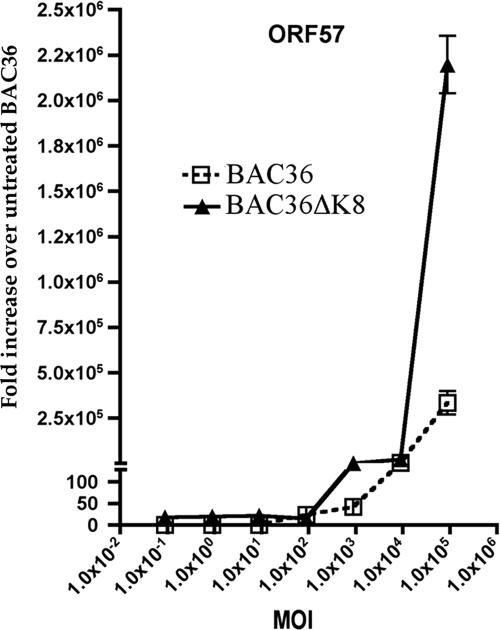
We also evaluated the mRNA accumulation of ORF57 in Ad50-treated BAC36- and BAC36ΔK8-containing cell lines. In transient assays, ORF57 expression was transactivated and upregulated by K-Rta, and the two proteins were shown to interact in infected and transfected cells (3, 5, 21, 24, 32). In addition, K-bZIP suppressed the K-Rta-mediated transcriptional activation of the ORF57 promoter (11). We infected BAC36ΔK8 cell lines using several different Ad50 MOIs and measured the relative amounts of ORF57 mRNA accumulation in each cell line. ORF57 accumulation at the highest MOI (4,500) resulted in an approximately 1,000-fold difference in ORF57 mRNA accumulation in BAC36ΔK8-containing cells (Fig. 4). To ensure that the levels of K-Rta expression were the same in both Ad50-infected cell lines, the relative amounts of K-Rta expression were measured by Western blotting in both cell lines at the three highest MOIs used. No difference in K-Rta expression was observed; hence, the increase in ORF57 mRNA was not due to an increase in K-Rta protein levels in BAC36ΔK8-containing cells (data not shown). These results indicated that in the absence of K-bZIP and under conditions where K-Rta is overexpressed, both virus replication and gene expression are greatly enhanced. This suggests that the replication function associated with K-bZIP is dispensable and can be compensated for by K-Rta.
FIG. 4.
Ad50 treatment induces a marked increase in the accumulation of ORF57 mRNA in BAC36ΔK8. BAC36 and BAC36ΔK8 cell lines were treated with TPA (25 ng/ml) and increasing amounts of Ad50. Total cellular RNA was extracted, and ORF57 mRNA accumulation was measured at 5 days posttreatment by qPCR. The data are shown as the increase over untreated BAC36. The error bars are the standard deviations from three separate experiments.
K-bZIP expression is required for viral replication when lytic cycle induction is performed with TPA/n-butyrate.
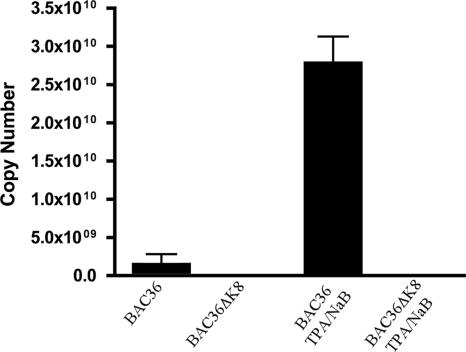
In light of the results from experiments where BAC-containing Vero cell lines were induced to enter the lytic cycle with Ad50, we wanted to investigate the growth characteristics under conditions using TPA/n-butyrate to induce the lytic cycle. In this case, the lytic cycle would be induced by the up regulation of K-Rta instead of the epigenetic expression of the transactivator. BAC36- and BAC36ΔK8-containing cell lines were treated with TPA/n-butyrate for 5 days, and supernatant virus was harvested and subjected to qPCR evaluation. No viral DNA was detected in supernatants harvested from BAC36ΔK8 cell lines by qPCR, whereas supernatants from BAC36 (wt)-containing cells showed over 2.5 × 1010 copies of viral DNA (Fig. 5). This experiment demonstrated that under the conditions traditionally used to induce HHV8 lytic replication, where much less K-Rta is produced, BAC36ΔK8 failed to replicate, indicating that K-bZIP was essential for virus growth under these treatment conditions.
FIG. 5.
BAC36ΔK8 is replication deficient when lytic replication is induced with TPA/n-butyrate. BAC36ΔK8 and BAC36 cell lines were treated with TPA/n-butyrate, and supernatant virus was collected, concentrated, and used for qPCR analysis as described in Materials and Methods. Shown is a representative graph from one experiment. The error bars are the standard deviations of viral copy numbers from three separate samples.
BAC36ΔK8 displays an aberrant gene expression pattern upon induction with TPA/n-butyrate.
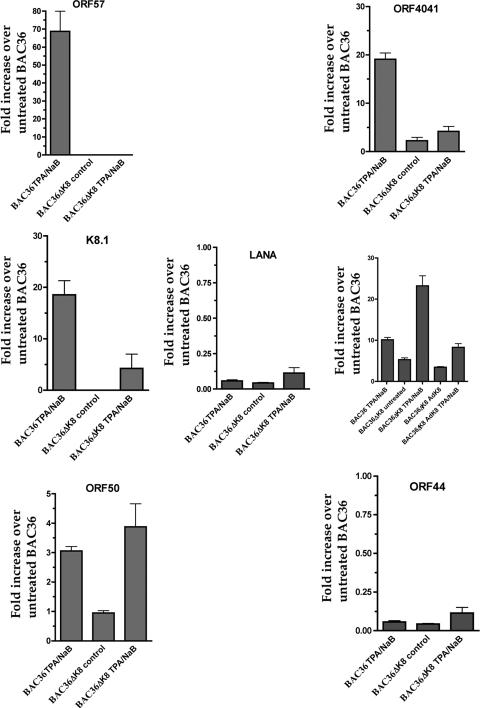
Once it was determined that virus production was impaired in BAC36ΔK8-harboring cell lines when the lytic cycle was induced using only TPA/n-butyrate, we examined the gene expression pattern from the mutant virus under these conditions. BAC36ΔK8 and BAC36 cell lines were induced with TPA/n-butyrate, and mRNA accumulation was measured for several HHV8-specific genes at 5 days posttreatment. The level of ORF50 (K-Rta) mRNA was slightly higher in TPA/n-butyrate-induced BAC36ΔK8 cell lines, and there was a small but measurable accumulation of K-Rta mRNA in uninduced cells versus that observed in BAC36 (wt)-containing cells (Fig. 6, ORF50 graph). Despite this apparent level of ORF50 mRNA accumulation, transcripts originating from the ORF57 locus were undetectable from BAC36ΔK8 cell lines (Fig. 6, ORF57 graph). In addition, RNA transcripts from the PAN locus were approximately 20-fold higher in TPA/n-butyrate-treated cells carrying BAC36ΔK8 than in those carrying BAC36 (Fig. 6, PAN graph). Upon treatment with AdK8, the PAN mRNA levels in BAC36ΔK8 returned to levels similar to those observed in BAC36 cell lines (Fig. 6, PAN graph). BAC36 displayed a fivefold increase in PAN RNA upon lytic cycle induction, consistent with an increase in K-Rta expression. mRNA accumulations of other virus-encoded transcripts, for example, ORF40/41 and K8.1, were much lower from BAC36ΔK8-containing cells than those observed from BAC36-containing cells (Fig. 6, ORF40/41 and K8.1 graphs). The levels of LANA mRNA were approximately the same in BAC36ΔK8- and BAC36-containing cell lines (Fig. 6, LANA graph). These results suggested that the lack of K-bZIP led to a general decrease in gene expression, most strikingly in ORF57, even in the environment where K-Rta was abundant; however, mRNA originating from the PAN locus accumulated to higher levels in BAC36ΔK8 cell lines with treatment, suggesting a deregulation of the PAN promoter when K-bZIP is not expressed.
FIG. 6.
qPCR analysis of BAC36ΔK8 cell lines induced with TPA/n-butyrate. Vero cell lines harboring BAC36 or BAC36ΔK8 were treated with TPA/n-butyrate, and total cellular RNA was extracted and subjected to qPCR analysis as described previously. Shown are graphs of qPCR data using cyclophilin as an internal standard. The data were normalized and compared to BAC36 untreated mRNA levels by the comparative threshold cycle method. The data are shown as the increase compared to the untreated BAC36 cell line. The error bars are the standard deviations from five separate experiments.
Expression of K-bZIP in trans complements and restores virus production in BAC36ΔK8-containing cells.
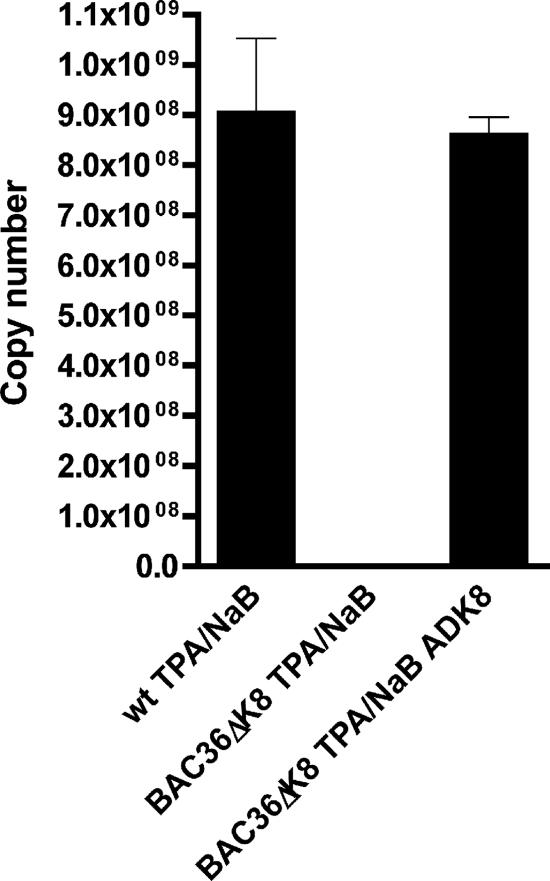
Since no virus replication was observed under conditions where the virus lytic cycle was induced using TPA/n-butyrate, we next sought to complement the replication-deficient mutant virus with the exogenous expression of K-bZIP. AdK8 was used to complement BAC36ΔK8 in the presence of TPA/n-butyrate. Under these conditions, wt virus production, as measured by qPCR, was observed when BAC36ΔK8-containing cells were treated with both TPA/n-butyrate and AdK8 (Fig. 7). This experiment strongly suggested that the null phenotype observed for BAC36ΔK8 was due to the lack of K-bZIP expression, and supplying K-bZIP in trans was sufficient to overcome the defect. These data indicated that K-bZIP can modulate gene expression in the context of the viral genome.
FIG. 7.
Restoration of wt virus production by complementation of BAC36ΔK8 with the epigenetic expression of K-bZIP. BAC36ΔK8 and BAC36 cell lines were treated with TPA/n-butyrate and AdK8 (MOI = 1,700) for 5 days. Supernatant virus was collected, concentrated, and used for qPCR analysis. Shown is a graph reporting the viral DNA copy number from supernatant virus. A representative experiment is shown. The error bars are the standard deviations from three sample replicates.
Disruption of the subcellular localization of LANA in the BAC36ΔK8 cell line.
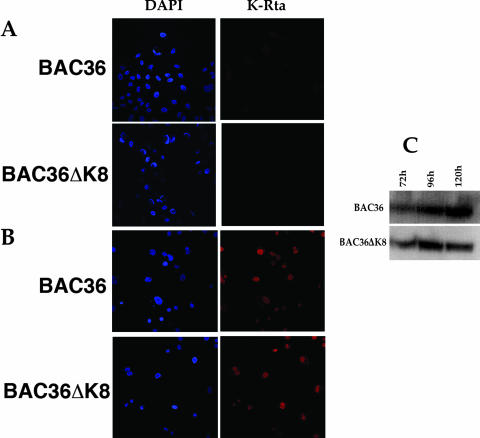
In order to further investigate the observed phenotype of the BAC36ΔK8 BACmid, we examined the subcellular distribution of LANA and K-Rta (ORF50) in BAC36ΔK8- and BAC36-containing cell lines. qPCR data suggested that K-Rta protein levels should be similar in BAC36ΔK8 and BAC36 cell lines, given that mRNA accumulations were similar in the two BACmids. No K-Rta-specific signal was detected in uninduced cell lines (Fig. 8A). However, when cell lines were induced with TPA/n-butyrate, a strong K-Rta-specific signal was detected within the nuclei of BACmid-containing cells at 5 days postinduction (Fig. 8B). No apparent difference in subcellular localization or intensity of the signal was observed between the two BACmid-containing cell lines (Fig. 8). We also measured the protein levels of K-Rta in each cell line treated with TPA/n-butyrate. Consistent with the qPCR data, similar K-Rta protein levels were observed in each BAC-containing cell line (Fig. 8C).
FIG. 8.
K-Rta subcellular localization is unchanged in BAC36ΔK8 cell lines. BAC36- and BAC36ΔK8-containing cell lines were fixed and reacted with anti-K-Rta antibody, followed by detection with Alexa Fluor 555. (A) Uninduced cell lines reacted with anti K-Rta. (B) TPA/n-butyrate-induced cell lines reacted with anti-K-Rta. (C) Levels of K-Rta protein in TPA/n-butyrate-induced cell lines. Shown is a Western blot of protein extracts from cell lines containing either BAC36 or BAC36ΔK8 DNA reacted with anti-K-Rta specific antibody. K-Rta protein levels were evaluated between 72 and 120 h postinduction. DAPI, 4′,6′-diamidino-2-phenylindole.
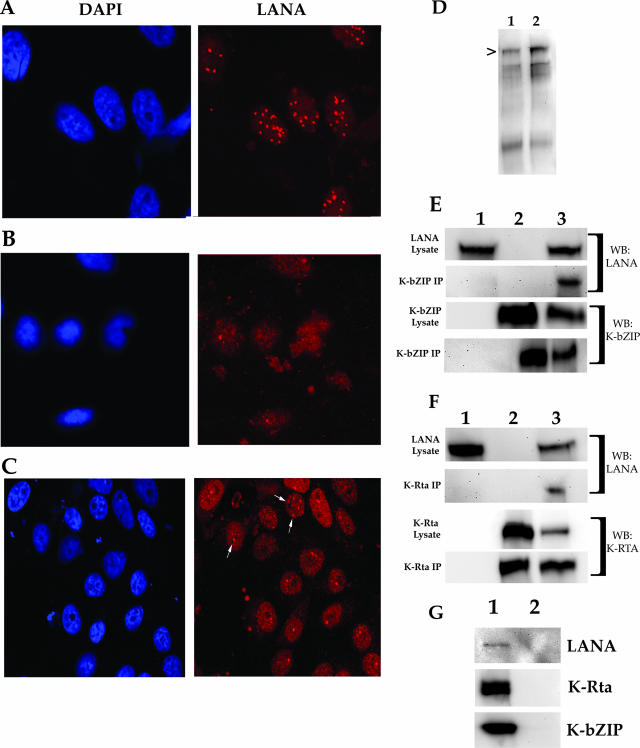
Uninduced cell lines were also subjected to immunofluorescence, using the anti-LANA antibody. Cell lines harboring BAC36 displayed a typical punctate nuclear staining of the LANA protein (Fig. 9A). However, in the cell line harboring BAC36ΔK8, LANA displayed a more diffuse pattern of staining and was not entirely localized within the cell nucleus (Fig. 9B). Transfection of the BAC36ΔK8 cell line with the K-bZIP expression construct, pCMVK8, was able to partially restore LANA subcellular localization to a more punctate nuclear staining, suggesting that even in cells that were not treated with TPA/n-butyrate, K-bZIP may influence the activity or localization of LANA (Fig. 9C). Examination of LANA protein levels by Western blotting showed similar protein expression levels of LANA in both BAC36ΔK8- and BAC36-containing cell lines (Fig. 9D, lanes 1 and 2, respectively).
FIG. 9.
LANA is dispersed in uninduced BAC36ΔK8 cell lines. BAC36- and BAC36ΔK8-containing cell lines were fixed and reacted with anti-LANA antibody, followed by detection with Alexa Fluor 555. The left column shows the cell nucleus stained with DAPI (4′,6′-diamidino-2-phenylindole). (A) Vero cell line harboring BAC36 DNA reacted with anti-LANA antibody (magnification, ×80). (B) Vero cell line harboring BAC36ΔK8 DNA reacted with anti-LANA antibody (magnification, ×80). (C) Vero cell line harboring BAC36ΔK8 DNA infected with AdK8 (MOI = 1,800) and reacted with anti-LANA antibody (magnification, ×80). The arrows show the punctate localization pattern of LANA protein. (D) LANA protein levels in BAC36- and BAC36ΔK8-containing cell lines. Lanes: 1, protein lysate from BAC36-containing cell lines; 2, protein lysates from BAC36ΔK8-containing cell lines. The arrowhead indicates the LANA protein band. (E) Coimmunoprecipitation of LANA and K-bZIP in transfected 293FT cells. The cells were transfected with a LANA expression plasmid and a K-bZIP expression plasmid. Protein complexes were coimmunoprecipitated using an anti-K-bZIP specific antibody. Immunoprecipitated protein complexes were separated through an SDS-PAGE gel, transferred to an Immobilon-P membrane, and reacted with either anti-LANA or anti-K-bZIP antibody. Lanes: 1, immunopre-cipitation from cells transfected with a LANA expression plasmid; 2, immunoprecipitation from cells transfected with a K-bZIP expression plasmid; 3, immunoprecipitation from cells transfected with LANA and K-bZIP expression plasmids. The blots were reacted with antibodies as indicated on the right. WB, Western blot. (F) Coimmunoprecipitation of LANA and K-Rta in transfected 293FT cells. The cells were transfected with a LANA expression plasmid and a K-Rta expression plasmid. Protein complexes were coimmunoprecipitated using an anti-K-Rta specific antibody. Immunoprecipitated protein complexes were separated through an SDS-PAGE gel, transferred to an Immobilon-P membrane, and reacted with either anti-LANA or anti-K-Rta antibody. Lanes: 1, immunoprecipitation from cells transfected with a LANA expression plasmid; 2, immunoprecipitation from cells transfected with a K-Rta expression plasmid; 3, immunoprecipitation from cells transfected with LANA and K-Rta expression plasmids. The blots were reacted with antibodies as indicated on the right. (G) Nonspecific antibody control. 293FT cells were cotransfected with plasmids expressing K-Rta, K-bZIP, and LANA, and protein lysates were prepared. The lysates were incubated with the anti-UL84 specific antibody MAb 84. Immunoprecipitated protein was separated on an SDS-PAGE gel, and Western blots were prepared. The blots were reacted with either anti-K-Rta, anti-K-bZIP, or anti-LANA specific antibodies. Lanes: 1, protein lysates prior to immunoprecipitation; 2, immunoprecipitated protein using MAb 84.
To show that K-bZIP can interact with LANA directly (in the absence of any other viral protein), we performed a series of experiments in which a LANA expression plasmid was cotransfected, along with a K-bZIP expression plasmid, with or without a K-Rta expression plasmid. Protein extracts were immunoprecipitated with anti-K-bZIP antibody, and Western blots of immunoprecipitated protein were reacted with anti-LANA or anti-K-bZIP specific antibodies. Immunoprecipitation of K-bZIP revealed that K-bZIP interacted with LANA directly and did not require the presence of K-Rta (Fig. 9E, lane 3, K-bZIP, WB:LANA). In control lanes where K-bZIP antibody was used to immunoprecipitate LANA alone, transfected cells failed to reveal a detectable band (Fig. 9E, lane 1, WB:LANA and WB:K-bZIP). As shown previously, K-Rta was also efficiently pulled down, along with LANA protein (Fig. 9F, lane 3, WB:LANA). These experiments demonstrated that LANA and K-bZIP can interact in the absence of K-Rta and that K-bZIP may influence the localization and activity of LANA. As an additional control to ensure that there was no nonspecific interaction in our cotransfection immunoprecipitation assays, we used an unrelated antibody (MAb 84) in an immunoprecipitation experiment with K-Rta-, K-bZIP-, and LANA-cotransfected protein lysates. In all cases, no protein was pulled down when an unrelated antibody was used (Fig. 9G, lane 2). Protein lysates showed that K-Rta and K-bZIP were all expressed efficiently (Fig. 9G, lane 1).
K-Rta can interact with oriLyt in the absence of K-bZIP.
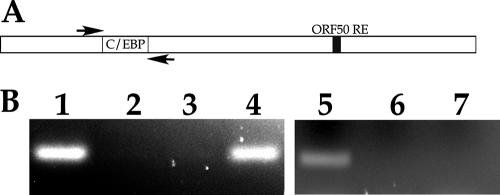
The current thinking on the dynamics of initiation of lytic DNA replication in HHV8 involves the interaction of K-bZIP with oriLyt. Recently, it was shown that K-Rta could also interact with oriLyt in the region where K-bZIP is known to bind (35). This region contains several C/EBPα consensus binding motifs. The interaction of K-Rta with oriLyt was mediated by K-bZIP. However, those studies used fragments of oriLyt DNA that contained mutations within the C/EBPα binding motifs; hence, a direct interaction of K-Rta with oriLyt at those sites could not be assessed. Consequently, it is not known whether K-Rta can interact with oriLyt directly through the C/EBPα sites in the context of the mutant virus that lacks K-bZIP and under conditions where K-Rta is abundant. We wanted to understand the reason why an environment where K-Rta is overexpressed is conducive to the complementation of BAC36ΔK8. The fact that we observed that the BACmid without K-bZIP could replicate when K-Rta is overexpressed suggests that K-Rta can perform a required replication function and therefore interact with oriLyt without the aid of K-bZIP. To test this theory, we infected the BAC36ΔK8 cell line with Ad50 and performed the ChIP assay using primers flanking the previously identified C/EBPα consensus binding motifs within oriLyt known to be a substrate for K-bZIP. Figure 10A is a schematic of the HHV8 oriLyt region showing the relative locations of the consensus C/EBPα sites and the ORF50 response element. The locations of PCR primers used in the ChIP assay are also shown. The ChIP assay revealed that K-Rta could interact with oriLyt in the absence of K-bZIP when immunoprecipitations were performed using anti-K-Rta specific antibody, as shown by the presence of a PCR-amplified product (Fig. 10B, lane 4). The control PCRs, consisting of no antibody or an anti-K-bZIP specific antibody, failed to show any amplified PCR product (Fig. 10B, lanes 2 and 3, respectively). Control primers were used that were complementary to the ORF45 locus. No PCR product was detected when PCR primers were used that amplified a region of DNA outside of oriLyt known not to interact with K-Rta (Fig. 10B, lanes 6 and 7). This experiment strongly suggests that K-bZIP may facilitate an interaction between K-Rta and oriLyt and that this ancillary function may be overcome by high concentrations of K-Rta, which provides an essential replication function without K-bZIP.
FIG. 10.
K-Rta can interact with oriLyt in the absence of K-bZIP. BAC36ΔK8-containing cell lines were infected with Ad50, and the ChIP assay was performed using anti-K-Rta or anti-K-bZIP antibodies to coimmunoprecipitate protein bound to DNA from the region of HHV8 oriLyt containing C/EBPα transcription factor binding sites. (A) Schematic of the HHV8 oriLyt region. The arrows depict the locations of the PCR primers used for amplification of immunoprecipitated protein-DNA complexes. (B) Ethidium bromide-stained gel of PCR amplification products from immunoprecipitations. Lanes: 1, input DNA (preimmunoprecipitation); 2, no-antibody control; 3, immunoprecipitation using anti-K-bZIP specific antibody; 4, immunoprecipitation using anti-K-Rta specific antibody; 5, input DNA for ORF45 primers (preimmunoprecipitation); 6, immunoprecipitation using isotype control antibody (same isotype as K-Rta antibody); 7, immunoprecipitation using K-Rta specific antibody.
DISCUSSION
The HHV8 K8 gene product, K-bZIP, is the proposed homolog to EBV Zta. On a functional level, however, the two proteins do not appear to have much in common. For EBV, Zta is the major transactivator and can reactivate latent virus, whereas K-bZIP is incapable of activating the lytic cycle and does not have a transactivation activity. One similarity is that Zta, like K-bZIP, is required for oriLyt-dependent DNA replication (1). Efficient oriLyt amplification using the cotransfection replication assay for HHV8 also required K-Rta, whereas in EBV, no such additional transactivator protein was needed (6). The most striking difference between Zta and K-bZIP is the association of K-bZIP with K-Rta and the repressive effects by K-bZIP on K-Rta-mediated transcriptional activation of certain HHV8 promoters. As for the replication function for K-bZIP, to date no known enzymatic or direct DNA binding activity has been identified for the protein. Apparently, K-bZIP interacts with oriLyt through binding with C/EBPα, and this protein complex binds to C/EBPα consensus binding sites within oriLyt (33). Therefore, it is logical to study the effects of a recombinant virus that lacks the K8 gene and is deficient for K-bZIP expression.
The most interesting observation from the study presented here was that K-bZIP was not required for viral gene expression, viral DNA replication, or virus production under conditions where K-Rta was overexpressed from an Ad vector. Surprisingly, the amount of virus produced was severalfold higher than that observed from the parent BAC36 under the same conditions. We initially used Ad50 to induce the lytic cycle because it was reported that this was a very efficient method to reactivate virus from cell lines harboring BACmid DNA (8). The BAC recombinant BAC36ΔK8 showed a marked increase in virus production as measured by the appearance of green cells indicative of infectious virus, as well as an increase in viral supernatant DNA. The growth-enhanced phenotype was unexpected, since K-bZIP is the proposed initiator protein for lytic DNA synthesis and therefore should be indispensable for growth. The use of a control Ad (AdTRACK) had no effect on BAC36ΔK8 gene expression or virus production, ruling out any complementation by an Ad protein (data not shown). Although viral mRNA accumulation for specific transcripts was somewhat higher in BAC36ΔK8, the increase in virus production could be due to disregulation at the protein level. For example, the activity or protein half-life of K-Rta may be affected by the absence of K-bZIP. One viral transcript that was markedly upregulated was ORF57. The ORF57 protein, which also interacts with K-Rta, is associated with regulation of gene expression via export of viral transcripts from the cytoplasm to the nucleus; all of these data are consistent with its being the proposed homolog to EBV MTA (24, 25). It was recently reported that expression of ORF57 was required for viral growth, as demonstrated using a BACmid with ORF57 deleted (8, 23). Overexpression of the ORF57 protein could account for an enhanced growth phenotype, given that its role is to enhance the expression of intronless viral mRNAs. Independent of the role of ORF57 in BAC36ΔK8-infected cells, it is apparent that the overexpression of K-Rta can compensate, directly or indirectly, for K-bZIP and complement a K-bZIP-deficient virus.
K-Rta was recently implicated as having a dual role in lytic DNA replication, in that it was shown to act as a transactivator (binding to an ORF50 response element), as well as interacting directly with oriLyt in a region that has not been identified as a K-Rta-responsive promoter (36). Also, K-Rta was shown to be part of the replication complex, and it has been proposed that it could act as a replication initiator protein for lytic DNA synthesis. The data presented here show that K-Rta is sufficient for initiating lytic virus replication in the absence of K-bZIP when K-Rta is present at high concentrations. It was previously shown that K-Rta binds to oriLyt via an interaction with K-bZIP, which subsequently binds to C/EBPα sites. This interaction is obviously not required in the context of the viral genome, since our data indicate the K-Rta can still mediate lytic DNA synthesis in the absence of K-bZIP in an environment where K-Rta is overexpressed. The ChIP assay confirmed that K-Rta interacts with oriLyt at the C/EBPα sites in the absence of K-bZIP. Therefore, another mechanism must exist by which K-Rta can interact with oriLyt, or it could be the case that K-Rta transactivation of the oriLyt promoter is sufficient to trigger lytic DNA synthesis. One other plausible explanation is that K-bZIP serves to amplify, augment, or target K-Rta to oriLyt, and consequently, it is K-Rta which performs an essential replication function. Hence, when the artificial condition of K-Rta overexpression is present, the ancillary role of K-bZIP is no longer needed. This would certainly explain why, in the absence of large amounts of K-Rta, for example, when TPA/n-butyrate was used to induce the lytic cycle, there was a lack of detectable virus production observed in the K-bZIP-negative mutant BACmid. One other scenario is that K-bZIP has a negative effect on initiation of DNA synthesis and “occupies” C/EBPα sites in oriLyt until a high concentration of intranuclear K-Rta displaces it, and subsequently, lytic replication ensues due to both the absence of K-bZIP and the presence of K-Rta interacting with the lytic origin.
Under conditions where lytic replication was induced using TPA/n-butyrate treatment, the BAC36ΔK8 virus failed to replicate or to produce infectious virus. One striking observation was the fact that PAN mRNA accumulation was approximately 25-fold higher in TPA/n-butyrate-induced BAC36ΔK8-containing cells than in untreated cells and 5-fold higher in TPA/n-butyrate-treated BAC36 samples. This is interesting, since in transient assays, K-bZIP was shown to have no influence on the PAN promoter with respect to repression of K-Rta-mediated transactivation. This suggests that, in the context of the viral genome, other factors may contribute to PAN mRNA expression or RNA half-life and K-bZIP may play some role in PAN regulation. Expression of K-bZIP in trans was able to restore PAN mRNA and virus replication in BAC36ΔK8-containing cell lines. This is clear evidence that the nature of the null virus phenotype for BAC36ΔK8 is due to the lack of K-bZIP expression.
BAC36ΔK8 preserved the polyadenylation site at nt 76714, and therefore, transcription of K-Rta and K8.1 was not disrupted. The deletion of the K-bZIP ORF left approximately 300 bp upstream of the K8.1 ORF. Although the promoter region has not been defined for the K8.1 ORF, there is apparently enough regulatory sequence present for adequate expression of this protein. The K8.1 protein is not required for viral replication, and it was postulated that the cis regulatory region controlling expression may be located within the K8 locus (22, 31). Our data here suggest that the cis regulatory region within the context of the viral genome for K8.1 expression is controlled either by the immediate upstream region from the ORF or by sequences that control the expression of ORF50 much further upstream from the K8.1 ORF. Another explanation for increased expression from this ORF could be a disruption of the native regulatory element, which led to an overexpression of K8.1 and the obvious overexpression of K-Rta.
We also investigated the subcellular distribution of K-Rta and LANA in BAC36ΔK8- and BAC36-containing cells. Since it is known that K-Rta interacts with LANA, we assumed that K-bZIP is part of this complex, and LANA was shown to have the ability to down regulate K-Rta-mediated transcriptional activation and the early expression of K-Rta was shown to contribute to the establishment of latency (15, 16). Hence, we were interested in the localization of LANA and K-Rta in cells containing BAC36ΔK8. The subcellular localization of K-Rta was apparently unchanged in BAC36ΔK8 cell lines compared to BAC36-containing cells. However, although only a slight increase in accumulation of LANA mRNA in the BAC36ΔK8 cell line was observed, there was a distinct difference in the subcellular distribution of LANA in the K-bZIP mutant BAC cell line compared to BAC36. This, however, did not translate into any apparent difference or difficulty in the establishment of latency with the mutant BAC. The change in subcellular localization of LANA in BAC36ΔK8 cell lines suggests that K-bZIP plays a role in latency, along with K-Rta. We did observe that K-bZIP was part of a complex with K-Rta and LANA in BCBL-1 cells. We further investigated if K-bZIP could directly interact with LANA by using cotransfection assays. The results of these assays revealed that LANA can directly interact with K-bZIP in the absence of any other viral protein. This is the first indication that K-bZIP is associated with LANA and may contribute to the activity of LANA.
It might at first seem unlikely that a protein that is known to be present during the lytic phase of replication can have an effect on LANA, a latent protein. However, the fact that K-bZIP was shown to be a component of the virion suggests that this lytic protein is present during initial and later stages of infection (2). Also, it was reported that the initial stages of virus infection appear to be lytic, with the expression of many lytic genes, including K-bZIP, followed by the subsequent establishment of latency (14, 16). This may also be a case where the initial stages following BAC transfection result in a short lytic replication cycle followed by a latent infection. It should be noted that neither K-Rta nor K-bZIP was shown to be expressed or present in our latently infected cell lines.
Another plausible explanation as to how a lytic protein can affect the distribution of LANA is that the initial expression of K-bZIP early in infection (or in this case BACmid transfection) may influence the subcellular localization and activity of LANA. Therefore, the localization of LANA can be impacted by the expression of K-bZIP at very early times after virus infections, as well as, in our BACmid-harboring cell lines, eliminating the need for continual expression during the latent phase. The fact that K-bZIP interacts with LANA allows the possibility that K-bZIP could modify or “target” LANA within the cell. The exact mechanism by which K-bZIP influence on the subcellular localization of LANA is disrupted in cells containing the mutant BAC is unknown and warrants further investigation of early events in the virus lytic cycle.
This is the first report to describe the function of K-bZIP in the context of the viral genome. K-bZIP is intriguing, since it appears to be a multifunctional protein but with no clear enzymatic or transactivation activity. The viral mutant described here, BAC36ΔK8, demonstrates that K-bZIP is dispensable under certain conditions and suggests that its role may be more regulatory with respect to modulating the activity of K-Rta. It appears that K-Rta is the elusive origin binding protein in HHV8 and K-bZIP is needed to facilitate this role. These results also emphasize the importance of evaluating replication-associated viral factors in the context of the viral genome, since transient assays like the cotransfection replication assay, powerful as they may be, should be used as a springboard to studies using whole virus in a cellular environment.
Acknowledgments
This work was supported by Public Health Service grant R01 CA085164.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318:542-555. [DOI] [PubMed] [Google Scholar]
- 2.Bechtel, J. T., R. C. Winant, and D. Ganem. 2005. Host and viral proteins in the virion of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, P. J., D. Shedd, and G. Miller. 2005. Two subclasses of Kaposi's sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J. Virol. 79:8750-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colletti, K. S., Y. Xu, S. A. Cei, M. Tarrant, and G. S. Pari. 2004. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 78:9203-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 6.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fixmann, E. D., G. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han, Z., and S. Swaminathan. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 80:5251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu, T. Y., Y. Chang, P. W. Wang, M. Y. Liu, M. R. Chen, J. Y. Chen, and C. H. Tsai. 2005. Reactivation of Epstein-Barr virus can be triggered by an Rta protein mutated at the nuclear localization signal. J. Gen. Virol. 86:317-322. [DOI] [PubMed] [Google Scholar]
- 10.Izumiya, Y., T. J. Ellison, E. T. Yeh, J. U. Jung, P. A. Luciw, and H. J. Kung. 2005. Kaposi's sarcoma-associated herpesvirus K-bZIP represses gene transcription via SUMO modification. J. Virol. 79:9912-9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izumiya, Y., S. F. Lin, T. J. Ellison, A. M. Levy, G. L. Mayeur, C. Izumiya, and H. J. Kung. 2003. Cell cycle regulation by Kaposi's sarcoma-associated herpesvirus K-bZIP: direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J. Virol. 77:9652-9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katano, H., K. Ogawa-Goto, H. Hasegawa, T. Kurata, and T. Sata. 2001. Human-herpesvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology 286:446-455. [DOI] [PubMed] [Google Scholar]
- 14.Krishnan, H. H., P. P. Naranatt, M. S. Smith, L. Zeng, C. Bloomer, and B. Chandran. 2004. Concurrent expression of latent and a limited number of lytic genes with immune modulation and antiapoptotic function by Kaposi's sarcoma-associated herpesvirus early during infection of primary endothelial and fibroblast cells and subsequent decline of lytic gene expression. J. Virol. 78:3601-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan, K., D. A. Kuppers, S. C. Verma, N. Sharma, M. Murakami, and E. S. Robertson. 2005. Induction of Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen by the lytic transactivator RTA: a novel mechanism for establishment of latency. J. Virol. 79:7453-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang, D., A. Birkmann, F. Neipel, W. Hinderer, M. Rothe, M. Ernst, and H. H. Sonneborn. 2000. Generation of monoclonal antibodies directed against the immunogenic glycoprotein K8.1 of human herpesvirus 8. Hybridoma 19:287-295. [DOI] [PubMed] [Google Scholar]
- 18.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luna, R. E., F. Zhou, A. Baghian, V. Chouljenko, B. Forghani, S. J. Gao, and K. G. Kousoulas. 2004. Kaposi's sarcoma-associated herpesvirus glycoprotein K8.1 is dispensable for virus entry. J. Virol. 78:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majerciak, V., N. Pripuzova, J. P. McCoy, S. J. Gao, and Z. M. Zheng. 2007. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8α, and K8.1 and the production of infectious virus. J. Virol. 81:1062-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik, P., D. J. Blackbourn, M. F. Cheng, G. S. Hayward, and J. B. Clements. 2004. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 85:2155-2166. [DOI] [PubMed] [Google Scholar]
- 25.Malik, P., and E. C. Schirmer. 2006. The Kaposi's sarcoma-associated herpesvirus ORF57 protein: a pleurotropic regulator of gene expression. Biochem. Soc. Trans. 34:705-710. [DOI] [PubMed] [Google Scholar]
- 26.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raab, M. S., J. C. Albrecht, A. Birkmann, S. Yaguboglu, D. Lang, B. Fleckenstein, and F. Neipel. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J. Virol. 72:6725-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 30.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang, S., and Z. M. Zheng. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 277:14547-14556. [DOI] [PubMed] [Google Scholar]
- 32.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 33.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, Y., H. Li, M. Y. Chan, F. X. Zhu, D. M. Lukac, and Y. Yuan. 2004. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: cis-acting requirements for replication and ori-Lyt-associated RNA transcription. J. Virol. 78:8615-8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y., Q. Tang, G. G. Maul, and Y. Yuan. 2006. Kaposi's sarcoma-associated herpesvirus ori-Lyt-dependent DNA replication: dual role of replication and transcription activator. J. Virol. 80:12171-12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Y., and Y. Yuan. 2007. Essential role of RBP-Jκ in activation of the K8 delayed-early promoter of Kaposi's sarcoma-associated herpesvirus by ORF50/RTA. Virology 359:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 79:3479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu, Y., A. Rodriguez-Huete, and G. S. Pari. 2006. Evaluation of the lytic origins of replication of Kaposi's sarcoma-associated virus/human herpesvirus 8 in the context of the viral genome. J. Virol. 80:9905-9909. [DOI] [PMC free article] [PubMed] [Google Scholar]