Abstract
Expression of a high-risk human papillomavirus (HPV) E7 oncoprotein is sufficient to induce aberrant centrosome duplication in primary human cells. The resulting centrosome-associated mitotic abnormalities have been linked to the development of aneuploidy. HPV type 16 (HPV16) E7 induces supernumerary centrosomes through a mechanism that is at least in part independent of the inactivation of the retinoblastoma tumor suppressor pRb and is dependent on cyclin-dependent kinase 2 activity. Here, we show that HPV16 E7 can concentrate around mitotic spindle poles and that a small pool of HPV16 E7 is associated with centrosome fractions isolated by sucrose density gradient centrifugation. The targeting of HPV16 E7 to the centrosome, however, was not sufficient for centrosome overduplication. Nonetheless, we found that HPV16 E7 can associate with the centrosomal regulator γ-tubulin and that the recruitment of γ-tubulin to the centrosome is altered in HPV16 E7-expressing cells. Since the association of HPV16 E7 with γ-tubulin is independent of pRb, p107, and p130, our results suggest that the association with γ-tubulin contributes to the pRb/p107/p130-independent ability of HPV16 E7 to subvert centrosome homeostasis.
Human papillomaviruses (HPVs) are small epitheliotrophic viruses that contain an approximately 8-kb double-stranded, closed circular DNA genome. Whereas there are more than 200 types of HPV, only a fraction of these types infect mucosal surfaces. These specific viruses are designated either “low risk,” if they cause benign warts, or “high risk,” if they cause premalignant lesions that have a propensity to progress towards malignancy. High-risk HPVs are associated with greater than 99% of cervical carcinomas and have also been associated with other anogenital tract carcinomas as well as oral carcinomas (reviewed in reference 47). During carcinogenic progression, the viral genome frequently integrates into a host cell chromosome, resulting in the cessation of the viral life cycle. Integration, however, gives rise to the persistent expression of viral E6 and E7 oncoproteins, and the deregulated expression of these proteins is necessary for the initiation and maintenance of the transformed phenotype (reviewed in reference 33). High-risk HPV E6 and E7 proteins interfere with key regulatory pathways, namely, the p53 and the retinoblastoma (pRb) tumor suppressor pathways, respectively (reviewed in reference 33). In order for an HPV-associated premalignant lesion to progress towards malignancy, however, further mutations in the host genome are required, and thus, malignant progression is a rather slow and altogether rare event. Consistent with a requirement for host cellular mutations, hallmarks of genomic instability are readily detected in HPV-immortalized cell lines and high-risk HPV-associated cervical neoplasias. High-risk HPV E6 and E7 oncoproteins can each independently disrupt genomic integrity (42). In contrast, low-risk HPV E6/E7 proteins do not subvert genomic integrity. Hence, the capacity of HPV E6/E7 proteins to induce genomic instability is considered key to the propensity of high-risk HPV-associated lesions to undergo malignant progression (reviewed in reference 11).
A common precursory step to the development of aneuploidy is the presence of supernumerary centrosomes (reviewed in references 3 and 35). Centrosomes are cellular organelles that normally duplicate exactly once per cell cycle and are responsible for the formation of a bipolar mitosis and, thus, equal chromosome segregation during cell division (reviewed in reference 22). Supernumerary centrosomes can result in the formation of multiple mitotic spindle poles and therefore contribute to the unequal segregation of chromosomes to daughter cells. Both numerical and structural centrosome abnormalities are readily detected in high-risk HPV-associated lesions (9), and in fact, multipolar mitoses are a diagnostic hallmark of high-risk HPV-expressing cervical lesions (4). The high-risk HPV E7 oncoprotein is able to uncouple centrosome duplication from the cell division cycle, and cells expressing HPV16 E7 display centrosome overduplication (7, 9, 17). HPV16 E7-induced supernumerary centrosomes require the deregulation of cyclin-dependent kinase 2 (cdk2) activity, linking this function of HPV type 16 (HPV16) E7 to its ability to degrade pRb/p107/p130 and the consequential disruption of the cell cycle (6, 8). Nevertheless, the induction of centrosome overduplication by HPV16 E7 is at least in part independent of HPV16 E7-mediated pRb degradation, as the expression of HPV16 E7 increases the incidence of supernumerary centrosomes in pRb/p107/p130−/− mouse embryo fibroblasts, cells that inherently mimic the degradation of the pRb family proteins by high-risk E7 (10). Interestingly, the pRb/p107/p130 binding-deficient HPV16 E7 Δ21-24 mutant that is unable to induce supernumerary centrosomes in normal cells (9) is still unable to do so in pRb/p107/p130−/− mouse embryo fibroblasts, suggesting that these sequences also contribute to the pRb/p107/p130-independent induction of centrosome abnormalities. Although it is clear that there are distinct mechanisms that contribute to high-risk E7-mediated supernumerary centrosomes, the molecular mechanisms by which the HPV16 E7 oncoprotein is able to induce centrosome overduplication remain elusive.
While the complex, dynamic architecture of the centrosome is still being investigated, various proteins have been identified as being centrosomal components that are critical for the normal function of the centrosome. Briefly, a centrosome contains a pair of centrioles surrounded by a pericentriolar matrix that contains numerous protein complexes, most notably the γ-tubulin ring complexes (γTuRCs) (reviewed in reference 38). γTuRCs are made up of γ-tubulin and associated proteins, and each “ring” is responsible for nucleating a single microtubule (31, 43). Whereas the role of γ-tubulin in γTuRCs has been studied extensively, modes of regulation as well as other functions of γ-tubulin have not been fully delineated. Up to 80% of the cellular pool of γ-tubulin is cytoplasmic, and there is a dynamic exchange with the centrosomal γ-tubulin pool (25, 32). Furthermore, γ-tubulin is recruited to the centrosome at different rates depending upon the cell cycle, with increased centrosomal association beginning at the onset of mitosis (25). Higher concentrations of centrosomal γ-tubulin are likely related to the increased capacity of centrosomes to nucleate microtubules during mitosis and to the organization of the mitotic spindle (reviewed in reference 43). The molecular mechanisms that regulate the fluctuation in γ-tubulin recruitment, as well as the biological significances, remain unclear but may involve the NEDD1 protein, as the depletion of this centrosomal protein interfered with the centrosomal targeting of γTuRCs and ultimately resulted in the inhibition of centriole duplication (18). More recently, it was reported that in mammary epithelial cells, the BRCA1 tumor suppressor protein regulates centrosome homeostasis as well as microtubule nucleation at centrosomes through the monoubiquitination of γ-tubulin (40, 41). The inhibition of BRCA1 function results in a rapid increase in centrosome numbers, suggesting that γ-tubulin also plays a key role in the regulation of centrosome duplication.
Since many regulators of centrosome duplication, such as cdk2, Aurora-A, mps1, NDR1, Id1, antizyme, and BRCA1, associate transiently and partially with centrosomes (12, 19, 21, 26, 27, 36, 46), and given the ability of the HPV16 E7 oncoprotein to induce aberrant centriole synthesis, we evaluated the subcellular localization of HPV16 E7. We found that a subpopulation of HPV16 E7 can concentrate around mitotic spindle poles and cofractionate with centrosome components upon sucrose density gradient centrifugation. Specific targeting of HPV16 E7 to centrosomes, however, was not sufficient for the induction of supernumerary centrosomes. Utilizing a recently described method to detect protein-protein interactions in vivo, we have discovered an association between HPV16 E7 and γ-tubulin that requires amino acid residues 21 to 24 of E7 but is independent of its ability to associate with pRb/p107/p130. We also tested whether HPV16 E7 expression alters the recruitment of γ-tubulin to the centrosome and used photobleaching experiments to show that γ-tubulin recovers at the centrosome more slowly in HPV16 E7-expressing cells. Thus, our results suggest that the expression of HPV16 E7 alters centrosome dynamics through a pRb/p107/p130-independent mechanism by interfering with the critical centrosome component γ-tubulin.
MATERIALS AND METHODS
Plasmids.
To construct the HPV16 E7/mCherry/μNS(471-721) fusions, forward primers containing an NheI site and a Kozak consensus sequence (5′-GCTAGGCTAGCCACCATGCATGGAGATACA-3′) and reverse primers containing an EcoRI site (5′-GGTACCGAATTCTGGTTTCTGAGAACAGATGGG-3′) were used to PCR amplify the E7 gene from the previously described vector CMV-E7, Δ21-24, C24G, or E26G (10, 16). The PCR products were restriction digested with NheI and EcoRI and ligated using T4 DNA ligase into NheI/EcoRI-cut, calf-intestinal alkaline phosphatase-treated pci-mCherry/μNS(471-721) (29). The inserts were sequenced and encode a fusion protein consisting of the HPV16 E7 sequence, followed by a linker sequence (EFTG), mCherry, and amino acid residues 471 to 721 of the reovirus μNS protein. The same process was used to construct the HPV6 E7/mCherry/μNS(471-721) fusion using forward primer 5′-GCT AGG CTA GCC ACC ATG CAT GGA AGA CAT-3′ and reverse primer 5′-GGT ACC GAA TTC GGT CTT CGG TGC GCA GAT-3′ for PCR amplification of the E7 gene.
For the construction of HPV16 E7 fused to the centrosome-targeting sequence of AKAP450 (15), we first PCR amplified a cDNA fragment corresponding to AKAP450 amino acid residues 3644 to 3808 from double-stranded PCR-ready HeLa cDNA (Quick-Clone; Clontech) using a forward primer encoding a BamHI site (5′-GCAGGATCCAACATTGAAGCCATCATTGCCTCTGAA-3′) and a reverse primer encoding a BglII site (5′-GCAAGATCTCTATGCACCTTGATTCAGTCCAAAGCCATC-3′). The amplified fragment was digested with BamHI and BglII and inserted into the BamHI site of plasmid pCMV-BamHI-neo (2) to generate pCMV-AKAP. In a second step, we cloned wild-type HPV16 E7 and the HPV16 E7 Δ21-24 mutant by digesting a PCR product, produced using forward primer 5′-CGAGGATCCACCATGCATGGAGATACACCTACA-3′ and reverse primer 5′-GCAGGATCCACCAGCCGCTGGTTTCTGAGAACAGAT-3′, with BamHI and ligating this fragment into BamHI-digested pCMV-AKAP. The inserts were fully sequenced and encode a 268-amino-acid fusion protein consisting of the full-length HPV16 E7 sequence followed by a linker sequence (AAGGS) and 165 amino acids corresponding to the AKAP450 centrosome-targeting sequence.
Other plasmids used in these studies were pcDNA3-γ-tubulin-GFP (25), a generous gift from Alexey Khodjakov, Wadsworth Center, Albany NY; pBudCE4.1 (Invitrogen); and poz-CE7 and poz-CE7Δ21-24 (23).
Cells.
NIH 3T3 mouse embryo fibroblasts, U2OS human osteosarcoma cells, and HPV-positive cervical carcinoma CaSki cells were obtained from the ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% calf serum, penicillin (50units/ml), and streptomycin (50 μg/ml). pRb/p107/p130−/− mouse embryo fibroblasts were previously described (5) and were maintained in DMEM supplemented with 10% fetal bovine serum, penicillin (50units/ml), and streptomycin (50 μg/ml).
NIH 3T3 cells with stable expression of either empty vector, poz-CE7, or poz-CE7Δ21-24 (23) were made via cotransfections with pBudCE4.1 (Invitrogen) at a 5:1 ratio after selection with 1 mg/ml zeocin (Invitrogen). Transfections were performed with FuGENE6 reagent (Roche Applied Science) according to the manufacturer's protocol. U2OS clones with stable expression of either empty vector or pCMV-E7 were described previously (9), as were U2OS cells with the stable expression of pcDNA3-cE7, which contains C-terminally Flag-tagged HPV16 E7 (16).
Immunofluorescence.
For immunofluorescence, cells were plated onto coverslips and fixed either with 4% paraformaldehyde in phosphate-buffered saline (PBS) or with methanol. For centrosomal staining, cells were extracted with cytoskeleton buffer [100 mM NaCl, 300 mM sucrose, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 3 mM MgCl2] prior to fixation, when indicated. Cells were blocked with 10% serum and 0.1% cold-water fish skin gelatin (Sigma) and permeabilized with 0.5% NP-40 in PBS. Primary antibodies used in these studies were anti-centrin (20H5; a generous gift from Jeffrey Salisbury, Mayo Clinic, Rochester, MN) (37), anti-E7 (ED19; generated by our laboratory), anti-ninein (ab4447; Abcam), anti-p107 (C-18; Santa Cruz Biotechnology), anti-pRb (G3-245; BD Pharmingen), anti-α-tubulin (11H10; Cell Signaling Technology), anti-β-tubulin (5H1; BD Pharmingen), and anti-γ-tubulin (ab11317 [Abcam] and H-183 [Santa Cruz Biotechnology]). Blocking solution and antibodies were diluted in PBS plus 1% (wt/vol) bovine serum albumin, 0.02% (wt/vol) saponin, and 0.05% (wt/vol) sodium azide. Secondary antibodies were goat Alexa Fluor 488 and Alexa Fluor 568 (Invitrogen). Nuclei were counterstained with Hoechst 33258 dye.
For experiments with μNS fusion proteins, U2OS cells were transfected with the corresponding μNS vector using FuGENE 6 (Roche Applied Science) 20 to 24 h prior to fixation. For transient transfection assays to assess centrosome overduplication, cells were transfected with the plasmid of interest together with a mitochondrial DsRED expression plasmid at a 10:1 ratio to specifically assess the percentage of transfected cells with abnormal centrosome numbers. After 48 h, cells were fixed and stained for γ-tubulin or centrin.
Immunofluorescence images were taken with either a laser scanning Zeiss LSM 510 microscope equipped with a photomultiplier tube or a laser scanning Zeiss Axioskop PCM2000.
Isolation of centrosomes and Western blotting.
To isolate centrosomes, we used sucrose density gradients as previously described (30). Briefly, cells were treated with 10 μg/ml nocodazole and 5 μg/ml cytochalasin B (Sigma) for 90 min before lysis in a buffer containing 1 mM Tris-HCl (pH 8.0), 0.5 mM MgCl2, 0.5% NP-40, 0.1% β-mercaptoethanol, and protease inhibitors (Complete EDTA-free tablets; Roche Diagnostics). Nuclei were pelleted, and the supernatant was layered onto a discontinuous sucrose gradient (40%, 50%, and 70% sucrose in buffer containing 10 mM PIPES, 1 mM EDTA, 10% Triton X-100, and 0.1% β-mercaptoethanol); some supernatant was kept to serve as a control for cytoplasmic lysates. The gradient was centrifuged in a Beckman L8-M ultracentrifuge using an SW28 rotor at 125,000 × g for 1 h. The fractions were collected from the bottoms of the tubes as 1-ml fractions (first 10 fractions) and then as approximately 2-ml fractions (fractions 11 to 25). To prepare nuclear extracts, pelleted nuclei were lysed using a buffer containing 0.01 M HEPES (pH 7.9), 15% glycerol, 0.4 M NaCl, 1.5 mM MgCl2, and 0.2 μM EDTA.
The fractions collected from the sucrose density gradients were analyzed by Western blotting. Fractions were run on sodium dodecyl sulfate-12% polyacrylamide gels next to 100-μg samples of cytoplasmic and nuclear extracts. After electrophoresis, proteins were electrotransferred onto a polyvinylidene difluoride membrane (Millipore). The membrane was probed with anti-Cdk2 (M2; Santa Cruz), anti-E7 (ED19), anti-Sp1 (07-124; Upstate Cell Signaling Solutions), and anti-γ-tubulin (GTU88; Sigma) antibodies. Secondary antibodies used were horseradish peroxidase-linked anti-mouse or anti-rabbit immunoglobulin G (GE Healthcare). Proteins were visualized using Western Lightning Chemiluminescence Reagent Plus (Perkin-Elmer Life Sciences, Inc.) or SuperSignal West Femto Maximum Sensitivity substrate (Pierce Biotechnology, Inc.) and exposed on Kodak BioMax XAR film or electronically acquired with a Kodak Image Station 4000R equipped with Kodak Imaging software, version 4.0.
FRAP.
For fluorescence recovery after photobleaching (FRAP) experiments, cells were transfected with pcDNA3-γ-tubulin-GFP, and prior to the experiment, culture medium was replaced with non-phenol-red-containing DMEM plus 25 mM HEPES (Invitrogen) supplemented with 10% calf serum. Centrosomes were photobleached using the MicroPoint laser system (Photonic Instruments), and images were taken using a Nikon TE2000U inverted microscope equipped with a spinning-disk confocal laser (Ultraview; Perkin-Elmer); a 100×, 1.4-numerical-perture Plan Apo objective (Nikon); and an Orca ER cooled charge-coupled-device camera (Hamamatsu). The coverslips were placed in a heated chamber set at 37°C for the duration of the experiments. Briefly, the pulse frequency was set between 40 and 50, and five pulses were used such that ∼80% of the green fluorescent protein (GFP) signal was lost after bleaching. For each individual experiment, images were captured in 5-s increments for the first 65 s (photobleaching occurred immediately prior to the third image capture) and then at 10-s intervals for the next 100 s and, lastly, at 1-min intervals for 5 to 10 min. All measurements were recorded using Metamorph (Molecular Devices), and intensity data were exported into Microsoft Excel. Background intensity was subtracted from the intensity values of the photobleached centrosome and corrected for overall photobleaching by dividing by the average intensity of a defined area within the same cell (that had reasonable distance from the photobleached centrosome). To determine the half-maximal fluorescence recovery (τ1/2) for the individual FRAP experiments, the data were curve fitted to the equation f(t) = A(1 − e−τt) using MATLAB software, where τ1/2 = ln0.5/(−τ), A is the plateau level of fluorescence recovery, and t is time.
RESULTS
HPV16 E7 can concentrate near mitotic spindle poles and cofractionate with centrosomal components.
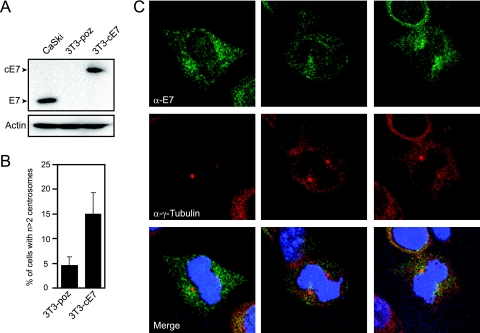
Many proteins that regulate centrosome duplication localize at least partially and transiently to centrosomes. Since HPV16 E7 induces centrosomal overduplication, we performed immunofluorescence experiments to examine the subcellular localization of E7 in comparison to centrosomal markers. We generated NIH 3T3 cells stably transfected with either empty vector (named 3T3-poz) or a C-terminally hemagglutinin (HA)/FLAG epitope-tagged HPV16 E7 (3T3-cE7) in order to examine specifically matched cell lines, which is not feasible when HPV-positive cervical cancer cells are used. The 3T3-cE7 cells express HPV16 E7 at levels similar to those of the HPV16-positive CaSki cervical cancer cell line (Fig. 1A). Furthermore, HPV16 E7 expression induces centrosome overduplication by about threefold (4.6% to 15.0%) in these noncancer cells (Fig. 1B), which is slightly higher than the approximately twofold increase previously described for U2OS osteosarcoma cells (10). This is likely due to the fact that U2OS cancer cells already contain an intrinsically higher level of supernumerary centrosomes, which may dampen the observed effect of E7 on centrosome overduplication in these cells. Immunofluorescence in stable E7-expressing NIH 3T3 cells revealed punctate and mostly cytoplasmic E7 staining during interphase, as has been detected in multiple HPV16 E7-expressing cell lines (23; our unpublished data). Nevertheless, in approximately 15% of mitotic cells, while the punctate staining still predominated, the HPV16 E7 signal was concentrated around the mitotic spindle poles (Fig. 1C). This quantification of 15% reflects a conservative estimation but may also indicate that E7 localization to mitotic spindle poles is confined to a specific phase of mitosis. Similar staining experiments in 3T3-poz control cells resulted in no signal (data not shown). To examine proteins that tightly associate with subcellular structures such as the cytoskeleton and/or centrosome, cells are often extracted prior to immunofluorescence to remove unbound pools of proteins that may otherwise obscure specific staining of such structures. We extracted the HPV16 E7-expressing cells to determine if we could detect E7 specifically associated with centrosome structures, but such treatment resulted in the complete loss of the E7 signal (data not shown).
FIG. 1.
HPV16 E7 can concentrate around mitotic spindle poles. (A) Western blot analysis of E7 expression levels in stable NIH 3T3, 3T3-poz (control), and 3T3-cE7 cells compared to HPV-positive CaSki cervical cancer cells. Actin is shown as a loading control. (B) Percentage of 3T3-cE7 cells with more than two centrosomes compared to control 3T3-poz cells. Bar graphs indicate averages of four experiments in which >150 cells were counted per experiment. Error bars indicate the standard errors between experiments. (C) Representative images of mitotic NIH 3T3 cells with stable expression of C-terminally HA- and FLAG-tagged HPV16 E7. Coimmunofluorescence was performed using an E7-specific antibody (α-E7) and a γ-tubulin antibody (α-γ-Tubulin) as a centrosomal marker. Nuclei were visualized with Hoechst 33258 DNA dye.
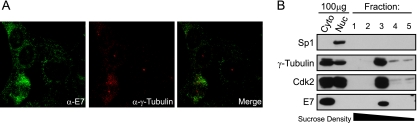
In order to further examine whether there is a centrosome-associated HPV16 E7 pool, we used sucrose density gradients specifically designed for the isolation of centrosomes (30). We utilized HPV16-positive cervical cancer CaSki cells that exhibit the same cytoplasmic distribution of E7 with concentration around mitotic spindle poles in some mitotic cells (Fig. 2A) that we observed in 3T3-cE7 cells (Fig. 1C). CaSki cytoplasmic lysates were layered onto a discontinuous sucrose gradient and centrifuged to separate centrosomes. Analysis of the fractions collected showed that a pool of E7 cofractionates with the centrosomal marker γ-tubulin as well as with cdk2, which was previously shown to partially localize to centrosomes (28) (Fig. 2B). Thus, a small pool of HPV16 E7 is associated with centrosomal material.
FIG. 2.
HPV16 E7 cofractionates with centrosomal components after sucrose density gradient centrifugation. (A) Immunofluorescence analysis of endogenous E7 and γ-tubulin expression in HPV16-positive CaSki cervical carcinoma cells. (B) Detection of HPV16 E7 in centrosome-containing fractions. Cytoplasmic lysates of the HPV16-positive CaSki cervical cancer cell line were subjected to sucrose density gradient centrifugation. Aliquots (150 μl) of individual fractions (fractions 1 to 5 of 25 fractions) along with 100-μg aliquots of total cytoplasmic (Cyto) and nuclear (Nuc) extracts were analyzed by Western blotting. E7 cofractionates with the centrosomal components γ-tubulin and cdk2 in fraction 3. Sp1 is shown as a nuclear marker, and the black bar indicates the direction of sucrose density. Consistent with its cytoplasmic localization, the majority of HPV16 E7 is contained in the later, cytoplasmic fractions (not shown).
Centrosomal targeting of HPV16 E7 is not sufficient for induction of supernumerary centrosomes.
Because HPV16 E7 concentrates around mitotic spindle poles and cofractionates with centrosome components, we investigated whether the localization of HPV16 E7 to centrosomes may be sufficient for the induction of centrosome overduplication. To target E7 directly to the pericentriolar matrix of the centrosome, we fused HPV16 E7 to the pericentrin-AKAP450 centrosomal targeting (PACT) domain of AKAP450 (15). Immunofluorescence analysis showed predominant centrosomal localization of the E7-AKAP protein (Fig. 3A). Transient expression of the E7-AKAP fusion protein in U2OS cells, however, did not result in an increased incidence of supernumerary centrosomes (Fig. 3B). Thus, targeting of HPV16 E7 to centrosomes is not sufficient to cause centrosome overduplication in this assay. The E7-AKAP fusion protein retains the ability to associate with pRb, demonstrating that the fusion does not globally interfere with the structural integrity of HPV16 E7 (data not shown).
FIG. 3.
Targeting HPV16 E7 to the centrosome is not sufficient for centrosome overduplication. (A) Coimmunofluorescence of E7 and γ-tubulin in a U2OS cell transiently transfected with HPV16 E7-AKAP. The merged image includes nuclear staining with Hoechst 33258 DNA dye (blue). (B) Quantitation of cells with abnormal centrosome numbers at 48 h after transfection with either empty pCMV Neo-Bam vector or E7-AKAP. Bar graphs show the means of counts done in triplicate where >200 cells were analyzed per experiment. Error bars indicate standard deviations.
HPV16 E7 associates with γ-tubulin.
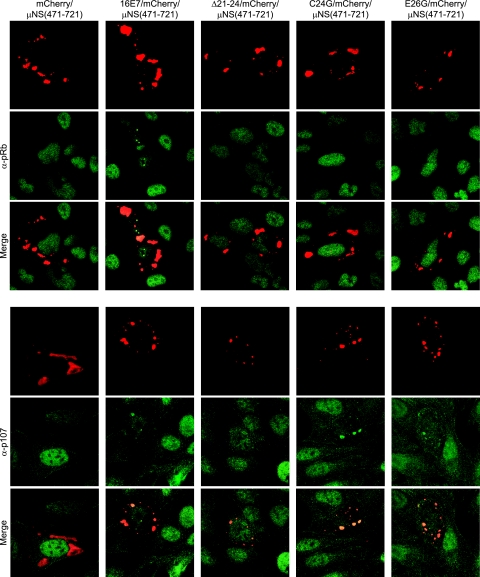
Despite the fact that a centrosome-targeted HPV16 E7 protein was unable to induce supernumerary centrosomes, the detection of HPV16 E7 near mitotic spindle poles and the association with centrosome structures observed by sucrose density centrifugation prompted us to determine if HPV16 E7 can interact with specific centrosomal components, which may account for these observations. Coimmunoprecipitation experiments with several antibodies specific for centrosomal components, such as centrin and γ-tubulin, however, failed to yield reproducible, conclusive results (data not shown), which may reflect a weak or low-level transient association that does not withstand detergent-based cell lysis. Hence, we utilized a recently described method that allows the visualization of protein-protein interactions in living cells (29). This method uses a fusion of reovirus μNS(471-721), the portion of the protein responsible for forming cytoplasmic inclusions during infection, to a fluorescent protein (mCherry) such that the inclusions can be readily visualized upon transfection (29). We then fused full-length HPV16 E7 as well as the Δ21-24, C24G, and E26G mutants to the N terminus of the mCherry/μNS(471-721) protein. The HPV16 E7 Δ21-24 mutant is pRb/p107/p130 binding deficient (34) and is defective in the induction of centrosome abnormalities even in cells that lack pRb/p107/p130 (10). The C24G and E26G mutants are pRb binding deficient but retain associations with p107 and p130 (16) and remain competent for the induction of centrosome abnormalities in cells with functional pRb/p107/p130 (10). To ensure that these various HPV16 E7/mCherry/μNS(471-721) fusion proteins function as expected, we analyzed them in U2OS cells for the relocalization of endogenous pRb and p107. As expected, the fusion of wild-type HPV16 E7, but not fusions of any of the HPV16 E7 mutants, was able to relocalize pRb to cytoplasmic inclusions, whereas fusions with wild-type HPV16 E7 as well as the C24G and E26G mutants, but not the Δ21-24 mutant, relocalized p107 as expected (Fig. 4).
FIG. 4.
HPV16 E7/mCherry/μNS(471-721) relocalizes pRb and p107. Shown is immunofluorescent analysis of U2OS cells expressing the indicated fusion proteins. Cells were transfected 20 to 24 h prior to fixation. The red fluorescent mCherry signal represents the inclusion bodies formed by μNS(471-721). Cells were stained with antibodies against either pRb or p107, shown in green, as indicated. Secondary-only controls showed no positive staining or bleed-through of the mCherry signal (data not shown).
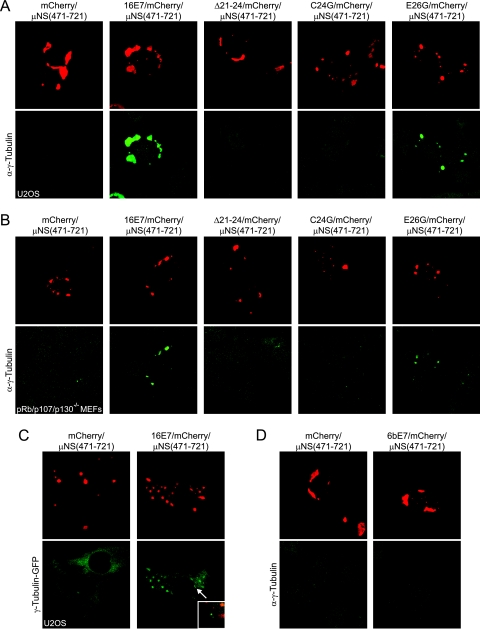
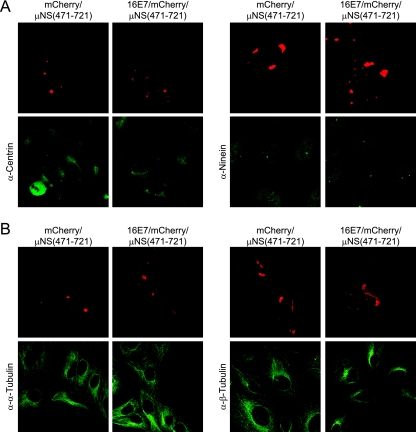
After validating the assay, we investigated whether these E7/mCherry/μNS(471-721) fusion proteins could relocalize centrosomal proteins by performing immunofluorescence experiments with various antibodies specific for known centrosome components. Expression of the 16E7/mCherry/μNS(471-721) fusion in U2OS cells caused a strong relocalization of endogenous γ-tubulin to inclusions in every cell examined (Fig. 5A). Furthermore, the Δ21-24 and C24G mutant fusions have greatly diminished capacities for relocalizing γ-tubulin, whereas the E26G mutant fusion remained competent for γ-tubulin relocalization (Fig. 5A). Given that the HPV16 E7 Δ21-24 mutant is also incompetent for pRb/p107/p130 binding, we wanted to determine whether the observed association of HPV16 E7 and γ-tubulin was dependent on pRb/p107/p130 integrity and similarly transfected triple-knockout pRb/p107/p130−/− mouse embryo fibroblasts (5). As we observed with U2OS cells, an mCherry/μNS(471-721) fusion of wild-type HPV16 E7 and the E26G mutant caused γ-tubulin relocalization, whereas the Δ21-24 and C24G mutants did not (Fig. 5B). We also observed a robust relocalization of γ-tubulin-GFP, demonstrating that the observed effect was not antibody specific (Fig. 5C). Interestingly, γ-tubulin was not relocalized by low-risk HPV6 E7 fused to mCherry/μNS(471-721) (Fig. 5D). Furthermore, we did not observe the relocalization of other centrosomal proteins such as centrin and ninein (Fig. 6A), and moreover, the centrosomal γ-tubulin signal was still observed in cells expressing the 16E7/mCherry/μNS(471-721) fusion protein (Fig. 5C, inset), suggesting that E7 may associate mostly with noncentrosomal γ-tubulin. We also tested whether HPV16 E7 may associate with other tubulin family members, but we did not detect an association of HPV16 E7 with α- or β-tubulin (Fig. 6B). Together, these results show that HPV16 E7 associates specifically with γ-tubulin but not with the other centrosomal proteins centrin and ninein or with other tubulin family members.
FIG. 5.
HPV16 E7/mCherry/μNS(471-721) relocalizes γ-tubulin to inclusions. (A) Immunofluorescent analysis of U2OS cells expressing the indicated fusion proteins as described in the legend of Fig. 4. The cells were stained for γ-tubulin as indicated. (B) As described above (A), pRb/p107/p130−/− cells were transfected with the indicated constructs and stained for γ-tubulin. (C) As described above (A), stable U2OS cells expressing γ-tubulin-GFP were transfected with the indicated constructs. The inset shows centrosomal γ-tubulin fluorescence. (D) As described above (A), U2OS cells were transfected with the indicated constructs and stained for γ-tubulin.
FIG. 6.
Relocalization of γ-tubulin by HPV16 E7/mCherry/μNS(471-721) is specific. (A) Immunofluorescence against the centrosomal proteins centrin (left) and ninein (right) was done on cells transfected with indicated plasmids as described in the legend of Fig. 4A. (B) Same as above (A). Cells were stained for α-tubulin (left) and β-tubulin (right). No relocalization of these proteins was detected.
Association with γ-tubulin may contribute to the pRb/p107/p130-independent ability of HPV16 E7 to induce centrosome overduplication.
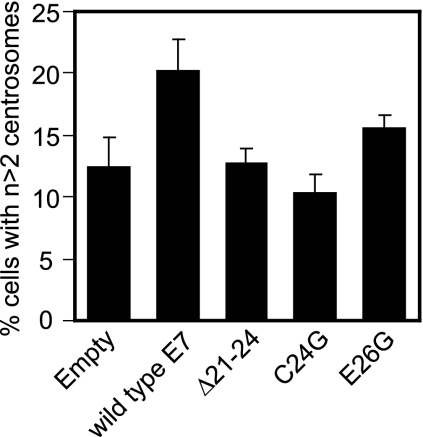
HPV16 E7 expression causes supernumerary centrosomes through both pRb/p107/p130-dependent and -independent mechanisms. Since we found that HPV16 E7 but not the HPV16 E7 Δ21-24 mutant interacts with γ-tubulin in pRb/p107/p130−/− cells, we investigated whether γ-tubulin associations might play a role in E7-mediated centrosome overduplication in a pRb/p107/p130-independent manner. We expected that the HPV16 E7 E26G mutant that retains γ-tubulin binding would be able to induce centrosome abnormalities in pRb/p107/p130−/− cells, whereas the γ-tubulin binding-defective HPV16 E7 C24G mutant would be inactive, similar to what was previously reported for the γ-tubulin binding-defective HPV16 E7 Δ21-24 mutant (10). Upon transient transfection of wild-type HPV16 E7 as well as the Δ21-24, C24G, and E26G mutants into pRb/p107/p130−/− mouse embryo fibroblasts, we found that HPV16 E7 induces an approximately 1.7-fold increase of cells with supernumerary centrosomes, whereas the HPV16 E7 Δ21-24 mutant was unable to do so (Fig. 7), as previously described (10). Consistent with our expectations, the HPV16 E7 C24G mutant, which has greatly diminished γ-tubulin binding, is also unable to cause centrosome overduplication, and the HPV16 E7 E26G mutant that retains γ-tubulin binding increases the percentage of cells with supernumerary centrosomes approximately 1.3-fold over control cells and 1.5-fold over cells transfected with the HPV16 E7 C24G mutant (Fig. 7).
FIG. 7.
Induction of supernumerary centrosomes in pRb/p107/p130-deficient mouse embryo fibroblasts by HPV16 E7. The bar graph shows the average percentages of cells with more than two centrosomes after expression of indicated constructs for 48 h. Counts were done in duplicate, where >100 cells were counted per experiment. Error bars indicate standard errors between experiments.
HPV16 E7 alters γ-tubulin recruitment to centrosomes.
It has been estimated that only 20% of the total cellular γ-tubulin pool is localized at centrosomes, with the remaining 80% localized in the cytoplasm (32). Moreover, γ-tubulin recruitment to the centrosomes is a dynamic process throughout the cell cycle (25). To gain insights into the functional consequences of the interaction between HPV16 E7 and γ-tubulin, we investigated whether E7 could affect the recruitment of cytoplasmic γ-tubulin to centrosomes. To experimentally address this issue, FRAP studies were performed on the above-mentioned stable 3T3-poz and 3T3-cE7 cells containing either empty vector or C-terminal HA/FLAG-tagged full-length HPV16 E7; we also used stable 3T3-Δ21-24 cells expressing the C-terminal HA/FLAG-tagged Δ21-24 mutant to levels similar to those seen in 3T3-cE7 cells (data not shown). The cells were transfected with γ-tubulin-GFP so that a single fluorescent centrosome could be discerned and photobleached (Fig. 8A). Fluorescence intensity of the centrosomal γ-tubulin signal in interphase cells was measured over time, and from this, we were able to extrapolate the amount of time it took to reach half-maximal fluorescence recovery (τ1/2). In vector-expressing cells, the observed average τ1/2 was 18.8 s. This value was increased to 26.1 s in E7-expressing cells (Fig. 8B). Therefore, the recruitment of γ-tubulin to the centrosome is approximately 40% slower in the E7-expressing cell line. Similar results were obtained when clonal populations of U2OS cells with stable expression of full-length, untagged HPV16 E7 were compared to control vector-transfected cells (data not shown). Interestingly, the average τ1/2 in cells expressing the HPV16 E7 Δ21-24 mutant, which showed diminished binding for γ-tubulin, decreased to 22.1 s (Fig. 8B). These results indicate that γ-tubulin dynamics may be altered in HPV16 E7-expressing cells, potentially due to an interaction between γ-tubulin and HPV16 E7.
FIG. 8.
Slower recovery of γ-tubulin to photobleached centrosomes in cells expressing HPV16 E7. (A) Representative images of a single FRAP experiment performed on a stable 3T3-Δ21-24 cell transiently transfected with γ-tubulin-GFP. The elapsed time at which each image was taken is indicated in the bottom right corner of the frame. The white arrow signifies the centrosome that was photobleached in this specific experiment where photobleaching occurred directly prior to the 10-s image. (B) Dot plot of the τ1/2 values calculated for each experiment. The τ1/2 represents the amount of time taken to reach half-maximal recovery in each trial. The averages of each cell line are shown as black bars, and numerical values are indicated.
DISCUSSION
Localization of HPV16 E7 near the centrosome in mitotic cells.
High-risk HPV E7 expression results in supernumerary centrosomes, a necessary step for the formation of multipolar mitoses, which in turn can lead to aneuploidy (3, 35). The ability of HPV16 E7 to induce aberrant centrosome duplication is at least in part independent of pRb/p107/p130 inactivation but requires cdk2 activity (6, 8, 10). In this study, we further investigated the mechanisms utilized by HPV16 E7 that may contribute to pRb/p107/p130-independent centrosome overduplication. As mentioned above, regulators of centrosome duplication often show at least partial centrosomal colocalization. Furthermore, proteins encoded by other viruses, including the Gag proteins of foamy virus, human immunodeficiency virus type 1, Jaagsiekte sheep retrovirus, and Mason-Pfizer monkey virus as well as human T-cell lymphotropic virus type 1 Tax and Epstein-Barr virus thymidine kinase, have also been shown to localize to the centrosome, although the consequences of these interactions have not been fully delineated (1, 14). Hence, we evaluated the subcellular localization of HPV16 E7. In HPV16 E7-expressing cells, we noticed that HPV16 E7 and γ-tubulin display similar punctate cytoplasmic stainings (Fig. 1 and 2) and that in approximately 15% of HPV16 E7-expressing mitotic cells, E7 concentrated around mitotic spindle pole bodies (Fig. 1). Coimmunofluorescence experiments, however, failed to provide conclusive evidence for a precise colocalization of E7 with various centrosome markers. Nevertheless, a fraction of HPV16 E7 cofractionated with centrosome components upon sucrose density gradient centrifugation (Fig. 2). These results suggest that HPV16 E7 may indeed be associated with cellular proteins, such as γ-tubulin, that localize to centrosomes. Our results cannot distinguish whether HPV16 E7 is loosely associated with centrosomes throughout the cell division cycle and/or whether the association may target centrosomal structures transiently during specific phases of cell division.
Generic targeting of HPV16 E7 to the centrosome alone, however, is not sufficient for the induction of supernumerary centrosomes (Fig. 3). Restricting the localization of other proteins that regulate centrosome duplication, such as the NDR1 kinase, to the centrosome has been shown to be sufficient for their regulatory functions (21), suggesting that E7 may disrupt the regulation of centrosome duplication through a distinct mechanism off of the centrosome. Furthermore, this finding is consistent with the notion that HPV16 E7 may preferentially target non-centrosome-associated pools of γ-tubulin. Nonetheless, these results do not rule out the possibility that E7 may also have an activity at the centrosome. Whereas the E7-AKAP fusion effectively localized to centrosomes, the PACT domain targets proteins to the pericentriolar matrix to sites shared with pericentrin (15). Therefore, it is possible that the PACT domain fails to target HPV16 E7 to the appropriate centrosomal structure, where it would productively interfere with specific regulators of centrosome function.
HPV16 E7 associates with γ-tubulin.
Since we observed a pool of E7 cofractionating with centrosome components on sucrose density gradients, we performed various coimmunoprecipitation experiments, which failed to yield compelling evidence for an association of HPV16 E7 with centrosomal proteins. Due to the high concentration of sucrose in centrosome fractions, we were unable to perform coimmunoprecipitation experiments specifically with centrosome-enriched materials. Similarly, E7 coimmunofluorescence experiments with purified centrosomes did not yield any interpretable results (data not shown). Hence, any potential interaction of HPV16 E7 with centrosomes may be transient and/or weak, or specific E7 epitopes may be blocked, which may bar us from detecting centrosomal localization or associations with centrosome components by immunoprecipitations. Therefore, we adapted a newly described method to visualize protein-protein interactions in vivo (29). Using a 16E7/mCherry/μNS(471-721) fusion, we show that HPV16 E7 can associate with γ-tubulin (Fig. 5). The detection of this association is specific, as the 16E7/mCherry/μNS(471-721) fusion was unable to relocalize the centrosomal proteins centrin, ninein, and pericentrin, nor was it able to relocalize α-tubulin and β-tubulin (Fig. 6 and data not shown). Moreover, E7 may preferentially associate with the noncentrosomal γ-tubulin pool, as centrosomal γ-tubulin staining remained intact in cells expressing 16E7/mCherry/μNS(471-721) (Fig. 5). This method does not allow us to distinguish whether E7 directly associates with γ-tubulin or whether an additional cellular protein(s) is necessary to mediate this association.
We also examined whether the expression of HPV16 E7 could alter the recruitment of γ-tubulin to the centrosome. In support of the hypothesis that an E7 association alters γ-tubulin dynamics, we found that γ-tubulin-GFP recovered to photobleached centrosomes at an approximately 40% slower rate in E7-expressing cells than in control cells (Fig. 8). Cells expressing the HPV16 E7 Δ21-24 mutant showed only a slight increase in the amount of time it took for γ-tubulin to recover to the centrosomes and was only approximately 20% slower than in control cells (Fig. 8). This result agrees with our above-mentioned data showing that the HPV16 E7 Δ21-24 mutant shows a greatly diminished, although not completely ablated, ability to bind γ-tubulin. Thus, it seems possible that the observed interaction between HPV16 E7 and γ-tubulin may alter the recruitment of γ-tubulin-GFP to the centrosome. While FRAP experiments do not with certainty demonstrate that the altered recruitment of γ-tubulin directly contributes to HPV16 E7-mediated centrosome overduplication, it is a provocative observation that warrants further study. Additionally, it will be interesting to examine whether the recruitment of other centrosomal regulators to the centrosome is altered upon E7 expression as well.
HPV16 E7 induces supernumerary centrosomes by both pRb-dependent and -independent mechanisms.
From all the published data, it is clear that the ability of HPV16 E7 to induce supernumerary centrosomes represents a congruence of several distinct mechanisms. Most strikingly, cdk2 activity is absolutely required for HPV16 E7 to induce supernumerary centrosomes, whereas it is dispensable for normal centrosome duplication (6, 8). HPV16 E7 can cause aberrant cdk2 activity through several mechanisms including the E2F-mediated activation of cyclin E and cyclin A expression, inactivation of the cdk inhibitors p21cip1 and p27kip1, and, potentially, direct binding to and activation of cdk2 (13, 20, 24, 44, 45). Our previous finding that ectopic expression of a dominant negative DP1 mutant, a heterodimerization partner required for the function of E2F, abrogates the ability of HPV16 E7 to induce supernumerary centrosomes suggests that aberrant E2F activation through the degradation of pRb/p107/p130 by HPV16 E7, which results in ectopic cyclin E and cyclin A expression, may be a critical step (9). However, there are also pRb/p107/p130-independent mechanisms that importantly contribute to the E7-mediated induction of supernumerary centrosomes. Most strikingly, HPV16 E7 can induce supernumerary centrosomes in pRb/p107/p130-deficient mouse embryo fibroblasts, and amino acid residues 21 to 24 are critical for this activity.
Several lines of evidence suggest that the association of HPV16 E7 with γ-tubulin may contribute to the pRb/p107/p130-independent mechanism of induction of supernumerary centrosomes. Our findings that HPV16 E7 associates with the centrosomal regulator γ-tubulin independent of pRb/p107/p130 and that amino acids 21 to 24 are essential for this association suggest that the E7-mediated subversion of γ-tubulin dynamics, in addition to aberrant cdk2 activation, may also contribute to the ability of E7 to induce aberrant centrosome synthesis. Furthermore, the HPV16 E7 C24G mutant that also exhibited diminished binding for γ-tubulin is also defective in the induction of centrosome overduplication in pRb/p107/p130-deficient cells (Fig. 6). We previously showed that the HPV16 E7 C24G mutant is competent for the induction of centrosome overduplication in U2OS cells which contain functional pRb/p107/p130. This is likely due to the fact that this mutant remains competent for p107/p130 degradation (16) and hence induces aberrant E2F activity, cyclin E and cyclin A overexpression, and cdk2 activation (10), which we hypothesize constitutes the major mechanism by which E7 induces supernumerary centrosomes. Thus, if the effects of an association with γ-tubulin contribute to E7-mediated centrosome overduplication to a lesser degree than the pRb/p107/p130- and/or cdk2-dependent mechanism, the lack of supernumerary centrosomes as a result of the failure of the HPV16 E7 C24G mutant to efficiently associate with γ-tubulin would indeed go unnoticed in pRb/p107/p130-expressing cells, as supernumerary centrosomes would result mostly from aberrant cdk2 activation. Nevertheless, since the E2F pathway is constitutively activated in pRb/p107/p130−/− cells, the finding that the γ-tubulin binding-defective HPV16 E7 C24G mutant does not induce additional supernumerary centrosomes in such cells is consistent with our model that the interaction between γ-tubulin and HPV16 E7 may contribute to centrosome overduplication in a pRb/p107/p130-independent manner. Our results showing that the low-risk HPV6 E7 protein, which is incompetent for the induction of centrosome abnormalities (9), does not associate with γ-tubulin (Fig. 5D) are also consistent with this model.
Proposed mechanisms of E7-mediated centrosome overduplication through targeting of γ-tubulin.
We envision two models of how E7 may interfere with γ-tubulin function. HPV16 E7 may target cytoplasmic noncentrosomal γ-tubulin and alter its functions or regulation away from centrosomes. Our finding that γ-tubulin recruitment to centrosomes is decelerated in HPV16 E7-expressing cells is consistent with this model. A second but not mutually exclusive model is that E7 targets specific sites on γ-tubulin and that the functional consequences of this interaction manifest themselves at the centrosome. In other words, centrosomal localization of E7 is indeed important for centrosome overduplication but only via an interaction with centrosomal γ-tubulin. Such an interaction may be weak, transient, and/or confined to a specific phase of the cell division cycle. The fact that HPV16 E7 concentrates around mitotic centrosomes as well as our finding that HPV16 E7 cofractionates with centrosomal components are consistent with both of these options, as centrosomal localization could be required or simply be coincidental with a γ-tubulin interaction. Additional experimentation will be required to clearly distinguish between these two possibilities. Nonetheless, as we have mentioned above, γ-tubulin has been implicated in the regulation of centrosome duplication, and an interaction between E7 and centrosomal and/or cytoplasmic γ-tubulin may interfere with the localization and/or modifications of γ-tubulin, such as monoubiquitination, that are important for its regulatory functions (reviewed in reference 39).
Additionally, an association of HPV16 E7 with γ-tubulin may subvert γ-tubulin activities that are not directly related to the regulation of centrosome duplication. It is interesting to note that γ-tubulin is involved in the nucleation of mitotic microtubules (38), and it remains to be determined whether E7 may affect this activity. Our previous analyses of mitotic abnormalities in HPV oncogene-expressing cells have revealed an increased incidence of lagging chromosomal material (9), a phenotype that may potentially be caused by hypotrophic centrosomes. The possibility that an E7/γ-tubulin interaction may play a role in centrosome overduplication and the possibility that this interaction interferes with nucleation are not mutually exclusive, and the abrogation of BRCA1-mediated γ-tubulin monoubiquitination has been shown to result in the induction of supernumerary centrosomes as well as to interfere with microtubule nucleation activity (40).
Here, we have identified γ-tubulin as being a novel interacting partner of HPV16 E7, and it will be interesting to determine how this association specifically alters various γ-tubulin functions. The viral E7 oncoprotein has proven to be a useful tool for examining normal cellular processes and, furthermore, how these processes are deregulated in cancer cells. Studies on E7-induced supernumerary centrosomes will hopefully shed more light on the complexities of centrosome duplication as well as give us insights into the role that centrosome abnormalities play in carcinogenic progression.
Acknowledgments
This work was supported by PHS grants R01 CA066980 (K.M.) and T32CA009031 (C.L.N.).
We thank A. Khodjakov, K. Kaye, and J. Salisbury for generously sharing reagents; J. Parvin for technical advice with centrosome preparations; J. Levitt for MATLAB programming; F. Wang for critical insights; and M. E. McLaughlin-Drubin and K. F. Bryant for critical comments and suggestions about the manuscript.
This paper is dedicated to the memory of K. Lerch.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Afonso, P. V., A. Zamborlini, A. Saib, and R. Mahieux. 2007. Centrosome and retroviruses: the dangerous liaisons. Retrovirology 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. [DOI] [PubMed] [Google Scholar]
- 3.Brinkley, B. R., and T. M. Goepfert. 1998. Supernumerary centrosomes and cancer: Boveri's hypothesis resurrected. Cell Motil. Cytoskel. 41:281-288. [DOI] [PubMed] [Google Scholar]
- 4.Crum, C. P., H. Ikenberg, R. M. Richart, and L. Gissman. 1984. Human papillomavirus type 16 and early cervical neoplasia. N. Engl. J. Med. 310:880-883. [DOI] [PubMed] [Google Scholar]
- 5.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duensing, A., Y. Liu, M. Tseng, M. Malumbres, M. Barbacid, and S. Duensing. 2006. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene 25:2943-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duensing, S., A. Duensing, C. P. Crum, and K. Munger. 2001. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 61:2356-2360. [PubMed] [Google Scholar]
- 8.Duensing, S., A. Duensing, D. C. Lee, K. M. Edwards, S. O. Piboonniyom, E. Manuel, L. Skaltsounis, L. Meijer, and K. Munger. 2004. Cyclin-dependent kinase inhibitor indirubin-3′-oxime selectively inhibits human papillomavirus type 16 E7-induced numerical centrosome anomalies. Oncogene 23:8206-8215. [DOI] [PubMed] [Google Scholar]
- 9.Duensing, S., L. Y. Lee, A. Duensing, J. Basile, S. Piboonniyom, S. Gonzalez, C. P. Crum, and K. Munger. 2000. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc. Natl. Acad. Sci. USA 97:10002-10007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duensing, S., and K. Munger. 2003. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J. Virol. 77:12331-12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duensing, S., and K. Munger. 2004. Mechanisms of genomic instability in human cancer: insights from studies with human papillomavirus oncoproteins. Int. J. Cancer 109:157-162. [DOI] [PubMed] [Google Scholar]
- 12.Fisk, H. A., C. P. Mattison, and M. Winey. 2003. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc. Natl. Acad. Sci. USA 100:14875-14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill, M. B., J. L. Kutok, and J. D. Fingeroth. 2007. Epstein-Barr virus thymidine kinase is a centrosomal resident precisely localized to the periphery of centrioles. J. Virol. 81:6523-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillingham, A. K., and S. Munro. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Münger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarguaglini, G., P. I. Duncan, Y. D. Stierhof, T. Holmstrom, S. Duensing, and E. A. Nigg. 2005. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol. Biol. Cell 16:1095-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haren, L., M. H. Remy, I. Bazin, I. Callebaut, M. Wright, and A. Merdes. 2006. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172:505-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasskarl, J., S. Duensing, E. Manuel, and K. Munger. 2004. The helix-loop-helix protein ID1 localizes to centrosomes and rapidly induces abnormal centrosome numbers. Oncogene 23:1930-1938. [DOI] [PubMed] [Google Scholar]
- 20.He, W., D. Staples, C. Smith, and C. Fisher. 2003. Direct activation of cyclin-dependent kinase 2 by human papillomavirus E7. J. Virol. 77:10566-10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hergovich, A., S. Lamla, E. A. Nigg, and B. A. Hemmings. 2007. Centrosome-associated NDR kinase regulates centrosome duplication. Mol. Cell 25:625-634. [DOI] [PubMed] [Google Scholar]
- 22.Hinchcliffe, E. H., and G. Sluder. 2001. “It takes two to tango”: understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 15:1167-1181. [DOI] [PubMed] [Google Scholar]
- 23.Huh, K. W., J. DeMasi, H. Ogawa, Y. Nakatani, P. M. Howley, and K. Munger. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. USA 102:11492-11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khodjakov, A., and C. L. Rieder. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lotti, L. V., L. Ottini, C. D'Amico, R. Gradini, A. Cama, F. Belleudi, L. Frati, M. R. Torrisi, and R. Mariani-Costantini. 2002. Subcellular localization of the BRCA1 gene product in mitotic cells. Genes Chromosomes Cancer 35:193-203. [DOI] [PubMed] [Google Scholar]
- 27.Mangold, U., H. Hayakawa, M. Coughlin, K. Munger, and B. R. Zetter. 30 July 2007. Antizyme, a mediator of ubiquitin-independent proteasomal degradation and its inhibitor localize to centrosomes and modulate centriole amplification. Oncogene doi: 10.1038/sj.onc.1210685. [DOI] [PubMed]
- 28.Matsumoto, Y., and J. L. Maller. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 306:885-888. [DOI] [PubMed] [Google Scholar]
- 29.Miller, C. L., M. M. Arnold, T. J. Broering, C. Eichwald, J. Kim, J. B. Dinoso, and M. L. Nibert. 2007. Virus-derived platforms for visualizing protein associations inside cells. Mol. Cell. Proteomics 6:1027-1038. [DOI] [PubMed] [Google Scholar]
- 30.Mitchison, T. J., and M. W. Kirschner. 1986. Isolation of mammalian centrosomes. Methods Enzymol. 134:261-268. [DOI] [PubMed] [Google Scholar]
- 31.Moritz, M., M. B. Braunfeld, J. W. Sedat, B. Alberts, and D. A. Agard. 1995. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature 378:638-640. [DOI] [PubMed] [Google Scholar]
- 32.Moudjou, M., N. Bordes, M. Paintrand, and M. Bornens. 1996. γ-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J. Cell Sci. 109:875-887. [DOI] [PubMed] [Google Scholar]
- 33.Münger, K., A. Baldwin, K. M. Edwards, H. Hayakawa, C. L. Nguyen, M. Owens, M. Grace, and K. Huh. 2004. Mechanisms of human papillomavirus-induced oncogenesis. J. Virol. 78:11451-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Münger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nigg, E. A. 2006. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer 119:2717-2723. [DOI] [PubMed] [Google Scholar]
- 36.Nishimura, T., M. Takahashi, H. S. Kim, H. Mukai, and Y. Ono. 2005. Centrosome-targeting region of CG-NAP causes centrosome amplification by recruiting cyclin E-cdk2 complex. Genes Cells 10:75-86. [DOI] [PubMed] [Google Scholar]
- 37.Paoletti, A., M. Moudjou, M. Paintrand, J. L. Salisbury, and M. Bornens. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109:3089-3102. [DOI] [PubMed] [Google Scholar]
- 38.Preble, A. M., T. M. Giddings, Jr., and S. K. Dutcher. 2000. Basal bodies and centrioles: their function and structure. Curr. Top. Dev. Biol. 49:207-233. [DOI] [PubMed] [Google Scholar]
- 39.Sankaran, S., and J. D. Parvin. 2006. Centrosome function in normal and tumor cells. J. Cell. Biochem. 99:1240-1250. [DOI] [PubMed] [Google Scholar]
- 40.Sankaran, S., L. M. Starita, A. C. Groen, M. J. Ko, and J. D. Parvin. 2005. Centrosomal microtubule nucleation activity is inhibited by BRCA1-dependent ubiquitination. Mol. Cell. Biol. 25:8656-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starita, L. M., Y. Machida, S. Sankaran, J. E. Elias, K. Griffin, B. P. Schlegel, S. P. Gygi, and J. D. Parvin. 2004. BRCA1-dependent ubiquitination of γ-tubulin regulates centrosome number. Mol. Cell. Biol. 24:8457-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White, A. E., E. M. Livanos, and T. D. Tlsty. 1994. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 8:666-677. [DOI] [PubMed] [Google Scholar]
- 43.Wiese, C., and Y. Zheng. 2006. Microtubule nucleation: gamma-tubulin and beyond. J. Cell Sci. 119:4143-4153. [DOI] [PubMed] [Google Scholar]
- 44.Zerfass, K., A. Schulze, D. Spitkovsky, V. Friedman, B. Henglein, and P. Jansen-Durr. 1995. Sequential activation of cyclin E and cyclin A gene expression by human papillomavirus type 16 E7 through sequences necessary for transformation. J. Virol. 69:6389-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zerfass-Thome, K., W. Zwerschke, B. Mannhardt, R. Tindle, J. W. Botz, and P. Jansen-Durr. 1996. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein. Oncogene. 13:2323-2330. [PubMed] [Google Scholar]
- 46.Zhou, H., J. Kuang, L. Zhong, W. L. Kuo, J. W. Gray, A. Sahin, B. R. Brinkley, and S. Sen. 1998. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat. Genet. 20:189-193. [DOI] [PubMed] [Google Scholar]
- 47.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]