Abstract
The Zaire ebolavirus protein VP24 was previously demonstrated to inhibit alpha/beta interferon (IFN-α/β)- and IFN-γ-induced nuclear accumulation of tyrosine-phosphorylated STAT1 (PY-STAT1) and to inhibit IFN-α/β- and IFN-γ-induced gene expression. These properties correlated with the ability of VP24 to interact with the nuclear localization signal receptor for PY-STAT1, karyopherin α1. Here, VP24 is demonstrated to interact not only with overexpressed but also with endogenous karyopherin α1. Mutational analysis demonstrated that VP24 binds within the PY-STAT1 binding region located in the C terminus of karyopherin α1. In addition, VP24 was found to inhibit PY-STAT1 binding to both overexpressed and endogenous karyopherin α1. We assessed the binding of both PY-STAT1 and the VP24 proteins from Zaire, mouse-adapted Zaire, and Reston Ebola viruses for interaction with all six members of the human karyopherin α family. We found, in contrast to previous studies, that PY-STAT1 can interact not only with karyopherin α1 but also with karyopherins α5 and α6, which together comprise the NPI-1 subfamily of karyopherin αs. Similarly, all three VP24s bound and inhibited PY-STAT1 interaction with karyopherins α1, α5, and α6. Consistent with their ability to inhibit the karyopherin-PY-STAT1 interaction, Zaire, mouse-adapted Zaire, and Reston Ebola virus VP24s displayed similar capacities to inhibit IFN-β-induced gene expression in human and mouse cells. These findings suggest that VP24 inhibits interaction of PY-STAT1 with karyopherins α1, α5, or α6 by binding within the PY-STAT1 binding region of the karyopherins and that this function is conserved among the VP24 proteins of different Ebola virus species.
Ebola virus (EBOV) and Marburg virus comprise the family Filoviridae. These viruses cause periodic outbreaks of severe hemorrhagic fever in humans and nonhuman primates, with case fatality rates as high as 90% (26, 36). Currently, no vaccines or antiviral therapies are approved for use against EBOV, although vaccines and additional strategies that protect nonhuman primates from lethal challenge have been described (16, 29, 50, 52).
Understanding of the molecular mechanisms underlying EBOV pathogenesis is incomplete. Several potential mechanisms that contribute to pathogenesis have been reviewed (36). These include viral glycoprotein-mediated cytotoxicity, dysregulation of the coagulation cascade due to the production of tissue factor, and the production of proinflammatory cytokines (10, 16, 17, 51, 54, 57). General immunosuppression also appears to be a characteristic of EBOV infection (7, 46), and inhibition of dendritic cell and macrophage activation are among the possible mechanisms of this suppression (5, 18, 38). These processes likely occur as a result of active viral replication. Therefore, the ability of the virus to evade the early antiviral response, including the interferon (IFN) response, is likely a critical step in pathogenesis. In fact, several studies have demonstrated the importance of the IFN response during EBOV infection. In particular, immunocompetent mice are resistant to lethal disease following infection with Zaire EBOV (ZEBOV) whereas type I IFN receptor or STAT1 knockout mice are susceptible to lethal EBOV infection (6).
The virulence of different EBOVs also appears to correlate with an ability to suppress innate immunity. Within the EBOV genus, ZEBOV appears to be the most pathogenic EBOV species (1, 28) while, in contrast, Reston EBOV (REBOV), which is lethal in nonhuman primates, seems to be attenuated in humans, having caused seroconversion without any apparent signs of illness in the only documented human infections (4, 27). When microarray analyses were performed on EBOV-infected Huh7 cells, REBOV was found to have a reduced ability compared with ZEBOV to inhibit expression of IFN-induced antiviral genes and thus evade the host antiviral response (30). Further evidence that suppression of IFN responses is important for EBOV virulence is the observation that adaptation of ZEBOV from a nonlethal to a lethal infection in mice required changes only in VP24 and nucleoprotein (NP), and these changes were shown to be associated with the ability of the virus to evade the type I IFN system in cultured mouse macrophages (14).
We have identified the EBOV proteins VP35 and VP24 as antagonists of IFN production and signaling, respectively (3, 45). The VP35 protein of EBOV inhibits the IFN-α/β system by blocking activation of IFN regulatory factor 3, a key transcription factor in IFN-α/β production (2, 3, 5, 9, 23, 24, 44), by targeting the RIG-I signaling pathway (9). VP24 inhibits IFN-induced gene expression and blocks nuclear accumulation of tyrosine-phosphorylated STAT1 (PY-STAT1). Consistent with this observed function of VP24, EBOV infection also inhibited IFN-induced gene expression and nuclear accumulation of PY-STAT1 (45). Moreover, VP24 was shown to interact with karyopherin α1 (45), the nuclear import receptor for PY-STAT1 (48).
Movement of molecules (∼50 kDa) into the nucleus in most cases occurs by active transport through the nuclear pore complex (35). In the classical pathway of nuclear import, proteins that contain a nuclear localization signal (NLS) bind to the karyopherin α/β heterodimer (reviewed in reference 49). Karyopherin α acts as an adaptor by binding both the NLS and karyopherin β, while karyopherin β mediates docking of the trimeric complex to the nuclear pore and subsequent translocation into the nucleus (20, 56). In humans, six members of the karyopherin α family have been identified. The family can be further divided into three distinct subfamilies based on sequence similarity (31), the Rch1 subfamily (karyopherin α2) (13, 55), Qip1 subfamily (karyopherin α3 and karyopherin α4) (31, 40, 42, 47), and the NPI-1 subfamily (karyopherin α1, karyophrein α5, and karyopherin α6) (12, 31, 32, 41, 43). Within the NPI-1 subfamily, karyopherin α1 shares greater than 80% sequence similarity with karyopherin α5 and karyopherin α6 (39).
In the present study we demonstrate that VP24 disrupts PY-STAT1-karyopherin α1 binding through an interaction within the region of karyopherin α1 required for PY-STAT1 binding. Binding within the PY-STAT1 binding region of the karyopherin enabled VP24 to disrupt the interaction of either endogenous or overexpressed PY-STAT1 with karyopherin α1. We also demonstrate that VP24 binds all three members of the NPI-1 subfamily of karyopherin α proteins, inhibiting their interaction with PY-STAT1, and we demonstrate that this function is a property common to the VP24 proteins from ZEBOV, mouse-adapted (MA) ZEBOV, and REBOV.
MATERIALS AND METHODS
Cells, plasmids, and antibodies.
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Derivatives of the plasmid pCAGGS were used for all expression studies. The antibodies used include monoclonal M2 anti-FLAG, anti-hemagglutinin (HA), anti-importin α5/7, rabbit polyclonal anti-FLAG and anti-HA (all from Sigma), and rabbit polyclonal PY-STAT1 (pY701; Cell Signaling Technology).
Coimmunoprecipitations and Western blotting.
293T cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as outlined by the manufacturer's protocol. At 1 day posttransfection, cells were harvested and washed once in ice-cold phosphate-buffered saline, lysed in extract buffer (50 mM Tris [pH 7.5], 280 mM NaCl, 0.5% Igepal, 0.2 mM EDTA, 2 mM EGTA, 10% glycerol, 1 mM dithiothreitol, 1 mM sodium vanadate, and protease inhibitors [Complete; Roche]) on ice for 10 min, and then centrifuged at 13,000 rpm for 10 min at 4°C in a microcentrifuge. For experiments with IFN-β treatment, 1 day posttransfection the cells were washed and then maintained in DMEM containing 0.3% bovine serum albumin with or without (mock treated) 1,000 U/ml recombinant human IFN-β (PBL Biomedical Laboratories, Piscataway, NJ) for no less than 30 min prior to being harvested and lysed. The supernatant was collected, and 25 μl of a 50% slurry of M2 (anti-FLAG) monoclonal antibody cross-linked to agarose beads (Sigma) was added and incubated for at least 1 h at 4°C with rotation. The M2 beads were then washed five times with extract buffer. The immunoprecipitated material and whole-cell extracts were boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were then transferred to a polyvinylidene difluoride membrane. The membrane was blocked in 5% milk and 0.1% Tween 20 in Tris-buffered saline and then probed with the aforementioned antibodies. Western blots were developed using a Western Lightning ECL kit (Perkin-Elmer, Boston, MA) and Kodak BioMax film (Kodak, Rochester, NY).
Reporter gene assays.
293T cells were transfected using Lipofectamine 2000 as described in the manufacturer's protocol. Cells (1.2 × 106) in suspension were transfected in six-well plates with 0.5 μg of an IFN-stimulated gene 54 (ISG54) promoter-chloramphenicol acetyltransferase (CAT) reporter construct, 0.1 μg of a constitutively expressing luciferase reporter construct (pCAGGS-luc), and the indicated (in the legend to Fig. 6) amounts of the relevant expression plasmids. Alternatively, an ISG54 promoter-firefly luciferase reporter plasmid was used, and measured firefly luciferase activity was normalized to the activity from a cotransfected, constitutively expressed Renilla luciferase reporter plasmid. Twenty-four hours posttransfection, cells were washed and maintained in DMEM containing 0.3% bovine serum albumin, with or without (mock-treated control) 1,000 U/ml of type I interferon (PBL Biomedical Laboratories, Piscataway, NJ). Sixteen hours post-IFN treatment, cells were harvested using reporter lysis buffer (Promega, Madison, WI) and analyzed for CAT and luciferase activities. The CAT activity was quantified by using a PhosphorImager and normalized to the luciferase activity. Alternatively, dual luciferase assays (Promega) were performed, and firefly luciferase activity was normalized to Renilla luciferase activity.
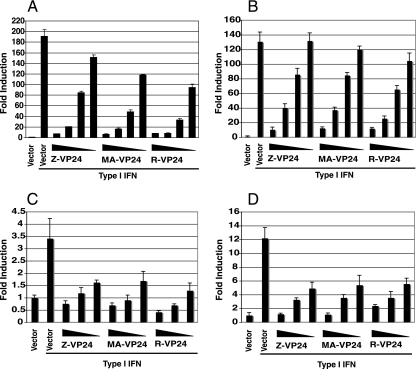
FIG. 6.
EBOV VP24 proteins inhibit reporter gene expression in human and murine cells. 293T (A), HuH7 (B), NIH-3T3 (C), or MEF (D) cells were transfected with the ISG54-CAT reporter plasmid, a constitutively expressed Renilla luciferase reporter plasmid, and either empty vector (vector) or the indicated plasmids expressing various concentrations (a, 500, 100, 20, and 4 ng; b, 500, 100, 20, and 4 ng; c, 500, 100, and 20 ng; and d, 500, 100, and 20 ng) of ZEBOV VP24 (Z-VP24), MA ZEBOV VP24 (MA-VP24), or REBOV VP24 (R-VP24). At 1 day posttransfection the cells were either mock treated or treated with 1,000 U/ml of universal type I IFN or human IFN-β (for 293T and HuH7 cells) or murine IFN-β (for NIH-3T3 cells and MEFs). At 16 h posttreatment the cells were harvested and assayed for reporter activities.
RESULTS
VP24 interacts with endogenous karyopherin α1.
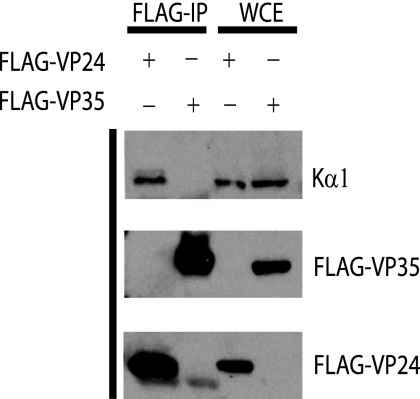
To determine if VP24 is able to interact with endogenous karyopherin α1, cells were transfected with a FLAG-tagged VP24 or VP35. One day posttransfection, immunoprecipitations were performed with an anti-FLAG monoclonal antibody, and the precipitated material was analyzed by Western blotting. Karyopherin α1 was coprecipitated with FLAG-VP24 but not with FLAG-VP35 (Fig. 1). This result indicates that EBOV VP24 is indeed able to interact with endogenous karyopherin α1.
FIG. 1.
HA-VP24 interacts with endogenous karyopherin α1. 293T cells were transfected with either FLAG-VP24 or FLAG-VP35, and 1 day posttransfection immunoprecipitation was performed with the anti-FLAG monoclonal antibody M2 bound to agarose beads. The immunoprecipitated material (FLAG-IP) and whole-cell extract (WCE) were then subjected to Western blot analysis with a monoclonal antibody against karyopherin α1 (Kα1) and an anti-FLAG monoclonal antibody.
VP24 and PY-STAT1 bind to overlapping regions of karyopherin α1.
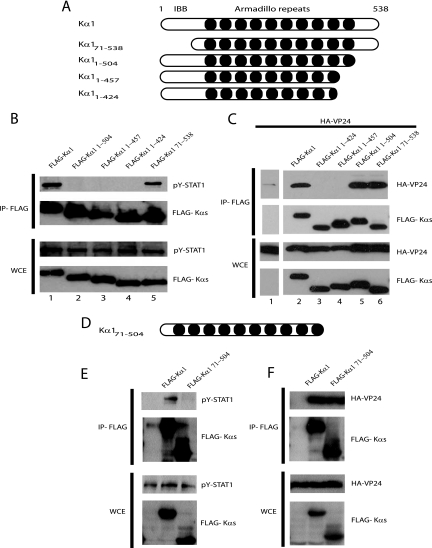
We previously demonstrated that EBOV VP24 binds karyopherin α1 and in doing so disrupts the interaction of an activated STAT1-green fluorescent protein (GFP) fusion protein with karyopherin α1 (45). PY-STAT1 interacts with the C-terminal armadillo (arm) repeats of karyopherin α1 for transport into the nucleus (39, 48); it was therefore of interest to determine if VP24 binds to a similar region of the karyopherin. Several karyopherin α1 truncation mutants were constructed (Fig. 2A) and tested for their ability to interact with PY-STAT1 and an HA-tagged VP24. Full-length FLAG-tagged karyopherin α1 (FLAG-Kα1) and the truncation mutants were transfected alone (Fig. 2B) or along with HA-VP24 (Fig. 2C). At 1 day posttransfection, immunoprecipitations were performed with an anti-FLAG monoclonal antibody, and the precipitated material was analyzed by Western blotting. In the absence of IFN-β treatment, STAT1 did not coprecipitate with FLAG-Kα1 (data not shown); however, upon IFN-β treatment STAT1 is readily coprecipitated with FLAG-Kα1 (Fig. 2B, lane 1). As previously reported (48), deletion of the importin beta domain of FLAG-Kα1 (yielding a karyopherin α1 comprised of residues 71 to 538 [FLAG-Kα171-538]) did not affect the interaction with PY-STAT1; in contrast, the C- terminal deletion mutants were unable to coprecipitate with PY-STAT1 (Fig. 2B). Interestingly, while FLAG-Kα1, FLAG-Kα11-504, and FLAG-Kα171-538 were able to coprecipitate HA-VP24 (Fig. 2C, lanes 2, 5, and 6), FLAG-Kα11-424 and FLAG-Kα11-457 were unable to bind (Fig. 2C, lanes 3 and 4). Based on these data we constructed another karyopherin α1, FLAG-Kα171-504 that lacks both the importin beta binding domain and the last 34 C-terminal amino acids (Fig. 2D). As expected, FLAG-Kα171-504 was unable to coprecipitate with PY-STAT1 (Fig. 2E) but clearly retained the ability to coprecipitate HA-VP24 (Fig. 2F). Thus, while PY-STAT1 requires amino acids 425 to 538 of karyopherin α1 for interaction, VP24 requires the region of karyopherin α1 that encompasses arm repeat 10, amino acids 458 to 504.
FIG. 2.
VP24 and PY-STAT1 bind to overlapping regions at the C terminus of karyopherin α1. (A and D) A schematic illustration of the full-length and truncation mutants of karyopherin α1. IBB, importin beta binding domain. Dark circles, arm repeats. (B and E) Coimmunoprecipitation of full-length and truncation mutants of FLAG-Kα1 with PY-STAT1. 293T cells were transfected with the indicated FLAG-Kα constructs. At 1 day posttransfection the cells were treated with 1,000 U/ml human IFN-β and subsequently immunoprecipitated with an anti-FLAG monoclonal antibody M2 bound to agarose beads. Immunoprecipitates were analyzed by Western blotting with a polyclonal rabbit antiserum recognizing PY-STAT1 and FLAG. (C and F) Coimmunoprecipitation of full-length and truncation mutant FLAG-Kα1 constructs with HA-VP24. 293T cells were transfected with HA-VP24 alone or along with the indicated FLAG-Kα constructs. One day posttransfection immunoprecipitation was performed as described above. Western blot analysis was then performed with polyclonal rabbit antiserum recognizing FLAG and a monoclonal antibody against HA. IP, immunoprecipitation. WCE, whole-cell extract.
Inhibition of the interaction of endogenous and overexpressed PY-STAT1 with karyopherin α1.
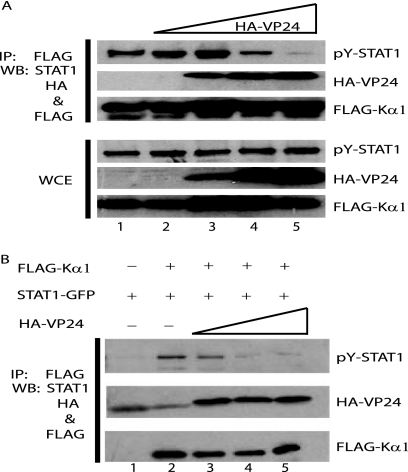
Our previous study demonstrated that VP24 could inhibit the interaction of karyopherin α1 with an overexpressed, activated STAT1-GFP fusion (45). To determine if VP24 can disrupt the interaction of FLAG-Kα1 with endogenous PY-STAT1, cells were transfected with FLAG-Kα1 alone or with increasing concentrations of HA-VP24. At 1 day posttransfection the cells were treated with IFN-β for 1 h, immunoprecipitation was performed, and the precipitated material was subjected to Western blot analysis. FLAG-Kα1 coprecipitated with PY-STAT1 in the absence and in the presence of low concentrations of HA-VP24 (Fig. 3A, lanes 1 to 3). However, as the concentration of HA-VP24 increased, the ability of FLAG-Kα1 to coprecipitate PY-STAT1 was clearly inhibited (Fig. 3A, IP: PY-STAT1), while HA-VP24 coprecipitated with FLAG-Kα1 (Fig. 3A, IP: HA-VP24).
FIG. 3.
Interaction of either endogenous or overexpressed PY-STAT1 with karyopherin α1 is inhibited by VP24. (A) 293T cells were transfected with FLAG-Kα1 either in the absence or the presence of increasing concentrations of HA-VP24. At 1 day posttransfection the cells were treated with 1,000 U/ml IFN-β for 1 h and then immunoprecipitated with anti-FLAG monoclonal antibody M2 bound to agarose beads. Western blot analysis of the immunoprecipitated material (IP) and whole-cell extract (WCE) was performed with a polyclonal rabbit antiserum recognizing PY-STAT1 and HA along with an anti-FLAG monoclonal antibody. (B) VP24 inhibits the interaction of PY-STAT1-GFP with karyopherin α1 in vitro. 293T cells were transfected with FLAG-Kα1, HA-VP24, or STAT1-GFP. One day posttransfection the STAT1-GFP-transfected cells were treated with 1,000 U/ml of IFN-β for 1 h, and cell lysates were subsequently prepared. The cell lysates were mixed as indicated and subjected to immunoprecipitation with the aforementioned anti-FLAG monoclonal antibody. Western blot analysis was performed with a polyclonal rabbit antiserum recognizing PY-STAT1 and anti-HA and FLAG monoclonal antibodies.
Because karyopherin α1 must shuttle into and out of the nucleus, it was possible that VP24 might prevent the interaction of PY-STAT1 with karyopherin α1 by sequestering karyopherin α1 in a subcellular compartment. This would, as a result, prevent access of karyopherin α1 to PY-STAT1. To determine whether VP24 could inhibit a karyopherin α1-PY-STAT1 interaction in vitro and, therefore, inhibit the interaction independently of subcellular localization, cell lysates containing FLAG-Kα1, PY-STAT1-GFP and HA-VP24 were separately prepared. The cell lysates were then combined in vitro, and coimmunoprecipitations were then performed. Cells were transfected with FLAG-Kα1, STAT1-GFP, or HA-VP24. One day posttransfection, the cells containing STAT1-GFP were treated with IFN-β for 1 h. Immunoprecipitation was then performed with an anti-FLAG monoclonal antibody, and the immunoprecipitated material was subjected to Western blot analysis. When mixed with the anti-FLAG antibody alone, PY-STAT1-GFP was not immunoprecipitated; however, when the FLAG-Kα1 lysate was added, PY-STAT1-GFP was coprecipitated (Fig. 3B, compare lanes 1 and 2). When increasing amounts of the HA-VP24 lysate were mixed with the PY-STAT1-GFP and FLAG-Kα1 lysate, the interaction of FLAG-Kα1 with PY-STAT1-GFP was inhibited (Fig. 3B, lane 3 to 5), while HA-VP24 coprecipitated with FLAG-Kα1 (Fig. 3B). Taken together, these data suggest that VP24 is able to disrupt the interaction of karyopherin α1 with endogenous PY-STAT1. Additionally, the in vitro experiments suggest that sequestration of karyopherin α1 in a subcellular compartment is not required for the inhibition to occur.
PY-STAT1 and HA-VP24 interact with karyopherins α1, α5, and α6.
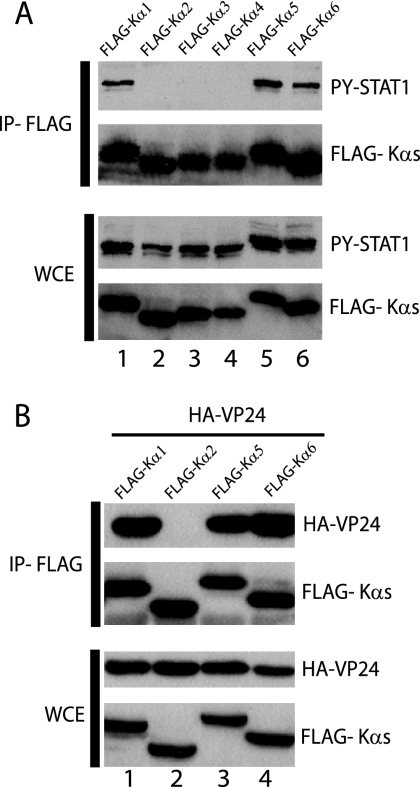
Karyopherins α1, α5, and α6 make up the NPI-1 subfamily of karyopherin α proteins. Because karyopherins α5 and α6 share a greater degree of amino acid identity to karyopherin α1 than does karyopherin α2, karyopherin α3, or karyopherin α4, we evaluated the interaction of both PY-STAT1 and VP24 with karyopherins α5 and α6. Cells were transfected with FLAG-tagged karyopherin α1, α2, α3, α4, α5, and α6 expression plasmids. At 1 day posttransfection the cells were treated with IFN-β for 30 min, and immunoprecipitation was performed. In contrast to previously published reports (39), PY-STAT1 coprecipitated exclusively with FLAG-Kα1, -Kα5, and -Kα6 (Fig. 4A). We next wanted to determine if VP24 was also able to interact with karyopherin α5 and α6. FLAG-Kα1, -Kα2, -Kα5, and -Kα6 were cotransfected with HA-VP24. At 1 day posttransfection the cells were lysed and subsequently immunoprecipitated as previously described. Similar to PY-STAT1, HA-VP24 coprecipitated with FLAG-Kα1, -Kα5, and -Kα6 but not with FLAG-Kα2 (Fig. 4B). Thus, PY-STAT1 and VP24 can interact with all three karyopherins in the NPI-1 subfamily.
FIG. 4.
PY-STAT1 and HA-VP24 interact with all three members of the NPI-1 subfamily of karyopherin α. (A) 293T cells were transfected with FLAG-Kα1, FLAG-Kα2, FLAG-Kα3, FLAG-Kα4, FLAG-Kα5, and FLAG-Kα6. One day posttransfection the cells were treated with 1,000 U/ml of IFN-β for 1 h, and then immunoprecipitation was performed with the anti-FLAG monoclonal antibody M2 bound to agarose. Immunoprecipitated (IP) material and whole-cell extract (WCE) were analyzed by Western blotting with polyclonal rabbit antiserum recognizing PY-STAT1 and FLAG. (B) 293T cells were cotransfected with HA-VP24 and either FLAG-Kα1, FLAG-Kα2, FLAG-Kα5, or FLAG-Kα6. One day posttransfection immunoprecipitation was performed as mentioned above, and Western blot analysis was performed with an anti-FLAG monoclonal antibody and polyclonal rabbit antiserum recognizing HA.
REBOV and MA ZEBOV VP24 also interact with karyopherins α1, α5, and α6 and disrupt PY-STAT1-karyopherin interaction.
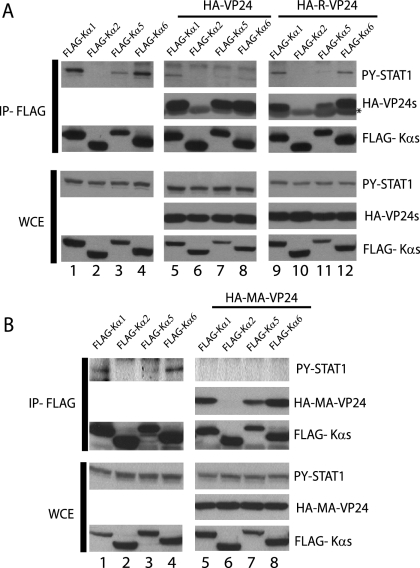
HA-VP24 has been shown to abrogate the interaction of PY-STAT1 with karyopherin α1 (Fig. 1). It was next of interest to determine if HA-VP24 could also abrogate the ability of PY-STAT1 to interact with karyopherin α5 and α6. Cells were transfected with FLAG-Kα1, -Kα2, -Kα5, and -Kα6 alone or along with HA-VP24. At 1 day posttransfection the cells were treated with IFN-β for 30 min and then subjected to immunoprecipitation. In the absence of HA-VP24, PY-STAT1 coprecipitated with FLAG-Kα1, -Kα5, and -Kα6 (Fig. 5A, lanes 1, 3, and 4). However, when HA-VP24 was cotransfected along with the karyopherins, the PY-STAT1-karyopherin interaction was inhibited (Fig. 5A, compare PY-STAT1 in lanes 1, 3, and 4 to lanes 5, 7, and 8). Because REBOV was found to inhibit IFN responses in infected Huh7 cells less efficiently than ZEBOV (30), it was of interest to determine whether REBOV VP24 retains the ability to interact with karyopherins α1, α5, and α6 and prevent PY-STAT1 binding. HA-tagged Reston VP24 was transfected along with FLAG-Kα1, -Kα2, -Kα5, and -Kα6. At 1 day posttransfection the cells were treated with IFN-β for 1 h and then immunoprecipitated as previously described. Similar to ZEBOV VP24, REBOV VP24 also coprecipitated with FLAG-Kα1, -Kα5, and -Kα6 and reduced the PY-STAT1-karyopherin interaction (Fig. 5A).
FIG. 5.
Reston and MA VP24 proteins interact with the NPI-1 subfamily of karyopherin α and inhibit the interaction of PY-STAT1 with karyopherin. (A) 293T cells were transfected with FLAG-Kα1, FLAG-Kα2, FLAG-Kα5, or FLAG-Kα6 alone or along with either HA-ZEBOV VP24 (HA-VP24), HA-REBOV VP24 (HA-R-VP24), or (B) HA MA ZEBOV (HA-MA-VP24). One day posttransfection the cells were treated 1,000 U/ml of IFN-β for 30 min and then subjected to immunoprecipitation with the anti-FLAG monoclonal antibody. The immunoprecipitated (IP) material and whole-cell extract (WCE) were then subjected to Western blot analysis with the polyclonal rabbit antiserum recognizing PY-STAT1, FLAG, and an anti-HA monoclonal antibody. The asterisk indicates the light chain of the anti-FLAG monoclonal M2 antibody used for the immunoprecipitation.
Adaptation of ZEBOV from a nonlethal to a lethal infection in adult mice resulted in a number of nucleotide changes in both noncoding and coding regions of the viral genome (22). Two amino acid changes were found to be critical for virulence in mice, one in NP and a second, a threonine-to-isoleucine change, in VP24 (14). However, our data demonstrate that MA VP24 still binds human FLAG-Kα1, -Kα5, and -Kα6 and prevents PY-STAT1 binding (Fig. 5B). Similarly, both ZEBOV and MA VP24s coimmunoprecipitated with similar efficiency to mouse karyopherins α1, α2, α3, α4, and α6 (data not shown). Thus, all three karyopherins in the NPI-1 subfamily interact with VP24s and prevent the interaction of PY-STAT1 with karyopherin.
ZEBOV, REBOV, and MA VP24 inhibit induction of reporter gene expression by IFN-β.
We have previously demonstrated that ZEBOV VP24 is able to inhibit IFN-β mediated activation of an IFN-responsive ISG54 promoter CAT reporter gene (45). Published data indicate that REBOV is less efficient than ZEBOV at inhibiting IFN responses in Huh7 cells (30). Similarly, MA ZEBOV was better able to replicate in the presence of parental ZEBOV in IFN-treated mouse macrophages (14). Because REBOV and MA VP24 both displayed the ability to disrupt a PY-STAT1-karyopherin interaction, it was of interest to determine if they were also able to inhibit IFN-induced gene expression. Two human cell lines, 293T and Huh7 cells, and two mouse cell lines, NIH 3T3 cells and transformed mouse embryo fibroblasts (MEFs), were cotransfected with an IFN-α/β-responsive ISG54 reporter along with either an empty vector expression plasmid or different amounts of ZEBOV, REBOV, or MA VP24 plasmids. At 1 day posttransfection the cells were either mock treated or treated with IFN-β, and 1 day posttreatment reporter assays were performed. IFN-β treatment of empty vector-transfected cells resulted in a striking up-regulation of reporter gene expression compared to untreated, empty vector-transfected cells (Fig. 6). As previously reported, ZEBOV VP24 inhibited reporter gene expression; similarly, both MA and REBOV VP24s were able to inhibit reporter gene expression in all cell lines tested (Fig. 6). Therefore, the ability to disrupt the PY-STAT1-karyopherin interaction and thus inhibit IFN-β-induced gene expression is shared by ZEBOV, REBOV, and MA VP24 proteins. However, differences in the capacity of these different VP24s to block IFN responses, at least when expressed in the absence of other viral proteins, do not appear to explain the different abilities of these viruses to block IFN responses.
DISCUSSION
We previously demonstrated that the VP24 of ZEBOV inhibits IFN-α/β and IFN-γ signaling (45). VP24 was found to block nuclear accumulation of PY-STAT1 and to interact with karyopherin α1, the NLS receptor for PY-STAT1 (45). These findings provided a molecular explanation for the observed EBOV-mediated impairment of IFN-α/β and IFN-γ responses (21). However, a number of questions related to this function remained, including how the interaction of VP24 with karyopherin α mediates the inhibition of STAT1 nuclear import and whether differences in the sequences of VP24s from different EBOV species result in different capacities to block IFN signaling.
Structurally, karyopherin α proteins contain a conserved N-terminal importin beta binding domain and a C-terminal export receptor domain, recognized by the cellular apoptosis susceptibility protein that mediates nuclear export (19, 25, 33, 56). Arm repeats that recognize the NLS on proteins destined for nuclear import make up the central portion of the karyopherin (11). Karyopherin (importin) β binds the importin beta binding domain of karyopherin α and mediates docking of karyopherin α-NLS-containing protein complex to the nuclear pore. The trimeric complex is subsequently transported through the pore and into the nucleus. In terms of PY-STAT1 nuclear import, PY-STAT1 has been reported to bind the C-terminal of karyopherin α1 between residues 425 to 538 (48), and subsequent studies have identified arm repeats 8 to 9 within this region as critical for PY-STAT1 interaction (39). This is distinct from classical monopartite and bipartite NLSs which bind arm repeats 2 to 4, 7, and 8 (11, 15). In the present study, mutational analysis with truncated forms of karyopherin α1 confirmed the requirement of karyopherin α1 residues 425 to 538 for interaction with endogenous PY-STAT1 (Fig. 2B). Our previous data suggested that VP24 might compete with PY-STAT1-GFP for binding to the karyopherin (45). Thus, VP24 and PY-STAT1 might be expected to bind to the same region of the karyopherin α as it has been shown in competition studies that a classic NLS coupled to bovine serum albumin was unable to compete with PY-STAT1 for binding to karyopherin α1 (48). Indeed, mutation analysis in our present study confirmed that VP24 bound the C terminus of karyopherin α1, displaying a requirement for residues 458 to 504, which comprise the arm repeat 10 region of the karyopherin, a region required for karyopherin α1 binding to PY-STAT1 (Fig. 2C). Interestingly, however, a karyopherin α1 mutant comprised of residues 71 to 504 (Fig. 2D) retained the ability to interact with VP24 and not PY-STAT1.
Coimmunoprecipitation studies confirmed that VP24 can prevent the interaction of endogenous as well as overexpressed PY-STAT1 with karyopherin α1 (Fig. 3A and B). First, VP24 displayed a dose-dependent inhibition of the PY-STAT1- karyopherin α1 interaction. In transfection studies with karyopherin α1 and VP24, expression of these proteins preceded IFN-β treatment and thus tyrosine-phosphorylation of STAT1. As a result, VP24-mediated inhibition of the PY-STAT1- karyopherin α1 interaction could have been explained by sequestration of karyopherin α1 prior to STAT1 activation. Thus, it was of interest to determine if VP24 could inhibit the interaction of PY-STAT1 with karyopherin α1 when the three proteins were mixed in vitro. In these studies VP24 again displayed the ability to bind the karyopherin and prevent PY-STAT1-GFP binding (Fig. 3B), suggesting that VP24 may competitively inhibit PY-STAT1 binding to the karyopherin αs.
Karyopherin α proteins are a multigene family that can be classified into three distinct subfamilies, the Rch1 subfamily (karyopherin α2), the Qip1 subfamily (karyopherin α3 and karyopherin α4), and the NPI-1 subfamily (karyopherin α1, karyopherin α5, and karyopherin α6) (31). The three subfamilies share up to 50% sequence similarity. However, within the NPI-1 subfamily, karyopherin α1 shares greater than 80% sequence similarity with karyopherin α5 and karyopherin α6 (39), suggesting that this subfamily may bind a similar set of NLS-containing proteins. Indeed, oncostatin M and IFN-γ stimulation can activate STAT1, leading to the PY-STAT1 interaction with karyopherin α1 and karyopherin α6 (34). In the present study we demonstrate that STAT1 activated by IFN-β treatment interacts with all three members of the NPI-1 subfamily, suggesting that any of these three karyopherin αs may mediate PY-STAT1 nuclear import (Fig. 4A). VP24 has previously been shown to interact specifically with karyopherin α1 and not members of the Rch1 and Qip1 subfamily (45). Similar to PY-STAT1, VP24 also interacts with all members of the NPI-1 subfamily (Fig. 4B). Thus, EBOV VP24 appears to have evolved to specifically interact with the karyopherin α subfamily utilized exclusively by PY-STAT1 for nuclear import.
Recently, in a transcriptional profiling study, ZEBOV-infected Huh7 cells were impaired in their ability to up-regulate antiviral and immune response-related genes. In contrast, REBOV infection resulted in comparatively greater up-regulation of antiviral and immune response-related genes (30). In comparison to REBOV, ZEBOV infection resulted in a more potent inhibition of ISG expression (30). In light of these studies it was of interest to assess the IFN antagonist function of REBOV VP24. In coimmunoprecipitation studies we were unable to detect a difference between ZEBOV and REBOV VP24 interactions with human karyopherin α proteins (Fig. 5A); similar results were obtained in reporter gene expression studies: REBOV inhibited reporter activation similar to ZEBOV VP24 in both 293T and Huh7 cells (Fig. 6). Our data suggest that REBOV VP24 functions similarly to ZEBOV VP24 under the conditions tested. Further studies will be required to determine whether the differences in the abilities of ZEBOV and REBOV to inhibit IFN responses can be attributed to VP24, for example, due to different kinetics of VP24 expression or to subtle differences in VP24 function.
The IFN system clearly plays a key role in the susceptibility of mice to lethal EBOV infection (6, 8, 37). While adult immunocompetent mice are resistant, type I IFN receptor or STAT1 knockout mice are susceptible to lethal EBOV infection (6). Additionally, treatment of immunocompetent mice with anti-IFN-α/β antibodies at the time of infection resulted in mice that were susceptible to lethal infection (6). Thus, in mice, the IFN system is critical for defense against lethal EBOV infection.
Changes in VP24 have been associated with adaptation of ZEBOV from a nonlethal to a lethal infection in animal models (14, 22, 53). In particular, in the mouse system, it has been demonstrated that two single amino acid changes, one in the NP and one in VP24, were sufficient to render ZEBOV lethal in mice (14). These changes also correlated with evasion of the type I IFN system (14). Because these data suggest that MA change to VP24 influences virus sensitivity to IFN-α/β responses, it was of interest to determine if nonadapted and MA ZEBOV VP24 would display different abilities to interact with human karyopherin α proteins. In coimmunoprecipitation studies we were unable to detect a difference in interaction with human karyopherin α between nonadapted ZEBOV and MA ZEBOV VP24 under the conditions tested (Fig. 5B). In reporter gene studies, MA VP24 inhibited reporter activation similar to nonadapted ZEBOV VP24 in two different mouse cell lines (Fig. 6). We were also unable to detect a difference in ZEBOV and MA VP24 interaction with mouse karyopherin αs (data not shown). (Human and mouse karyopherin α1 are greater than 95% similar in overall sequence at the amino acid level.) The possibility exists that our experimental conditions were not sensitive enough to detect more subtle yet meaningful differences in VP24 function. Alternatively, adaptive changes in VP24 may not be related to karyopherin α1 binding, and inhibition of PY-STAT1 nuclear import might reflect an additional function of VP24 during EBOV infection. However, further studies will be needed to determine whether the VP24-karyopherin α interaction influences the replication of EBOV in different species.
Acknowledgments
This work was supported by grant AI059536 to C.F.B. from the National Institutes of Health.
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Anonymous. 2003. Outbreak(s) of Ebola hemorrhagic fever, Congo and Gabon, October 2001 to July 2002. Can. Commun. Dis. Rep 29:129-133. [PubMed] [Google Scholar]
- 2.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, S., H. Feldmann, C. Will, and W. Slenczka. 1992. Evidence for occurrence of filovirus antibodies in humans and imported monkeys: do subclinical filovirus infections occur worldwide? Med. Microbiol. Immunol. 181:43-55. [DOI] [PubMed] [Google Scholar]
- 5.Bosio, C. M., M. J. Aman, C. Grogan, R. Hogan, G. Ruthel, D. Negley, M. Mohamadzadeh, S. Bavari, and A. Schmaljohn. 2003. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J. Infect. Dis. 188:1630-1638. [DOI] [PubMed] [Google Scholar]
- 6.Bray, M. 2001. The role of the type I interferon response in the resistance of mice to filovirus infection. J. Gen. Virol. 82:1365-1373. [DOI] [PubMed] [Google Scholar]
- 7.Bray, M., and T. W. Geisbert. 2005. Ebola virus: the role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int. J. Biochem. Cell. Biol. 37:1560-1566. [DOI] [PubMed] [Google Scholar]
- 8.Bray, M., J. L. Raymond, T. Geisbert, and R. O. Baker. 2002. 3-Deazaneplanocin A induces massively increased interferon-alpha production in Ebola virus-infected mice. Antivir. Res. 55:151-159. [DOI] [PubMed] [Google Scholar]
- 9.Cardenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 80:5168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, S. Y., M. C. Ma, and M. A. Goldsmith. 2000. Differential induction of cellular detachment by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J. Gen. Virol. 81:2155-2159. [DOI] [PubMed] [Google Scholar]
- 11.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 12.Cortes, P., Z. S. Ye, and D. Baltimore. 1994. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl. Acad. Sci. USA 91:7633-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuomo, C. A., S. A. Kirch, J. Gyuris, R. Brent, and M. A. Oettinger. 1994. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. USA 91:6156-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebihara, H., A. Takada, D. Kobasa, S. Jones, G. Neumann, S. Theriault, M. Bray, H. Feldmann, and Y. Kawaoka. 2006. Molecular determinants of Ebola virus virulence in mice. PLOS Pathog. 2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontes, M. R., T. Teh, and B. Kobe. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 297:1183-1194. [DOI] [PubMed] [Google Scholar]
- 16.Geisbert, T. W., L. E. Hensley, P. B. Jahrling, T. Larsen, J. B. Geisbert, J. Paragas, H. A. Young, T. M. Fredeking, W. E. Rote, and G. P. Vlasuk. 2003. Treatment of Ebola virus infection with a recombinant inhibitor of factor VIIa/tissue factor: a study in rhesus monkeys. Lancet 362:1953-1958. [DOI] [PubMed] [Google Scholar]
- 17.Geisbert, T. W., H. A. Young, P. B. Jahrling, K. J. Davis, E. Kagan, and L. E. Hensley. 2003. Mechanisms underlying coagulation abnormalities in Ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. J. Infect. Dis. 188:1618-1629. [DOI] [PubMed] [Google Scholar]
- 18.Gibb, T. R., M. Bray, T. W. Geisbert, K. E. Steele, W. M. Kell, K. J. Davis, and N. K. Jaax. 2001. Pathogenesis of experimental Ebola Zaire virus infection in BALB/c mice. J. Comp. Pathol. 125:233-242. [DOI] [PubMed] [Google Scholar]
- 19.Gorlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 20.Gorlich, D., F. Vogel, A. D. Mills, E. Hartmann, and R. A. Laskey. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377:246-248. [DOI] [PubMed] [Google Scholar]
- 21.Harcourt, B. H., A. Sanchez, and M. K. Offermann. 1999. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1β, in endothelial cells. J. Virol. 73:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart, M. K. 2003. Vaccine research efforts for filoviruses. Int. J. Parasitol. 33:583-595. [DOI] [PubMed] [Google Scholar]
- 23.Hartman, A. L., J. E. Dover, J. S. Towner, and S. T. Nichol. 2006. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J. Virol. 80:6430-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartman, A. L., J. S. Towner, and S. T. Nichol. 2004. A C-terminal basic amino acid motif of Zaire Ebola virus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology 328:177-184. [DOI] [PubMed] [Google Scholar]
- 25.Herold, A., R. Truant, H. Wiegand, and B. R. Cullen. 1998. Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 143:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Society for Infectious Diseases. 2005. Marburg hemorrhagic fever—Angola, archive no. 20051108.3269. International Society for Infectious Diseases, Brookline, MA.
- 27.Jahrling, P. B., T. W. Geisbert, D. W. Dalgard, E. D. Johnson, T. G. Ksiazek, W. C. Hall, and C. J. Peters. 1990. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet 335:502-505. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, K. M., J. V. Lange, P. A. Webb, and F. A. Murphy. 1977. Isolation and partial characterisation of a new virus causing acute haemorrhagic fever in Zaire. Lancet 1:569-571. [DOI] [PubMed] [Google Scholar]
- 29.Jones, S. M., H. Feldmann, U. Stroher, J. B. Geisbert, L. Fernando, A. Grolla, H. D. Klenk, N. J. Sullivan, V. E. Volchkov, E. A. Fritz, K. M. Daddario, L. E. Hensley, P. B. Jahrling, and T. W. Geisbert. 2005. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 11:786-790. [DOI] [PubMed] [Google Scholar]
- 30.Kash, J. C., E. Muhlberger, V. Carter, M. Grosch, O. Perwitasari, S. C. Proll, M. J. Thomas, F. Weber, H. D. Klenk, and M. G. Katze. 2006. Global suppression of the host antiviral response by Ebola- and Marburgviruses: increased antagonism of the type I interferon response is associated with enhanced virulence. J. Virol. 80:3009-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler, M., S. Ansieau, S. Prehn, A. Leutz, H. Haller, and E. Hartmann. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104-108. [DOI] [PubMed] [Google Scholar]
- 32.Kohler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Gorlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutay, U., F. R. Bischoff, S. Kostka, R. Kraft, and D. Gorlich. 1997. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90:1061-1071. [DOI] [PubMed] [Google Scholar]
- 34.Ma, J., and X. Cao. 2006. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cell Signal. 18:1117-1126. [DOI] [PubMed] [Google Scholar]
- 35.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol Rev. 65:570-594, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahanty, S., and M. Bray. 2004. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect. Dis. 4:487-498. [DOI] [PubMed] [Google Scholar]
- 37.Mahanty, S., M. Gupta, J. Paragas, M. Bray, R. Ahmed, and P. E. Rollin. 2003. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology 312:415-424. [DOI] [PubMed] [Google Scholar]
- 38.Mahanty, S., K. Hutchinson, S. Agarwal, M. McRae, P. E. Rollin, and B. Pulendran. 2003. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 170:2797-2801. [DOI] [PubMed] [Google Scholar]
- 39.Melen, K., R. Fagerlund, J. Franke, M. Kohler, L. Kinnunen, and I. Julkunen. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193-28200. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto, Y., N. Imamoto, T. Sekimoto, T. Tachibana, T. Seki, S. Tada, T. Enomoto, and Y. Yoneda. 1997. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J. Biol. Chem. 272:26375-26381. [DOI] [PubMed] [Google Scholar]
- 41.Moroianu, J., G. Blobel, and A. Radu. 1995. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc. Natl. Acad. Sci. USA 92:2008-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nachury, M. V., U. W. Ryder, A. I. Lamond, and K. Weis. 1998. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc. Natl. Acad. Sci. USA 95:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill, R. E., and P. Palese. 1995. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology 206:116-125. [DOI] [PubMed] [Google Scholar]
- 44.Reid, S. P., W. B. Cardenas, and C. F. Basler. 2005. Homo-oligomerization facilitates the interferon-antagonist activity of the Ebolavirus VP35 protein. Virology 341:179-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, 4th ed., vol. 1. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 47.Seki, T., S. Tada, T. Katada, and T. Enomoto. 1997. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem. Biophys. Res. Commun. 234:48-53. [DOI] [PubMed] [Google Scholar]
- 48.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart, M. 2007. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell. Biol. 8:195-208. [DOI] [PubMed] [Google Scholar]
- 50.Sullivan, N. J., T. W. Geisbert, J. B. Geisbert, L. Xu, Z. Y. Yang, M. Roederer, R. A. Koup, P. B. Jahrling, and G. J. Nabel. 2003. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature 424:681-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sullivan, N. J., M. Peterson, Z. Y. Yang, W. P. Kong, H. Duckers, E. Nabel, and G. J. Nabel. 2005. Ebola virus glycoprotein toxicity is mediated by a dynamin-dependent protein-trafficking pathway. J. Virol. 79:547-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 53.Volchkov, V. E., A. A. Chepurnov, V. A. Volchkova, V. A. Ternovoj, and H. D. Klenk. 2000. Molecular characterization of guinea pig-adapted variants of Ebola virus. Virology 277:147-155. [DOI] [PubMed] [Google Scholar]
- 54.Volchkov, V. E., V. A. Volchkova, E. Muhlberger, L. V. Kolesnikova, M. Weik, O. Dolnik, and H. D. Klenk. 2001. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science 291:1965-1969. [DOI] [PubMed] [Google Scholar]
- 55.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 56.Weis, K., U. Ryder, and A. I. Lamond. 1996. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 15:1818-1825. [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Z. Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886-889. [DOI] [PubMed] [Google Scholar]