Abstract
All hepatitis B viruses replicate by protein-primed reverse transcription, employing a specialized reverse transcriptase, P protein, that carries a unique terminal protein (TP) domain. To initiate reverse transcription, P protein must bind to a stem-loop, ɛ, on the pregenomic RNA template. TP then provides a Y residue for covalent attachment of the first nucleotide of an ɛ-templated DNA oligonucleotide (priming reaction) that serves to initiate full-length minus-strand DNA synthesis. ɛ binding requires the chaperone-dependent conversion of inactive P protein into an activated, metastable form designated P*. However, how P* differs structurally from P protein is not known. Here we used an in vitro reconstitution system for active duck hepatitis B virus P combined with limited proteolysis, site-specific antibodies, and defined P mutants to structurally compare nonactivated versus chaperone-activated versus primed P protein. The data show that Hsp70 action, under conditions identical to those required for functional activation, transiently exposes the C proximal TP region which is, probably directly, involved in ɛ RNA binding. Notably, after priming and ɛ RNA removal, a very similar new conformation appears stable without further chaperone activity; hence, the activation of P protein is triggered by energy-consuming chaperone action but may be completed by template RNA binding.
Hepatitis B viruses (HBVs), or hepadnaviruses, are small enveloped DNA-containing viruses (12, 34) that replicate through protein-primed reverse transcription of a pregenomic RNA (pgRNA) intermediate (37). This unusual mechanism (4) is reflected in the structural organization of their reverse transcriptase (RT), P protein, which in addition to the conserved RT and RNase H (RH) domains (45) contains a unique terminal protein (TP) domain of about 200 amino acids not found in any other RT (Fig. 1A). TP is connected to the RT domain by a functionally dispensable spacer. Binding of P protein to a 5′-proximal stem-loop, ɛ (Dɛ for duck HBV [DHBV]), on the pgRNA triggers both encapsidation of the pgRNA and initiation of reverse transcription. Using the hydroxyl group of a specific Y residue in TP (Y63 in HBV [24], Y96 in DHBV [44, 47]) rather than a tRNA 3′ end as starting point, P protein generates a 3- to 4-nucleotide, ɛ-templated DNA oligonucleotide (“priming”; Fig. 1B). After this initiation phase, ɛ is replaced as a template by the 3′-proximal DR1* element on pgRNA and, primed by the TP-linked DNA oligonucleotide, minus-strand DNA is elongated from there. The end product of reverse transcription is a partially double-stranded, relaxed circular DNA in which the minus strand is still covalently linked to TP. This mechanism appears to hold for all hepadnaviruses, but most current knowledge is derived from DHBV (32), whose P protein, in contrast to that of HBV (17), is capable of performing the priming reaction in appropriate cell-free systems (2, 6, 16, 19, 41).
FIG. 1.
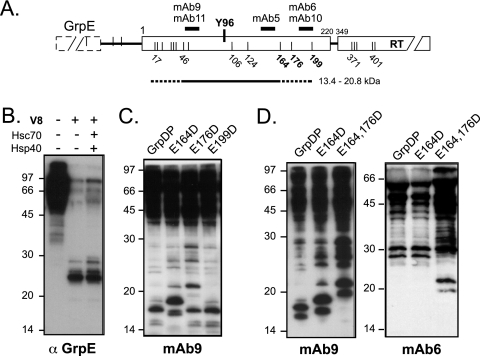
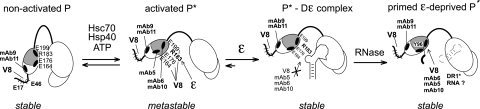
P-protein domain structure and activation. (A) Wild-type DHBV P protein (wt P) and the recombinant P proteins (rec P) used in the present study. Like all hepadnaviral P proteins, DHBV P protein consists of conserved RT and RH domains plus an N-terminal TP domain and a functionally dispensable spacer. Y96 in TP serves as attachment site for the 5′ end of the DNA primer oligonucleotide. Numbers represent amino acid positions. In the recombinant proteins the spacer is replaced by a short SPG linker and the C-terminal amino acids past position 761 are deleted (6); the heterologous domains (GrpE or NusA) enhance solubility. (B) Schematic representation of primed P protein. Bound ɛ RNA undergoes a conformational alteration, which opens up the upper ɛ stem (3) and appropriately positions the Y residue in TP and the template region in ɛ so as to allow the RT domain to copy the DNA primer (thick curved line connected to TP). DNA synthesis ceases until ɛ is replaced as a template by the DR1* element close to the opposite end of the pgRNA. (C) Stepwise formation of primed P protein. Inactive P is transiently activated into P* by the ATP-consuming activity of Hsc70 and Hsp40; Hsp90 plus Hop, or Bag-1S, can further stimulate P* formation. P* is metastable, and its steady-state concentration is determined by the rates of P* production and decay. Only P* can bind ɛ RNA, forming a stable initiation-competent complex. If provided with deoxynucleoside triphosphates, the complex carries out synthesis of the primer oligonucleotide.
Initial studies with P protein in vitro-translated in rabbit reticulocyte lysate (RL) revealed a strict dependence of the RT on cellular chaperones for gaining ɛ RNA binding and, consequently, priming competence. Based on some similarities to steroid hormone receptor activation (29), both the heat shock protein 70 (Hsp70; or its constitutive form Hsc70) and Hsp90, together with their respective cochaperones such as Hsp40 and Hop, were proposed to be essential for P activation (18, 20). Recently developed in vitro reconstitution systems (2, 19) consisting of bacterially expressed DHBV P protein, fused to solubility enhancing heterologous domains, and purified chaperones seemed to support this notion for a glutathione S-transferase fusion protein (19); however, using two different fusion partners (NusA and GrpE), we found strong evidence that Hsc70 plus Hsp40 plus ATP are sufficient (2, 5). New work from our laboratory has corroborated this conclusion and, in addition, has shown that Hsp90 plus Hop, as well as the Hsp70 nucleotide exchange factor Bag-1S (7, 38), can further stimulate, by four- to fivefold, Hsc70-mediated activation, most likely by increasing the quantity, not the quality, of priming-competent P molecules (35).
However, how chaperone action activates P protein is largely unknown. Given their role as folding catalysts (27, 46), the chaperones are likely to modulate P protein structure (Fig. 1C) from a state where the binding site for Dɛ RNA is occluded (P) to another one where it is accessible (P*). P* is metastable, and maintaining steady-state levels of it requires continuous, ATP-consuming chaperone activity; upon ATP depletion, the activated molecules decay within minutes. However, once Dɛ RNA is bound, the resulting P-Dɛ complexes are stable over hours (2).
As yet, no direct structural information is available on any P protein; this holds particularly for the TP domain, whose sequence dissimilarity to all current database entries even prevents homology-based molecular modeling. Moreover, the structure of P protein must be highly dynamic to cope with the various nucleic acid transformations during reverse transcription. For instance, once P protein has bound ɛ RNA, the TP-borne Y-residue must be placed in the RT active site; then TP must give way to the growing DNA primer and, with the bound primer, must translocate to DR1*. Eventually, there will be 3 kb of DNA between TP linked to the 5′ end of the minus-strand DNA and the RT domain elongating the 3′ end toward the end of the pgRNA template. However, none of these events can occur without the initial, chaperone-dependent activation step, which is in the focus of the present study.
Limited proteolysis is a powerful method for gaining information about dynamic structural changes (10, 11, 22). Applied to RL-translated DHBV P protein, it provided first evidence that upon P-Dɛ complex formation, not only the RNA (3) but also the protein is structurally altered (39, 40). However, resolution was limited by the multitude of factors in RL that could affect protease accessibility, the chaperone-activated P* intermediate could not directly be analyzed, and the available tools did not allow more explicit statements than that ɛ binding stabilized a large fragment of the RT domain contained between residues 265 and 652.
We combined here limited proteolysis with several recently developed tools for a higher-resolution analysis of structural alterations accompanying P-protein activation. The in vitro reconstitution system allowed us to test the effects of defined combinations of purified chaperones, and ATP, on P-protein structure and to correlate them with functional P activation; moreover, the initial chaperone activation step could directly be monitored in the absence of Dɛ RNA. Second, several monoclonal antibodies (MAbs) against different regions in the TP domain (8) and site-specifically mutated P proteins allowed us to assign proteolytic fragments to specific sequence segments. Third, the known location of the priming Y residue allowed us to identify TP-associated changes in enzymatically active, primed P protein. Finally, application of the limited proteolysis assay to an RNA-binding-defective TP point mutant, R183G, allowed us to demonstrate that the ablated Dɛ RNA binding capacity was not caused by an aberrant structure but rather by a direct involvement of R183 in enabling RNA binding. All of the data suggest that transient exposure of a Dɛ RNA-binding region involving TP is a key event in the chaperone activation of P protein.
MATERIALS AND METHODS
Recombinant DHBV P proteins.
GrpDP is a recombinant fusion protein containing the P protein from DHBV16 (26) fused, via a tag-containing linker, to the C terminus of the Escherichia coli GrpE protein. The P protein part lacks the functionally dispensable spacer region (amino acids [aa] 221 to 348) and the 25 C-terminal amino acids; NusDP is a homologous fusion with the bacterial NusA protein; both fusion proteins can be activated for priming in RL and in vitro (2). The indicated point mutations were introduced via PCR using a mega-primer protocol (28). A construct encoding GrpDP including the authentic spacer was obtained by replacing an appropriate restriction fragment in the GrpDP expression vector by the corresponding authentic fragment from plasmid pCD16 (30), which carries a terminally redundant 1.1-mer DHBV16 genome.
Purification of proteins.
Recombinant P proteins and Hsc70, Hsp40, Bag-1S, and Hop were expressed in E. coli BL21 Codon Plus cells as described previously (2, 35). Purified human Hsp90β was a gift from J. Buchner (Technical University of Munich).
MAbs.
Five MAbs with approximately known epitope locations in the DHBV P protein TP domain (8) were kindly provided by J. Tavis (St. Louis University). Using the PepScan technique (13) and cellulose membrane immobilized TP-derived 15-mer peptides, each overlapping the previous one by 13 amino acids, we confirmed and refined the mapping data previously obtained by using a set of truncated DHBV P protein derivatives (8). Operationally, we defined an epitope as the common sequence present in usually three to five overlapping peptides that reacted strongly with the respective MAb (the previously reported mapping data are given in parentheses): MAb9 and MAb11, aa 53 to 59 (aa 46 to 77); MAb5, aa 141 to 147 (aa 138 to 160); and MAb6 and MAb10, aa 191 to 197 (aa 182 to 202). We noted that both MAb9 and MAb11 reacted well with peptides 47-61 and 53-67 but less well with peptides 49-63 and 51-65, possibly reflecting some conformational contributions to epitope formation.
Limited proteolysis and immunoblotting.
Chaperone activation reactions containing 150 ng of the appropriate recombinant P protein, 3 μg of Hsc70, 2 μg of Hsp40 and, where indicated, 0.5 μg of Hop plus 0.5 μg of Hsp90 or 1.2 μg of Bag-1S were set up in TKD buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 2 mM dithiothreitol) in a final volume of 10 μl, supplemented with a 1/10 volume of an ATP regenerating system (50 mM ATP, 25 mM MgCl2, 250 mM creatine phosphate, 100 U of phosphocreatine kinase/ml), and incubated at 30°C for 30 min. In some experiments ATP was depleted by the addition of glucose (final concentration, 25 mM) and 0.1 U of hexokinase (Sigma)/μl for 5 min at room temperature; controls were performed identically except that hexokinase was replaced by bovine serum albumin. For limited proteolysis, the samples were incubated for 15 min at room temperature with 0.25 U of endoproteinase Glu-C (Sigma) or with 50 ng of endoproteinase Asp-N (Roche Diagnostics). Reactions were stopped by adding an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, followed by boiling for 5 min. Reaction products were separated by SDS-PAGE in discontinuous 14% acrylamide gels run in Tris-glycine buffer (23) or in continuous 15% acrylamide gels run in Tris-Tricine buffer (31). After electroblotting onto polyvinylidene difluoride membranes, TP-containing fragments were detected by incubation with anti-TP MAb, followed by the addition of anti-mouse peroxidase or alkaline phosphatase conjugates and ECL+ (GE Healthcare) or CDP-Star (Roche Diagnostics) as chemiluminescent substrates. Signals were recorded on X-ray film or by using an LAS-3000 imager (Fuji). Efficient priming requires specialized buffer conditions, and therefore proteolysis experiments with primed P-ɛ complexes (see below) were performed in the presence of priming buffer. To exclude that the different buffer conditions affected the V8 cleavage patterns of chaperone-activated but nonprimed P protein, control experiments were performed under identical buffer conditions as with primed P protein (see below) except that no Dɛ RNA and no deoxynucleoside triphosphates were added. However, in both buffer systems the same chaperone-dependent small fragments were produced.
Limited proteolysis of primed P-ɛ complexes.
Reconstitution of P-ɛ complexes and subsequent labeling of P protein by incorporation of [α-32P]dATP during the priming reaction were essentially carried out as previously reported (2). Briefly, reconstitution reactions, set up as described above, were supplemented with 1 μM Dɛ RNA and 1 U of RNasin RNase inhibitor (Ambion)/μl. P-ɛ complex formation was allowed to proceed for 3.5 h at 30°C. After the addition of an equal volume of 2× priming buffer (20 mM Tris-HCl [pH 8.0], 20 mM NH4Cl, 12 mM MgCl2, 4 mM MnCl2, 0.4% [vol/vol] NP-40, 20 mM β-mercaptoethanol, 1 mM spermidine, 0.1 mM dGTP, 0.1 mM dTTP) supplemented with 0.3 μCi of [α-32P]dATP/μl, the reactions were further incubated for 1 h at 37°C to allow DNA primer synthesis to proceed. To account for the increased reaction volume after priming (20 μl) and the different buffer conditions, 0.9 U of V8 protease per reaction was used for proteolysis. The incubation time and temperature were the same as described above. ATP depletion was done as described above. If appropriate, Dɛ RNA was removed by the addition of 0.1, 1, or 10 μg of RNase A per sample and allowing the cleavage to proceed for 5 min. Reactions were stopped by the addition of sample buffer and immediately freezing the samples at −20°C or by boiling for 5 min at 100°C. Reaction products were separated by SDS-PAGE and visualized by phosphorimaging or autoradiography.
Immunoprecipitations.
Reconstitution reactions were set up as described above and incubated at 30°C for 30 min. The reactions were split, 0.7 μg of the respective MAb was added per 150 ng of recombinant P protein, and the samples were further incubated for 2 h at 30°C. Chaperone-activated P molecules continuously generated during this period would then immediately be sequestered into P-antibody complexes. Thereafter, antibodies and bound proteins were adsorbed to GammaBind Plus Sepharose (GE Healthcare) suspended in 600 μl of TNN (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% NP-40) overnight at 4°C. The beads were washed four times with 1 ml of TNN, and the immunocomplexes were disrupted by boiling them in sample buffer. The samples were separated by SDS-PAGE in 10% polyacrylamide gels, and P protein was detected by Western blotting with MAb9.
Immunoprecipitation of V8 fragments from primed, ɛ-deprived GrpDP and its E164D and E164,176D mutants.
Primed P-ɛ complexes were generated and subjected to RNase A and V8 treatment as described above. The reactions were split and incubated overnight at 4°C with MAb9 or MAb6 or an irrelevant anti-green fluorescent protein (GFP) MAb immobilized on Gammabind Plus-Sepharose beads suspended in 0.5 ml of radioimmunoprecipitation assay (RIPA) buffer (8). Beads were washed four times with 1 ml each of RIPA buffer. Bound proteins were released by boiling in sample buffer, and the samples were subjected to SDS-PAGE using the Tris-Tricine buffer system. The gel was dried, and 32P-labeled fragments were visualized by autoradiography.
Dɛ RNA binding by P protein.
RNA-binding competence was assessed as previously described (1, 21). In brief, wild-type and the indicated mutant P proteins were in vitro translated in RL in the presence of [35S]methionine (Amersham) to label the protein and in the presence of internally 32P-labeled in vitro-transcribed Dɛ RNA. After incubation for 3 h, the proteins were immobilized to Ni-nitrilotriacetic acid (NTA) agarose beads via their C-terminal His tags, and the beads were thoroughly washed. Thereafter, protein and potentially protein-bound RNA were released by boiling in sample buffer, and the eluates were analyzed by SDS-PAGE and autoradiography.
Priming inhibition by MAbs.
Reconstitution reactions including 150 ng of GrpDP and 1 μM Dɛ RNA were allowed to proceed for 3.5 h, and then priming was initiated, in the presence of [α-32P]dATP, as described above. MAb5, MAb6, or MAb10 (0.7 μg per reaction), or an irrelevant MAb to GFP, or no antibody was added immediately after mixing the reconstitution components (t = 0), after 2 h of P-Dɛ complex formation (t = 2 h), or immediately before switching to priming conditions (t = 3.5 h). Products of the individual reactions were separated by SDS-PAGE. The amounts of 32P-labeled P protein were quantified by phosphorimaging using MacBas software (Fuji).
RESULTS
Hsc70/Hsp40 plus ATP induce a specific conformational rearrangement involving the TP domain.
Deriving region-specific information on conformationally different states of a protein by limited proteolysis requires suitable conditions for inducing the alternative conformation(s), and the presence of appropriate recognition sites for the given protease. Our previous work had established conditions under which recombinant DHBV P protein, fused to GrpE (GrpDP) or NusA (NusDP), is reproducibly converted into a Dɛ RNA-binding and priming-competent state by Hsc70 plus Hsp40 plus ATP (2). This reconstitution system was used here. Of several initially tested proteases, V8 protease (endoproteinase Glu-C; cleaving specifically after E residues) (15) proved most suitable because it robustly produced distinctive fragments that were appropriately sized for analysis by SDS-PAGE, were well detectable by immunoblotting with MAb9 (recognizing an epitope comprising TP residues 53 to 59), and reproducibly occurred only in the presence of specific components in the reconstitution reactions. V8 protease was therefore used in most experiments.
After optimizing proteolysis conditions with respect to V8 concentration and incubation time, we analyzed different combinations of reconstitution components for their effects on the proteolysis pattern (Fig. 2A). The GrpDP protein (∼100 kDa) was, under all conditions, efficiently cleaved into a major TP-containing product with an apparent mass of about 66 kDa (resulting from cleavage between the GrpE and the P protein part; see below), plus a series of less-abundant smaller fragments. Most importantly, two fragments of about 14 kDa (doublet, sometimes triplet) and 18 kDa were prominently enriched only in reactions containing Hsc70, Hsp40, and ATP, i.e., all of the components also required for P activation (2). The same chaperone-dependent fragment patterns were observed when V8 proteolysis was performed under conditions identical to those used with the 32P-labeled primed P protein preparations described below.
FIG. 2.
Hsc70 and Hsp40 plus ATP induce a specific conformational change involving the TP domain. (A) V8 fragments (14 to 18 kDa) from GrpDP are only formed in the presence of Hsc70 plus Hsp40 plus ATP. GrpDP was incubated with the indicated factors and then subjected to limited V8 proteolysis. Products were analyzed by SDS-PAGE and immunoblotting, with MAb9 recognizing an epitope involving TP residues 53 to 59. Numbers on the left show the positions of marker proteins of the indicated sizes in kilodaltons. (B) Chaperone-induced 14- to 18-kDa fragments occur independently of the heterologous fusion partner and of the spacer region. NusDP, containing NusA instead of GrpE as a fusion partner, and GrpDP including the authentic P-protein spacer region were incubated with Hsc70, Hsp40 plus ATP, or not and then subjected to V8 digestion as in panel A. (C) Limited proteolysis with Asp-N protease also detects a chaperone-dependent conformational alteration involving TP. GrpDP was incubated with Hsc70, Hsp40 plus ATP, or not and then subjected to proteolysis with Asp-N protease. Reactions were analyzed as in panel A. (D) Hsp90/Hop and Bag-1S do not induce detectable additional conformational changes in TP. GrpDP was incubated with Hsc70 and Hsp40 plus ATP as in panel A, but in addition with Hsp90/Hop or Bag-1S. Reactions were analyzed as in panel A.
To test whether the heterologous GrpE part, or the spacer deletion in our standard constructs, affected generation of these fragments, we performed analogous experiments with NusDP (P protein fused to NusA instead of GrpE) and a GrpDP derivative containing the authentic spacer. Despite some differences in the higher-molecular-weight range, both proteins yielded Hsc70/Hsp40 plus ATP-dependent small fragments with mobilities identical to that of the GrpDP protein (Fig. 2B). Hence, generation of the 14- to 18-kDa fragments was an intrinsic property of P protein, and the fragment borders resided inside the TP sequence. A chaperone-dependent conformational change was also detectable with Asp-N protease; a band at around the 22-kDa position was strongly reduced, and a band at about the 14-kDa position was strongly enhanced (Fig. 2C).
Finally, we tested whether the additional presence of Bag-1S, as well as Hsp90 plus Hop, influenced the V8 proteolysis pattern (Fig. 2D). Again, fragments of 14 to 18 kDa were strongly enhanced. The pattern of the other bands did not show major differences from that obtained with only Hsc70/Hsp40 plus ATP. Hence, neither Bag-1S nor Hsp90/Hop induced the formation of a conformationally distinguishable form of activated P protein with respect to the TP domain.
The conformational rearrangement in the TP domain is metastable.
A hallmark of P protein activation is the metastability of the activated state P*, which is maintained only by constant ATP-consuming chaperone activity (2). We therefore tested whether the conformationally altered form of P protein was also metastable. GrpDP was mixed with Hsc70, Hsp40, and ATP, and then the reaction was split. From one-half ATP was depleted by hexokinase plus glucose treatment; from the other half it was not. Both samples were further incubated for 3 h. At different time points aliquots were removed, subjected to V8 cleavage, and analyzed as before (Fig. 3). In the nondepleted samples, the amounts of the characteristic small fragments did not significantly change, whereas ATP depletion caused a rapid reduction already in the first few minutes. As estimated using a luminescent image analyzer, the band intensities were reduced to ca. 25% after 5 min and to ca. 20% after 10 min. A minor fraction of the fragments declined more slowly, however, with very little (ca. 5%) left at the end of the incubation period. Hence, the bulk of P molecules in the conformationally altered state was metastable.
FIG. 3.
The chaperone-induced conformation of P protein is metastable. GrpDP was incubated in the presence of Hsc70, Hsp40, and ATP as in Fig. 2, and then the reaction mixture was split. One-half was treated with hexokinase and glucose for ATP depletion (panel −ATP); the other half was not (panel +ATP). The reactions were further incubated at 30°C for 3 h, and aliquots were taken at the indicated time points, subjected to V8 protease cleavage, and analyzed by immunoblotting. Note that the chaperone-induced 14- to 18-kDa fragment (arrowheads) remained present at essentially at the same concentration in the presence of ATP and yet disappeared upon ATP depletion; most of the other proteolysis products remained unaffected.
Chaperone action exposes the C proximal part of TP.
Obviously, the cleavages producing the chaperone-specific 14- to 18-kDa fragments must have occurred at E residues bracketing the MAb9 epitope (Fig. 4A). This suggested that the numerous N-proximal E residues (E17, E19, E26, E34, E35, E36, and E46; E55 overlaps already with the MAb9 epitope) and either E164, E176, or E199, but not E106, E124, or E244 (corresponding to E371 in the presence of the authentic spacer) as likely V8 target sites. N-proximal cleavage should clip off the GrpE domain. This was corroborated by subjecting GrpDP to V8 proteolysis, in the absence or presence of Hsc70/Hsp40 plus ATP, and probing it with an MAb against GrpE. The major product, in both cases, had an apparent mass of around 25 kDa (Fig. 3B), a finding in line with the calculated mass of GrpE (26 kDa, including the vector-encoded linker). Hence, the region connecting GrpE and TP is highly sensitive to protease, regardless of chaperone action. Exactly which E residue constitutes the major cleavage site could not yet be assigned; however, the fine structure of the band pattern suggests that several of the first seven E residues between positions E17 and E46 could be involved. To define the C proximal cleavage site(s), we individually replaced the three E residues in question by D residues (variants E164D, E176D, and E199D). V8 cleavage, under the conditions used, is highly specific for E (15); on the other hand, the E→D exchange is so subtle that it should preserve overall structure. This was confirmed in that all variants displayed wild-type-like priming activities (see below). Next, the mutant proteins were subjected to V8 proteolysis. As with wild-type GrpDP, no specific small fragments were seen without chaperones (not shown). In the presence of Hsc70/Hsp40 and ATP, the major 18-kDa product from the E164D mutant appeared almost unaffected when analyzed using the common Tris-glycine gel system (not shown); however, in a Tris-Tricine gel system (31) both the 14-kDa doublet and the 18-kDa band migrated distinctly slower (Fig. 4C), indicating that E164 represents a major V8 cleavage site. Weak bands at the position of the dominant E164D product were also present in the wild-type GrpDP and E199D samples but not in E176D, suggesting that they had arisen from cleavage at E176. In addition, a weak band shortly above the 20-kDa marker position was slightly enhanced with the E164D mutant and more pronouncedly with the E176D mutant. This difference in relative intensity might relate to slightly different efficiencies of proteolysis in the different reactions; however, the absence of a corresponding band in the E199D sample would be consistent with E199 representing another chaperone-dependent V8 target site. To substantiate these conjectures, we next analyzed the double mutant E164,176D, which produced a further upward shift of the small fragments to positions corresponding to about 20 and 22 kDa (Fig. 4D). This indicated that, in the absence of both E164 and E176, cleavage occurred at E199 and produced stable fragments. If so, the upward-shifted fragments should contain the epitope of MAb6 (around residues 191 to 197). This MAb, indeed, recognized only the 20- to 22-kDa fragment from the double mutant (Fig. 4D, right panel).
FIG. 4.
Mapping the borders of the chaperone-induced 14- to 18-kDa V8 fragments. (A) Potential V8 cleavage sites in and around the TP part of GrpDP. TP is shown as a bar, flanked by the GrpE and the RT domain. Each vertical line represents an E residue. The positions of some of the E residues, of Y96, and of the MAb epitopes are indicated. The candidate region producing the 14- to 18-kDa fragments is represented by the straight line with the minimal (solid part) and maximal (dashed extensions) segments plus their calculated molecular masses. (B) Cleavage between GrpE and TP is chaperone independent. GrpDP was incubated with the indicated chaperones and then subjected to V8 proteolysis. Products were monitored by immunoblotting with an anti-GrpE antibody. GrpE was efficiently clipped off with or without chaperones. (C) E164 is involved in the C-terminal V8 cleavage. GrpDP and the indicated mutants were incubated with Hsc70, Hsp40, and ATP and then subjected to V8 proteolysis. Products were separated by SDS-PAGE in a Tris-Tricine gel system and analyzed by immunoblotting with MAb9. Note the mobility shift for the 14- to 18-kDa fragments from mutant E164D. (D) E176 and E199 are also exposed by chaperone action. GrpDP and the indicated mutants were treated as described above, and then the products were detected by using MAb9 (left panel) or MAb6 (right panel). Note the upward shift of the fragments in question with the double mutant and the exclusive recognition of these upshifted fragments by MAb6.
Hence, the N-proximal part of TP between E17 and E46 is continuously accessible to protease, the central part encompassing E106 and E124 is permanently inaccessible, and the C proximal region comprising E164 to E199 is transiently exposed by the ATP-consuming chaperone activity of Hsc70 plus Hsp40.
Efficient immunoprecipitation of P protein by MAbs against the C-terminal TP part requires chaperones.
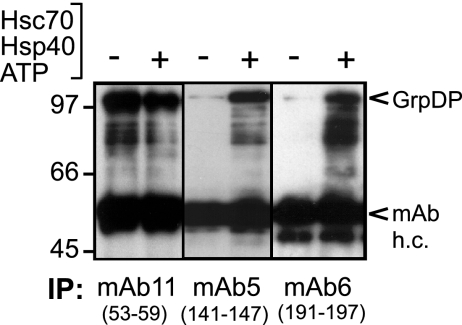
We next used immunoprecipitation with site-specific MAbs to monitor chaperone effects on epitope accessibility. An epitope close to or within the region that is becoming surface exposed should only be recognized in the presence of the proper chaperones. In addition to MAb11 (recognizing the same TP region as MAb9 around positions 53 to 59) and MAb6 (epitope around positions 191 to 197), we included MAb5, whose epitope encompasses residues 141 to 147. GrpDP was incubated either without or with Hsc70/Hsp40 and ATP; the reactions were then split, and the respective MAbs were added. The reactions were allowed to proceed for another 2 h to sequester P molecules with potentially chaperone-exposed MAb epitopes generated during this time into antibody complexes. These were subsequently immobilized onto Gammabind Plus-Sepharose beads, and the immunopellets were analyzed by Western blotting with MAb9 (Fig. 5). MAb11 precipitated GrpDP with comparable efficiency in the absence or presence of the chaperones. In contrast, the precipitation efficiency of GrpDP by MAb5 and MAb6 was poor without chaperones but strongly enhanced in their presence. Hence, in accord with the proteolysis data, exposure of the TP region around positions 53 to 59 was independent, and that of the region around positions 191 to 197 was dependent on chaperone activity. The data with MAb5 extend this region to include the residues around positions 141 to 147.
FIG. 5.
Efficient immunoprecipitation of GrpDP by MAbs recognizing the C-proximal TP part requires chaperone activity. GrpDP was incubated with or without chaperones and immunoprecipitated with MAb5, MAb6, or MAb11. Bound proteins were analyzed by immunoblotting with MAb9. Bands corresponding to GrpDP and to the heavy chains of the immunoprecipitation (IP) MAbs (MAb h.c.) are indicated on the right.
Conformational changes in primed P protein.
To substantiate the functional relevance of the chaperone-induced conformational change, we applied limited V8 proteolysis to in vitro-primed P protein. Here, fragments containing the [32P] DNA-labeled Y96, and therefore originating exclusively from enzymatically active molecules, can specifically be detected by autoradiography. Due to the proximity of Y96 to the epitope of MAb9, a V8 pattern might be expected that is similar to that obtained by Western blotting with MAb9. However, as shown below, the chaperone-dependently exposed TP region also contains residues that are crucial for Dɛ RNA binding; hence, bound RNA could affect protease accessibility. Equal aliquots from an in vitro priming reaction with GrpDP were therefore either left untreated or incubated with increasing concentrations of RNase A, and then all of the samples were subjected to V8 proteolysis. As shown in Fig. 6A, the untreated primed P protein was nearly quantitatively cleaved to yield one major fragment with an apparent molecular mass of around 45 kDa, and very few smaller products. This is consistent with the previously proposed stabilization of a relatively large domain in P protein by bound Dɛ RNA (39, 40).
FIG. 6.
A conformation similar to that transiently induced by chaperones is stabilized in primed, RNA-deprived P protein. (A) Primed wild-type GrpDP protein. GrpDP was incubated with Hsc70, Hsp40, ATP, and Dɛ RNA and then subjected to priming conditions in the presence of [α-32P]dATP. Prior to V8 proteolysis, some samples were treated with increasing amounts of RNase A (0.1, 1.0, or 10 μg per reaction), as indicated by “+,” “++,” or “+++.” Proteolysis products were detected by SDS-PAGE, followed by phosphorimaging. (B) V8 fragments (14 to 20 kDa) from primed RNA-deprived P protein are stable without ATP-consuming chaperone activity. Reconstituted primed P-Dɛ complexes were subjected to ATP depletion or not; both samples were then treated with RNase and further incubated. Over a time course of 2 h aliquots were removed and probed by V8 proteolysis. Products were analyzed by SDS-PAGE, followed by phosphorimaging. The signal intensities of the 14- to 20-kDa fragments after 2 h were still ca. 70% as high as those immediately after RNase treatment, irrespective of ATP depletion. (C) Mutants E164D and E164,176D are functionally active. The mutant proteins were analyzed in parallel to wild-type GrpPD as in panel A. No significant differences in priming signal intensities and formation of stable 45-kDa V8 fragments were detectable in the absence of RNase treatment. (D) E164 and E176 are involved in generating the chaperone-specific small V8 fragments from primed P protein. In vitro-primed P proteins were treated with 10 μg of RNase A per reaction and subjected to V8 proteolysis as in panel A, followed by immunoprecipitation with either MAb9 or MAb6. Precipitated proteins were analyzed by SDS-PAGE using the Tris-Tricine system. Control lanes on the left show primed untreated P protein, RNA-deprived V8-treated P protein without immunoprecipitation, and primed untreated P protein incubated with an anti-GFP MAb. wt, wild type.
RNase treatment prior to V8 proteolysis drastically and dose dependently changed the fragment pattern in that, at the cost of the 45-kDa product, three new smaller products in the 14- to 20-kDa range appeared, similar to those observed by Western blotting in the chaperone-activated nonprimed samples. Direct evidence that these fragments arise, indeed, from cleavage at the same E164, E176, and E199 residues is presented below.
These data were compatible with two explanations: either primed RNA-deprived P protein stably maintained a conformation similar to that initially induced by chaperone action, or it readopted the conformation of nonactivated P protein and was subsequently converted into the chaperone-specific conformation by the Hsc70, Hsp40, and ATP still present in the reactions. Therefore, a reconstitution reaction containing primed GrpDP was split, and from one-half ATP was depleted, while from the other half it was not. Both were then treated with RNase A, and aliquots taken over a time course of 2 h were subjected to V8 proteolysis. In contrast to the rapid loss of chaperone-dependent fragments from nonprimed GrpDP protein upon ATP depletion (Fig. 3), the corresponding fragments from the primed GrpDP molecules declined only slowly, with 60 to 70% of the initial signal left after 2 h of incubation, as estimated by phosphorimaging. Importantly, there was no significant difference between the ATP-depleted and nondepleted samples (Fig. 6B). This suggested that the primed molecules stably adopted a conformation similar to the one that is transiently and ATP-dependently induced in nonprimed P protein.
Despite the generally similar pattern of the small proteolysis fragments obtained from activated P protein (Fig. 2) versus activated plus primed P protein (Fig. 6A), the band mobilities were not exactly identical. Although this could relate to the attached primer oligonucleotide, we sought to directly test whether the same cleavage sites were involved. Therefore, we applied the same assay to the E164D and E164,176D mutants. Both proteins displayed wild-type-like priming activity (Fig. 6C, left three lanes), and both yielded, in the absence of RNase A, one major 45-kDa product. RNase A treatment caused, again, the appearance of smaller fragments which, especially for the double mutant, were detectably upward shifted when analyzed in the Tris-glycine gel system (not shown). To further enhance specificity of the assay, we subjected the proteolysis samples to an additional immunoprecipitation step using MAb9 (epitope aa 53 to 59) and MAb6 (aa 191 to 197), or an irrelevant anti-GFP MAb, and resolved the precipitated proteins using the Tris-Tricine system (Fig. 6D). MAb9 precipitated 32P-labeled fragments from wild-type and both mutant proteins, and these showed the characteristic shifts expected if cleavage at E164 or at E164 plus E176 is prevented by mutation. Furthermore, MAb6 precipitated only the topmost fragment from the double mutant but not the smaller fragments from the wild-type and E164D mutant protein, as predicted if the former but not the latter two contain its epitope. Therefore, the corresponding fragments from primed, RNase A-treated wild-type GrpDP resulted from cleavages involving E164 and/or E176, as in chaperone-activated but nonprimed P protein.
MAbs against the chaperone-exposed TP region involving residues 140 to 200 inhibit P-Dɛ complex formation.
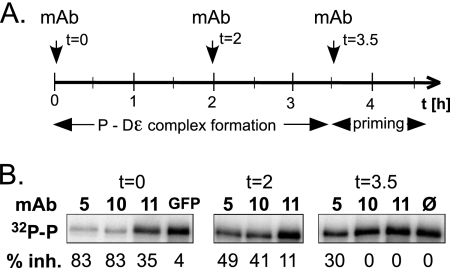
If bound Dɛ RNA blocked protease access to the C-terminal TP region (Fig. 6A and B), then, conversely, MAbs binding to this region might block access of the RNA and consequently inhibit priming. We therefore monitored the effects on in vitro priming efficiency of MAb5 (aa 141 to 147), MAb10 (aa 191 to 197), and MAb11 (aa 53 to 59). P-Dɛ complex formation was initiated by mixing GrpDP with Hsc70/Hsp40, ATP, and Dɛ RNA and allowed to proceed for 3.5 h. The reactions were then adjusted to priming conditions and further incubated for 1 h. The MAbs, and as controls an irrelevant anti-GFP MAb or no antibody, were added immediately at the initiation of complex formation (t = 0) or after 2 h (t = 2 h) or at the start of the priming period (t = 3.5 h). Samples were analyzed for 32P-labeled GrpDP by SDS-PAGE and autoradiography (Fig. 7). The addition, at t = 0, of MAb5 and MAb10 strongly (>80%) and of MAb11 modestly (35%) reduced the priming signals compared to the irrelevant MAb control. When the antibodies were added at 2 h, inhibition was less pronounced for MAb5 and MAb6 (40%-50%) and essentially absent for MAb11. At 3.5 h, only MAb5 showed a residual inhibitory activity (30%) compared to the no-antibody control. These results are expected if MAb5 and MAb10 compete strongly and MAb11 competes weakly, if at all, with RNA binding by the chaperone-activated P protein. One caveat was that some components in the MAb preparations, such as denatured antibody arising from repeated freeze-thaw cycles, could have reduced chaperone activity, and consequently priming efficiency. This was excluded by in vitro luciferase refolding assays which quantitatively assess chaperoning activity (25), in this case of Hsc70/Hsp40 plus ATP. None of the MAb preparations used had any significant negative impact (data not shown).
FIG. 7.
Selective inhibition of P-Dɛ complex formation and priming by MAbs recognizing the C proximal TP region. (A) Experimental setup. GrpDP was incubated with Hsc70, Hsp40, ATP, and Dɛ RNA for 3.5 h and then subjected to priming conditions in the presence of [α-32P]dATP. MAbs directed against different epitopes in TP and against a GFP antibody or no antibody as controls were added at the indicated time points. (B) Effects of MAbs on the formation of primed P protein. Samples from the priming reactions were analyzed by SDS-PAGE and autoradiography. Signals for 32P-labeled P protein were quantified by phosphorimaging. Relative inhibition (percent inhibition values) was derived by normalizing the signal intensities to that of the reaction without added antibody (0% inhibition).
Evidence that the chaperone-exposed TP region directly contributes to Dɛ RNA binding.
Because of the large size of intact antibody molecules, the inhibitory effects of the MAbs described above did not necessarily derive from a direct competition with Dɛ RNA for a common binding site. As a more site-specific tool we used GrpDP with point mutations in the center of the chaperone-exposed region, encompassing the so-called T3 motif, a short stretch of amino acids (A177GILYKR183) that is highly conserved between the TP sequences of avian and mammalian hepadnaviruses (8). In addition to K182 and R183, there are four more basic (three K and one R) amino acids, but only one acidic (E176), in close proximity. As previously shown, double replacement of R183 and K186 by A essentially abolished in vitro Dɛ RNA binding and priming (42), and similar effects were seen with double mutants with the K182R183 motif replaced by AA (33) or EE or TT (39, 40). However, the latter two KR mutations also caused drastically altered MAb accessibility profiles compared to priming-competent variants (8), demonstrating that they had altered structures.
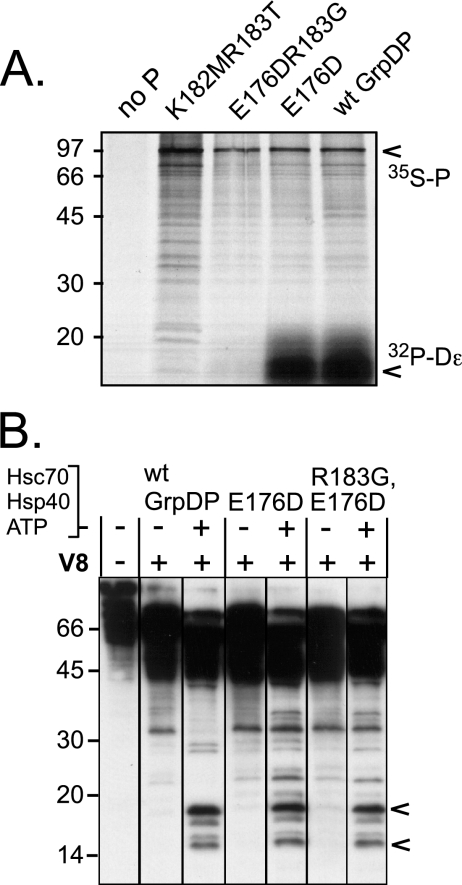
From a library of partially randomized TP constructs we had isolated a double mutant (K182MR183T) that showed no priming activity in RL, and the same priming deficiency was seen for an R183G mutation isolated in the functionally innocuous E176D context. To test for RNA binding, the corresponding GrpDP proteins were 35S labeled by in vitro translation in RL, incubated with 32P-labeled Dɛ RNA, and immobilized to Ni-NTA resin via their C-terminal His tags. Protein and RNA were then simultaneously detected by SDS-PAGE and autoradiography (Fig. 8A). Both the double and the single KR motif mutation caused a complete loss of the RNA signal, although the proteins were present in similar amounts. Hence, these data confirmed the previous reports and further showed that replacement of R183 alone is sufficient to abolish Dɛ RNA binding.
FIG. 8.
RNA-binding deficiency of P mutant R183G is not caused by a gross structural alteration. (A) The single R183G mutation ablates Dɛ RNA binding as efficiently as a K182MR183T double mutation. The indicated proteins were in vitro translated in RL in the presence of [35S]methionine and 32P-labeled Dɛ RNA and immobilized via their His tags to Ni-NTA agarose. Protein and potentially bound RNA were analyzed by SDS-PAGE and autoradiography. The positions of 35S-labeled P protein and of 32P-labeled RNA are indicated by arrowheads. (B) The RNA-binding-deficient R183G mutant undergoes the same chaperone-induced conformational change as RNA-binding-competent P protein. The indicated proteins were incubated with Hsc70, Hsp40, and ATP, or not and subjected to limited V8 proteolysis, followed by immunoblotting with MAb9. All samples were run on the same gel, except that the lanes have been reordered for ease of comparison between chaperone-treated and nontreated samples from one protein. The R183G mutant produced essentially the same chaperone-induced fragments as wild-type GrpDP and the parental, RNA-binding-competent variant E176D. wt, wild type.
To test for potential structural alterations by the R183G mutation, we next used the V8 proteolysis assay to determine whether the conformation of the R183G mutant was chaperone regulated similarly to that of RNA-binding competent P protein. As shown in Fig. 8B, the patterns for the E176DR183G mutant were essentially indistinguishable from those produced by the parental E176D mutant and wild-type GrpDP, with prominent small fragments in the 14- to 18-kDa range occurring only in the presence of Hsc70, Hsp40, and ATP. Hence, the RNA-binding-defective R183G protein can undergo the same chaperone-induced conformational alteration as active P protein, suggesting that its loss of RNA binding relates directly, and not indirectly, to the R183G mutation.
DISCUSSION
We provide here first insights into the molecular mechanism of the chaperone-dependent activation of a hepadnaviral RT. All data are compatible with a model whereby a major function of Hsc70 plus Hsp40 plus ATP is the transient, energy-consuming exposure of a TP-borne template RNA-binding site as a crucial prerequisite for initiation complex formation; Dɛ RNA binding and/or priming may then be sufficient to keep the P protein in an enzymatically active state.
Specificity of the chaperone-induced conformational change in the TP domain.
Limited proteolysis of a protein in distinct conformational states should result in distinct fragmentation patterns. This criterion fully applies to our data, which showed the highly reproducible formation, by V8 protease, of 14- to 18-kDa P-protein fragments in a Hsc70-plus-Hsp40-plus-ATP-dependent fashion. These fragments were formed regardless of the heterologous fusion protein partner or of the presence or absence of the authentic spacer region (Fig. 2). Hence, the capacity for their formation is an inherent property of P protein.
Protease target site mutants demonstrated that the fragments arose from exposure of C-proximal TP residues E164, E176, and (if these were blocked by mutation) E199. An exact assignment of the N proximal V8 cleavage sites was not yet possible. However, the fine structure of the bands in the 14-kDa region (often two, sometimes three closely spaced bands) suggests an initial efficient cleavage event at one of the most upstream E residues, E17 or E19, and, possibly secondary, cleavages at one of the following E residues down to E46. These cleavages occurred independently of chaperone action (Fig. 4B). Hence, the N proximal TP region is constantly solvent exposed. Fragments indicative of cleavage at the centrally located E106 and E124 residues were never observed; hence, this region is always inaccessible.
These data were fully corroborated, and extended to the region around position 140, by the chaperone-plus-ATP dependence of GrpDP immunoprecipitation by MAb5 and MAb6 (with epitopes around residues 141 to 147 and residues 191 to 197, respectively), whereas the epitope of MAb11 and MAb9 (aa 53 to 59) was accessible regardless of the presence of chaperones (Fig. 5). This finding resembles the immunoprecipitation data for vitro-translated P protein (8), where MAb5 and MAb6, but not MAb9, required RIPA buffer for efficient precipitation. Hence, superficially, detergent appears to have structural effects similar to that of specific chaperone action. However, a fundamental difference is that RIPA buffer dissociates P-Dɛ complexes (8), probably by global unfolding of the protein, whereas chaperone-induced exposure of these epitopes likely reflects specific domain movements enabling complex formation; in severely truncated P-protein-glutathione S-transferase fusions this requirement can apparently be substituted for by mild detergent, although not by RIPA buffer (43). Another seeming difference is the poor immunoprecipitation efficiency of MAb5 and MAb6 in RL (8) despite its high chaperone content. Most likely, the comparatively strong signals we obtained in the reconstitution system are due to the presence of the antibodies, in soluble form, during the chaperone incubation period when new activated P molecules are constantly produced. In contrast, in the previous report the RL was at least 25-fold diluted before addition of the immobilized antibodies. P activation requires high concentrations of chaperones and ATP, and even 10-fold dilution efficiently blocks activation in the reconstitution system (2). Given the short half-life of P* (2), the number of activated P molecules in strongly diluted RL is expected to drastically drop within minutes.
In essence, a TP region involving residues from about positions 140 to 200 is specifically exposed by the concerted action of Hsc70, Hsp40, and ATP. No further changes in this region were detectably induced by Bag-1S and by Hsp90 plus Hop, which both can further stimulate Hsp70 activation (35). This finding supports our conclusion that neither Bag-1S nor Hsp90/Hop act by creating a distinct form of activated P protein.
Functional relevance of the chaperone-induced structural alteration.
Several lines of evidence support our conclusion that the chaperone-induced conformational alteration in the TP domain is a structural correlate to functional P protein activation. First, the chaperone requirements for inducing the structural change were absolutely congruent with those enabling P protein to bind Dɛ RNA and initiate DNA synthesis (Fig. 2A). Second, the altered conformation was metastable (Fig. 3), as is the case for P* (2). Third, enzymatically active primed P protein, after digestion of the bound RNA, produced a very similar proteolysis pattern involving similar cleavage events. Finally, the TP region becoming exposed by chaperone action appears to directly contribute to Dɛ RNA binding.
As for functional P protein activation, the combined presence of Hsc70, Hsp40, and ATP was required for the formation of the chaperone-specific V8 fragments. Only minute amounts of similarly sized fragments were produced if one of these factors was omitted (Fig. 2A). Likewise, the amounts of the conformationally altered P-protein molecules rapidly decreased, by 70 to 80%, within the first 10 min of ATP depletion (Fig. 3). Hence, the majority of molecules in the chaperone-altered state is metastable. Hsc70 with bound ADP (Hsc70ADP) binds strongly to substrate, and substrate release requires exchange of the bound ADP for ATP (27). Possibly, then, the small fraction of molecules remaining in the altered state in the absence of ATP may stably be complexed with Hsc70ADP, but this explanation requires further experimental confirmation.
Proteolysis of primed P protein revealed similarities but also distinct differences from the chaperone-activated but nonprimed molecules. Dɛ RNA-containing complexes were nearly quantitatively converted into one major 45-kDa fragment (Fig. 6A), indicating that the bound RNA, and/or the DNA primer, conferred to this fragment a compact protease-resistant structure. If we assume a similarly efficient N proximal cleavage as in nonprimed P protein, the C-proximal cleavage site would be located in a region downstream of the RT active-site motif YMDD (positions 511 to 514 in the full-length protein nomenclature) that encompasses numerous E residues (at positions 538, 555, 565, 571, 572, and 577), as well as many other charged and polar residues, as often observed for surface-exposed regions. This is close to the proposed C-terminal border of the protease-resistant fragments obtained, in the presence of Dɛ RNA, from RL-translated P protein (39, 40). The suggested N-terminal cleavage site(s) in the spacer is deleted in our constructs. This, and the use of proteases with different specificities (papain and trypsin) is likely to account for the differences in fragment size. However, our data fully support the Dɛ RNA-induced generation of a highly protease-resistant P fragment encompassing about two-thirds of the RT domain.
Upon RNase A treatment, small fragments were generated that were similar in size to those generated by chaperone action only, and their generation involved the same C-terminal residues E164, E176, and E199 (Fig. 6D). Hence, in primed RNA containing P molecules the chaperone-exposed V8 sites were occluded but were reexposed upon RNA digestion. However, in contrast to nonprimed P protein, the altered conformation was stable in the absence of ATP and intact Dɛ RNA, with a half-life exceeding 2 h (Fig. 6B). Hence, prior priming stabilized a new structure similar to the one transiently induced by chaperone action. Whether this stabilization is mediated by additional rearrangements in other domains of the protein, or residual bound RNA fragments from incomplete nuclease digestion, and/or the covalently linked DNA primer remains to be determined. Similarly, it is currently not known which fraction of the P molecules with a chaperone-altered conformation binds ɛ RNA and which fraction of these becomes eventually competent for DNA synthesis.
However, a causal connection between the chaperone-induced exposure of the C-terminal TP region and P-protein activation is further supported by the selective inhibition of priming activity by MAbs binding in this region (MAb5 and MAb10) when they were added early during P-Dɛ complex formation (Fig. 7). Thus, the MAbs and Dɛ RNA compete for the transiently chaperone-exposed TP region. We note that these data are not fully congruent with those reported for RL-derived P protein (8), where a stronger priming inhibition (80 to 90%) by MAbs binding to the N-proximal TP part than by MAb5 and MAb6 (40 to 60%) was observed. However, both studies agree that the latter MAb epitopes are not continuously exposed; it is therefore likely that the partial inhibition by these MAbs in RL occurs, as in our reconstitution system, via the fraction of chaperone-activated P molecules. The more effective inhibition by MAbs against the N proximal TP region may relate to a more efficient sequestration of P protein (activated or not) into antibody complexes due to the much higher ratio of MAbs to P (3 μg of MAb per 20 μl in vitro translation reaction which, in our experience, would yield ca. 20 ng of P protein) compared to our experiments (0.7 μg of MAb per 150 ng of P protein). More important is the seeming discrepancy in interpretation. Cao et al. (8) concluded that occlusion of the epitopes around positions 140 and 190 is a characteristic of active P proteins and that their exposure, as in the KR double mutants, is a characteristic of inactive P proteins. This is only superficially contradictory to our data showing that transient exposure of this region is essential for P activation. As outlined below, we consider it likely that at some point during activation the Dɛ RNA binds to this region; however, prior to and after Dɛ binding the T3 motif comprising the two basic residues could well fulfill its proposed “molecular contact point” function (8) with other partners. Furthermore, from both data sets it is highly conceivable that constant, chaperone-independent exposure of the epitopes around aa 140 and 190 is a hallmark of inactive, or possibly more correctly, inactivatable P proteins.
Is the chaperone-exposed C-proximal TP region part of the Dɛ RNA binding site of P protein?
That TP residue Y96 must be close to the template region in ɛ for priming is obvious; however, the evidence for a direct role of another specific TP region in ɛ RNA binding was mostly indirect. Priming inhibition with some of the MAbs suggested, but did not prove, a direct competition for a narrowly defined target site, given the dimensions of an antibody and that of an ∼60-nucleotide RNA stem-loop such as ɛ (9, 14). Simultaneous exchanges of two of the various basic residues in the C-proximal TP region had previously been reported to ablate RNA binding (33, 39, 40), but EE and TT double mutations of the K182R183 motif also altered P-protein structure (8). In contrast, the single R183G mutant used here, although completely defective for RNA binding (Fig. 8A), shared all structural features, in the presence or absence of chaperones, with functionally active P protein (Fig. 8B). Hence, R183 is either in direct contact with the RNA, or its replacement by G exerts a locally restricted, but not a global, effect on nearby residues that are essential for RNA binding. We therefore consider it likely that the region around R183 is directly involved in forming part of the Dɛ RNA-binding site of P protein, which probably requires additional contacts to the RT domain.
Model for chaperone-mediated hepadnaviral P protein activation.
A model for P protein activation, incorporating the data from the present study, is shown in Fig. 9. The initial key event is the Hsc70-, Hsp40-, and ATP-dependent transient conversion of inactive P protein into an ɛ RNA-binding-competent state, P*. P* differs structurally from P protein by exposure of the C-proximal TP region including MAb epitopes, V8 protease sites, and important parts of the ɛ RNA-binding site (see legend to Fig. 9 for details). ɛ binding reoccludes the MAb epitopes and V8 sites and leads to the formation of stable, priming-competent P-ɛ complexes. RNA removal after priming re-exposes these sites; however, in primed P protein this altered structure, termed P′ for distinction from P*, is maintained without chaperones and ATP. It is tempting to speculate that P′ might be similar to active P protein, after the switch from initiation to elongation mode that occurs in virtually all polymerases (36), and that for P protein is likely to happen after ɛ has been removed and replaced as a template by the 3′-proximal part of the pgRNA. There is, in fact, circumstantial evidence supporting this view by way of the so-called trans-reaction, i.e., template-dependent reverse transcription of exogenous RNA by DHBV P protein. Accepting the exogenous RNA template required prior digestion of the endogenous nucleic acids, including ɛ, and yet occurred efficiently only when P protein had been translated in the presence of ɛ (39, 40), suggesting the prior encounter with ɛ had imprinted some functionally important alteration on P protein.
FIG. 9.
Structural model for hepadnaviral P protein activation. Initially, P protein is in a stable nonactivated state, in which the C-proximal TP region including E164, E176, and E199 and the epitopes for MAb5, MAb6, and MAb10 is occluded, whereas the N-proximal E residues in TP and the epitopes for MAb9 and MAb11 are, and remain, accessible. Occlusion might involve the RH domain, as suggested by the partial chaperone independence of P proteins with large C-terminal truncations (2, 43). ATP-consuming Hsc70 plus Hsp40 action transiently exposes the C-proximal sites, including R183, which is involved in RNA binding. In this metastable activated state P*, the corresponding MAbs, and ɛ can compete for binding to the C-proximal TP region. ɛ binding generates a stable priming-competent P*-Dɛ complex in which the RNA blocks the access of V8 protease and the respective MAbs. RNase A treatment creates a new state P′ in which the same sites are exposed as in chaperone-activated P*; however, different from P*, P′ remains stable without ATP-consuming chaperone activity. P′, here artificially created by RNA digestion, may resemble P protein in transition from ɛ-dependent initiation to elongation mode, which likely accompanies the replacement of ɛ by DR1* as the RNA template.
Hence overall, hepadnaviral RT activation appears to follow a “double hit and then run” mechanism. The first hit is transient chaperone activation into an ɛ binding-competent state which, however, has no sustained consequences unless the second hit, i.e., ɛ RNA binding, occurs. Thereafter, P protein might have gained autonomous polymerase activity and be able to complete genome replication, possibly without further chaperone assistance.
Acknowledgments
We gratefully acknowledge J. Tavis for providing MAbs and J. Buchner for providing purified Hsp90. We thank Kerstin Semmler for excellent technical assistance.
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG Na154/7-2) and is part of the activities of the VIRGIL European Network of Excellence on Antiviral Drug Resistance sponsored by the European Commission (LSHM-CT-2004-503359).
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Beck, J., and M. Nassal. 1996. A sensitive procedure for mapping the boundaries of RNA elements binding in vitro translated proteins defines a minimal hepatitis B virus encapsidation signal. Nucleic Acids Res. 24:4364-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck, J., and M. Nassal. 2003. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis B virus reverse transcriptase, an assumed Hsp90 client protein. J. Biol. Chem. 278:36128-36138. [DOI] [PubMed] [Google Scholar]
- 3.Beck, J., and M. Nassal. 1998. Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 18:6265-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, J., and M. Nassal. 2007. Hepatitis B virus replication. World J. Gastroenterol. 13:48-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, J., and M. Nassal. 2004. In vitro reconstitution of epsilon-dependent duck hepatitis B virus replication initiation. Methods Mol. Med. 95:315-325. [DOI] [PubMed] [Google Scholar]
- 6.Beck, J., and M. Nassal. 2001. Reconstitution of a functional duck hepatitis B virus replication initiation complex from separate reverse transcriptase domains expressed in Escherichia coli. J. Virol. 75:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehmer, D., S. Rüdiger, C. S. Gässler, D. Klostermeier, L. Packschies, J. Reinstein, M. P. Mayer, and B. Bukau. 2001. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8:427-432. [DOI] [PubMed] [Google Scholar]
- 8.Cao, F., M. P. Badtke, L. M. Metzger, E. Yao, B. Adeyemo, Y. Gong, and J. E. Tavis. 2005. Identification of an essential molecular contact point on the duck hepatitis B virus reverse transcriptase. J. Virol. 79:10164-10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flodell, S., M. Petersen, F. Girard, J. Zdunek, K. Kidd-Ljunggren, J. Schleucher, and S. Wijmenga. 2006. Solution structure of the apical stem-loop of the human hepatitis B virus encapsidation signal. Nucleic Acids Res. 34:4449-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontana, A., P. P. de Laureto, B. Spolaore, E. Frare, P. Picotti, and M. Zambonin. 2004. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 51:299-321. [PubMed] [Google Scholar]
- 11.Fontana, A., P. Polverino de Laureto, V. De Filippis, E. Scaramella, and M. Zambonin. 1997. Probing the partly folded states of proteins by limited proteolysis. Fold. Des. 2:R17-R26. [DOI] [PubMed] [Google Scholar]
- 12.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA. [Google Scholar]
- 13.Geysen, H. M., S. J. Rodda, T. J. Mason, G. Tribbick, and P. G. Schoofs. 1987. Strategies for epitope analysis using peptide synthesis. J. Immunol. Methods 102:259-274. [DOI] [PubMed] [Google Scholar]
- 14.Girard, F. C., O. M. Ottink, K. A. Ampt, M. Tessari, and S. S. Wijmenga. 2007. Thermodynamics and NMR studies on duck, heron, and human HBV encapsidation signals. Nucleic Acids Res. 35:2800-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houmard, J., and G. R. Drapeau. 1972. Staphylococcal protease: a proteolytic enzyme specific for glutamoyl bonds. Proc. Natl. Acad. Sci. USA 69:3506-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, J., and D. Anselmo. 2000. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 74:11447-11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., and M. Boyer. 2006. Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 80:2141-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, J., and C. Seeger. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 93:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, J., D. Toft, D. Anselmo, and X. Wang. 2002. In vitro reconstitution of functional hepadnavirus reverse transcriptase with cellular chaperone proteins. J. Virol. 76:269-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, J., D. O. Toft, and C. Seeger. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 16:59-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu, K., J. Beck, and M. Nassal. 2004. SELEX-derived aptamers of the duck hepatitis B virus RNA encapsidation signal distinguish critical and non-critical residues for productive initiation of reverse transcription. Nucleic Acids Res. 32:4377-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard, S. J. 1998. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta 1382:191-206. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 24.Lanford, R. E., L. Notvall, H. Lee, and B. Beames. 1997. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71:2996-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, G. J., and E. Vierling. 2000. A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol. 122:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandart, E., A. Kay, and F. Galibert. 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J. Virol. 49:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer, M. P., and B. Bukau. 2005. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol. Life Sci. 62:670-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picard, V., E. Ersdal-Badju, A. Lu, and S. C. Bock. 1994. A rapid and efficient one-tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 22:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pratt, W. B., M. D. Galigniana, Y. Morishima, and P. J. Murphy. 2004. Role of molecular chaperones in steroid receptor action. Essays Biochem. 40:41-58. [DOI] [PubMed] [Google Scholar]
- 30.Protzer, U., M. Nassal, P. W. Chiang, M. Kirschfink, and H. Schaller. 1999. Interferon gene transfer by a hepatitis B virus vector efficiently suppresses wild-type virus infection. Proc. Natl. Acad. Sci. USA 96:10818-10823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 32.Schultz, U., E. Grgacic, and M. Nassal. 2004. Duck hepatitis B virus: an invaluable model system for HBV infection. Adv. Virus Res. 63:1-70. [DOI] [PubMed] [Google Scholar]
- 33.Seeger, C., E. H. Leber, L. K. Wiens, and J. Hu. 1996. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology 222:430-439. [DOI] [PubMed] [Google Scholar]
- 34.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl, M., M. Retzlaff, M. Nassal, and J. Beck. 2007. Chaperone activation of the hepadnaviral reverse transcriptase for template RNA binding is established by the Hsp70 and stimulated by the Hsp90 system. Nucleic Acids Res. doi: 10.1093/nar/gkm628. [DOI] [PMC free article] [PubMed]
- 36.Steitz, T. A. 2006. Visualizing polynucleotide polymerase machines at work. EMBO J. 25:3458-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 38.Takayama, S., and J. C. Reed. 2001. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3:E237-E241. [DOI] [PubMed] [Google Scholar]
- 39.Tavis, J. E., and D. Ganem. 1996. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J. Virol. 70:5741-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavis, J. E., B. Massey, and Y. Gong. 1998. The duck hepatitis B virus polymerase is activated by its RNA packaging signal, epsilon. J. Virol. 72:5789-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, G. H., and C. Seeger. 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71:663-670. [DOI] [PubMed] [Google Scholar]
- 42.Wang, G. H., F. Zoulim, E. H. Leber, J. Kitson, and C. Seeger. 1994. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 68:8437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., X. Qian, H. C. Guo, and J. Hu. 2003. Heat shock protein 90-independent activation of truncated hepadnavirus reverse transcriptase. J. Virol. 77:4471-4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber, M., V. Bronsema, H. Bartos, A. Bosserhoff, R. Bartenschlager, and H. Schaller. 1994. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68:2994-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, J. C., V. R. Agashe, K. Siegers, and F. U. Hartl. 2004. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell. Biol. 5:781-791. [DOI] [PubMed] [Google Scholar]
- 47.Zoulim, F., and C. Seeger. 1994. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68:6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]