Abstract
The feline junctional adhesion molecule A (fJAM-A) is a functional receptor for feline calicivirus (FCV). fJAM-A is a member of the immunoglobulin superfamily (IgSF) and consists of two Ig-like extracellular domains (D1 and D2), a membrane-spanning domain, and a short cytoplasmic tail. To identify regions of fJAM-A that interact with FCV, we purified recombinant fJAM-A ectodomain and D1 and D2 domains. We found that preincubation of FCV with the ectodomain or D1 was sufficient to inhibit FCV infection in plaque reduction assays. In enzyme-linked immunosorbent assays, FCV binding to fJAM-A ectodomain was concentration dependent and saturable; however, FCV bound D1 alone weakly and was unable to bind D2. To characterize FCV binding to surface-expressed fJAM-A, we transfected truncated and chimeric forms of fJAM-A into a nonpermissive cell line and assayed binding by flow cytometry. Only D1 was necessary for FCV binding to cells; all other domains could be replaced. Using a structure-guided mutational approach, we identified three mutants of fJAM-A within D1 (D42N, K43N, and S97A) that exhibited significantly decreased capacities to bind FCV. In contrast to our finding that D1 mediated FCV binding, we found that all domains of fJAM-A were necessary to confer susceptibility to FCV infection. Furthermore, surface expression of fJAM-A was not sufficient to permit FCV infection by all of the isolates we investigated. This indicates that (i) other cellular factors are required to permit productive FCV infection and (ii) individual FCV isolates differ in the factors they require.
Members of the Caliciviridae are small, nonenveloped viruses that carry a positive-stranded RNA genome of ∼7 to 8 kb. Despite including important human and animal pathogens, the family Caliciviridae is relatively understudied. Feline caliciviruses (FCVs), members of the genus Vesivirus, are highly contagious pathogens that cause a variety of mild to severe disease syndromes in cats. Recently, feline junctional adhesion molecule A (fJAM-A) was identified as a functional receptor for FCV, the first proteinaceous receptor identified for any member of the Caliciviridae (28).
Virus entry often requires multiple interactions between virus particles and cell surface receptors. These interactions are critical determinants of productive virus entry and are often important factors in viral pathogenesis. Increasingly, it is being found that viral entry is not a passive process but often involves activation of distinct cellular signaling pathways that program correct endocytic uptake of the viral particle and/or prime the cell for viral replication (32). Both a carbohydrate (α-2,6 sialic acid) and the cell surface glycoprotein fJAM-A have been shown to be involved in FCV binding to cells (28, 48). The roles these molecules play in FCV entry have not been determined.
JAM-A is a type I transmembrane glycoprotein (mass, 36 to 41 kDa) and a member of the immunoglobulin superfamily (IgSF). It consists of an N-terminal signal peptide, an extracellular domain (composed of two Ig-like domains, a membrane-distal D1 and a membrane-proximal D2), a transmembrane domain, and a short cytoplasmic domain (13, 31). JAM-A localizes to intercellular tight junctions of endothelial and epithelial cells in humans and mice and is expressed on the surfaces of platelets, leukocytes, and erythrocytes (27, 30, 34, 39). In solution, JAM-A forms homodimers that are stabilized by ionic and hydrophobic interactions between residues in the D1 domain dimerization motif. Homodimer formation is critical in establishing a regulated tight-junctional barrier between cells, and it is thought that homophilic interactions between JAM-A dimers on opposing cell surfaces are important for forming intercellular tight junctions (27, 30, 34, 39, 51). JAM-A is also believed to play an important role in modulating leukocyte diapedesis and platelet aggregation (1, 2, 23, 29, 34).
Several different virus families use IgSF cell surface proteins as receptors. The σ1 attachment protein of mammalian orthoreoviruses (Reoviridae) engages residues present within the dimerization motif of the human JAM-A (hJAM-A) D1 domain (4, 8, 15, 19). Coxsackie B viruses (CVB) (Picornaviridae) and most adenovirus (Adenoviridae) subgroups utilize the coxsackievirus and adenovirus receptor (CAR) (6). Like JAM-A, CAR has two extracellular Ig-like domains, localizes to tight junctions in polarized epithelial cells or to sites of cell-cell contact, and forms homodimers mediated by a dimerization motif within its D1 domain dimerization motif (9, 34). Comparable to the interaction of the reovirus σ1 protein with JAM-A, the adenovirus fiber knob protein also interacts with residues found mostly within the dimer interface of the CAR D1 domain (7, 25). CVB interact with the distal end of the CAR D1 domain via surface residues in the canyon region of the capsid that surrounds the fivefold icosahedral axes (20). Other Picornaviridae, including human rhinovirus type 14 (HRV14) and HRV16; coxsackie A virus 21; and poliovirus type 1 (PV1), PV2, and PV3, also use IgSF receptors (ICAM-I, ICAM-I, and PVR, respectively) (44).
Makino et al. showed that expression of fJAM-A cDNA in nonpermissive hamster and human cell lines conferred susceptibility to FCV infection and that infection could be blocked with fJAM-A antiserum (28). However, a direct interaction between the virus and receptor was not investigated. In the present study, we used both sequence comparisons and the structures of the human and murine JAM-A molecules to guide mutational analyses of the domains and residues of fJAM-A required for FCV binding and infection. We show that FCV binds to the membrane-distal D1 domain of fJAM-A close to the linker region between the D1 and D2 domains. Replacing two specific fJAM-A residues (Asp 42 and Lys 43) with the corresponding human sequence substantially diminished virus binding and infection. Although the D1 domain was necessary for FCV binding, it was insufficient for infection. We found that both the D1 and D2 domains of fJAM-A were required for saturable binding of FCV; the cytoplasmic and transmembrane domains were required, in addition to the extracellular domain, for infection of nonpermissive CHO cells. Lastly, we found that expression of fJAM-A in nonpermissive cell lines was not sufficient for productive infection by all of the FCV isolates we investigated.
MATERIALS AND METHODS
Cells and viruses.
Crandell-Reese feline kidney (CRFK; ATCC CCL-94) and HeLa (ATCC CCL-2) cells were grown in Eagle's minimal essential medium (EMEM) (CellGro) supplemented with 5% fetal bovine serum (HyClone), 100 U ml−1 of penicillin, 100 μg ml−1 streptomycin, 0.25 μg ml−1 amphotericin B, 1 mM sodium pyruvate, and nonessential amino acids (CellGro). Adherent Chinese hamster ovary (CHO-K1; ATCC CCL-61) cells were grown in Ham's F12 (HyClone) supplemented with 10% fetal bovine serum, 100 U ml−1 of penicillin, 100 μg ml−1 streptomycin, 0.25 μg ml−1 amphotericin B, 1 mM sodium pyruvate, and nonessential amino acids. Suspension Chinese hamster ovary (CHO-S) cells (Invitrogen) were grown in CHO-serum-free medium (Gibco). Flp-In T-REx 293 (Invitrogen) cells were maintained in Dulbecco's modified essential medium (HyClone) supplemented with 10% fetal bovine serum, 100 U ml−1 of penicillin, and 100 μg ml−1 streptomycin. Chinese hamster lung (CHL; ATCC CRL-1935) and Vero (ATCC CCL-81) cells were grown in EMEM supplemented with 10% fetal bovine serum, 100 U ml−1 of penicillin, 100 μg ml−1 streptomycin, 0.25 μg ml−1 amphotericin B, 1 mM sodium pyruvate, and nonessential amino acids.
The F9 vaccine strain (VR-782) of FCV was obtained from the ATCC. The viral isolates FCV-5, Deuce, Kaos, 127, and 131 were previously characterized (37). Third-passage viral stocks were prepared from twice-plaque-purified viruses amplified in CRFK cells.
Reverse transcription-PCR, 3′- and 5′-random amplification of cDNA ends, and sequencing of fJAM-A.
A cDNA of fJAM-A was prepared from total RNA isolated from CRFK cells using reverse transcription-PCR and 5′ and 3′ random amplification of cDNA ends. This cDNA was sequenced for correctness (Cornell University Life Sciences Core Laboratories Center) and cloned into the pCI-neo vector (Promega) to create the pCI-fJAM-A plasmid. Details of the primers used for amplification and the cloning strategy are available upon request.
Generation and purification of recombinant fJAM-A-GST fusion proteins.
The fJAM-A extracellular domain (amino acid residues 26 to 232) and the single Ig-like domains D1 (residues 26 to 127) and D2 (residues 132 to 232) were amplified by PCR from the pCI-fJAM-A plasmid using primers that incorporated BamHI and SalI restriction sites (Table 1). Amplified fragments were cloned into a modified pET-41A vector (Novagen) that had a 3C protease cleavage site introduced following the N-terminal glutathione S-transferase (GST) and six-His tags. Escherichia coli (BL21) was grown at 25°C, and protein expression of the recombinant plasmids was induced by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacteria were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS) (137 mM NaCl, 3 mM KCl, 8 mM Na2HPO4 [pH 7.5]) containing 0.2% Triton X-100, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride, and lysed by sonication. Recombinant proteins were purified by affinity chromatography using GSTrap Fast Flow columns and Glutathione Sepharose 4 Fast Flow beads (GE Healthcare). GST was expressed and purified from the empty, modified pET41A vector as described above. When GST-free proteins were desired, GST and His tags were cleaved from the recombinant protein using HRV 3C protease (Novagen) and removed from the fJAM-A-containing fraction by passing the samples back over GSTrap columns. The cleaved proteins contained 4 additional nonnative N-terminal amino acids (Gly-Pro-Arg-Gly) and 17 additional nonnative C-terminal amino acids, including a His tag (Val-Asp-Lys-Leu-Ala-Ala-Ala-Leu-Glu-His-His-His-His-His-His-His-His).
TABLE 1.
GST fusion primers
| Domain | Orientation | Sequence |
|---|---|---|
| Ectodomain | Sense | 5′-TAGGATCCGGCAGGGGCGCAGTG-3′ |
| Antisense | 5′-AGGTCGACCTCCGCGGCTTCCAT-3′ | |
| D1 | Sense | 5′-TAGGATCCGGCAGGGGCGCAGTG-3′ |
| Antisense | 5′-AGGTCGACCACAGTGAGCTGGAC-3′ | |
| D2 | Sense | 5′-TAGGATCCTCCAAGCCCACGGTC-3′ |
| Antisense | 5′-AGGTCGACCTCCGCGGCTTCCAT-3′ |
Generation of rabbit fJAM-A-specific antiserum.
Endotoxin was removed from the purified fJAM-A ectodomain using an AffinityPak Detoxigel endotoxin-removing gel (Pierce). Antibody was produced in a rabbit inoculated three times at 3-week intervals with the purified ectodomain (Cornell Center for Research Animal Resources). Preimmune samples were tested by both enzyme-linked immunosorbent assay (ELISA) and immunofluorescence to verify lack of reactivity. Antiserum was aliquoted and stored at −20°C.
Electrophoresis and immunoblot analysis of recombinant fJAM-A.
Soluble, recombinant fJAM-A full-length ectodomain and single Ig-like loop (D1 and D2) GST fusion and 3C-cleaved proteins were separated on 4 to 15% gradient sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis gels (Bio-Rad). The gels were stained with Coomassie blue or transferred to nitrocellulose membranes. The membranes were blocked in PBS with 5% nonfat milk powder and then incubated with the fJAM-A rabbit antiserum diluted in blocking buffer. After being washed with PBS-0.1% Tween 20, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Jackson Laboratories). After the membranes were washed, SuperSignal enhanced-chemiluminescence substrate (Pierce) was used to detect immunoreactive bands. The membranes were stripped (Restore Western Blot Stripping Buffer; Pierce), reblocked with blocking buffer, and then incubated with an HRP-conjugated anti-GST antibody (GE Healthcare) diluted in blocking buffer. Detection of immunoreactive bands was performed as described above.
Detection of surface fJAM-A in transiently transfected CHO cells.
CHO-K1 cells (3 × 105 per well) in six-well plates containing 18-mm glass coverslips were transiently transfected with pCI-fJAM-A or empty vector using FuGENE 6 (Roche) according to the manufacturer's directions. At 24 h posttransfection (p.t.), coverslips were washed with PBS, incubated with the fJAM-A rabbit antiserum (diluted in PBS plus 1% bovine serum albumin [BSA]) for 1 h on ice, washed, fixed in 2% paraformaldehyde in PBS for 15 min, washed, and then incubated with Alexa 594-conjugated goat anti-rabbit IgG (Invitrogen) and DAPI (4′-6′-diamidino-2-phenylindol; Invitrogen) for 1 h. The coverslips were mounted on slides using Prolong (Invitrogen). A Nikon TE2000 inverted microscope equipped with a 60× 1.4-numerical-aperture oil objective linked to a Coolsnap HQ charge-coupled-device camera (Roper) and Openlab software (Improvision) were used to collect fluorescence and phase-contrast images. The images were then prepared for publication using Photoshop and Illustrator software (Adobe Systems).
Specificity of fJAM-A antiserum.
CRFK and HeLa cells were seeded in six-well plates containing coverslips as described above and incubated overnight. Cells on coverslips were washed and then incubated with either fJAM-A rabbit antiserum or a mouse monoclonal antibody (MAb) against hJAM-A (BV-16; Santa Cruz Biotechnology). The cells were washed and fixed before being incubated with Alexa-594-conjugated goat anti-rabbit or goat anti-mouse IgG and DAPI. The coverslips were mounted on slides with Prolong. Images were obtained as described above.
fJAM-A antiserum plaque reduction.
Tenfold dilutions of fJAM-A rabbit antiserum in PBS plus 1% BSA were incubated with CRFK cells for 1 h on ice. Control wells were incubated with PBS plus 1% BSA or a 1:10 dilution of rabbit preimmune serum in PBS plus 1% BSA. Virus was adsorbed (∼20 PFU/well, as titered on CRFK cells) to cell monolayers for 1 h at room temperature. The monolayers were then overlaid with EMEM containing 5% fetal bovine serum and 1% Bacto Agar. After incubation at 37°C for 48 h in humidified 5% CO2, the overlay was removed and the cells were fixed with 10% buffered formalin, and stained with 1% (wt/vol) crystal violet solution. The plaques were counted, and percent reduction from controls was calculated.
fJAM-A ectodomain, D1, and D2 plaque reduction.
Twofold dilutions of purified fJAM-A ectodomain, D1, D2, or GST were incubated with FCV-5 (∼20 PFU/well) in Dulbecco's modified essential medium plus 0.1% BSA for 1 h on ice; control samples were incubated with medium only. The virus-receptor mixture was adsorbed to CRFK monolayers for 1 h at room temperature. The monolayers were then overlaid with 3 ml of EMEM containing 5% fetal bovine serum and 1% Bacto Agar. After incubation at 37°C for 48 h in humidified 5% CO2, the overlay was removed and the plaques were stained as described above. The plaques were counted, and percent reduction from controls was calculated.
ELISA binding of FCV to fJAM-A ectodomain, D1, and D2.
Purified fJAM-A ectodomain, D1, or D2 (5 μM solutions) in carbonate/bicarbonate buffer (Sigma) was bound to 96-well ELISA plates at 4°C overnight. The wells were blocked with PBS containing 0.05% Tween 20 plus 0.5% BSA for 1 h at room temperature and washed three times with PBS containing 0.05% Tween 20. Two dilutions of FCV-5 (initial dilution, 1:10) were prepared, and 50 μl of each dilution was bound to plates for 1 h at room temperature. After the plates were washed, bound virus was detected with rabbit anti-FCV-5, followed by HRP-conjugated goat anti-rabbit IgG. Antibodies diluted in blocking buffer were incubated with the plates for 1 h at room temperature. After the plates were washed, substrate, 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (Sigma) in a citric acid-sodium phosphate buffer was added to each well and incubated for 15 min. Absorbance was measured at 595 nm on a Microplate Biokinetics Reader (BioTek Instruments).
Creation of fJAM-A deletion, chimeric, and point mutants.
Deletion and chimeric mutants were prepared using a general strategy of amplifying individual domains, introducing unique restriction sites on either end, and then joining the domains together in the expression vector pCI-Neo to create the desired constructs. fJAM-A deletion mutants are named according to the Ig-like domain deleted. The deletion mutants were ΔD1/D2 (fJAM-A amino acid residues 1 to 27 and 128 to 298; fJAM-A residue 129 changed from a valine to a glycine) and D1/ΔD2 (fJAM-A 1 to 127 and 233 to 298). Chimeric constructs were named to indicate the sources of the Ig-like domains from N to C termini (fJAM-A, fJ; hJAM-A, hJ; or human CAR [hCAR], hC). The chimeric mutants created were hJ/fJ (hJAM-A 1 to 128 and fJAM-A 128 to 298), hC/fJ (hCAR 1 to 144 and fJAM-A 128 to 298), fJ/hJ (fJAM-A 1 to 131, hJAM-A 133 to 234, and fJAM-A 234 to 298), and fJ/hC (fJAM-A 1 to 127, hCAR 138 to 227, and fJAM-A 234 to 298). The GPI-fJAM-A chimeric construct was created by coupling the fJAM-A ectodomain to the glycosylphosphatidylinositol (GPI) anchor of human decay accelerating factor (hDAF) (fJAM-A 1 to 231 and hDAF 345 to 364; an additional alanine was added between the segments). The full-length open reading frames of hCAR and hJAM-A were also cloned into pCI-Neo. The PCR primers used to generate the deletion and chimeric mutants are listed in Table 2. fJAM-A D1 point mutants were generated using the Quick Change site-directed mutagenesis strategy (Stratagene). The PCR primers used to generate the point mutants are listed in Table 3. Common molecular-cloning techniques were utilized to create the constructs discussed above. Details of the amplification conditions and cloning strategies are available upon request. All constructs were sequenced for correctness.
TABLE 2.
Chimeric/deletion mutant primers
| Construct | Template | Orientation | Sequence (5′-3′)a |
|---|---|---|---|
| fJAM-A | fJAM-A | Sense | TAGCTAGCATCGCCAATGGGGACCGA |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| ΔD1/D2 | fJAM-A | Sense | ACACGTACGGGGAGGTCAGCGTCCAGCTCACTGTGTTGAATGTGGGGGGCATT |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| D1/ΔD2 | fJAM-A | Sense | TAGCTAGCATCGCCATGGGGACCGA |
| Antisense | TAACCTAGGCGGCCCAACGCCAGGGA | ||
| fJAM-A | Sense | ATCCTAGGTCCTCCATCCAAGCCCACG | |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| hJAM-A | hJAM-A | Sense | TAGCTAGCATCGCCATGGGGACAAAG |
| Antisense | ATCTCGAGGGCCTCACACCAGGAATGA | ||
| hCAR | hCAR | Sense | ATGCTAGCGCCACCATGGCGCTCCT |
| Antisense | CTCTCGAGCTATACTATAGACCCATCC | ||
| fJ/hJ | hJAM-A | Sense | ATCGTACGGGGAGGTCAGCGTCCAGCTCACTGTGCTTGTGCCTCCATCCAAG |
| Antisense | ATGCGGCCGCCACAATGCCCCCCACATTCCGCTCCACAGCTTCCAT | ||
| fJAM-A | Sense | AAGCGGCCGCACTTGTCACACTCATTCTCCT | |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| fJ/hC | hCAR | Sense | ATCGTACGGGGAGGTCAGCGTCCAGCTCACTGTGCTTGTTAAGCCTTCAGGT |
| Antisense | ATGCGGCCGCCACAATGCCCCCCACATTTAGACGCAACAGGCACT | ||
| fJAM-A | Sense | AAGCGGCCGCACTTGTCACACTCATTCTCCT | |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| hJ/fJ | hJAM-A | Sense | TAGCTAGCATCGCCATGGGGACAAAG |
| Antisense | TAGGTACCACGATGAGCTTGACCTT | ||
| fJAM-A | Sense | ATGGTACCTCCATCCAAGCCCA | |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| hC/fJ | hCAR | Sense | ATGCTAGCGCCACCATGGCGCTCCT |
| Antisense | TAGGTACCGCACCTGAAGGCTTAACAAG | ||
| fJAM-A | Sense | ATGGTACCTCCATCCAAGCCCA | |
| Antisense | TAACGCGTGGGACCAGGGTCACACCAG | ||
| GPI-fJAM-A | fJAM-A | Sense | TAGCTAGCATCGCCATGGGGACCGA |
| Antisense | TAATGCGGCCGCTTCCATGCGCACAGCCTCT | ||
| hDAF | Sense | TAATGCGGCCGCTCCAAATAAAGGAAGTGGAACC | |
| Antisense | ATTACGCGTTAAGTCAGCAAGCCCATG |
Restriction sites are underlined.
TABLE 3.
fJAM-A D1 point mutants
| Mutation | Orientation | Primer sequence (5′-3′)a |
|---|---|---|
| Y31H | Sense | CAGGGGCGCAGTGCATACTTCTGAGCCCG |
| Antisense | CGGGCTCAGAAGTATGCACTGCGCCCCTG | |
| D36E | Sense | GTGTATACTTCTGAGCCCGAGGTCAGGGTACCTGAGGAC |
| Antisense | GTCCTCAGGTACCCTGACCTCGGGCTCAGAAGTATACAC | |
| E41Q | Sense | CCCGATGTCAGGGTACCTCAGGACAAACCCGCCAAGTTG |
| Antisense | CAACTTGGCGGGTTTGTCCTGAGGTACCCTGACATCGGG | |
| D42N | Sense | CCCGATGTCAGGGTACCTGAGAACAAACCCGCCAAGTTG |
| Antisense | CAACTTGGCGGGTTTGTTCTCAGGTACCCTGACATCGGG | |
| D42K | Sense | GCCCGATGTCAGGGTACCTGAGAAGAAACCCGCCAAGTTG |
| Antisense | CAACTTGGCGGGTTTCTTCTCAGGTACCCTGACATCGGGC | |
| D42A | Sense | CCCGATGTCAGGGTACCTGAGGCCAAACCCGCCAAGTTG |
| Antisense | CAACTTGGCGGGTTTGGCCTCAGGTACCCTGACATCGGG | |
| K43N | Sense | CCCGATGTCAGGGTACCTGAGGACAACCCCGCCAAGTTG |
| Antisense | CAACTTGGCGGGGTTGTCCTCAGGTACCCTGACATCGGG | |
| N56S | Sense | CGGGCTTCTCCAGCCCGCGCGTGGAG |
| Antisense | CTCCACGCGCGGGCTGGAGAAGCCCG | |
| K75N | Sense | CACCAGCCTCGTTTGTTATAATAACAAGATCACGGCCT |
| Antisense | AGGCCGTGATCTTGTTATTATAACAAACGAGGCTGGTG | |
| A83E | Sense | GATCACGGCCTCATATGAAGACCGAGTCACCTTCTCG |
| Antisense | CGAGAAGGTGACTCGGTCTTCATATGAGGCCGTGATC | |
| S89L | Sense | GCAGACCGAGTCACCTTCAAGCACAGTGGCATCACTTTC |
| Antisense | GAAAGTGATGCCACTGTGCTTGAAGGTGACTCGGTCTGC | |
| H90P | Sense | CGAGTCACCTTCTCGCCCAGTGGCATCACTTTC |
| Antisense | GAAAGTGATGCCACTGGGCGAGAAGGTGACTCG | |
| H96K | Sense | CGCACAGTGGCATCACTTTCAAGTCGGTGACGCGTAAAGACACG |
| Antisense | CGTGTCTTTACGCGTCACCGACTTGAAAGTGATGCCACTGTGCG | |
| H96E | Sense | GGCATCACTTTCGAGTCGGTGACGCGTAAAGACACGGGGACG |
| Antisense | CGTCCCCGTGTCTTTACGCGTCACCGACTCGAAAGTGATGCC | |
| S97A | Sense | GGCATCACTTTCCATGCGGTGACGCGTAAAGACACGGG |
| Antisense | CCCGTGTCTTTACGCGTCACCGCATGGAAAGTGATGCC | |
| K101E | Sense | CCATTCGGTGACGCGTGAAGACACGGGGACGTACAC |
| Antisense | GTGTACGTCCCCGTGTCTTCACGCGTCACCGAATGG | |
| D113E | Sense | CGTACACTTGCATGGTGTCTGACGAGGGCGGCAACACATACGGG |
| Antisense | CCCGTATGTGTTGCCGCCCTCGTCAGACACCATGCAAGTGTACG |
Restriction sites are underlined.
Virus-binding assay by flow cytometry.
CHO-S cells (106 cells/sample) were transiently transfected with plasmid DNA constructs using FuGENE 6. Cells were incubated at 37°C in an 8% CO2 humidified atmosphere on a shaker set to 125 rpm for 24 h. The cells were washed once in PBS and incubated with FCV (multiplicity of infection [MOI] = 5) in PBS plus 1% BSA on ice for 30 min. After being washed twice, the cells were incubated with an antibody for the appropriate receptor (fJAM-A rabbit antiserum, anti-hJAM-A mouse MAb, or anti-hCAR mouse MAb [Ambion]) and an antibody against the virus (anti-FCV mouse MAb [Custom Monoclonal Antibodies International] or rabbit anti-FCV antiserum) in PBS plus 1% BSA for 30 min on ice. The cells were washed and then fixed in fluorescence-activated cell sorter (FACS) fix buffer (PBS plus 1% paraformaldehyde and 0.05% NaN3). After being washed, the cells were incubated with the appropriate secondary antibody, either Alexa 647-conjugated goat anti-rabbit or goat anti-mouse IgG for the receptor and either Alexa 488-conjugated goat anti-mouse or goat anti-rabbit IgG for the virus, in PBS plus 1% BSA for 30 min on ice. The cells were washed, suspended in FACS fix, and analyzed with a FACSCalibur (Becton Dickinson). Transfected cells were gated on positive surface receptor expression, and ∼10,000 gated cells were analyzed for each sample.
Virus infection assay by flow cytometry.
CHO-S cells (106/sample) were transfected with receptor constructs using FuGENE 6. After overnight incubation, the cells were washed in cold PBS and virus was adsorbed (MOI = 0.5 in PBS plus 1% BSA) on ice for 30 min. Immediately following virus adsorption, the cells were washed to remove unbound virus. Samples to assay virus binding were then incubated on ice with fJAM-A rabbit antiserum in PBS plus 1% BSA, washed, and fixed. Samples to be tested for infection were resuspended in 2 ml CHO-serum-free medium and incubated at 37°C for 24 h with shaking. After 24 h, infected samples were washed in cold PBS, incubated with the fJAM-A antiserum as described above, and then washed and fixed. All samples were then incubated with an anti-FCV mouse MAb in PBS plus 0.1% Triton X-100 and 10% normal goat serum, followed by Alexa 647-conjugated goat anti-rabbit and Alexa 488-conjugated goat anti-mouse IgGs in PBS plus 0.1% Triton X-100 and 10% normal goat serum. After being washed, the cells were suspended in FACS buffer and analyzed with a FACSCalibur cytometer. Transfected cells were gated on positive surface receptor expression, and ∼10,000 gated cells in each sample were analyzed for expression of FCV capsid antigen.
Creation of a stably fJAM-A-transduced CHO-K1 cell line.
CHO-K1 cells were transfected with pCI-fJAM-A using FuGENE 6. At 24 h p.t., G418 (1.2 mg/ml; Cellgro) was added to the growth medium. The cells were maintained and passaged in the selective medium, and clones were selected using cloning cylinders (Fisher). Clonal populations were maintained in 400 μg/ml G418.
Virus infection plaque assay.
CHO-K1, Flp-In T-REx 293, Vero, HeLa, or CHL cells were seeded in six-well plates at a concentration of 3 × 105 cells per well in 2 ml of appropriate growth medium. The next day, the cells were transfected using FuGENE 6 and 1 μg DNA of either pCI-fJAM-A or the empty vector pCI-Neo; the cells were incubated for 24 h to allow protein expression. CRFK, nontransfected CHO-K1, and stably transfected CHO-K1 cells expressing fJAM-A were seeded in six-well plates at a concentration of 4 × 105 cells per well in appropriate growth medium 16 h before infection. Before virus inoculation, the cells were washed in PBS and then incubated with selected FCV isolates (MOI = 0.5) diluted in PBS plus 1% BSA for 1 h at room temperature. After virus adsorption, the cells were washed with PBS, and prewarmed growth medium was added. Samples of each virus were frozen immediately at −80°C to calculate input titers; the remaining samples were incubated at 37°C for 24 or 48 h and then frozen at −80°C and subjected to three freeze-thaw cycles. Plaque assays were performed as previously described (37). The change in plaque titer was calculated by subtracting the log10 titer of input virus from the log10 titer at 24 or 48 h. The mean and standard deviation of three replicates of a representative experiment are shown.
Statistical analyses.
The Analyze-it (Analyze-it Software) statistical analysis add-in for Microsoft Excel was used to perform analysis of variance (ANOVA) where necessary. Graphs were prepared using Kaleidagraph (Synergy Software). Protein images were prepared using PyMOL (DeLano Scientific) (12).
RESULTS
Expression and purification of the fJAM-A ectodomain and D1 and D2 subdomains.
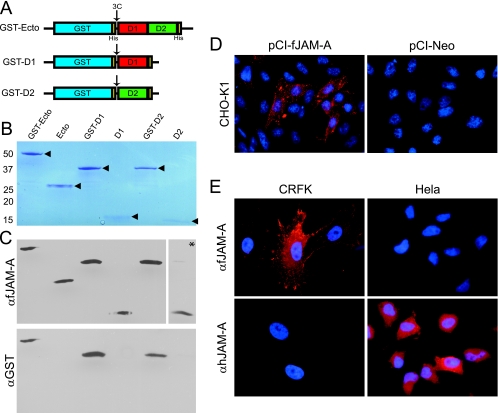
To determine if FCV binds directly to the ectodomain of fJAM-A and to identify which of the two Ig-like domains FCV interacts with, we prepared the ectodomain (amino acids 26 to 232) and the individual Ig-like D1 (amino acids 26 to 127) and D2 (amino acids 132 to 232) domains of fJAM-A for expression in bacteria. The fJAM-A domains were expressed as GST fusion proteins in a modified pET41a plasmid that had a hepatitis C virus 3C protease site inserted in frame at the fusion site (Fig. 1A). After purification by affinity chromatography, the expressed GST fusion proteins were of the expected masses by SDS-polyacrylamide gel electrophoresis analysis (GST-fJAM-A, 53 kDa; GST-D1, 41 kDa; and GST-D2, 41 kDa) (Fig. 1B). In addition, when the fusion proteins were cleaved with 3C protease and the GST portion was removed (see Materials and Methods), the ectodomain, D1, and D2 proteins were of the expected masses (ectodomain, 25 kDa; D1, 13.5 kDa; and D2, 13.5 kDa) (Fig. 1B).
FIG. 1.
Characterization of recombinant fJAM-A ectodomain and fJAM-A-specific rabbit antisera. (A) GST-Ecto, GST-D1, and GST-D2 were expressed in E. coli (BL21). Soluble recombinant proteins were purified by affinity chromatography. (B) The fJAM-A regions of the recombinant proteins were released by 3C protease cleavage, and the predicted sizes of the fusion and cleaved proteins were verified by SDS-polyacrylamide gel electrophoresis. (C) A rabbit polyclonal antiserum against soluble fJAM-A ectodomain was prepared, and its capacity to specifically recognize the purified fusion and cleavage proteins was confirmed by immunoblotting; a higher concentration of antibody was used to detect the cleaved D2 (indicated by an asterisk). Membranes were also probed with anti-GST. (D) Surface expression of fJAM-A in CHO cells by fluorescence microscopy. CHO-K1 cells were transfected with pCI-fJAM-A to express full-length fJAM-A or the empty vector, pCI-Neo. The cells were fixed at 24 h p.t. and immunostained with fJAM-A rabbit antiserum, followed by Alexa 594-conjugated goat anti-rabbit IgG. Nuclei were stained with DAPI. (E) The fJAM-A rabbit antiserum recognizes JAM-A on feline CRFK but not human HeLa cells. CRFK or HeLa cells were immunostained with either anti-fJAM-A or anti-hJAM-A (MAb BV16), followed by Alexa 594-conjugated goat anti-rabbit or Alexa 594-conjugated goat anti-mouse IgG; nuclei were stained with DAPI.
Detection of fJAM-A.
To detect expression of fJAM-A cDNA, we prepared a rabbit antiserum against the purified fJAM-A cleaved ectodomain. This antiserum detected both GST-fused and cleaved fJAM-A proteins in immunoblots, while an anti-GST antibody recognized only the GST fusion proteins (Fig. 1C). To test the specificity of the fJAM-A antiserum, we expressed the full-length fJAM-A cDNA in CHO-K1 cells, which do not form coherent tight junctions or express endogenous JAM-A (34, 38). We detected surface expression of fJAM-A in CHO-K1 cells transfected with pCI-fJAM-A, but not in cells transfected with empty vector (Fig. 1D). As reported for expression of murine and human JAM-A, we found that fJAM-A was distributed throughout the plasma membrane (30, 34). To further test the specificity of the antiserum, CRFK and HeLa cells were stained with either the fJAM-A antiserum or a mouse MAb against hJAM-A (Fig. 1E). The anti-fJAM-A bound to CRFK but not HeLa cells, while anti-hJAM-A bound HeLa but not CRFK cells. We also tested CHL and Flp-In T-REx 293 cells and, as expected, found that the fJAM-A antiserum did not recognize any cell surface antigen (data not shown).
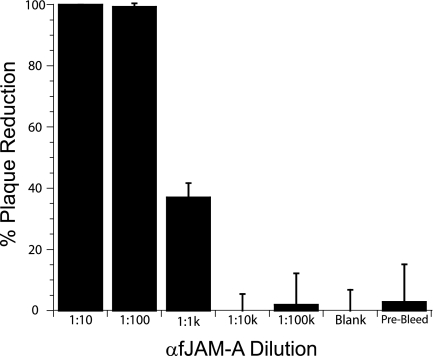
A rabbit antiserum against fJAM-A inhibits FCV plaque formation in CRFK cells.
Makino et al. showed that fJAM-A mouse antiserum partially inhibited FCV binding and blocked infection in permissive CRFK cells (28). We therefore tested the fJAM-A rabbit antiserum for its capacity to neutralize FCV infection of CRFK cells. We found that preincubation of cells with a 1:100 dilution of fJAM-A antiserum almost completely inhibited plaque formation (Fig. 2). In contrast, preincubation of monolayers with a 1:10 dilution of preimmune serum had no substantial effect on the number of plaques formed (Fig. 2). A 1:100 dilution of the fJAM-A antiserum also inhibited infection by other FCV isolates (F9, Kaos, Deuce, 127, and 131) (data not shown). Based on these findings, we conclude that the fJAM-A rabbit antiserum can inhibit FCV infection of CRFK cells.
FIG. 2.
Inhibition of FCV infection of CRFK cells by fJAM-A antiserum. Serial dilutions of anti-fJAM-A or a 1:10 dilution of rabbit preimmune serum was preincubated with monolayers of CRFK cells for 1 h on ice. The cells were then inoculated with ∼20 PFU of FCV-5 and incubated for an additional 1 h at room temperature. The cells were then overlaid with EMEM-5% fetal bovine serum and 1% Bacto Agar and cultured at 37°C for 48 h. The plaques in each well were counted, and the results were expressed as the percentage of plaque reduction from infected monolayers that were untreated. The results shown are the mean plaque reductions plus standard deviations of six replicate wells of a single representative experiment.
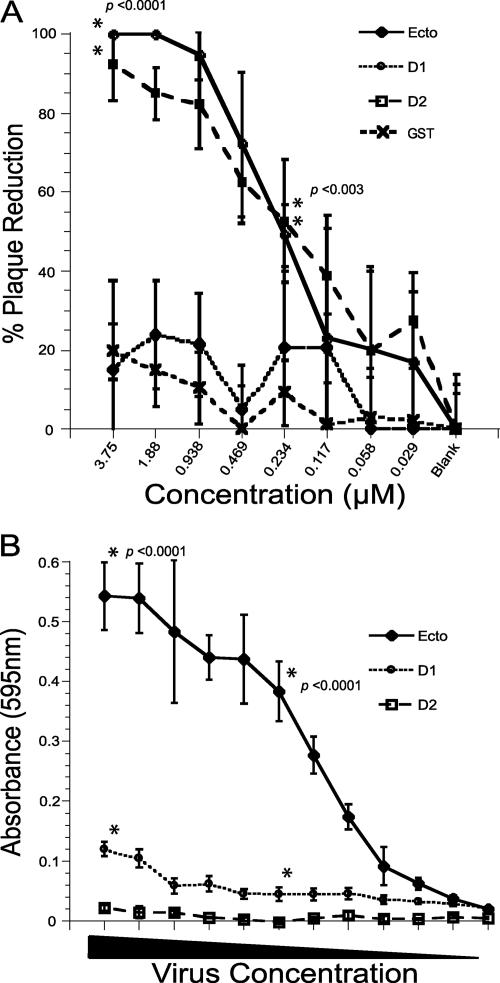
Purified soluble fJAM-A ectodomain and D1 domain neutralize FCV infection.
Neutralization of virus following incubation with soluble receptor has been demonstrated for a number of viruses (18, 22, 33). Therefore, we used plaque reduction assays to assess the capacity of the fJAM-A ectodomain to neutralize FCV-5 infection. We found that preincubation of FCV-5 with fJAM-A ectodomain reduced viral infectivity by ∼50% at 234 nM fJAM-A ectodomain and abolished infectivity at concentrations of >938 nM (Fig. 3A). The fJAM-A ectodomain consists of two Ig-like domains: the membrane-distal D1 and membrane-proximal D2. We found that preincubation of FCV-5 with purified D1 domain inhibited viral infectivity at concentrations similar to that of fJAM-A ectodomain (Fig. 3A). In contrast, preincubation of FCV-5 with purified D2 domain or GST had no effect on infectivity at any of the concentrations we tested (Fig. 3A). Plaque reduction by the ectodomain and D1 domain was statistically greater than that by the D2 domain at two of the three concentrations (3.75 and 0.234 μM) we analyzed by ANOVA (P < 0.0001 and P < 0.003, respectively). Preincubation of FCV-5 with the GST fusion proteins inhibited viral infectivity to the same extent as the cleaved forms of the proteins (data not shown). In addition, we found that addition of purified fJAM-A to cells after virus was adsorbed for 1 h on ice had no effect on virus infectivity (data not shown). We conclude from these results that the full-length fJAM-A ectodomain and the membrane-distal D1 domain can inhibit FCV-5 infection of CRFK cells when preincubated with virus, most likely by preventing virus binding to the fJAM-A receptors present on the surfaces of the cells.
FIG. 3.
Binding of fJAM-A ectodomain and the D1 and D2 Ig-like domains to FCV and their effects on infectivity. (A) FCV-5 was incubated with purified fJAM-A ectodomain (Ecto), D1, D2, or GST for 1 h on ice and then adsorbed to a monolayer of CRFK cells for 1 h at room temperature. The cells were then overlaid with EMEM, 5% fetal bovine serum, and 1% Bacto Agar and incubated at 37°C for 48 h. The plaques in each well were counted, and the data were expressed as the percentage of plaque reduction relative to monolayers infected with untreated virus. The data shown are the means of six replicates ± standard deviations from one representative experiment. ANOVA was performed on three concentrations (3.75, -0.234, and 0.029 μM) to determine statistical differences; significant concentrations are indicated by asterisks. (B) ELISA plates were coated with 5 μM solutions of soluble fJAM-A ectodomain, D1, or D2. Serial dilutions of FCV-5 were incubated with the immobilized proteins for 1 h, and then the plates were washed extensively. Bound FCV-5 was detected with rabbit anti-FCV serum, followed by HRP-conjugated goat anti-rabbit IgG. Colorimetric HRP substrate was added, and the amount of bound FCV-5 was quantified by absorbance at 595 nm. The means and standard deviations are shown; n = 3. As for panel A, ANOVA was performed on three concentrations; significant concentrations are indicated by asterisks.
Purified FCV-5 particles directly bind to purified fJAM-A ectodomain and the D1 domain by ELISA.
Our findings thus far strongly suggested that FCV directly interacted with fJAM-A ectodomain. To test this hypothesis, we used an ELISA to detect direct binding of viral particles to the fJAM-A ectodomain and D1 and D2 domains bound to 96-well plates. We found saturable concentration-dependent binding of FCV-5 to plate-bound fJAM-A ectodomain (Fig. 3B). Higher concentrations of FCV bound the plate-bound fJAM-A at levels similar to those with the highest concentration of FCV shown in Fig. 3B (data not shown). We also found that FCV-5 bound to the D1 domain at low levels (Fig. 3B); however, this binding was nonsaturable and did not significantly change with increasing concentrations of virus. In contrast, FCV did not bind the D2 domain at any of the concentrations we tested (Fig. 3B). ANOVA analysis of three concentrations showed significantly more viral binding to the ectodomain and D1 domain than to the D2 domain at all but the lowest concentration of virus (P < 0.0001). We also assayed binding of fJAM-A ectodomain or D1 or D2 domain to viral particles when virus was bound to the plate. Under these conditions, although we detected saturable concentration-dependent binding of soluble fJAM-A to virus bound to the solid phase, we were unable to detect any substantial binding of either the D1 or D2 domain to plate-bound virus using the fJAM-A rabbit antiserum to detect the bound proteins (data not shown). We hypothesized that our difficulty in detecting binding of the D1 domain to virus bound to the plate was because the fJAM-A antiserum competed with the virus for binding of D1. As the purified fJAM-A ectodomain and D1 and D2 proteins all possessed a C-terminal His tag (Fig. 1A), we also used a rabbit anti-His polyclonal antibody to detect binding of fJAM-A ectodomain, D1, or D2 to plate-bound virus. Using the anti-His antibody, our findings were similar to the results shown in Fig. 3B; we were able to detect concentration-dependent saturable binding of fJAM-A ectodomain and low levels of D1 binding, but no detectable binding of D2 to the plate-bound virus (data not shown). We conclude that FCV-5 can bind directly to the fJAM-A D1 and full-length ectodomain but that saturable concentration-dependent binding requires the D2 domain. In addition our results suggest that the fJAM-A antiserum inhibits the interaction between purified D1 and virus particles, perhaps by competing for binding to regions of the fJAM-A ectodomain that are involved in FCV binding.
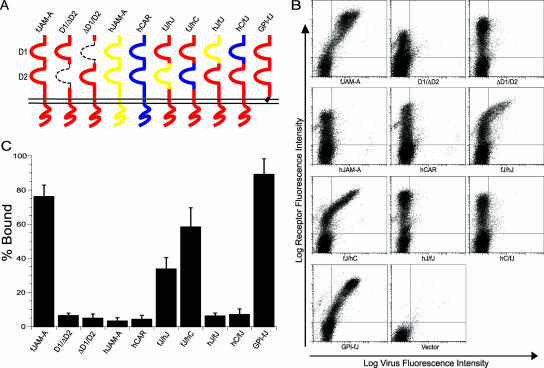
FCV binding to fJAM-A deletion and chimeric mutants.
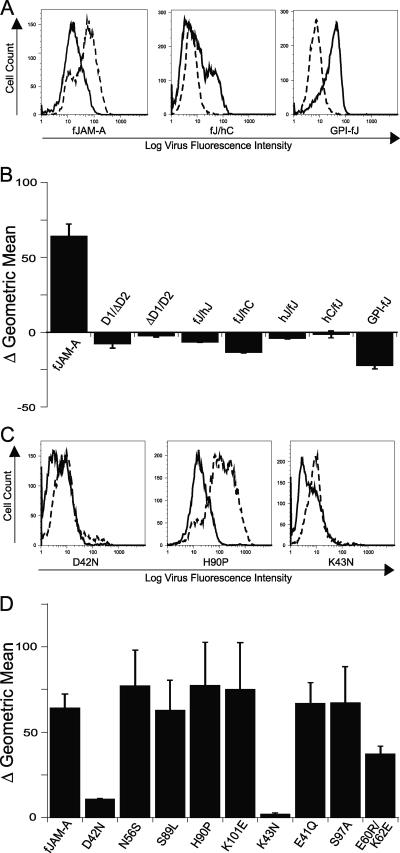
To analyze the regions of fJAM-A responsible for virus binding, we prepared deletion and chimeric mutants of fJAM-A for expression in CHO-S cells (Fig. 4A). The D1 and D2 domains were deleted or replaced with those of the related IgSF molecules, hJAM-A or hCAR. Binding of virus to CHO-S cells expressing the different surface receptor molecules was detected by flow cytometry (Fig. 4B). We found that virus bound to nearly 80% of cells that expressed cell surface full-length fJAM-A. However, FCV-5 did not bind above control levels to cells that expressed the single D1 or D2 Ig-like loops (Fig. 4C). In addition, virus did not bind to native hJAM-A or hCAR. In contrast, FCV-5 bound to cells expressing chimeric receptors that contained the fJAM-A D1 fused with the hJAM-A or hCAR D2 domain, albeit at lower levels than wild-type receptor (∼25% and 60%, respectively) (Fig. 4C). We observed only background levels of virus binding to cells that expressed chimeric receptors that consisted of the D1 from hJAM-A or hCAR in the fJAM-A background. FCV bound to CHO-S cells expressing a GPI-anchored form of fJAM-A at levels similar to those of cells expressing wild-type receptor. The background level of virus binding to cells transfected with empty vector was 3% (Fig. 4B). We also investigated the binding of the FCV isolates Deuce and F9 to the full panel of chimeric receptors by immunofluorescence (data not shown) and found levels of binding similar to that of FCV-5. The binding of the FCV isolates Deuce, F9, 127, 131, and Kaos to both the full-length fJAM-A receptor and the GPI-anchored form was detected by flow cytometry (data not shown). We noted similar binding levels for all isolates. We conclude that the fJAM-A D1 domain was necessary for FCV binding and that the cytosolic and transmembrane domains were not required. However, it appears that the context in which the D1 domain is presented on the cell surface is important, as FCV was unable to bind the fJAM-A D1 when it was expressed alone, possibly because the viral particle cannot access the domain when it lies close to the cell membrane.
FIG. 4.
FCV binding to CHO cells expressing fJAM-A deletion and chimeric mutants. (A) A panel of fJAM-A deletion and chimeric mutants was created to investigate FCV binding. Chimeric receptors were generated by exchanging single Ig-like loops from the IgSF proteins fJAM-A (red), hJAM-A (yellow), and hCAR (blue). Deletion constructs lacking single Ig-like domains, as well as a construct in which the transmembrane and cytoplasmic domains were replaced with a GPI anchor, were generated. (B) CHO-S cells were transfected with each construct. At 24 h p.t., FCV was adsorbed to the cells on ice for 30 min. After being washed with cold PBS, the bound virus and cell surface fJAM-A were detected with mouse anti-FCV MAb and rabbit anti-fJAM-A antibodies, followed by Alexa 488-conjugated goat anti-mouse IgG and Alexa 647-conjugated goat anti-rabbit IgG. Virus binding and receptor expression were analyzed by flow cytometry. (C) Virus binding was measured by determining the percentage of receptor-positive cells that were positive for virus. The means (n ≥ 3) (1 × 104 cells) and standard deviations are shown.
FCV binding to fJAM-A D1 point mutants.
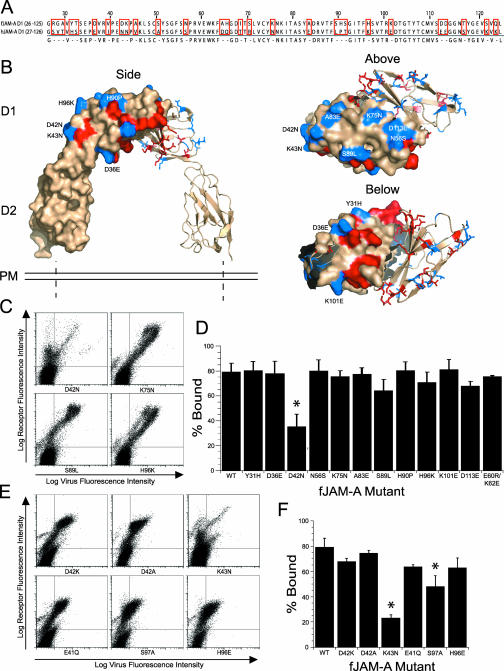
To identify individual regions of fJAM-A D1 required for FCV binding, we compared the amino acid sequences of the human and feline D1 domains and identified nonidentical residues (Fig. 5A). Dissimilar surface-exposed residues are indicated on a model of the hJAM-A structure (Protein Data Bank identification no. 1NBQ) (Fig. 5B). We prepared 11 point mutants by replacing selected feline residues with the corresponding residues of hJAM-A. In addition, we prepared a mutant (E60R/K62E) that reversed the charges on two of the four residues predicted to be involved in fJAM-A dimer formation. Mutations to the charged residues involved in salt bridge formations have been shown to prevent hJAM-A dimerization (19, 24). Each construct was transiently transfected into CHO-S cells, and FCV-5 binding was detected by flow cytometry (Fig. 5C). The level of FCV-5 binding to cells expressing the various constructs is presented as the percentage of receptor-positive cells that bound virus (Fig. 5D). Of the initial panel of mutant receptors we examined, only cells expressing fJAM-A D42N were bound by FCV-5 less effectively than the wild type (∼2.2-fold fewer cells bound virus) (Fig. 5D). FCV-5 bound all of the other mutants to levels not significantly different than those with the wild-type fJAM-A (Fig. 5C and D). To further investigate the role of fJAM-A residue 42 in binding, we prepared two additional fJAM-A mutants in which the charge on residue 42 was reversed (D42K) or removed (D42A); neither of these changes significantly decreased FCV-5 binding compared to binding to the wild-type receptor (Fig. 5E and F). As residue 43 of JAM-A also differed between feline (lysine) and human (asparagine) sequences, we examined binding of FCV-5 to cells expressing fJAM-A K43N. We found that FCV-5 bound 3.5-fold fewer cells expressing this mutant than those expressing the wild-type receptor (Fig. 5E and F). We prepared three other mutants that had changes to charged residues that were positioned close to residues 42 and 43 in the hJAM-A structure. Two of these mutants (E41Q and H96E) bound levels of FCV-5 similar to those with wild-type receptor. However, mutant fJAM-A S97A bound ∼1.7-fold less virus than wild-type fJAM-A (Fig. 5E and F). The D1 point mutants D42N, K43N, and S97A were all bound by significantly less virus than the wild-type fJAM-A, as determined by ANOVA (P < 0.0001). One caveat to these observations is that the cell surface expression of the D42N and K43N mutants was lower than that of the wild-type fJAM-A. Thus, it is possible that some of the lower FCV cell binding could be attributed to lower levels of expression; however, it is clear that those cells that did express lower levels of these mutants still bound far less virus than a comparable population of wild-type-expressing cells. The binding of FCV isolates F9 and Deuce to selected mutants (D42N, S89L, D42K, K43N, E41Q, and E60R/K62E) was also measured, with similar binding levels noted (data not shown). Taken together, these results suggest that residues D42, K43, and S97 of the fJAM-A D1 are likely involved in interactions with FCV virions.
FIG. 5.
FCV binding to fJAM-A D1 point mutants. (A) The amino acid sequences of the D1 domain of fJAM-A (residues 26 to 125) and hJAM-A (27 to 126) were aligned, and nonidentical residues were identified (highlighted by red boxes). (B) Identified residues were mapped on the hJAM-A crystal structure (Protein Data Bank identification no. 1NBQ; red and blue residues), and 11 surface-exposed residues were selected to mutate to the hJAM-A sequence (blue residues; images were created in PyMOL [Delano Scientific]). A dimerization mutant (E60R/K62E) that reverses the charges on two of the four charged residues in the dimerization motif was also created. (C) CHO-S cells were transfected with each of the constructs. At 24 h p.t., cells were incubated with FCV-5 on ice for 30 min, followed by immunostaining to detect surface expression of receptor and FCV binding, and then analyzed by flow cytometry. (D) After gating was performed for receptor-positive cells, virus binding was measured and expressed as a percentage. The means of at least three replicates (1 × 104 cells each) plus standard deviations are shown. To further investigate the decreased binding observed with the point mutant D42N, additional mutants were made to reverse or eliminate the charge on residue 42, as well as to alter other charged residues in the vicinity of residue 42. (E and F) Virus binding to cells expressing the constructs was measured by flow cytometry, and virus binding was determined as for panel D (F). The means (n ≥ 3; 1 × 104 cells each) plus standard deviations are shown. Constructs indicated by asterisks were bound by significantly lower levels of virus, as determined by ANOVA (P < 0.0001).
Capacity of FCV to infect nonpermissive CHO cells expressing fJAM-A or deletion and chimeric mutants of fJAM-A.
We used flow cytometry to examine the capacity of low multiplicities of FCV F9 to infect CHO-S cells transiently expressing fJAM-A and the panel of fJAM-A deletion and chimeric mutants. Controlling for levels of input capsid bound to cells, we quantified the change in the geometric mean fluorescence associated with viral capsid protein levels at 24 h postinoculation (Fig. 6A). In this way, infection was assayed by increased intracellular levels of viral capsid protein resulting from viral replication. In cells expressing the full-length fJAM-A molecule, we found significantly increased expression of viral capsid protein, indicating successful virus entry and infection (Fig. 6B). All of the mutant constructs that were unable to bind FCV (D1/ΔD2, ΔD1/D2, hJ/fJ, and hC/fJ) (Fig. 4C) were also unable to mediate infection (Fig. 6B). Although virus bound to cells expressing the D2 exchange mutant constructs (fJ/hJ and fJ/hC) and the GPI-anchored construct (GPI-fJAM), we were unable to detect infection in cells expressing these constructs (Fig. 6B). We also prepared a mutant construct of fJAM-A that lacked three conserved C-terminal residues of the cytosolic tail (Phe, Leu, and Val) known to be required for binding of PDZ domain-containing proteins (5). Both viral binding and infection of CHO-S cells expressing this construct were comparable to that mediated by the wild-type receptor construct (data not shown). From these results, we conclude that all the domains (D1, D2, transmembrane, and cytoplasmic) of the fJAM-A receptor are necessary for infection but that interactions with proteins that contain a PDZ domain are not required.
FIG. 6.
FCV infection of cells expressing chimeric, deletion, or select D1 point mutant constructs. (A) F9 was bound to two sets of cells expressing chimeric or deletion constructs for 30 min on ice. One set of F9-inoculated cells was resuspended in growth medium and placed at 37°C for 24 h; the other set of cells was washed and immediately immunostained for the virus and receptor to determine background binding of virus. After 24 h, the infected cells were washed and immunostained for virus and receptor as described above. Bound (solid lines) and 24-h-incubated (dashed lines) samples were then analyzed by flow cytometry. (B) Infectivity was measured by determining the change in mean log virus fluorescence intensity between bound and incubated samples. The averages of the changes in the geometric means (n ≥ 4) plus standard errors are shown. (C) Cells expressing select D1 point mutant constructs were prepared and analyzed in the same fashion. (D) The change in the mean virus fluorescence intensity was used as a measure for infection, and the averages of the changes in geometric means (n ≥ 4) plus standard errors are shown.
FCV infection of fJAM-A D1 point mutants.
We similarly analyzed the fJAM-A D1 point mutants for the capacity to confer susceptibility to FCV infection in CHO-S cells. We found that the two fJAM-A mutants with decreased capacity to bind virus (D42N and K43N) (Fig. 5D and F) also showed decreased capacity to confer susceptibility to infection on CHO-S cells relative to the wild-type receptor (Fig. 6C and D). The other point mutations investigated (N56S, S89L, H90P, K101E, and E41Q) conferred susceptibility to infection at levels similar to that of the wild-type receptor. The mutant S97A, which demonstrated a significant decrease in virus binding (Fig. 5F), was also able to confer levels of susceptibility to infection similar to those of the wild-type receptor. Lastly, the dimerization mutant (E60R/K62E) also conferred susceptibility to CHO-S cell infection by F9.
Isolate variation in the capacity to infect fJAM-A-expressing cells.
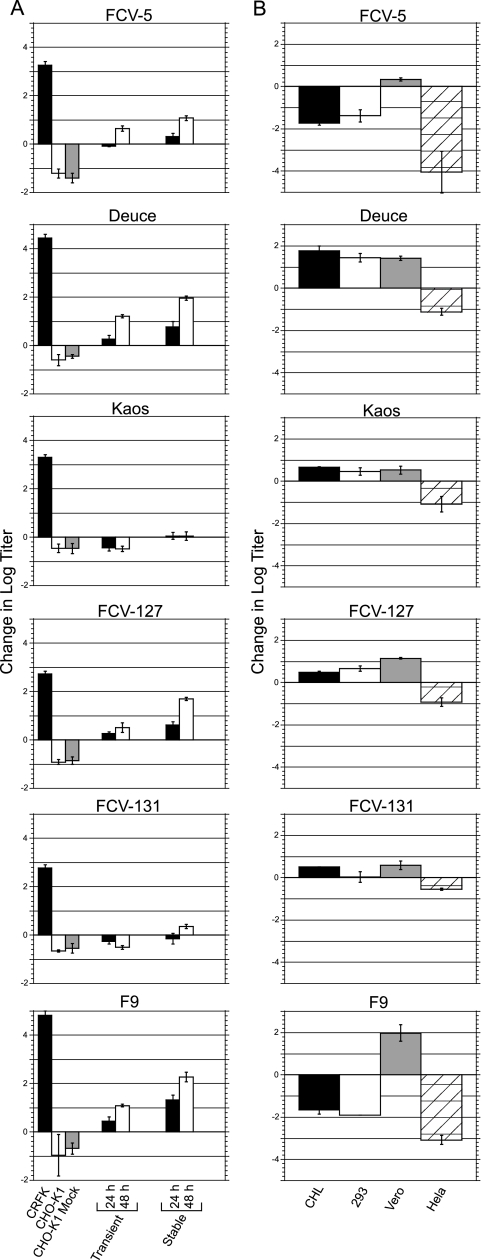
In the above-mentioned experiments, we used the F9 vaccine strain to assay for infection. However, we found that not all of the FCV isolates we examined were able to infect CHO-S cells expressing the wild-type fJAM-A receptor (data not shown). We selected six FCV isolates (including the vaccine strain, F9) to investigate the capacities of different isolates to infect transiently transfected adherent CHO-K1 cells (Fig. 7A). We found that four isolates (FCV-5, Deuce, 127, and F9) were able to productively infect CHO-K1 cells transiently expressing fJAM-A, with yields ranging from 0.5 to 1.2 log units greater than the input titer. The viral yields, however, were considerably lower than the yields observed in CRFK cells incubated for 24 h (2.8 to 4.8 log units). Two isolates, Kaos and 131, were unable to productively infect the transfected CHO-K1 cells. Incubation of these isolates with the fJAM-expressing CHO-K1 cells resulted in a net loss of titer similar to that observed following incubation with CHO-K1 controls.
FIG. 7.
FCV isolate infectivity in nonpermissive cell lines expressing fJAM-A. (A) FCV isolates (MOI = 0.5) were incubated with monolayers of CHO-K1 cells expressing fJAM-A (transiently or stably) for 24 or 48 h. Virus was also incubated for 24 h with empty CHO-K1 cells, CHO-K1 cells transfected with the empty vector (CHO K1 Mock), and CRFK cells. The infectivities of the samples were determined by plaque assay, and the change in titer from virus-bound-only samples was calculated. The mean log10 changes in titers of three replicates ± standard deviations from a representative experiment are shown. (B) Monolayers of four additional nonpermissive cell lines (CHL, Flp-In T-REx 293, Vero, and HeLa) were transiently transfected with fJAM-A and incubated with virus (MOI = 0.5) for 24 (CHL, Vero, and HeLa) or 48 (Flp-In T-REx 293) hours. The infectivities of the samples were determined by plaque assay, and the changes in titers from virus-bound-only samples were calculated. The mean log10 titers of three replicates ± standard deviations are shown.
The process of transfection using cationic liposomes has been shown to induce a robust type I interferon response (40), and FCV infection is sensitive to the presence of type I interferon (3, 17, 36). To eliminate the potential effects of a transfection-induced antiviral state, we created a CHO-K1 cell line stably expressing fJAM-A and then repeated the infection assay in the fJAM-A stable cells. The four isolates able to productively infect the transiently transfected cells demonstrated enhanced infectivity in the stable cell lines, with yields 1.1 to 2.3 log units higher than the input titer following 48 h of incubation. However, infection of FCV isolates 131 and Kaos was still greatly restricted in the stably transfected cell line. Neither isolate displayed a net increase in the viral yield after 24 h; after 48 h, the titer of FCV 131 had increased 0.35 log units while Kaos showed no change in titer over the bound virus sample.
To further investigate the abilities of FCV isolates to infect cells expressing fJAM-A, we transfected four additional cell lines with the receptor and incubated the cells for 24 or 48 h with the selected FCV isolates (Fig. 7B). Results varied by both cell line and FCV isolate. Transfected Vero cells were the only cell line identified that supported a productive infection by all FCV isolates tested. In contrast, transfected HeLa cells were unable to support a productive infection by any of the FCV isolates investigated. Transfected CHL cells supported infection by Deuce, Kaos, FCV-127, FCV-131, and F9 (following 48 h of incubation) (data not shown), but not FCV-5; Flp-In T-REx 293 cells supported infection by Deuce, Kaos, and FCV-127, but not isolates FCV-5, FCV-131, and F9. The FCV isolates Deuce and 127 were able to productively infect all of the transfected cell lines we tested, other than HeLa cells; isolates FCV-5, Kaos, FCV-131, and F9 were more restricted and showed differential capacities to infect the other transfected cell lines. All isolates were capable of productive infection in at least one fJAM-A-expressing nonpermissive cell line.
DISCUSSION
Makino et al. showed that fJAM-A is a receptor for FCV (28). Here, we have confirmed and extended those results. We have shown that binding of FCV to fJAM-A requires the membrane-distal D1 domain; furthermore, mutations to three residues within D1 (D42, K43, and S97) decreased viral binding. The D1 domain of fJAM-A is predicted to contain two antiparallel β-sheets (strands ABED and GFCC′C"). Structural and biochemical analyses of human and murine JAM-A molecules revealed dimer formation through extensive interactions between the GFCC′ faces of two D1 domains (dimerization interface); at the center of the dimer interface are several charged residues critical in mediating JAM-A-JAM-A dimer formation by forming four salt bridges (dimerization motif) (19, 43). Residues D42 and K43 are predicted to reside in a short turn between β-strands A and B on the β-sheet opposite the dimerization face of hJAM-A, in close proximity to the short linker region between D1 and D2. Interestingly, these residues are also contained within a peptide that blocks the binding of MAb (F11) to human JAM-A (F11R) on the surfaces of platelets. This MAb induces platelet secretion and aggregation, and these stimulatory activities are inhibited by a peptide that contains residues 28 to 50 of human JAM-A (1).
We hypothesize that the decreased binding observed in mutations of fJAM-A residues D42 and K43 is due to the importance of these residues in the FCV-fJAM-A interaction. However, the possibility exists that the mutations D42N and K43N introduce N-linked glycosylation sites in the fJAM-A receptor. Although the mutated fJAM-A constructs do not contain a classical Asn-X-Ser/Thr glycosylation motif at the position of the mutation, we cannot rule out atypical glycosylation. The presence of a glycan, and not the actual residue, could then be responsible for the decreased binding. Another possibility that we cannot completely exclude is that the lower levels of surface-expressed fJAM-A mutants D42N and K43N than of the wild-type fJAM-A (Fig. 5C and E) were responsible for the decreased virus binding. However, a distinct population of CHO-S cells expressed these mutants at levels comparable to those in cells expressing the wild-type receptor, yet these cells bound significantly less virus (Fig. 5C and E). Furthermore, mutating residue 97, which resides on a loop between β-strands E and F in very close proximity to residues 42 and 43, to an alanine resulted in significantly reduced viral binding, with expression levels similar to those of the wild-type fJAM-A. Thus, taken together, our data support the hypothesis that FCV binds to the D1 domain of fJAM-A in the vicinity of these residues.
Makino et al. reported that the FCV isolates they examined (which included the vaccine strain F9) could bind human 293T and monkey Vero cell lines and that anti-hJAM-A antibodies decreased that binding (28). Confounding this observation, Stuart and Brown (48) showed negligible binding of radiolabeled F9 virions to Vero and 293T cells. In agreement with the findings of Stuart and Brown, we were unable to detect an interaction between FCV and hJAM-A expressed on the surfaces of CHO cells. Furthermore, we did not detect an interaction between FCV and soluble recombinant hJAM-A ectodomain by ELISA or surface plasmon resonance studies (data not shown). Therefore, the binding detected by Makino et al. seems unlikely to have been mediated by hJAM-A.
The mammalian reovirus σ1 protein interacts with hJAM-A via residues E61 and K63 present at the dimer interface of D1, and it is the charge on these residues that is important for binding (19). Mutations to just one of the residues of the dimerization motif prevented the formation of hJAM-A-hJAM-A dimers (19). In contrast, FCV binding to cells expressing the E60R/K62E dimerization mutant of fJAM-A was similar to that of the wild-type fJAM-A (79% and 75%, respectively). The E60R/K62E mutations reverse the charges of two residues involved in forming the four predicted salt bridges important in dimerization (R58-E60, E60-R58, K62-E120, and E120-K62) (43). Thus, we conclude that the charges of fJAM-A residues E60 and K62 (corresponding to hJAM-A E61 and K63) are not important in FCV binding. In addition, we predict that the E60R/K62E mutant is unable to form fJAM-A-fJAM-A dimers. Therefore, the observed binding of FCV to the dimerization mutant suggests that FCV can interact with both monomeric and dimeric forms of fJAM-A.
The FCV-fJAM-A receptor interaction is also different from the adenovirus-CAR interaction. The adenovirus attachment fiber protein is similar to the reovirus σ1 protein. The fiber protein is a trimer with a globular C-terminal knob that engages the CAR at its amino-terminal D1 domain (16). Like the σ1 head domain, the fiber knob attaches at the dimer interface to residues important in CAR-CAR interactions at the dimer interface (7, 46, 47).
Similar to caliciviruses, coxsackieviruses (Picornaviridae) are small, nonenveloped, positive-strand RNA viruses. CVB utilize CAR as a host cell receptor (6, 20, 35). CVB also engage the amino-terminal Ig-like domain D1 of CAR. However, unlike adenoviruses, the CVB interaction with CAR does not involve critical residues in the dimerization motif, although the binding site is on the distal end and the lateral side (A-G face) of CAR D1 and lies close to the dimer interface (20). Residues D42, K43, and S97, which we found were important for FCV binding, are predicted to also be part of the orthologous lateral A-G face of fJAM-A; however, these residues lie much closer to the D1-D2 linker than the dimerization interface.
The ectodomain of fJAM-A was sufficient to mediate FCV binding, but when expressed on the cell surface with a GPI anchor, it could not mediate infection. In contrast, a GPI-anchored CAR mediated CVB and adenovirus binding and infection (50). This finding suggests that the transmembrane and/or the cytosolic domain of fJAM-A is required for productive FCV infection of nonpermissive cells. We first hypothesized that signaling via the consensus C-terminal type II PDZ domain-binding motif at the carboxyl terminus of the cytoplasmic tail of fJAM-A might be required for infection; however, a construct that lacked the predicted PDZ binding motif (the last three amino acids of fJAM-A: Phe-Leu-Val) was as efficient as the wild-type receptor in mediating FCV infection of CHO cells (data not shown). Taken together, our data suggest a model in which FCV binding to the fJAM-A ectodomain transduces signals through its transmembrane and cytosolic domains that are required for infection. Evidence suggesting such a model for CVB was reported by Coyne and Bergelson, where interaction of CVB with the cellular receptor DAF activates kinases that trigger actin rearrangements and phosphorylation events necessary for mediating viral infection (10).
Makino et al. (28) reported that fJAM-A expression on nonpermissive hamster lung cells conferred susceptibility to FCV infection. Here, we show that expression of fJAM-A on the surfaces of five different cell lines was not necessarily sufficient to support productive infection for all FCV isolates. Not all of the cell lines that we investigated supported productive infection by all of the isolates; furthermore, the abilities of FCV isolates to productively infect each cell line tested varied. Only Vero cells were able to support productive infection by all of the FCV isolates we examined. While native nontransfected Vero cells have been reported to support infection by some FCV isolates (28), none of the isolates we tested were capable of infecting Vero cells in the absence of fJAM-A.
The capacities of different FCV isolates to infect CHO-K1 cells that expressed fJAM-A varied. Indeed, we found that some isolates (FCV-131 and Kaos) were unable to productively infect these cells. Stuart and Brown reported that cell binding and infection by FCV are partially mediated by an N-linked glycoprotein containing α-2,6-linked, but not α-2,3-linked, sialic acids (48). CHO cells lack the sialyltransferase necessary for generating α-2,6-sialic acid linkages and express predominantly α-2,3-linked sialic acid glycans (26). Therefore, our results imply that at least a subset of FCV isolates are capable of binding and infecting fJAM-A-expressing cells independently of α-2,6-sialic acid linkages. fJAM-A contains a single putative N-linked glycosylation site at residue N184 in D2. The glycosylation status of this site, as well as the role of glycosylation in FCV binding and infection, warrants further investigation. It is also possible that there is an additional surface receptor for FCV. The findings that not all FCV isolates were capable of infecting nonpermissive cell lines expressing fJAM-A lends support to the idea that fJAM-A alone is not sufficient for FCV infection by all isolates and that additional coreceptors may be required.
In the last 10 years, there have been sporadic reports of highly virulent outbreaks of FCV disease in cats (11, 21, 41, 45). The virus isolates responsible for these outbreaks have been termed virulent systemic (VS) FCV (21). The pathology associated with VS-FCV outbreaks indicates breakdown of the epithelial and endothelial barrier functions (41, 42). These observations may reflect the capacity of FCV to target fJAM-A, as it is known that JAM-A is required for maintenance of epithelial and endothelial tight junctions. It is possible that FCV can disrupt the homophilic interactions that occur between JAM-A molecules on apposing cells and thus disrupt tight junctions and barrier integrity, an effect proposed for secreted adenovirus fiber protein (49).
JAM-A is also expressed on platelets, including feline platelets (T. Stokol and J. S. L. Parker, unpublished data). In some cases of VS-FCV, thrombocytopenia has been noted, together with disseminated intravascular coagulation (14, 21, 41, 45). It is possible that large amounts of FCV in the blood might disrupt normal platelet function. A monoclonal antibody (F11) that binds hJAM-A induces platelet secretion and aggregation. Binding of F11 to the surfaces of platelets is blocked by a peptide that incorporates the first 23 residues of hJAM-A D1 (residues 28 to 50) (1). As we have shown, FCV likely interacts with residues D42 and K43 on fJAM-A. Therefore, it is possible that FCV binding to fJAM-A might induce secretion and aggregation of feline platelets and that this in turn might be responsible for some of the pathogenic sequelae seen with VS-FCV disease. We are currently examining these and other possibilities.
Acknowledgments
We thank Kristen Guglielmi, Meg Crapster-Pregont, Stephen Campbell, and Patricia Pesavento for excellent technical assistance. We thank Karin Hoelzer for advice on statistical analysis. We thank Terry Dermody and Doug Lublin for the generous gift of reagents.
This work was supported by grants from The Cornell Feline Health Center and the Winn Feline Foundation. R.J.O. is the recipient of a scholarship from Cornell University.
We dedicate this paper to the memory of James Richards, a tireless advocate for feline research.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Babinska, A., M. H. Kedees, H. Athar, T. Ahmed, O. Batuman, Y. H. Ehrlich, M. M. Hussain, and E. Kornecki. 2002. F11-receptor (F11R/JAM) mediates platelet adhesion to endothelial cells: role in inflammatory thrombosis. Thromb. Haemost. 88:843-850. [PubMed] [Google Scholar]
- 2.Babinska, A., M. H. Kedees, H. Athar, T. Sobocki, M. B. Sobocka, T. Ahmed, Y. H. Ehrlich, M. M. Hussain, and E. Kornecki. 2002. Two regions of the human platelet F11-receptor (F11R) are critical for platelet aggregation, potentiation and adhesion. Thromb. Haemost. 87:712-721. [PubMed] [Google Scholar]
- 3.Baldwin, S. L., T. D. Powell, K. S. Sellins, S. V. Radecki, J. John Cohen, and M. J. Milhausen. 2004. The biological effects of five feline IFN-alpha subtypes. Vet. Immunol. Immunopathol. 99:153-167. [DOI] [PubMed] [Google Scholar]
- 4.Barton, E. S., J. C. Forrest, J. L. Connolly, J. D. Chappell, Y. Liu, F. J. Schnell, A. Nusrat, C. A. Parkos, and T. S. Dermody. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441-451. [DOI] [PubMed] [Google Scholar]
- 5.Bazzoni, G., O. M. Martinez-Estrada, F. Orsenigo, M. Cordenonsi, S. Citi, and E. Dejana. 2000. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J. Biol. Chem. 275:20520-20526. [DOI] [PubMed] [Google Scholar]
- 6.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 7.Bewley, M. C., K. Springer, Y. B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, J. A., P. Schelling, J. D. Wetzel, E. M. Johnson, J. C. Forrest, G. A. Wilson, M. Aurrand-Lions, B. A. Imhof, T. Stehle, and T. S. Dermody. 2005. Junctional adhesion molecule A serves as a receptor for prototype and field isolate strains of mammalian reovirus. J. Virol. 79:7967-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen, C. J., J. T. Shieh, R. J. Pickles, T. Okegawa, J. T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coyne, C. B., and J. M. Bergelson. 2006. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124:119-131. [DOI] [PubMed] [Google Scholar]
- 11.Coyne, K. P., B. R. Jones, A. Kipar, J. Chantrey, C. J. Porter, P. J. Barber, S. Dawson, R. M. Gaskell, and A. D. Radford. 2006. Lethal outbreak of disease associated with feline calicivirus infection in cats. Vet. Rec. 158:544-550. [DOI] [PubMed] [Google Scholar]
- 12.Delano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, Pala Alto, CA.
- 13.Ebnet, K., C. U. Schulz, M. K. Meyer Zu Brickwedde, G. G. Pendl, and D. Vestweber. 2000. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J. Biol. Chem. 275:27979-27988. [DOI] [PubMed] [Google Scholar]
- 14.Foley, J., K. Hurley, P. A. Pesavento, A. Poland, and N. C. Pedersen. 2006. Virulent systemic feline calicivirus infection: local cytokine modulation and contribution of viral mutants. J. Feline Med. Surg. 8:55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forrest, J. C., J. A. Campbell, P. Schelling, T. Stehle, and T. S. Dermody. 2003. Structure-function analysis of reovirus binding to junctional adhesion molecule 1. Implications for the mechanism of reovirus attachment. J. Biol. Chem. 278:48434-48444. [DOI] [PubMed] [Google Scholar]
- 16.Freimuth, P., K. Springer, C. Berard, J. Hainfeld, M. Bewley, and J. Flanagan. 1999. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J. Virol. 73:1392-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulton, R. W., and L. J. Burge. 1985. Susceptibility of feline herpesvirus 1 and a feline calicivirus to feline interferon and recombinant human leukocyte interferons. Antimicrob. Agents Chemother. 28:698-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodfellow, I. G., D. J. Evans, A. M. Blom, D. Kerrigan, J. S. Miners, B. P. Morgan, and O. B. Spiller. 2005. Inhibition of coxsackie B virus infection by soluble forms of its receptors: binding affinities, altered particle formation, and competition with cellular receptors. J. Virol. 79:12016-12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guglielmi, K. M., E. Kirchner, G. H. Holm, T. Stehle, and T. S. Dermody. 2007. Reovirus binding determinants in junctional adhesion molecule-A. J. Biol. Chem. 282:17930-17940. [DOI] [PubMed] [Google Scholar]
- 20.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hurley, K. E., P. A. Pesavento, N. C. Pedersen, A. M. Poland, E. Wilson, and J. E. Foley. 2004. An outbreak of virulent systemic feline calicivirus disease. J. Am. Vet. Med. Assoc. 224:241-249. [DOI] [PubMed] [Google Scholar]
- 22.Kaplan, G., M. S. Freistadt, and V. R. Racaniello. 1990. Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol. 64:4697-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khandoga, A., J. S. Kessler, H. Meissner, M. Hanschen, M. Corada, T. Motoike, G. Enders, E. Dejana, and F. Krombach. 2005. Junctional adhesion molecule-A deficiency increases hepatic ischemia-reperfusion injury despite reduction of neutrophil transendothelial migration. Blood 106:725-733. [DOI] [PubMed] [Google Scholar]
- 24.Kostrewa, D., M. Brockhaus, A. D'Arcy, G. E. Dale, P. Nelboeck, G. Schmid, F. Mueller, G. Bazzoni, E. Dejana, T. Bartfai, F. K. Winkler, and M. Hennig. 2001. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 20:4391-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law, L. K., and B. L. Davidson. 2005. What does it take to bind CAR? Mol. Ther. 12:599-609. [DOI] [PubMed] [Google Scholar]
- 26.Lee, E. U., J. Roth, and J. C. Paulson. 1989. Alteration of terminal glycosylation sequences on N-linked oligosaccharides of Chinese hamster ovary cells by expression of beta-galactoside alpha 2,6-sialyltransferase. J. Biol. Chem. 264:13848-13855. [PubMed] [Google Scholar]
- 27.Liu, Y., A. Nusrat, F. J. Schnell, T. A. Reaves, S. Walsh, M. Pochet, and C. A. Parkos. 2000. Human junction adhesion molecule regulates tight junction resealing in epithelia. J. Cell Sci. 113:2363-2374. [DOI] [PubMed] [Google Scholar]
- 28.Makino, A., M. Shimojima, T. Miyazawa, K. Kato, Y. Tohya, and H. Akashi. 2006. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 80:4482-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malergue, F., F. Galland, F. Martin, P. Mansuelle, M. Aurrand-Lions, and P. Naquet. 1998. A novel immunoglobulin superfamily junctional molecule expressed by antigen presenting cells, endothelial cells and platelets. Mol. Immunol. 35:1111-1119. [DOI] [PubMed] [Google Scholar]
- 30.Mandell, K. J., I. C. McCall, and C. A. Parkos. 2004. Involvement of the junctional adhesion molecule-1 (JAM1) homodimer interface in regulation of epithelial barrier function. J. Biol. Chem. 279:16254-16262. [DOI] [PubMed] [Google Scholar]
- 31.Mandell, K. J., and C. A. Parkos. 2005. The JAM family of proteins. Adv. Drug Deliv. Rev. 57:857-867. [DOI] [PubMed] [Google Scholar]
- 32.Marsh, M., and A. Helenius. 2006. Virus entry: open sesame. Cell 124:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, S., J. M. Casasnovas, D. E. Staunton, and T. A. Springer. 1993. Efficient neutralization and disruption of rhinovirus by chimeric ICAM-1/immunoglobulin molecules. J. Virol. 67:3561-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Padura, I., S. Lostaglio, M. Schneemann, L. Williams, M. Romano, P. Fruscella, C. Panzeri, A. Stoppacciaro, L. Ruco, A. Villa, D. Simmons, and E. Dejana. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J. Cell Biol. 142:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martino, T. A., M. Petric, H. Weingartl, J. M. Bergelson, M. A. Opavsky, C. D. Richardson, J. F. Modlin, R. W. Finberg, K. C. Kain, N. Willis, C. J. Gauntt, and P. P. Liu. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology 271:99-108. [DOI] [PubMed] [Google Scholar]
- 36.Mochizuki, M., H. Nakatani, and M. Yoshida. 1994. Inhibitory effects of recombinant feline interferon on the replication of feline enteropathogenic viruses in vitro. Vet. Microbiol. 39:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ossiboff, R. J., A. Sheh, J. Shotton, P. A. Pesavento, and J. S. Parker. 2007. Feline caliciviruses (FCVs) isolated from cats with virulent systemic disease possess in vitro phenotypes distinct from those of other FCV isolates. J. Gen. Virol. 88:506-527. [DOI] [PubMed] [Google Scholar]
- 38.Ostermann, G., K. S. Weber, A. Zernecke, A. Schroder, and C. Weber. 2002. JAM-1 is a ligand of the β2 integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 3:151-158. [DOI] [PubMed] [Google Scholar]
- 39.Ozaki, H., K. Ishii, H. Horiuchi, H. Arai, T. Kawamoto, K. Okawa, A. Iwamatsu, and T. Kita. 1999. Cutting edge: combined treatment of TNF-α and IFN-γ causes redistribution of junctional adhesion molecule in human endothelial cells. J. Immunol. 163:553-557. [PubMed] [Google Scholar]
- 40.Park, M. S., M. L. Shaw, J. Munoz-Jordan, J. F. Cros, T. Nakaya, N. Bouvier, P. Palese, A. Garcia-Sastre, and C. F. Basler. 2003. Newcastle disease virus (NDV)-based assay demonstrates interferon-antagonist activity for the NDV V protein and the Nipah virus V, W, and C proteins. J. Virol. 77:1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedersen, N. C., J. B. Elliott, A. Glasgow, A. Poland, and K. Keel. 2000. An isolated epizootic of hemorrhagic-like fever in cats caused by a novel and highly virulent strain of feline calicivirus. Vet. Microbiol. 73:281-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pesavento, P. A., N. J. MacLachlan, L. Dillard-Telm, C. K. Grant, and K. F. Hurley. 2004. Pathologic, immunohistochemical, and electron microscopic findings in naturally occurring virulent systemic feline calicivirus infection in cats. Vet. Pathol. 41:257-263. [DOI] [PubMed] [Google Scholar]
- 43.Prota, A. E., J. A. Campbell, P. Schelling, J. C. Forrest, M. J. Watson, T. R. Peters, M. Aurrand-Lions, B. A. Imhof, T. S. Dermody, and T. Stehle. 2003. Crystal structure of human junctional adhesion molecule 1: implications for reovirus binding. Proc. Natl. Acad. Sci. USA 100:5366-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossmann, M. G., Y. He, and R. J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324-331. [DOI] [PubMed] [Google Scholar]
- 45.Schorr-Evans, E. M., A. Poland, W. E. Johnson, and N. C. Pedersen. 2003. An epizootic of highly virulent feline calicivirus disease in a hospital setting in New England. J. Feline Med. Surg. 5:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiradake, E., H. Lortat-Jacob, O. Billet, E. J. Kremer, and S. Cusack. 2006. Structural and mutational analysis of human Ad37 and canine adenovirus 2 fiber heads in complex with the D1 domain of coxsackie and adenovirus receptor. J. Biol. Chem. 281:33704-33716. [DOI] [PubMed] [Google Scholar]
- 47.Stehle, T., and T. S. Dermody. 2004. Structural similarities in the cellular receptors used by adenovirus and reovirus. Viral Immunol. 17:129-143. [DOI] [PubMed] [Google Scholar]
- 48.Stuart, A. D., and T. D. Brown. 2007. α2,6-Linked sialic acid acts as a receptor for feline calicivirus. J. Gen. Virol. 88:177-186. [DOI] [PubMed] [Google Scholar]
- 49.Walters, R. W., P. Freimuth, T. O. Moninger, I. Ganske, J. Zabner, and M. J. Welsh. 2002. Adenovirus fiber disrupts CAR-mediated intercellular adhesion allowing virus escape. Cell 110:789-799. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X., and J. M. Bergelson. 1999. Coxsackievirus and adenovirus receptor cytoplasmic and transmembrane domains are not essential for coxsackievirus and adenovirus infection. J. Virol. 73:2559-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams, L. A., I. Martin-Padura, E. Dejana, N. Hogg, and D. L. Simmons. 1999. Identification and characterisation of human Junctional Adhesion Molecule (JAM). Mol. Immunol. 36:1175-1188. [DOI] [PubMed] [Google Scholar]