Abstract
Proteins of the capsid proximal tegument are involved in the transport of incoming capsids to the nucleus and secondary envelopment after nuclear egress. Homologs of the essential large capsid proximal tegument protein pUL36 are conserved within the Herpesviridae. They interact with another tegument component, pUL37, and contain a deubiquitinating activity in their N termini which, however, is not essential for virus replication. Whereas an internal deletion of 709 amino acids (aa) within the C-terminal half of the alphaherpesvirus pseudorabies virus (PrV) pUL36 does not impair its function (S. Böttcher, B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter, J. Virol. 80:9910-9915, 2006), deletion of the very C terminus does (J. Lee, G. Luxton, and G. A. Smith, J. Virol. 80:12086-12094, 2006). For further characterization we deleted several predicted functional and structural motifs within PrV pUL36 and analyzed the resulting phenotypes in cell culture and a mouse infection model. Extension of the internal deletion to encompass aa 2087 to 2981 exerted only minor effects on virus replication but resulted in prolonged mean survival times of infected mice. Any additional extension did not yield viable virus. Deletion of an N-terminal region containing the deubiquitinating activity (aa 22 to 248) only slightly impaired viral replication in cell culture but slowed neuroinvasion in our mouse model, whereas a strong impairment of viral replication was observed after simultaneous removal of both nonessential domains. Absence of a region containing two predicted leucine zipper motifs (aa 748 to 991) also strongly impaired virus replication and spread. Thus, we identify several domains within the PrV UL36 protein, which, though not essential, are nevertheless important for virus replication.
Herpesvirus virions consist of four morphologically distinct structures: core, capsid, tegument, and envelope. Of these, the tegument is the most complex in terms of protein content and the least understood concerning its structure and function (39, 40). Whereas major parts of the tegument are released from the incoming virus particle after fusion of the virion envelope with the cellular plasma membrane, several proteins have been shown to remain associated with nucleocapsids on their transport along microtubules to the nuclear pore (17, 34). In the alphaherpesvirus pseudorabies virus (PrV) these include pUS3, pUL37, and pUL36 (17, 34). It has been suggested that the last two are components of the inner part of the tegument (40). Capsid assembly and packaging of newly replicated viral DNA occur in the nucleus, and mature capsids are subsequently translocated to the cytoplasm for final morphogenesis by an envelopment/deenvelopment process, which includes acquisition of a primary envelope by budding at the inner nuclear membrane followed by fusion of the primary envelope with the outer nuclear membrane (40). Tegumentation in the cytoplasm may nucleate at two separate viral structures, the translocated capsid and the future envelopment site. At the future envelopment site in trans-Golgi vesicles, glycoproteins interact with outer tegument proteins, whereas addition of inner tegument proteins to nucleocapsids appears to take place in juxtanuclear regions (5, 11, 29, 42). In PrV, a complex of pUL48 and pUL3.5 may link the two subassemblies, so that complete virus particles are subsequently released from the cell by exocytosis (12, 13, 18). Ultrastructural studies (1, 19, 50) also supported the distinction between a well-organized, at least partially icosahedral inner tegument and a less ordered envelope-proximal portion.
Tegument proteins homologous to pUL36 of herpes simplex virus type 1 (HSV-1) (36, 37) represent the largest herpesvirus-encoded gene products, ranging in size from 2,241 amino acids (aa) in human cytomegalovirus (HCMV) (7) to 3,534 aa in equine herpesvirus 4 (46). Deletion of pUL36 from PrV or HSV-1 resulted in abrogation of virus replication (8, 14). Although these proteins are conserved throughout the Herpesviridae, sequence homology is low (27, 43) and confined to only a few regions within these proteins. These include the N-terminal part, which harbors a deubiquitinating activity that has recently been identified in pUL36 orthologs of all three herpesvirus subfamilies (21, 43, 44). However, this activity is not required for virus replication in cell culture (32, 48). An internal, carboxy-terminally located region of 709 aa could also be deleted from PrV pUL36 without impairment of viral replication (4). In contrast, removal of or in-frame deletions within the highly conserved C-terminal 62 aa of PrV pUL36 resulted in a complete abrogation of productive virus replication (4, 32).
Based on studies using different mutants several functions have been proposed for pUL36. The temperature-sensitive HSV-1 mutant tsB7, whose defect had been mapped to the UL36 locus, failed to release viral DNA into the nucleus after entry at the nonpermissive temperature, whereas a temperature shift allowed the uncoating of the viral genome (2, 28). Marker rescue experiments indicated that ca. 300 nucleotides (nt) within the pUL36 gene open reading frame (ORF) might be responsible for this phenotype (2). Ultrastructural analysis of an HSV-1 pUL36 deletion mutant, KΔUL36, revealed an accumulation of unenveloped nucleocapsids in the cytoplasm, with no obvious defects in DNA packaging or nuclear egress (8). A similar phenotype has been described for the PrV pUL36 deletion mutant PrV-ΔUL36F (14). Thus, both studies indicate a key role for pUL36 in secondary envelopment. However, due to the necessity to propagate these mutant viruses on complementing cell lines, an effect of the pUL36 deletion on early events in virus replication could not be investigated. Although a recent study using cell fractionation and capsid purification suggested that pUL36 and pUL37 may already be associated with intranuclear HSV-1 capsids (6), neither of the analyzed pUL36 null mutants in either HSV-1 or PrV exhibited a nuclear phenotype (8, 14).
Herpesvirus virion formation requires numerous protein-protein interactions to ensure correct maturation of nascent particles (reviewed in reference 40). Besides the well-characterized interaction of pUL36 with the pUL37 tegument protein (3, 14, 19a, 27, 47), a tight association of pUL36 with the capsid has been proposed using either differential extraction (45), coimmunoprecipitation (36), or three-dimensional image reconstruction after cryoelectron microscopical analyses (50). Further interactions of pUL36 homologs with viral (47) or cellular (41) proteins have been proposed, but these remain to be verified and assayed for their biological relevance.
In PrV, pUL36 is the only tegument protein analyzed so far which is strictly essential for viral replication (14). Thus, we are interested in dissecting this huge protein and identifying functional domains. In this study, based on computational analyses of PrV pUL36, we deleted a region containing two predicted leucine zipper motifs, removed the deubiquitinating domain within the N terminus of pUL36, and enlarged the previously described deletion of a proline/alanine-rich region within the C terminus to delineate its borders to essential functional domains within PrV pUL36 (4).
MATERIALS AND METHODS
Viruses and cells.
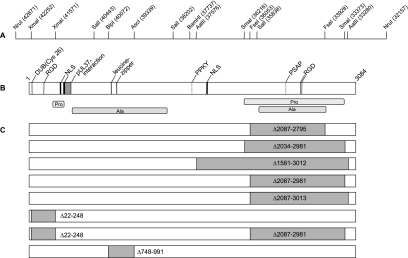
The wild-type PrV strain Kaplan (PrV-Ka) (20) was used in all experiments as the parental strain. Viruses were propagated in rabbit kidney cells (RK13) and porcine kidney cells (PSEK). A UL36 deletion mutant, PrV-ΔUL36F, lacking nearly the complete UL36 coding region, was propagated on RK13-UL36 cells which contain a 10.5-kb NruI fragment of the viral genome encompassing the UL36 promoter and complete ORF (14, 27) (Fig. 1). PrV-UL36ΔFseI lacking codons 2087 to 2795 of UL36 is able to replicate in noncomplementing cells (4) and is referred to here as PrV-UL36Δ2087-2795.
FIG. 1.
Predicted motifs and deletions in PrV pUL36. (A) UL36 ORF with relevant cleavage sites and corresponding nucleotide numbers (GenBank accession no. BK001744). (B) Protein motifs such as nuclear localization signals (NLS), the leucine zipper, and RGD motifs as well as two putative L-domain motifs (PPKY and PSAP) are shown, as predicted by Prosite release 19.24 (http://www.expasy.org/prosite/), PSORTII (http://psort.hgc.jp/), and ELM (http://elm.eu.org/) (4). The pUL37 binding domain was identified by yeast two-hybrid screening (27), and the active site for deubiquitinating activity (DUB) has been described recently (21, 43). Proline- and alanine-rich regions are indicated. (C) Deletions within mutated PrV pUL36 are highlighted in gray. Numbers show the extent of deletion in amino acid positions.
Plasmid constructs.
The complete UL36 gene of PrV-Ka had been cloned in plasmid pcDNA-UL36 (4, 27) (Fig. 1). For convenient exchange of 5′- and 3′-terminal parts of the UL36 ORF, pcDNA-UL36IV was constructed by inactivation of the BamHI site within the polylinker region, thus leaving only one BamHI site approximately in the middle of the UL36 ORF. Relevant cleavage sites used for cloning are indicated in Fig. 1A. For enlargement of the C-terminal deletion of the recently described pUL36ΔFseI (4), a 5.5-kb BamHI/KpnI fragment comprising the 3′-terminal half of UL36 was cloned in pBluescriptSK(+) (Stratagene), resulting in plasmid pBS-UL36C-2. To generate pUL36Δ2034-2981, pBS-UL36C-2 was cleaved by SmaI (nt 36216 and 33373) and an EcoRI linker (5′-CGGAATTCCG-3′; New England Biolabs) was inserted, cleaved, and blunt ended by Klenow polymerase, resulting in the in-frame insertion of five codons. To construct pUL36Δ1581-3012, pBS-UL36C-2 was cleaved by AatII (nt 37576 and 33280) and religated. After cleavage with BamHI and KpnI the resulting 2.7-kb and 1.2-kb fragments were inserted into BamHI/KpnI-cleaved pcDNA-UL36IV, thereby replacing the wild-type 3′ part of UL36 to obtain pcDNA-UL36Δ2034-2981 and pcDNA-UL36Δ1581-3012.
For isolation of pUL36Δ2087-2981, pcDNA-UL36Δ2034-2981 was cleaved by AseI, which cuts within the EcoRI linker at nt 33373 and assures subsequent in-frame ligation, and XbaI, which cuts within the multiple cloning site of pcDNA3. The resulting 1.2-kb fragment was inserted into pcDNA-UL36 that had been cleaved with FseI (nt 36053) and XbaI. To isolate pUL36Δ2087-3013, pcDNA-UL36IV was cleaved by AatII (nt 33280) and XbaI, followed by insertion of the resulting 1.1 kb into pcDNA-UL36 that had been cleaved by FseI (nt 36053) and XbaI. To restore the UL36 ORF, noncompatible overhangs were blunt ended, resulting in the insertion of three additional codons in pUL36Δ2087-2981. Correct in-frame cloning was verified by sequencing using primer UL36REV (5′-CACAGAATTCCAATTTATTAACCGAGAATC-3′).
To construct pUL36Δ22-248, an 1.2-kb HindIII/SphI fragment from pUC-UL36 was subcloned into pUC19 (New England Biolabs) and the resulting plasmid was cleaved by XmaI (nt 41571 and 42252), religated, and sequenced by using primer UL362FOR (5′-CACAGGTACCTTCAGCCATG-3′). Subsequent reconstitution of the full-length UL36 gene in pcDNA3 resulted in pcDNA-UL36Δ22-248. For deletion of a central region comprising a predicted leucine zipper region, a SalI fragment (nt 40443 and 38202) was subcloned in pUC19, cleaved with BlpI (nt 40072) and AscI (nt 39339), blunt ended with Klenow polymerase, and ligated. The resulting plasmid was sequenced using primers M13 and M13Rev to verify correct in-frame cloning. Subsequent reconstitution of the full-length UL36 gene in pcDNA3 resulted in pcDNA-UL36Δ748-991.
To combine the deletions within the 5′ and 3′ regions of UL36, pcDNA-UL36Δ22-248Δ2087-2981 was generated by replacement in pcDNA-UL36Δ22-248 of a 2.8-kb wild-type BamHI/XbaI fragment with a corresponding fragment lacking codons 2087 to 2981.
Generation of PrV-UL36 in-frame deletion mutants.
All pcDNA-UL36 constructs were cotransfected with genomic DNA of PrV-ΔUL36F into RK13 cells using calcium phosphate coprecipitation (16). Transfection progeny was plated onto RK13 cells, and single plaques were picked and purified to homogeneity. DNA from several single plaque isolates for each of the mutant viruses was analyzed by restriction enzyme digestion and Southern blot hybridization, which yielded the expected fragment patterns. Moreover the affected regions were amplified by PCR from viral DNA using the following primers: for PrV-UL36Δ2087-2981, primers UL36REV (5′-CACAGAATTCCAATTTATTAACCGAGAATC-3′) and sb_sf (5′-CTGGAGGCGGCTCTCGGC-3′); for PrV-UL36Δ22-248, primers UL362FOR (5′-CACAGGTACCTTCAGCCATG-3′) and sb_UL36rev (5′-ACCGTGTGCGTGCTGCTCAGG-3′); for PrV-UL36Δ748-991, primers sb_LZFOR (5′-GCTTCGACGCCATGGACGC-3′) and sb_LZREV (5′-GCCGAAGAGCGACCCGATG-3′). For PrV-UL36Δ22-248Δ2087-2981 two PCR products were generated with primers UL362FOR and sb_UL36rev as well as UL36REV and sb_sf. PCR fragments were eluted and sequenced with one of the primers used for PCR. All sequenced regions showed correct in-frame fusions. By all of these criteria, DNAs from the mutant viruses contained only the intended deletions.
Since the single-plaque isolates all exhibited the expected mutations, individual isolates were randomly chosen and designated PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, PrV-UL36Δ748-991, and PrV-UL36Δ22-248Δ2087-2981. No infectious progeny could be isolated from cotransfections of PrV-ΔUL36F and pcDNA-UL36ΔΔ2034-2981, pcDNA-UL36Δ1581-3012, and pcDNA-UL36Δ2087-3013.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis.
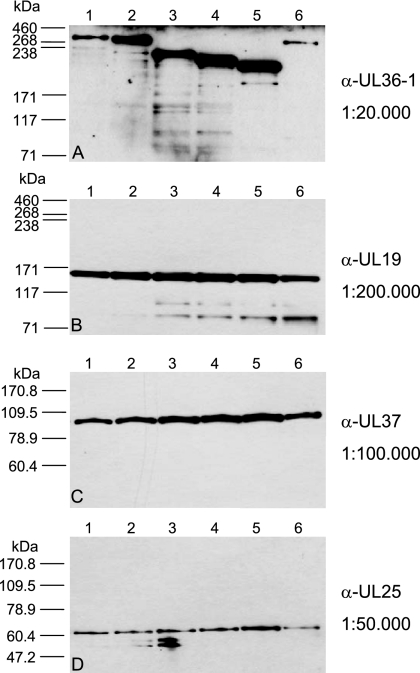
To assay for virion incorporation of truncated pUL36, cells were infected at a multiplicity of infection (MOI) of 0.1 with PrV-Ka and mutant PrV-UL36Δ2087-2795, PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, PrV-UL36Δ748-991, or PrV-UL36Δ22-248Δ2087-2981 and incubated until complete cytopathic effect developed. Remaining intact cells were lysed by freezing (−70°C) and thawing (37°C), cellular debris was removed by low-speed centrifugation, and the virus-containing supernatant was precleared by centrifugation through a 35% sucrose cushion. The resulting pellet was resuspended in phosphate-buffered saline (PBS) and layered onto a discontinuous sucrose gradient of 30, 40, and 50% sucrose. Virions which accumulated at the boundary between 40 and 50% sucrose were harvested by aspiration, pelleted, and resuspended in PBS. Virion lysates were loaded onto 6% discontinuous polyacrylamide gels (30), and proteins were electrophoretically separated for 1.5 h at 200 V. Blotting to nitrocellulose membrane was performed for 1.5 h at 100 V, followed by incubation with anti-UL36-1 serum (1:20.000) (27). After incubation with peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany), bound antibody was detected by chemiluminescence (Super Signal; Pierce, Bonn, Germany) recorded on X-ray film. To control for the presence of other virion components, parallel blots were probed with antisera against tegument protein pUL37 (1:100.000) (24), major capsid protein pUL19 (1:200.000) (26), or capsid-associated protein pUL25 (1:50.000) (25).
In vitro replication studies.
For analysis of one-step growth kinetics, RK13 cells were infected with PrV-Ka or PrV-UL36Δ2087-2795, PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, PrV-UL36Δ748-991, and PrV-UL36Δ22-248Δ2087-2981 at an MOI of 10, and incubated on ice for 1 h. Prewarmed medium was then added, and cells were further incubated for 1 h at 37°C. Thereafter, nonpenetrated virus was inactivated by low-pH treatment (38), and cells were scraped into the medium immediately (0 h), as well as after 4, 8, 12, 24, and 36 h, and frozen at −70°C. After all samples had been harvested, they were thawed at 37°C and centrifuged for 2 min at 13,000 rpm, and combined titers of virus in the supernatant and infectious virus released by freezing and thawing were determined on RK13 cells.
For determination of plaque size RK13 cells in six-well culture dishes were infected with 300 PFU/well under plaque assay conditions. The cells were fixed 2 days postinfection (p.i.) with 5% formaldehyde and stained with crystal violet. Plaque diameters were measured microscopically. For each virus 30 plaques were measured in three independent experiments, and values were calculated by comparison to the plaque size of PrV-Ka, which was set at 100%.
Electron microscopy.
For analysis of ultrathin sections, RK13 cells were infected at an MOI of 1 with PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, PrV-UL36Δ748-991, or PrV-UL36Δ22-248Δ2087-2981 and incubated for 14 h at 37°C. Fixation and embedding were done as described previously (18). Ultrathin sections were counterstained with uranyl acetate and lead salts and examined with a model 400T electron microscope (Philips, Eindhoven, The Netherlands).
In vivo studies.
To determine the relevance of the deleted pUL36 regions for the neurovirulence of PrV, 10 8-week-old CD1 mice each were infected by bilateral intranasal instillation of 104 PFU of either PrV-Ka, PrV-UL36Δ2087-2795, PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, or PrV-UL36Δ748-991. The animals were observed three times a day, and mean times to death (MTDs) were determined (22, 23).
RESULTS
Construction of UL36 plasmids and isolation of PrV pUL36 mutants.
Two functional domains, both located in the N terminus, had previously been described for pUL36: the pUL37 interaction domain (27) and a new class of deubiquitinating activity (21, 43). Neither domain is essential for virus replication, at least in cell culture (14, 32, 48), nor is a 709-aa proline/alanine-rich stretch in the C-terminal part (4). In contrast, the very C terminus of PrV-UL36 could not be deleted without loss of pUL36 function (4, 32). To further investigate the relevance of the proline/alanine-rich region within the C-terminal part of PrV pUL36 and to delineate more exactly the boundaries of the internal nonessential region, we enlarged the 709-aa (Δ2087-2795) internal in-frame deletion to 1432, 948, 927, and 895 aa, resulting in pcDNA-UL36Δ1581-3012, pcDNA-UL36Δ2034-2981, pcDNA-UL36Δ2087-3013, and pcDNA-UL36Δ2087-2981, respectively, for expression of the mutated pUL36 in eukaryotic cells (Fig. 1C). Correct in-frame cloning was verified by sequencing, and expression was demonstrated by indirect immunofluorescence of transiently transfected cells (data not shown). To transfer the mutant UL36 ORFs into the viral genome, viral DNA of PrV-ΔUL36F was cotransfected with pUL36 expression constructs into RK13 cells for marker rescue. Although all constructs transiently expressed the expected mutated pUL36 after transfection (data not shown), only pUL36Δ2087-2981 was able to complement the defect in PrV-ΔUL36F to yield PrV-UL36Δ2087-2981. Thus, the 5′ end of the internal deletion could not be expanded, whereas in the 3′ direction codons for an additional 186 aa could be deleted without loss of function.
For deletion of the deubiquitinating motif, aa 22 to 248, comprising the catalytic triad (21), were removed, resulting in pcDNA-UL36Δ22-248 (Fig. 1C). After cotransfection with PrV-ΔUL36F DNA, rescued virus PrV-UL36Δ22-248 could be isolated. Since the deletions in the N and C terminus were apparently tolerated individually, we also removed them simultaneously, resulting in PrV-UL36Δ22-248Δ2087-2981 (Fig. 1C).
Computational sequence analysis of PrV pUL36 revealed two putative leucine zipper motifs at aa 779 to 800 and 827 to 848 (4) (Fig. 1B). These motifs are also present in pUL36 homologs of other members of all three herpesvirus subfamilies except in human herpesvirus 6 (HHV-6), HHV-7, and Epstein-Barr virus (EBV) (4). To remove these leucine zipper motifs, an internal deletion from aa 748 to 991 was introduced, resulting in pcDNA-UL36Δ748-991 and rescued virus PrV-UL36Δ748-991 (Fig. 1C).
Expression and virion incorporation of truncated pUL36.
The authentic pUL36 of PrV consists of 3,084 aa, resulting in a calculated mass of 324 kDa. In contrast, the engineered truncated pUL36 comprise 2,857 aa (pUL36Δ22-248), 2192 aa (pUL36Δ2087-2981), 1965 aa (pUL36Δ22-248Δ2087-2981), or 2840 aa (pUL36 Δ748-991), with calculated masses of 300 kDa, 235 kDa, 211 kDa, and 298 kDa, respectively. Western blot analysis of purified virions using our anti-UL36-1 serum showed that the truncated proteins were incorporated into virus particles (Fig. 2A). Moreover, they exhibited the predicted decrease in molecular mass (Fig. 2A, lanes 2 and 4 to 6) compared to wild-type pUL36 (Fig. 2A, lane 1). PrV-UL36Δ2087-2795, which specifies a 255-kDa truncated pUL36 (Fig. 2A, lane 3), was also included. Neither deletion impaired incorporation of the interaction partner pUL37, as judged by the Western blots shown in Fig. 2, indicating that the pUL37-binding domain was not affected (Fig. 2C). Detection of pUL19 and pUL25 was used for control (Fig. 2B and D). The different intensities of the pUL36 bands, which decrease with increasing molecular mass, are most likely due to differences in the efficiencies of transfer of the high-molecular-mass proteins from the gel to the nitrocellulose membrane.
FIG. 2.
Western blot analysis of purified virions. Purified virions of PrV-Ka (lane 1), PrV-UL36Δ22-248 (lane 2), PrV-UL36Δ2087-2795 (lane 3), PrV-UL36Δ2087-2981 (lane 4), PrV-UL36Δ22-248Δ2087-2981 (lane 5), and PrV-UL36Δ748-991 (lane 6) were separated by electrophoresis on 6% (A and B) or 10% (C and D) polyacrylamide gels. Parallel blots were probed with the indicated antisera against pUL36 (A), pUL19 (B), pUL37 (C), and pUL25 (D) used at the indicated dilutions. The locations of molecular mass marker proteins (high-molecular-weight marker and Benchmark; Invitrogen) are indicated on the left. α, anti.
In vitro replication of pUL36 truncation mutants.
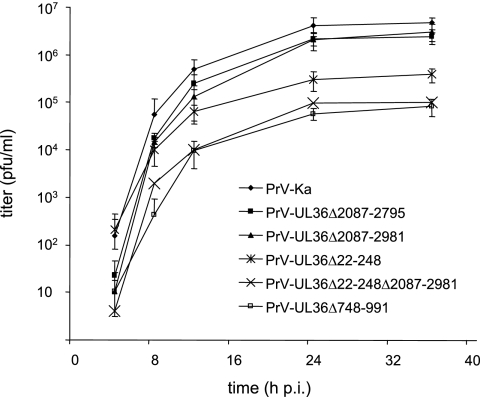
To reveal possible effects on virus replication, plaque formation of pUL36 mutant viruses on RK13 cells was investigated and one-step growth analysis was performed. In one-step growth analysis, PrV-UL36Δ2087-2981 replicated to similar titers as PrV-Ka and PrV-UL36Δ2087-2795. In contrast, PrV-UL36Δ22-248 showed ca. 15-fold-decreased final titers, and mutants PrV-UL36Δ22-248Δ2087-298 and PrV-UL36Δ748-991 exhibited ca. 100-fold-reduced final titers compared to PrV-Ka (Fig. 3).
FIG. 3.
One-step growth kinetics. Replication of PrV-Ka and respective mutant viruses was assessed after infection of RK13 cells. Average values and standard deviations of three independent experiments are shown.
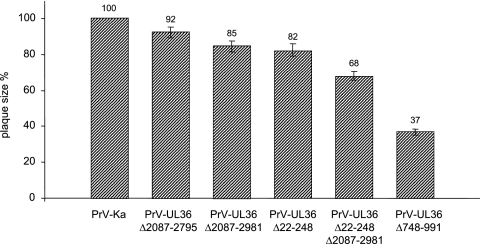
Concomitant with the one-step growth defect, plaque formation of PrV-UL36Δ748-991 lacking two predicted leucine zipper motifs was drastically decreased (37%) compared to PrV-Ka, set as 100% (Fig. 4). In contrast, deletion of only the N-terminal (Δ22-248) or C-terminal (Δ2087-2981) region had little effect on plaque size (82 and 85%, respectively), whereas simultaneous removal of both domains reduced plaque size to 68%. Thus, single deletions of the N- or C-terminal domain affected plaque formation of PrV to a much lesser extent than simultaneous deletion of both.
FIG. 4.
Plaque size determination. Plaques of PrV-Ka and respective mutant viruses on RK13 cells were measured, and mean plaque sizes of three independent experiments were calculated as percentages of plaques induced by PrV-Ka. Standard deviations are indicated.
Ultrastructural analysis.
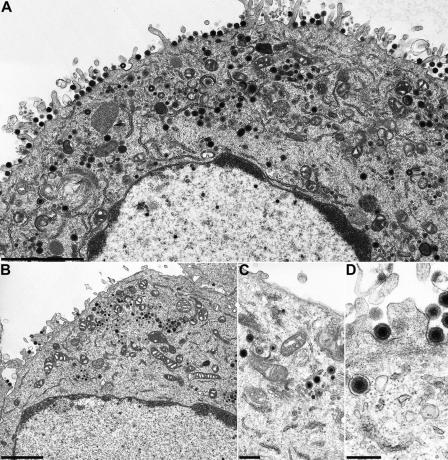
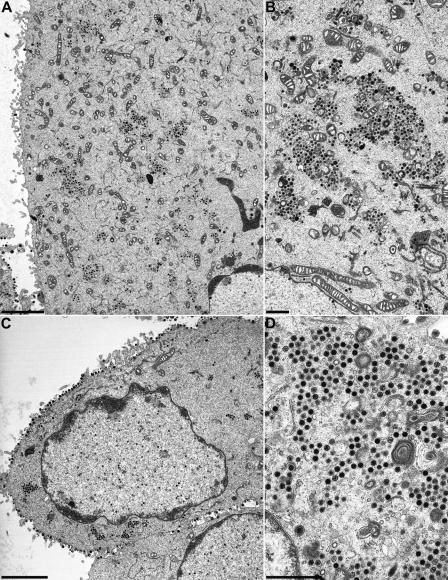
To assay for the effects of the engineered deletions in pUL36 on virion formation, cells infected with PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, PrV-UL36Δ22-248Δ2087-2981, and PrV-UL36Δ748-991 were analyzed at 14 h p.i. by electron microscopy. Apparently normal virion maturation could be observed in cells infected with PrV-UL36Δ22-248 (Fig. 5A) or PrV-UL36Δ2087-2981 (Fig. 5B-D). In contrast, cells infected with PrV-UL36Δ22-248Δ2087-2981 (Fig. 6A and B) or PrV-UL36Δ748-991 (Fig. 6C and D) displayed distinct cytoplasmic accumulations of nonenveloped nucleocapsids (Fig. 6B and D), whereas nuclear egress was not impaired. Only a few extracellular mature virus particles could be detected on PrV-UL36Δ22-248Δ2087-298- and PrV-UL36Δ748-991-infected cells, whereas capsidless L particles were released in abundance (Fig. 6C).
FIG. 5.
Ultrastructural analysis of PrV-UL36Δ22-248 and PrV-UL36Δ2087-2981. RK13 cells were infected at an MOI of 1 and analyzed 14 h after infection. (A) Overview of a PrV-UL36Δ22-248-infected cell. (B) Overview of a PrV-UL36Δ2087-2981-infected cell. (C and D) Higher magnifications of the panel B image showing secondary envelopment (C) and extracellular mature virions (D). Bars, 2.4 μm (A), 2 μm (B), 500 nm (C), and 300 nm (D).
FIG. 6.
Ultrastructural analysis of PrV-UL36Δ22-248Δ2087-2981, and PrV-UL36Δ748-991. RK13 cells were infected at an MOI of 1 and analyzed 14 h after infection. (A) Overview of a PrV-UL36Δ22-248Δ2087-2981-infected cell. (B) Enlargement of capsid aggregations in the cytoplasm. (C) Overview of a PrV-UL36Δ748-991-infected cell. (D) Higher magnification on cytoplasmic capsid aggregations. Bars, 3 μm (A and C) and 1 μm (B and D).
Animal infections.
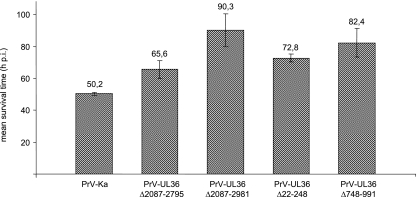
Since PrV is a neurotropic herpesvirus, we assayed with our small-animal infection model, i.e., intranasal infection of mice (22, 23), whether the engineered mutations within the pUL36 protein influence neurovirulence. To this end, 10 8-week-old CD1 mice were intranasally infected with the different PrV UL36 mutants and mean survival times were determined (Fig. 7). Mice infected with PrV-Ka developed their first clinical symptoms on day 2 p.i. and died at an average time of 50 h p.i. Although PrV-UL36Δ2087-2981 showed no obvious defects in culture, the infected mice had a considerably longer mean survival time of 90 h, whereas infection with PrV-UL36Δ22-248 or PrV-UL36Δ2087-2795 resulted in MTDs of 73 and 65 h, respectively. In contrast, mice infected with PrV-UL36Δ748-991 showed depression and a hunched position at 55 h p.i. and died after clinical signs like pruritus, severe dyspnea, and extensive scratching of the facial skin at an average time of 82 h p.i., which was unexpectedly low in view of the drastic replication defects in cell culture. Interestingly, PrV-Ka, PrV-UL36Δ2087-2795, and PrV-UL36Δ2087-2981 exhibited similar phenotypes in cell culture, whereas in the animal remarkably prolonged mean survival times of PrV-UL36Δ2087-2981-infected mice were observed. Thus, the C-terminal extension in PrV-UL36Δ2087-2981 of the deletion in PrV-UL36Δ2087-2795 might impair regulatory pathways exclusively available in vivo.
FIG. 7.
Mean survival times of mice. Ten CD-1 mice each were intranasally infected with PrV-Ka, PrV-UL36Δ2087-2795, PrV-UL36Δ2087-2981, PrV-UL36Δ22-248, PrVUL36Δ22-248Δ2087-2981, and PrV-UL36Δ748-991, and mean survival times were calculated.
DISCUSSION
In this study we engineered PrV mutants lacking different regions within pUL36 to analyze their role in virus replication in more detail. The salient findings are as follows. (i) A previously described deletion of 709 aa within the C-terminal part of pUL36 could be increased to 895 aa without significant impairment of viral in vitro replication. (ii) Further increase of this deletion to 927 aa did not yield replication-competent viruses. (iii) Deletion of a 227-aa region within the N terminus of pUL36 including the putative deubiquitinating activity was also tolerated, although viral titers decreased. (iv) Simultaneous deletion of the N- and C-terminal regions strongly impaired viral replication in vitro. (v) Deletion of a predicted leucine zipper region also impaired pUL36 function. Thus, we identified several nonessential regions within pUL36 whose deletion produced variable effects on protein function.
Previously, we isolated PrV-UL36Δ2087-2795, which expresses a pUL36 with an internal deletion of 709 aa, whereas deletions affecting the very C terminus were not tolerated (4). This correlates with another report which suggested that only the extreme C-terminal 6 aa of pUL36 could be deleted without impairment of PrV replication (32). We show here that the internal deletion can be extended towards the C terminus to include a total of 895 aa with only minor effects on PrV replication in vitro. Final titers of PrV-UL36Δ2087-2981 were similar to those of PrV-UL36Δ2087-2795 and were only slightly lower than those of PrV-Ka, as was plaque size. This deletion covers a region that is not conserved between herpesvirus pUL36 homologs. It is rich in proline and alanine residues and, therefore, is supposed to display a disordered secondary structure (4). Proline-rich regions can be recognized by SH3 domains, which are present in many eukaryotic proteins and which are involved in signal transduction or mediate protein-protein interactions of cytoskeletal proteins (33). Whatever function this region has for PrV, it is apparently not required for productive replication in RK13 cells.
Surprisingly, there were distinct differences in vivo between PrV-Ka, PrV-UL36Δ2087-2795, and PrV-UL36Δ2087-2981, resulting in MTDs of mice after intranasal infection of 50, 65, and 90 h. This is in stark contrast to the in vitro phenotypes, where no drastic alterations between the two deletion mutants could be detected and there were only minor differences from the wild type, PrV-Ka. In ultrastructural analyses neither accumulations of nucleocapsids in the cytoplasm, which are characteristic of the impairment of secondary envelopment, nor a visibly reduced release of mature virus particles was observed, indicating that increased survival times are not simply due to a basic defect in viral replication. Thus, apparently the larger deletion specifically impaired pUL36 functions important for infection in vivo.
In contrast, strong in vitro phenotypes were observed for PrV-UL36Δ748-991, which lacks a putative leucine zipper domain, and PrV-UL36Δ22-248Δ2087-2981, carrying a deletion of 227 aa in the N terminus encompassing the active site of the deubiquitinating enzymatic activity and also lacking 895 aa in the C terminus. Viral titers of both mutants were decreased ca. 100-fold, but plaque sizes were affected differently. Whereas plaques produced by PrV-UL36Δ22-248Δ2087-2981 reached 68% of the wild-type diameter, those produced by PrV-UL36Δ748-991 amounted to only 37% of the wild-type size. Surprisingly, the strong reduction in plaque formation was not reflected in the survival times of mice infected intranasally, yielding an MTD of 82 h after infection with PrV-UL36Δ748-991. No information is available yet on the in vivo phenotype of PrV-UL36Δ22-248Δ2087-2981.
In PrV-UL36Δ22-248 the predicted active site of a conserved deubiquitinating activity (21, 43, 44) has been deleted. The resulting mutant virus showed only a minor reduction in plaque size compared to the wild type, PrV-Ka, whereas final titers were reduced by ca. 15-fold. However, ultrastructurally no specific defect in viral replication could be pinpointed. Furthermore, mean survival times of mice infected intranasally with PrV-UL36Δ22-248 were prolonged only to 73 h p.i. compared to 50 h p.i for PrV-Ka-infected mice. These data on PrV pUL36 correlate with those obtained by analysis of an HCMV mutant carrying an exchange of the catalytic cysteine residue (C24I) (48), which did not exhibit strikingly altered growth characteristics. However, they differ from results for a PrV mutant expressing pUL36 with an N-terminal deletion of aa 6 to 225, which showed a substantial decrease in single-step growth (32). This difference could conceivably be attributed to the different parental viruses and cell systems used or to the different amino termini of the deletions. This could indicate that amino acids between 6 and 22 are important for pUL36 function. However, Lee et al. (32) also report that their deletion impaired pUL37 expression and virion incorporation, which are known to interfere with efficient viral replication (9, 24). Thus, an effect of impaired pUL37 expression on the observed phenotype could not be ruled out. In our mutant virus, neither expression nor virion incorporation of pUL37 was detectably affected, indicating that this portion of PrV pUL36 can indeed be deleted without affecting pUL37 and without a strong impairment of viral replication.
The described amino-terminal deletion has also been associated with a defect in egress of capsids in axons of sensory neurons, whereas retrograde transport was indistinguishable from that for wild-type PrV (32). In our animal model survival times of PrV-UL36Δ22-248-infected mice were only moderately increased, which may correlate with a somewhat slower neuroinvasion. However, mice still succumbed to the infection, exhibiting the classical symptoms of PrV infection. Thus, although our deletion resulted in a delay in neuroinvasion, infection of the nervous system apparently still occurred and neurovirulence was retained.
Although we did not factually demonstrate the presence of a deubiquitinating activity in PrV pUL36 by an enzymatic assay, overall sequence homology (21) and analyses of pUL36 homologs in murine cytomegalovirus, EBV (43), and HCMV (48) indicate that the deubiquitinating activity is also present in PrV pUL36.
Thus, the exact biological role of this activity remains unclear. Ubiquitination has been shown to be involved in cell cycle control, NF-κB regulation, and proteasomal degradation of major histocompatibility complex class I proteins (15), and many pathogens hijack these pathways. However, in PrV the deubiquitinating activity is not required for replication either in cell culture or in a murine host.
Despite the nonessential nature of the N-terminal and C-terminal amino acids deleted in the mutants, simultaneous absence of both in PrV-UL36Δ22-248Δ2087-2981 drastically impaired viral replication. Ultrastructural analysis revealed the presence of large accumulations of nonenveloped nucleocapsids in the cytoplasm (Fig. 6), whereas nuclear egress was not affected (data not shown). The presence of only few mature virus particles on the cell surface correlates with the 100-fold-lower final titers compared to those of PrV-Ka. Accumulations of seemingly tegument-free nucleocapsids in the cytoplasm have been observed in PrV-ΔUL37 (24), which lacks pUL37, and PrV-UL36BSF, a pUL36 mutant which lacks the pUL37 binding site (14). However, in cells infected with these virus mutants, cytoplasmic nucleocapsid accumulations appeared in geometrically ordered clusters. This pattern differs from the accumulation observed in PrV-UL36Δ22-248Δ2087-2981-infected cells, which appeared more random.
Thus, whereas single deletions of either the N- or C-terminal region exerted only minor effects on pUL36 function, simultaneous removal of both resulted in an additive or even synergistic effect. This could indicate that both regions function independently of each other. On the other hand, the simultaneous deletion might interfere with pUL36 overall structure, resulting in impairment of possible pUL36 interactions with other viral tegument components. An effect on pUL36 structure by the other introduced mutations could also not be excluded, although it is clear that interaction with pUL37 is not abolished by any of them. Interestingly, in this double-mutant virus all three RGD motifs in PrV pUL36 (Fig. 1B), which may serve for interaction with cellular integrins, have also been deleted, which demonstrates that, if they are active at all, they are certainly not absolutely required for pUL36 function.
It has been suggested that, besides the described interaction of pUL36 with the major capsid protein pUL19 (36) and the tegument protein pUL37 (3, 14, 19a, 27, 47), other viral (47) or cellular proteins (41) bind to pUL36. Furthermore, the inner tegument proteins are prime candidates for interaction with molecular motors like dynein or dynactin (10, 49) in the transport of incoming capsids towards the nuclear pore. Leucine zipper motifs which mediate DNA-protein as well as protein-protein interactions (31) have been predicted for pUL36 orthologs of members of all herpesvirus families except HHV-6, HHV-7, and EBV (4, 27). PrV-UL36Δ748-991, which lacks the region containing both predicted leucine zippers, exhibited strong impairment of one-step growth and, even more drastically, plaque formation. In ultrastructural analysis accumulations of capsids in the cytoplasm similar to those in cells infected with PrV-UL36Δ22-248Δ2087-2981 were observed concomitant with an increase in the formation and release of capsidless L particles. In contrast, and unexpectedly, neurovirulence was only moderately affected, as mean survival times of infected mice amounted to 82 h.
In a recent study, a pUL36 with a deletion of an alanine-rich region from aa 397 to 1293 failed to function in cell culture (32), indicating that regions which flank the sequence removed in this study might be important for virus replication. Based on the accumulation of nucleocapsids observed by electron microscopy we hypothesize that the region containing two leucine zipper motifs is required for continuing tegumentation and may play a role in linking capsids carrying inner tegument proteins with outer tegument proteins assembled at the envelopment site. Alternatively, removal of these protein-protein interaction motifs could impair a potential pUL36 homo- oligomerization, which may be required for tegumentation.
In summary, we show that regions of PrV pUL36 containing either a highly conserved deubiquitinating activity or predicted leucine zipper motifs as well as a large internal deletion within the C terminus are not essential for viral replication either in vitro or in vivo. However, they may nevertheless be important for virus replication in cell culture and infection of animals. It remains to be analyzed whether the observed effects correlate with impairment of specific functions of the protein during PrV replication. In general, our data on these and previous pUL36 mutants are not congruent with a defect in nuclear egress in the absence of pUL36 (35) but rather support the notion that pUL36 is important for tegumentation of capsids in the cytoplasm.
So far the exact roles for PrV pUL36 remain unclear. However, we have now identified 45% of the protein sequence as being nonessential for viral replication and constructed a functional pUL36 simultaneously lacking a total of 1,122 aa. Despite this huge deletion, the morphology of mature virus particles released from cells infected by any of the mutant viruses does not differ significantly from that of wild-type virions, which gives testimony to the amazing flexibility in herpesvirus morphogenesis. Based on our findings, the remaining, apparently essential part of pUL36 can now be analyzed with more ease, shedding more light on the multiple roles of this impressively huge virion component.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/8-1).
We thank Petra Meyer, Kathrin Müller, and Diana Werner for expert technical assistance and Mandy Jörn for photographic help.
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Baines, J. D., C. E. Hsieh, E. Wills, C. Mannella, and M. Marko. 2007. Electron tomography of nascent herpes simplex virus virions. J. Virol. 81:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W. D., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bechtel, J. T., and T. Shenk. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol. 76:1045-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Böttcher, S., B. G. Klupp, H. Granzow, W. Fuchs, K. Michael, and T. C. Mettenleiter. 2006. Identification of a 709-amino-acid internal nonessential region within the essential conserved tegument protein (p)UL36 of pseudorabies virus. J. Virol. 80:9910-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucks, M. A., K. J. O'Regan, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, and J. A. Martignetti. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 8.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai, P., G. L. Sexton, J. M. McCaffery, and S. Person. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dohner, K., A. Wolfstein, U. Prank, C. Echeverri, D. Dujardin, R. Vallee, and B. Sodeik. 2002. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13:2795-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, W., H. Granzow, B. G. Klupp, A. Karger, K. Michael, C. Maresch, R. Klopfleisch, and T. C. Mettenleiter. 2007. Relevance of the interaction between the alphaherpesvirus UL3.5 and UL48 proteins for virion maturation and neuroinvasion. J. Virol. 81:9307-9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., H. Granzow, H., B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 2004. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J. Virol. 78:11879-11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao, G., and H. Luo. 2006. The ubiquitin-proteasome pathway in viral infections. Can. J. Physiol. Pharmacol. 84:5-14. [DOI] [PubMed] [Google Scholar]
- 16.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 17.Granzow, H., B. G. Klupp, and T. C. Mettenleiter. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J. Virol. 79:3200-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 19a.Harmon, M. E., and W. Gibson. 1996. Proc. 15th Annu. Meet. Am. Soc. Virol., abstr. W35-4, p.144.
- 20.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 21.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 22.Klopfleisch, R., B. G. Klupp, W. Fuchs, M. Kopp, J. P. Teifke, and T. C. Mettenleiter. 2006. Influence of pseudorabies virus proteins on neuroinvasion and neurovirulence in mice. J. Virol. 80:5571-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klopfleisch, R., J. P. Teifke, W. Fuchs, M. Kopp, B. G. Klupp, and T. C. Mettenleiter. 2004. Influence of tegument proteins of pseudorabies virus on neuroinvasion and transneuronal spread in the nervous system of adult mice after intranasal inoculation. J. Virol. 78:2956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knipe, D. M., W. Batterson, C. Nosal, B. Roizman, and A. Buchan. 1981. Molecular genetics of herpes simplex virus. VI. Characterization of a temperature-sensitive mutant defective in the expression of all early viral gene products. J. Virol. 38:539-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, and T. C. Mettenleiter. 2004. Simultaneous deletion of pseudorabies virus tegument protein UL11 and glycoprotein M severely impairs secondary envelopment. J. Virol. 78:3024-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA-binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 32.Lee, J. I., G. W. Luxton, and G. A. Smith. 2006. Identification of an essential domain in the herpesvirus VP1/2 tegument protein: the carboxy terminus directs incorporation into capsid assemblons. J. Virol. 80:12086-12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, S. S. 2005. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem. J. 15:641-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luxton, G. W., S. Haverlock, K. E. Coller, S. E. Antinone, A. Pincetic, and G. A. Smith. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. USA 102:5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luxton, G. W., J. I.-H. Lee, S. Haverlock-Moyns, J. M. Schober, and G. A. Smith. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J. Virol. 80:201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 37.McNabb, D. S., and R. J. Courtney. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 66:7581-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 39.Mettenleiter, T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167-180. [DOI] [PubMed] [Google Scholar]
- 40.Mettenleiter, T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163-169. [DOI] [PubMed] [Google Scholar]
- 41.Ogawa-Goto, K., S. Irie, A. Omori, Y. Miura, H. Katano, H. Hasegawa, T. Kurata, T. Sata, and Y. Arao. 2002. An endoplasmic reticulum protein, p180, is highly expressed in human cytomegalovirus-permissive cells and interacts with the tegument protein encoded by UL48. J. Virol. 76:2350-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Regan, K. J., M. A. Bucks, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192-200. [DOI] [PubMed] [Google Scholar]
- 43.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpes virion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Telford, E. A., M. S. Watson, J. Perry, A. A. Cullinane, and A. J. Davison. 1998. The DNA sequence of equine herpesvirus-4. J. Gen. Virol. 79:1197-1203. [DOI] [PubMed] [Google Scholar]
- 47.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfstein, A., C. H. Nagel, K. Radtke, K. Dohner, V. J. Allan, and B. Sodeik. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227-237. [DOI] [PubMed] [Google Scholar]
- 50.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]