Abstract
Human cytomegalovirus (CMV), a ubiquitous human pathogen, is a leading cause of congenital infections and represents a serious health risk for the immunosuppressed patient. A vaccine against CMV is currently not available. CMV is characterized by its large genome and by multiple genes modulating the immunity of the host, which cluster predominantly at genome termini. Here, we tested whether the deletion of gene blocks rich in immunomodulatory genes could be used as a novel concept in the generation of immunogenic but avirulent, herpesvirus vaccines. To generate an experimental CMV vaccine, we selectively deleted 32 genes from the mouse cytomegalovirus (MCMV) genome. The resulting mutant grew to titers similar to that of wild-type MCMV in vitro. In vivo, the mutant was 10,000-fold attenuated and well tolerated, even by highly susceptible mice deficient for B, T, and NK cells or for the interferon type I receptor. Equally relevant for safety concerns, immune suppression did not lead to the mutant's reactivation from latency. Immunization with the replication-competent mutant, but not with inactivated virus, resulted in protective immunity, which increased over time. Vaccination induced MCMV-specific antibodies and a strong T-cell response. We propose that a targeted and rational approach can improve future herpesvirus vaccines and vaccine vectors.
The human cytomegalovirus (HCMV), a betaherpesvirus subfamily member, is a ubiquitous human pathogen that causes congenital infections and also represents a major morbidity risk for immune-suppressed or immunodeficient patients (56).
HCMV carries approximately 200 genes, which represent a large antigenic potential. However, despite previous efforts, (59, 60), no effective vaccine has been generated so far (67). Although the Towne strain was shown in numerous studies to be a safe and immunogenic vaccine (1, 29, 30), its immunogenicity was lower than that of the wild-type (WT) virus, and it failed to confer immune protection against infection by natural contact (2). Several features of HCMV infection make vaccine development uniquely difficult. A large number of HCMV genes modulate the innate and adaptive host immune responses to the advantage of the pathogen (43, 50, 74). Natural CMV infection provides only partial protection, and reinfection can cause congenital CMV disease even in infants of mothers with preconceptional immunity (5, 22). Moreover, persistence of the virus in state of latency, with the possibility of reactivation over the course of the patient's life, represents a safety concern when a live attenuated herpesvirus vaccine is used. Finally, cytomegaloviruses are characterized by strict species specificity, and there is no animal model for direct studies of HCMV infection and immunity in vivo.
The infection of mice with mouse CMV (MCMV) represents a widely used in vivo model of CMV infection (40). Studies with the MCMV model revealed that protective immunity against CMV infection requires both B and T cells (31-33, 64). Therefore, it is not surprising that subunit immunization strategies, which induced either cellular (24, 52) or humoral (20, 69) immunity, provided only partial immune protection against a challenge with virulent MCMV. Approaches using DNA immunization followed by formalin-inactivated virus have shown promising results (53). Live vaccines resulted in a much stronger protection (23, 47, 51), yet their application is connected with the concern for virus latency and virus reactivation in immunocompromised hosts. Moreover, cellular immunity against CMV follows kinetics not seen in most other viral infections. The number of CD8+ memory-effector cells directed against immunodominant HCMV or MCMV peptides expands over time (28, 34), and low-level transcription of viral genes during latency (25) has been implied as the underlying mechanism (35). MCMV recombinants that expressed heterologous immunodominant peptides during latency induced protective immunity against lymphocytic choriomeningitis and influenza (35). Therefore, live attenuated CMVs are attractive vaccine candidates if their pathogenicity can be lowered without affecting their immunogenicity and if the risk of reactivation in the immunodeficient host can be excluded.
A prototypical live attenuated CMV vaccine or a CMV-based vaccine vector should possess the following properties. (i) For easy production, the vaccine should grow to high titers in cell culture. (ii) The virus should be severely attenuated in vivo, even in immune-compromised hosts. (iii) It should elicit a strong immune response that protects against a challenge to the same extent as or better than that of an infection with the WT virus.
A rational approach to the generation of such a vaccine is the targeted deletion of genes modulating the immune response, because this should expose the virus to the immune system and thereby decrease its fitness and increase its immunogenicity. This approach has become technically achievable with novel advances in herpesvirus genetics. The cloning of herpesvirus genomes as bacterial artificial chromosomes (BACs) in Escherichia coli (49) and the subsequent improvements in BAC technology (72) have allowed quick and reliable targeted deletion of cytomegalovirus genes or regions encompassing multiple genes.
To date, the full range of viral functions that modulate host immunity is unknown for both HCMV and MCMV. However, conserved and essential genes tend to cluster in the middle of the genome, whereas genes involved in the modulation of host functions accumulate at the genome termini. In MCMV, the three major histocompatibility complex class I (MHC-I) modulating genes (m04 [37], m06 [66], and m152 [43, 74]) and the five genes known to regulate NK-cell function (m144 [21], m145 [42], m152 [41], m155 [45], and m157 [3, 9]) are located at either end of the genome. This is also true for m147.5, which affects dendritic cells by downregulating CD86 surface expression (46).
Here, we describe an MCMV recombinant lacking 31.2 kbp at the left and right genome termini of the WT genome, corresponding to 32 genes altogether. The recombinant replicated to WT levels in vitro but was severely attenuated in vivo, even in immunodeficient mice. The recombinant virus induced cellular and humoral immunity and protected mice from a challenge with infection by WT-MCMV.
MATERIALS AND METHODS
Cells, viruses, and mice.
NIH 3T3 (ATCC no. CRL-1658), IC-21 (ATCC no. TIB-186), SVEC4-10 (ATCC no. CRL-2181), and M2-10B4 (ATCC no. CRL-1972) were grown as previously described (48). Ana-1 mouse macrophages (14) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal calf serum (FCS). C127 mouse epithelial cells (ATCC no. CRL-1616) were maintained in DMEM-10% FCS. Mouse embryonic fibroblasts (MEFs) were prepared and maintained as described previously (61).
Wild-type MCMV (WT-MCMV) was derived from pSM3fr (71), a molecular clone with in vivo growth properties comparable to those of the MCMV Smith strain (71). Δm01-17+m144-158-MCMV was generated as described below. All viruses were propagated on M2-10B4 cells, and virus stocks were purified on a sucrose cushion as described previously (48).
Female BALB/c, C.B-17/IcrHsd-Prkcdscid (SCID), and C.B-17/IcrHsd-PrkcdscidLystbg (SCID/bg) mice were purchased from Harlan-Winkelmann (Borchen, Germany). Interferon (IFN) type I receptor−/− mice on a 129 background (IFN-αβR−/−) were bred under specific pathogen-free conditions at the Vaccine and Gene Therapy Institute Vivarium. All mice were infected at 8 to 10 weeks of age and kept in barrier housing throughout the experimental procedures. Animal experiments were approved by the responsible office of the state of Bavaria (approval no. 211-2531-38/99), by the ethics committee at the University of Rijeka, or by the institutional animal care and use committee at Oregon Health and Science University.
Generation of mutant virus by MCMV BAC mutagenesis.
The recombinant MCMV BAC pΔm01-17+m144-158, lacking 31.2 kb of the MCMV genome (nucleotide [nt] positions 480 to 16949 and 203001 to 217800, according to Rawlinson et al. [63]) was generated by three subsequent mutagenesis steps with the parental MCMV BAC pSM3fr (71), essentially as described previously (70), except that the redα, redβ, and redγ recombinases were expressed by the plasmid pKD46 (15). In brief, (i) a linear DNA fragment containing the kanamycin resistance gene, flanked by 34-bp FRT sites and sequences homologous to the MCMV genome target site (nt positions 449 to 479 and 16952 to 16992), was generated using primers 5-m01 (5′-CAC GTG TTA GCA TAG GAA TCC AGA CGC GCG CTC GCC TGA GCG TCG TGG AAT GCC TTC GAA TTC-3′) and 3-m17 (5′-CTT TGA AAT CGG ACG ACC GAT CAG AAC GTC CGC CTT CGA GAA CAA GGA CGA CGA CGA CAA GTA A-3′) and the plasmid pSLFRTKn (4) as template. This PCR fragment was inserted into pSM3fr by homologous recombination in E. coli to delete the gene region m01 to m17. (ii) The kanamycin resistance gene was excised by transiently expressed FLP recombinase, as described previously (12). (iii) Next, genes m144 to m158 were deleted. For this step, a PCR fragment containing a kanamycin marker flanked by mutant FRT sites (5′-GAA GTT CCT ATT CTT CAA AAG GTA TAG GAA CTT C-3′) and MCMV homologies to the up- and downstream sequences of genes m144 and m158 (nt positions 202960 to 203000 and 217800 to 217844), respectively, was generated using primers 5-mFRT-m144 (5′-AGC GCC GTC CGA ACC TCC TAC GCG TCT TCC TCT GTT CCT TGC CCC GAA AAG TGC CAC CTG CAG AT-3′) and 3-mFRT-m158 (5′-CTA CCG TCT CCT CGA ATG GCA GAG AGG CGA TCT CGT ACC TCT AAC GTG ACA CAG GAA CAC TTA ACG GCT GA-3′) and plasmid pOri6k-F5 (7) as the template. This fragment was inserted into the MCMV BAC deficient for genes m01 to m17. Recombinant virus from the resulting MCMV BAC pΔm01-17+m144-158 was reconstituted as described previously (70).
Southern blotting analysis of viral DNA.
Isolation of MCMV BAC DNA from E. coli cultures was performed by using a Nucleobond kit (Macherey-Nagel, Dueren, Germany). Viral DNA was isolated from ∼106 M210B4 cells, which were infected for 24 h with 3 × 106 PFU of WT and mutant MCMV at passage seven after reconstitution from the respective BACs, using a DNeasy kit (QIAGEN, Germany) according to the manufacturer's instructions. Southern blotting analysis was carried out as described previously (71) by applying PCR-synthesized probes for hybridization, which were amplified from the pSM3fr template with primers Bfor (GTGAAGCGATTCACAGATGTCT) and Brev (CGACAGAGGATAAACGGTAATC), using a digoxigenin probe synthesis kit (Roche, Germany) according to the manufacturer's instructions. For the experiment shown in Fig. 1c, 6 μg of each DNA purified from the infected cells in parallel with 1 μg of pΔm01-17+m144-158 and 0.5 μg of the pSM3fr BAC DNA were digested with PciI overnight and separated by agarose gel electrophoresis prior to the blotting.
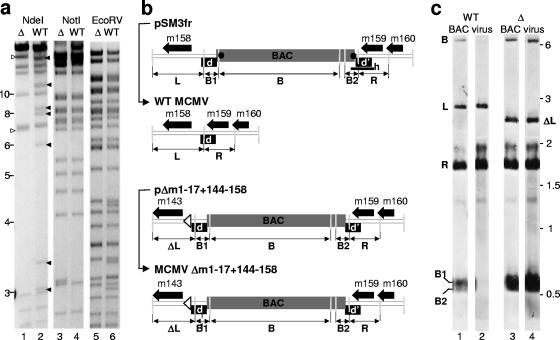
FIG. 1.
Analysis of the BAC-cloned genomes pSM3fr and Δm01-17+m144-158 and of the corresponding viral DNA after virus reconstitution. (a) The pΔm01-17+m144-158 (Δ) and the pSM3fr (WT) BACs were analyzed by restriction endonuclease digests using NdeI, NotI, and EcoRV. Filled arrowheads indicate NdeI fragments which are lost by the deletion of the regions comprising the genes m01 to m17 and m144 to m158. Open arrowheads indicate two new fragments generated by mutagenesis. Analysis of NotI and EcoRV restriction patterns excluded unwanted mutagenesis that occurred in other areas of the mutant genome. (b) Schematic representation of the m158 gene locus in the pSM3fr (WT-MCMV BAC) and pΔm01-17+m144-158-BAC plasmids and upon reconstitution of the respective virus (BAC and virus maps are connected with thin black arrows). The viral ORFs are indicated by thick black arrows; the BAC vector sequences are represented by a gray box; the repeat region, which is duplicated at both the BAC/viral DNA joints, are shown as black boxes (d and d′); the loxP sites at the ends of the BAC vector sequences are indicated by black dots. The open arrowhead indicates the site at which the region m144 to m158 was deleted from the mutant genome. The probe used for the Southern blotting shown in panel c was amplified on the pSN3fr template and is indicated in the uppermost map by the thick horizontal (h) bar. PciI restriction sites are indicated by the light gray vertical bars. PciI fragments recognized by the Southern probe and corresponding to the signals indicated in panel c are indicated with bold capital letters below each map (L, ΔL, R, B, B1, and B2). (c) Southern blotting analysis of the BAC vector sequence in pΔm01-17+m144-158 and in the Δm01-17+m144-158-MCMV genome. DNA isolated from M2-10B4 cells and infected either with WT (lane 2) or Δm01-17+m144-158-MCMV (lane 4) was cleaved by PciI, separated by electrophoresis, and hybridized with the probe “h,” indicated in panel b (lanes 2 and 4). pSM3fr and pΔm01-17+m144-158-BAC plasmid DNA was used for control (lanes 1 and 3). The specifically recognized fragments are indicated by the letters on the left side of the panel corresponding to the maps in panel b. To visualize the separation of fragments B1 and B2, only half of the pSM3fr BAC DNA was loaded (lane 1). Values shown are given in kilobases.
Infectious virus replication and quantification.
Cells were infected at a multiplicity of infection (MOI) of 0.1 or 1. The inoculum was removed 1 h postinfection (p.i.), and cells were washed with phosphate-buffered saline (PBS). The infected cell cultures were then incubated in appropriate complete media. Supernatants from infected cells were harvested every 24 h from day 0 (input virus) through day 5 after infection and were stored at −80°C until quantification. Virus titers were determined by a standard plaque assay with MEFs, as described previously (61, 65).
For the determination of virus replication in vivo, BALB/c, SCID, or SCID/bg mice were infected by an intravenous (i.v.) or a subcutaneous (s.c.) route. Organs were collected at the indicated time points and assayed by plaque assay with centrifugal enhancement, essentially as described previously (65), with the following modifications: organs were homogenized on 100-μm-pore-sized strainers (BD-Falcon) and resuspended in DMEM-10% FCS. Thirty minutes of centrifugal enhancement at room temperature was followed by a 30-min incubation at 37°C, after which the monolayers were washed and overlaid with DMEM supplemented with 10% carboxymethylcellulose (Sigma, Munich, Germany).
Immunization experiments.
BALB/c mice were immunized by s.c. injection of a 100-μl solution of virus that had been diluted in PBS to equal 106 PFU/mouse or, where indicated, to 105 PFU/mouse. Where indicated, virus was inactivated with overnight exposure to UV light, and the loss of infectivity was confirmed by a plaque assay of an aliquot of the inactivated virus. Mock-treated mice received the same volume of PBS. Booster injections were carried out 14 days p.i. After mice were infected, their weight loss was monitored daily. At 16 weeks p.i., titers of virus-specific antibodies in sera of immunized mice were determined by indirect enzyme-linked immunosorbent assay (ELISA), as described previously (18). In brief, plates were coated with lysates from MCMV-infected MEF as the source of antigen or with uninfected MEF as the background control. Background optical density values were subtracted prior to analysis. CD8 T cells specific for the MCMV pp89-derived YPHFMPTNL peptide were quantified by flow cytometry, essentially as described previously (34). In brief, 50 μl of blood collected from the tail vein was stained with fluorescein isothiocyanate (FITC)-αCD8 monoclonal antibodies (BD-Pharmingen) and phycoerythrin (PE)-conjugated H-2Ld-YPHFMPTNL tetramers (kindly provided by P. Klenerman, Oxford, United Kingdom), incubated for 5 min in NH4Cl buffer, washed, and run in a Coulter EPICS XL flow cytometer.
Mice were challenged by an i.p. injection of 106 PFU of tissue culture-derived WT-MCMV. On day 5 postchallenge, mice were sacrificed, and lungs, liver, and spleen were collected under sterile conditions and frozen at −80°C. Organ homogenates were analyzed for infectious virus by standard plaque assays with MEFs, as described above. Each experiment was repeated at least two times.
Peptide stimulation and intracellular cytokine staining.
Blood cells obtained at day 7 p.i. were depleted of erythrocytes and incubated at 37°C in the presence of purified YPHFMPTNL peptide (10−6 M) for 6 h in RPMI medium supplemented with FCS (10% [vol/vol]), β-mercaptoethanol, and brefeldin A. Subsequently, cells were surface stained with anti (α)-CD4-Pacific Blue and α-CD8-PerCP-cy5.5, treated for 5 min with IC Fixation Buffer and Permeabilization Buffer (both from eBioscience, San Diego, CA), and stained with α-IFN-γ-allophycocyanin and α-tumor necrosis factor (TNF)-α-PE antibodies.
Reactivation experiments.
BALB/c mice infected i.p. with 5 × 105 PFU of WT or mutant MCMV were immunosuppressed at 9 months p.i. by an injection of polyclonal α-asialo antibodies (WAKO, Osaka, Japan) and monoclonal αCD4 and αCD8 (13) antibodies (1 mg each), followed by whole-body gamma irradiation with 7 Gy 1 day later. Spleen explants were layered on MEF monolayers at day 11 postirradiation. MEFs were subcultured 5 days later and monitored for virus reactivation for an additional 3 weeks.
RESULTS
Generation of an MCMV recombinant lacking 32 genes.
The majority of the known genes that modulate the immune evasion of NK and T cells belong to the m02 or the m145 gene families (63), which cluster, respectively, at the left end of the MCMV genome or between the m144 and the m158 gene. Therefore, we reasoned that targeted deletion of these gene clusters might allow us to test a novel concept in herpesvirus vaccinology and generate an MCMV mutant that would be immunogenic and selectively attenuated in vivo but not in vitro.
A mutant lacking genes m02 to m16 showed no replication defect in vitro but was severely attenuated in vivo (55). In a previous study, we found the m143 gene to be essential (48). Therefore, by homologous recombination of the MCMV genomes maintained as BAC in E. coli (see Materials and Methods), we deleted the m01 to m17 and m144 to m158 gene regions to generate the recombinant MCMV BAC pΔm01-17+m144-158. The analysis of four restriction enzyme patterns using NdeI, NotI, and EcoRV confirmed the expected recombinant genome (Fig. 1a).
MEFs were transfected with pΔm01-17+m144-158 DNA, and virus progeny, referred to as Δm01-17+m144-158-MCMV, were grown. Thus, the 32 deleted genes are dispensable for virus growth in vitro. To test the stability of the significantly shortened genome, we analyzed viral DNA in infected fibroblasts after several cell culture passages. Two independently transfected clones of Δm01-17+m144-158-MCMV were passaged at least five times with M2-10B4 cells prior to DNA isolation. EcoRI restriction enzyme patterns of isolated virus DNA showed that the genome of the deletion mutant had the same restriction pattern as its parental BAC (data not shown). We also analyzed the stability of the locus in which the BAC vector sequence is inserted. WT-MCMV genomes carrying the BAC sequence are believed to be oversized and thus less efficient at packaging. A 500-bp sequence belonging to m159 is repeated at both ends of the BAC vector (Fig. 1b), which allows the homologous recombination and removal of the BAC vector sequences by natural selection after a few passages (71) (Fig. 1b, compare the first two linear maps). In line with the prediction that MCMV are selected for their genome size, the BAC vector sequence was not excised from Δm01-17+m144-158-MCMV, as revealed by Southern blotting upon seven passages of the mutant on M2-10B4 cells (Fig. 1b and c). The probe used for Southern blotting was amplified by PCR on the pSN3fr template and recognized the repeated sequence and the 3′ end of the BAC vector, including the loxP sites. The BAC DNA and viral DNA were treated with PciI restriction endonuclease, which cleaves within the repeat and the 3′ end of the BAC. Therefore, there are five fragments which are recognized specifically by this probe: the BAC sequence (Fig. 1b, fragment B), the upstream and the downstream repeated sequences connected to the 5′ or the 3′ end of the BAC vector (Fig. 1b, B1 and B2), the left genomic fragment, which contains the m158 gene open reading frame (ORF) (Fig. 1b, L), and the right genomic fragment, which contains the m159 gene ORF (Fig. 1b, R). Due to the deletion of the m144 to m158 region, the L-derived fragment in the mutant (Fig. 1b, ΔL) shifts down from the original 2.8 kbp to 2.4 kbp. While the BAC-derived B, B1, and B2 fragments become undetectable after passaging of the reconstituted WT construct (Fig. 1c, lanes 1 and 2), the virus stocks derived from the BAC pΔm01-17+m144-158 still maintain these fragments (Fig. 1c, lanes 3 and 4). Thus, the retention of the BAC vector sequences indicated that, in contrast to WT-MCMV, the deletion mutant virus was not selected for shorter genome size.
Δm01-17+m144-158-MCMV replicates to WT levels in fibroblasts and endothelial and epithelial cells.
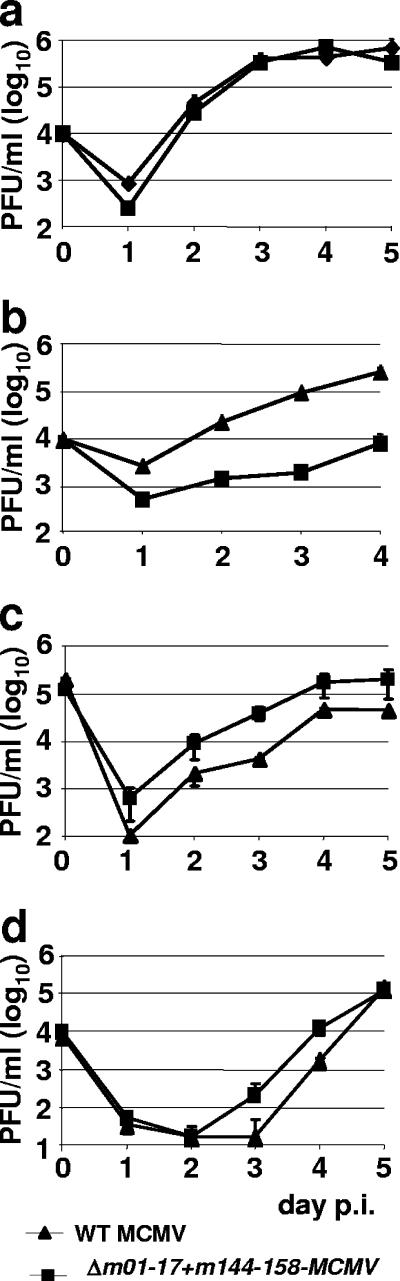
To test Δm01-17+m144-158-MCMV replication in cell culture, we compared it to WT-MCMV in a multistep growth kinetics assay of NIH 3T3 cells. The mutant replication was comparable to that of WT-MCMV (Fig. 2a). We also tested the growth of the recombinant virus on the macrophage cell line Ana-1, the endothelial cell line SVEC4-10, and the mammary epithelial cell line C127 (Fig. 2b, c, and d). Δm01-17+m144-158-MCMV replicated like WT-MCMV in endothelial and epithelial cell lines but was about 15-fold attenuated in Ana-1 macrophages. A similar attenuation was seen in another macrophage line, IC-21, but this growth deficit was overcome at an MOI of 1 (data not shown). The mutant was also not attenuated in mHTC-K2 cells, a hepatocyte cell line (data not shown). Therefore, with the exception of a moderate growth deficit in macrophages, the mutant and the WT-MCMV showed similar growth kinetics in different cell lines. We concluded from these experiments that the lack of 32 genes and the presence of the BAC cassette had little, if any, effect on virus replication in vitro.
FIG. 2.
Replication of Δm01-17+m144-158-MCMV and WT-MCMV in vitro. NIH 3T3 fibroblasts (a), Ana-1 macrophages (b), SVEC4-10 endothelial cells (c), and C127 mammary epithelial cells (d) were infected in triplicate with Δm01-17+m144-158-MCMV or WT-MCMV at an MOI of 0.1. Virus in the supernatant was harvested every 24 h, from day 0 (input virus) to day 4 or 5 p.i., and infectious virus titers were determined by plaque assay with MEF. Values shown are means ± standard deviations.
Δm01-17+m144-158-MCMV is severely attenuated in vivo.
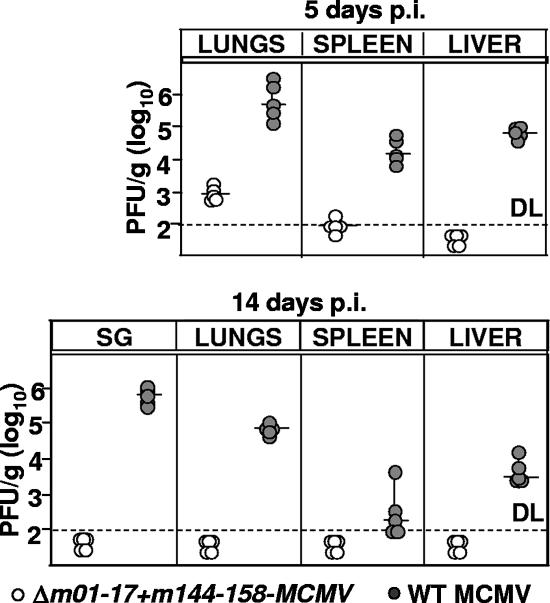
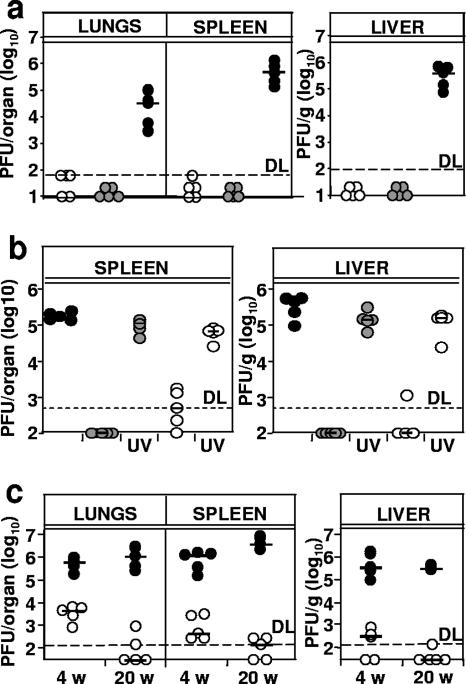
BALB/c mice, a mouse strain susceptible to MCMV (41), was used to test the virulence of Δm01-17+m144-158-MCMV in vivo. Infectious WT and mutant titers in lungs, spleen, liver, and salivary glands at days 5 and 12 p.i. were compared. Mutant virus growth was severely attenuated in all organs at both time points (Fig. 3a and b), although some virus detected at day 5 p.i. in the lungs and the spleen of mice infected with Δm01-17+m144-158 indicated that the virus was still able to colonize the organs and to replicate in vivo. Moreover, infection with the mutant did not result in weight loss, a rough indicator of compromised health (data not shown).
FIG. 3.
Δm01-17+m144-158-MCMV is severely attenuated in BALB/c mice. Groups of five BALB/c mice were infected i.v. with 5 × 105 PFU of the indicated viruses, and virus titers in lungs, spleen, liver, and salivary glands were determined at days 5 and 14 p.i. by plaque assay with MEFs. Virus titers in the salivary gland (SG), spleen, and lungs are shown per organ, and virus titers in the liver are shown per gram. Each circle represents a mouse; horizontal lines show median values; DL, detection limit.
Compromised Δm01-17+m144-158-MCMV reactivation from latency.
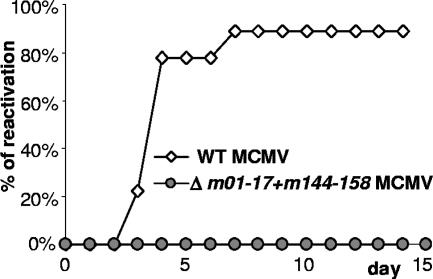
A hallmark of CMV infection is the ability to reactivate its productive viral infection from latency upon immune suppression. Therefore, there is concern that even an attenuated live CMV vaccine might reactivate and cause disease under immune suppressive conditions. We tested Δm01-17+m144-158-MCMV reactivation upon immune suppression. Nine months after mice received primary infection with mutant virus or WT-MCMV, they were immune suppressed by antibody depletion of T-cell subsets, followed by gamma irradiation. Eleven days later, spleen explants were cocultivated with MEFs to detect virus reactivation. Virus could be detected in mice infected with WT-MCMV but not in mice infected with the mutant virus (Fig. 4). In an independent experiment, no reactivation of Δm01-17+m144-158-MCMV was observed for the liver and spleen of mice that were immune suppressed by gamma irradiation at 28 days p.i. and sacrificed 7 days later, although WT-MCMV reactivated in all of the control mice (data not shown). Thus, replicating Δm01-17+m144-158-MCMV could not be isolated upon immune suppression.
FIG. 4.
No reactivation of Δm01-17+m144-158-MCMV. BALB/c mice were i.p. infected with 5 × 105 PFU of WT-MCMV (n = 9) or Δm01-17+m144-158-MCMV (n = 4). Nine months later, mice were immune suppressed by CD8 and CD4 depletion and whole-body gamma irradiation with a dose of 7 Gy. Eleven days after immune suppression, spleen explants were placed on MEFs. Cells were kept for 3 weeks, and virus reactivation in individual animals (y axis) was monitored daily by the occurrence of MCMV-specific cytopathic effects.
Δm01-17+m144-158-MCMV induces humoral and cellular immunity.
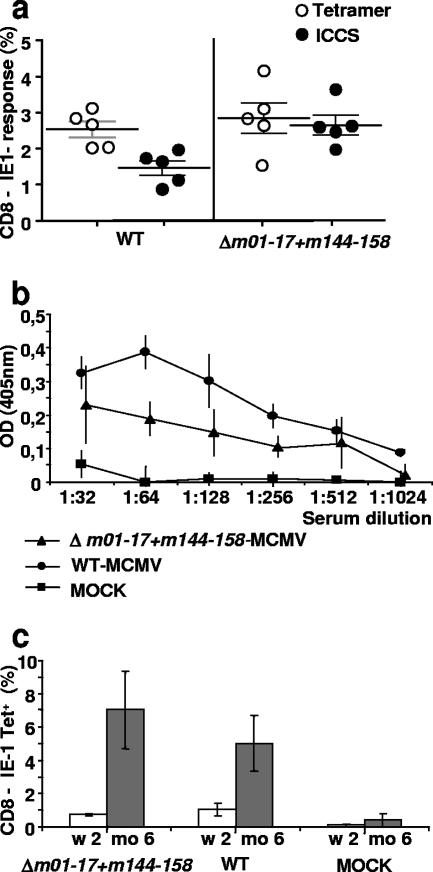
To test the cellular immune response to Δm01-17+m144-158-MCMV, blood cytotoxic T lymphocytes (CTL) with specificity for the immunodominant YPHFMPTNL peptide (encoded by the IE-1 gene [16]) were quantified by tetramer staining or by peptide stimulation, followed by intracellular cytokine staining (ICCS). Seven days after infection, the mutant and the WT induced similar CD8 responses, as observed by using tetramer staining (Fig. 5a). Remarkably, ICCS showed that the functional response to peptide stimulation was significantly (P = 0.007, Mann-Whitney U test) stronger in mice infected with the mutant than those infected with the WT virus (Fig. 5a). Taken together, these results indicated that Δm01-17+m144-158-MCMV infection induced cellular responses that were functionally even stronger than the ones induced by WT virus.
FIG. 5.
Immunity to MCMV elicited by Δm01-17+m144-158-MCMV. (a) Blood CD8 T cells from mice infected with 5 × 105 PFU of the indicated virus were analyzed for specificity of the immune response to the immunodominant YPHFMPTNL peptide. Cells from individual mice were split into two aliquots, which were subjected to tetramer staining (○) or to peptide stimulation, followed by ICCS for TNF-α and IFN-γ induction (•). Symbols represent percentage values of tetramer+ (○) or IFNg+ TNFa+ (•) cells in CD8+-gated lymphocytes of individual mice. Horizontal bars indicate average values, error bars indicate standard errors of the means. (b) Groups of mice (n = 4) were primed and 14 days later were boosted by s.c. injection of 106 PFU of the indicated virus or of sterile PBS (MOCK). At week 16 p.i., sera were collected, and titers of antibodies specific for MCMV were determined by ELISA. (c) Blood cells from mice immunized with 5 × 105 PFU of the indicated MCMV taken at 2 weeks (white bars) or 6 months (gray bars) p.i. were stained with antiCD8-FITC and YPHFMPTNL-MHC-IKd tetramers. CD8+ lymphocytes were gated, and the percentage of tetramer-positive cells was determined. Histograms indicate the means and standard deviations from at least four mice. OD, optical density.
To test the long-term immunogenicity of Δm01-17+m144-158-MCMV, mice were bled at 16 weeks after a prime/boost immunization, and antibody titers to MCMV were determined by ELISA. WT-MCMV-infected mice served as positive controls and mock-immunized mice as negative controls. Δm01-17+m144-158-MCMV infection induced antiviral antibodies (Fig. 5b), although optical density values and titers were not as high as those seen in the positive control. The cellular immune response to CMV infection is characterized by inflated kinetics, expanding over time, and is deemed to be a reflection of persistent stimulation by latently expressed antigen. To define whether Δm01-17+m144-158-MCMV follows this kinetic pattern, we compared the responses to the IE-1 peptide at an early and a late point after infection. Two weeks p.i., 1% of the total peripheral CD8+ cells were specific for this peptide in animals infected with Δm01-17+m144-158-MCMV or with WT-MCMV. At 6 months p.i., this percentage increased to ∼4 to 8% in both the mutant- and the WT-MCMV-infected mice (Fig. 5c). In mice subjected to a prime/boost protocol, we observed a similar proportion of tetramer-positive cells at 18 weeks postpriming (data not shown). Therefore, in contrast to the antibody response, Δm01-17+m144-158-MCMV induced a specific CTL response that was comparable to that induced by WT-MCMV with respect to kinetics and strength.
Δm01-17+m144-158-MCMV induces protective immunity.
To test whether the immune response correlated with protection, we challenged mice with a sublethal dose of WT-MCMV at 20 weeks after priming. At the expected peak of virus productivity (47), virus titers were determined in organs. The comparison of virus titers in immunized mice to those in mock-infected ones served to evaluate the protective immunity. Both the Δm01-17+m144-158-MCMV and the WT-MCMV immunization strongly suppressed MCMV replication on challenge, whereas mock-immunized controls showed high virus titers in all of the organs tested (Fig. 6a). Thus, immunization with Δm01-17+m144-158-MCMV and with WT-MCMV led to comparable degrees of protection for mice against a challenge with WT-MCMV.
FIG. 6.
Immune protection upon Δm01-17+m144-158-MCMV infection. (a) Mice that were s.c. primed and boosted with 106 PFU of Δm01-17+m144-158-MCMV (white circles) or WT-MCMV (gray circles) or mock immunized (black circles) were challenged at 20 weeks postpriming with 106 PFU of i.p.-delivered WT-MCMV. Five days later, infectious virus titers in lungs, spleen, and livers were determined by plaque assay on MEFs. (b) Mice were immunized with 105 PFU of Δm01-17+m144-158-MCMV (white circles) or WT-MCMV (gray circles) or were mock immunized with sterile PBS (black circles). Where indicated, an equal amount of UV-inactivated virus was used. Challenge with 106 PFU of WT-MCMV was performed i.p. at 4 weeks p.i., and virus was quantified 5 days later. (c) Mice were immunized with 105 PFU of Δm01-17+m144-158-MCMV (white circles) or PBS (black circles). At 4 and 20 weeks p.i., mice were challenged, and virus was titrated in the indicated organs, as described for panel a. Short horizontal bars indicate medians. Circles represent individual mice, and horizontal bars mark the median values. DL, detection limit.
The poor replication of Δm01-17+m144-158-MCMV in vivo (Fig. 3) raised the question of whether the induction of protective immunity required virus replication at all. Therefore, UV-inactivated virus as a source of antigen was included. Mice were immunized with one single 105-PFU dose of Δm01-17+m144-158-MCMV or with an equal amount of UV-inactivated virus. Control groups were mock immunized or immunized with WT-MCMV. To monitor for immune protection, mice were challenged with WT-MCMV, and viral growth was assessed. As expected, high viral titers were observed for the mock-treated group but not for the mice immunized with replication-competent WT-MCMV or Δm01-17+m144-158-MCMV (Fig. 6b). Mice that received UV-inactivated virus showed high virus titers, equal to those of mock-immunized controls. Thus, viral gene expression was required for protective immunity.
The comparison of virus growth shown in Fig. 6a and b indicated that the immunization with a single 105-PFU dose of Δm01-17+m144-158-MCMV provided less protection against challenge at 4 weeks p.i. than a prime/boost immunization with 106 PFU at 20 weeks p.i. It was not clear whether this difference reflected the long-term increase of antigen-specific CTL, as shown in Fig. 5c, or simply the difference in the vaccine dose. To investigate whether the immune protection increases over time, we immunized mice with a single dose of 105 PFU of Δm01-17+m144-158-MCMV and challenged them at 4 or at 20 weeks p.i. with WT-MCMV. The challenge resulted in significantly lower MCMV titers in immunized mice than in mock-immunized controls, but this difference was more pronounced at week 20 than at week 4 (Fig. 6c). Therefore, immunization with Δm01-17+m144-158-MCMV resulted in a CD8+ T-cell response and protection against a WT challenge that increased over time.
Δm01-17+m144-158-MCMV is attenuated in immunodeficient mice.
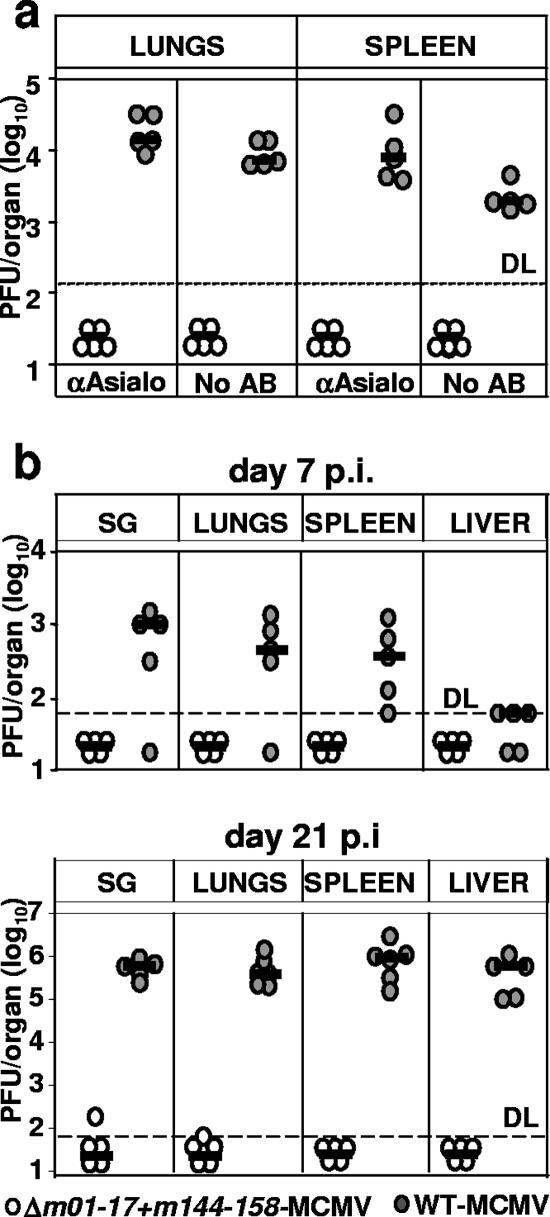
Although we observed no Δm01-17+m144-158-MCMV reactivation from latency upon severe immune suppression (Fig. 4), the sustained increase in antigen-specific immunity after the resolution of productive virus infection (Fig. 3 and 5c) indicated that the recombinant may persist in the host and express antigens. Therefore, it was possible that the virus may reactivate upon alternative immune suppressive protocols and become harmful to the host. To address this possibility, we injected infectious Δm01-17+m144-158-MCMV in constitutively immunodeficient mice and assessed its fitness and virulence. SCID mice, which lack T and B cells but still contain NK cells, were injected i.p. with 104 PFU of mutant MCMV or with WT-MCMV. NK cells were depleted in half of the animals from each group. All mice infected with WT-MCMV had detectable virus titers, whereas infectious Δm01-17+m144-158-MCMV could not be isolated from spleen, lungs (Fig. 7a), liver, and salivary glands (not shown) of any of the mice at day 12 p.i. NK-cell depletion increased the WT-MCMV titers but did not help to detect the mutant virus. To confirm and extend these results, we infected SCID/bg mice, a mouse strain which constitutively lacks B, T, and NK cells. Mice received s.c. injections of 104 PFU of mutant or WT-MCMV, and virus titers were determined at days 7 and 21 p.i. WT virus was found in all organs and found to increase over time (Fig. 7b). Mutant virus titers were below the detection limit in most of the mice, but infectious virus isolated in rare occasions at day 21 p.i. indicated that the infection with Δm01-17+m144-158-MCMV was not abortive. In conclusion, our data argued that even if Δm01-17+m144-158-MCMV could reactivate upon severe immune suppression, it would still be attenuated in immune compromised hosts.
FIG. 7.
Δm01-17+m144-158-MCMV is severely attenuated in immunodeficient mice. (a) Groups of SCID mice were infected i.p. with 104 PFU of the indicated virus. Where indicated, NK cells were depleted with α-asialo antibody. Virus titers in lungs and spleen at day 14 p.i. are shown. Each circle represents the value of an individual mouse; short horizontal bars indicate median values. DL, detection limit. (b) Groups of SCID/bg mice were s.c. infected with 104 PFU of the indicated virus. Virus titers in lungs, spleen, livers, and salivary glands (SG) at 7 and 21 days p.i. are shown. Virus titers in the spleen and lungs are shown per organ, and virus titers in the liver are shown per gram.
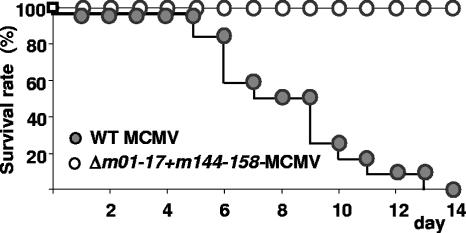
The high degree of attenuation suggested that Δm01-17+m144-158-MCMV should not harm even the most severely immunocompromised host. To test this, we infected IFN-αβR−/− mice with an equivalent of a lethal dose. These mice are 1,000-fold more susceptible to MCMV infection than the parental mouse strain (62). Mice were infected with an i.v. injection of 5 × 104 PFU of mutant or WT-MCMV, a dose corresponding to 10 times the 50% lethal dose for IFN-αβR−/− mice. All mice infected with WT-MCMV succumbed by day 7 p.i. (Fig. 8), whereas all mutant-infected mice survived the infection during the subsequent test period of 3 months p.i. Therefore, we concluded that Δm01-17+m144-158-MCMV presents a low risk, even in immunodeficient animals.
FIG. 8.
Δm01-17+m144-158-MCMV is avirulent in immunodeficient mice. Groups (n = 10) of IFN-αβR−/− mice were infected i.v. with 5 × 104 PFU of Δm01-17+m144-158-MCMV (white circles) or WT-MCMV (gray circles), and the survival rate was monitored daily.
DISCUSSION
The introduction of BAC-based technologies into herpesvirus genetics (49) has allowed reliable site-directed mutagenesis of the large herpesvirus DNA genomes (reviewed in reference 72). Several attenuated MCMV mutants with single or multiple site-directed gene deletions have been generated (see references 48 and 70). Here, we show for the first time that the MCMV genome tolerates the deletion of 32 genes, equal to 31.3 kb of genomic sequence.
The mutant Δm01-17+m144-158-MCMV is able to replicate, and restriction fragment analysis did not reveal any genome instability upon multiple passages in cell culture. Short HCMV-based amplicons of 15 kb in length are disposed to package concatemers to the preferred size of 210 kb into HCMV capsids (6), which matched the size of the Δm01-17+m144-158-MCMV genome with the retained BAC. Such a genome size preference suggested replication problems for viral mutants with undersized genomes. In this context, it should be noted that the deletion of 101 kb from the genome of Epstein-Barr virus (EBV), a gammaherpesvirus, results in viral mutants which can be packaged into infectious virions in the presence of a helper EBV but are unable to lytically replicate (36). The fact that the BAC cassette was maintained in the genomes was suggestive for a genome size preference, but further studies are required to explore the minimal size constraint of the CMV genome. The apparent size preference opens the potential to exchange large fragments of the genome and provides perspectives for the use of CMV-based gene vectors.
Only a minority of CMV genes are essential (17), and a previous report showed that a mutant MCMV lacking a block of 15 genes in the genomic area ranging from m02 to m16 was not attenuated in fibroblasts but was severely attenuated in immune-competent mice (55). Here, we extend this finding by showing that the loss of 32 genes, including the genes m02 to m16, did not affect the replication of the mutant MCMV in fibroblasts and epithelial and endothelial cell lines. We observed a moderate but consistent attenuation in cell culture at a low MOI in macrophage cell lines. We explain the growth deficit in macrophage lines as a cell type-specific function of one of the 32 genes, which have not yet been studied in detail.
Several mutants lacking individual immune-modulating MCMV genes have been described previously. MCMV mutants that lack genes which inhibit MHC-I-mediated peptide presentation (70) or that lack genes which evade the antiviral activity of NK cells (41) or lack the genomic regions comprising several genes with homologies to MHC-I genes (55) are all attenuated in the immunocompetent host. Nevertheless, all these mutants can be rescued to the WT level by immune suppression of the host. Therefore, these mutants are not attenuated in the immunodeficient host. We present here for the first time an MCMV mutant with a new quality. It grows like WT-MCMV in vitro but is severely attenuated even in the SCID/bg mouse, which lacks NK, B, and T cells and which is, to date, the mouse strain most sensitive to MCMV (38). Accordingly, we also observed a loss of virulence of Δm01-17+m144-158-MCMV in mice deficient for the type I IFN receptor. Finally, CMV disease could not be provoked by the most severe immune suppression (33). These data indicated that Δm01-17+m144-158-MCMV attenuation cannot be attributed exclusively to the loss of known immune-evasive genes suppressing the antiviral function of CD8 and NK cells. In line with this, at least one of the genes missing from Δm01-17+m144-158-MCMV affects the function of dendritic cells (46). Therefore, the attenuation in immunodeficient mice was likely caused by the loss of other viral functions, i.e., viral dissemination, the modest loss of tropism for macrophages (Fig. 2d), or the suppression of innate immune functions. This is an important consideration in the design of safe CMV vaccines, because CMV vaccine strains rendered apathogenic by the exclusive deletion of genes affecting antigen presentation may still become harmful in the event of an episode of immune suppression. Altogether, our data show for the first time that a CMV mutant can be engineered to be fully replication competent in vitro and, nevertheless, to represent a safe CMV vaccine or CMV-based vaccine vector in vivo.
Immunization with Δm01-17+m144-158-MCMV prevented virulent MCMV replication and induced a robust CTL response against the IE-1-derived epitope YPHFMPTNL. Remarkably, a comparison of responding cells identified by tetramer staining or by peptide stimulation revealed that many of the peptide-specific CD8+ T cells in WT (but not in mutant) infection do not respond to peptide stimulation. Therefore, our data suggested that WT-MCMV may affect the functionality of the cellular immune response, while CTL clones induced by Δm01-17+m144-158-MCMV perform better in terms of cytokine induction. CTL clones directed against the IE-1-derived epitope YPHFMPTNL (16) have been shown to mediate a strong but not sterilizing immunity (31, 54). Similarly, CTL specific for the HCMV IE-1 protein define immune protection against HCMV in patients who have undergone transplantation (10). A comparison of the numbers of peptide-specific CD8 T cells at 2 weeks and at 6 months p.i. with Δm01-17+m144-158-MCMV showed an increase comparable to that observed after WT-MCMV infection (Fig. 5c and references 28 and 34). The antibody titer induced by the recombinant MCMV was lower than that induced by WT-MCMV, which is most likely a consequence of the fast clearance of the mutant virus (Fig. 3) and the low antigenic load during lytic infection, although it may also have been caused by the loss of hitherto unknown CD4 and/or antibody epitopes. Although it is unclear to what extent the protection against challenge with WT-MCMV was mediated by T cells, the degree of protection also appeared to increase over time.
Previous HCMV vaccine concepts were based on virus attenuation in cell culture (19, 58, 59). However, these viruses acquired mutations at random (11), and their attenuation was associated with a 20-fold loss of immunogenicity and the inability to confer adequate immune protection (2). The HCMV strains used as vaccines retained the majority of known immune-evasive genes (44), which may have interfered with the immune recognition of the vaccine. Here, we provided proof for the principle that targeting immune-evasive genes for deletion results in a prototypical vaccine that induces even stronger cytokine responses in CD8 cells than WT-MCMV, a strong loss of pathogenicity, and vigorous immune protection that increases over time. Moreover, the Towne and AD-169 vaccine strains showed a loss of tropism for important cell types, i.e., endothelial or hemopoietic cells (57, 68). In contrast Δm01-17+m144-158-MCMV was still able to infect all cell types tested. The sites of CMV latency are still a matter of debate, but several reports indicate endothelial cells and hemopoietic precursor cells as sites of CMV latency (26, 39). An attenuated virus which is able to infect these cells might still be able to establish latency, and abortive reactivation events in these cells might suffice to maintain a virus-specific CTL response needed for sustained protective immunity (25). In line with this hypothesis the UV-inactivated virus was not able to induce a protective immune response.
How long can an attenuated mutant like Δm01-17+m144-158-MCMV persist after the resolution of lytic infection? Although we could not reactivate the virus and did not get definitive data in respect to genome persistence, we assume that the virus genome persists at a very low level, sufficient to undergo abortive reactivation events and express immunodominant antigens (25).
BAC cloning has made herpesvirus genomes accessible to rational mutant design. It is possible to delete any combination of genes in a herpesvirus genome. Thus, the key issue in herpesvirus genetics has shifted from the construction of mutants to the selection of genes to be targeted for deletion. We had constructed additional mutants which lack other gene blocks, and those that showed growth deficits in vitro were not considered further (data not shown). Mutants that grow poorly in cell culture are not suitable for vaccine preparation. Testing of the natural host showed that severe CMV attenuation is not necessarily linked with a loss of immunogenicity. Thus, our approach showed the feasibility of generating CMV vaccine strains with the desired properties by reverse genetics. The mapping of essential genes in the HCMV genome (17) and the mapping of genes governing cell tropism (8, 27, 48, 73), as well as those mediating immune evasion, facilitate the decision about which genes or gene blocks to keep and which to remove from the genome of an HCMV vaccine candidate. Therefore, the data presented here may represent the first and decisive steps in the generation of live attenuated CMV vaccines or vaccine vectors.
Acknowledgments
L.C.S. was supported by a DFG fellowship, CI 129/1-1. U.H.K. and Z.R. were supported by the DFG SFB 455. I.B and S.J. were supported by the Croatian Ministry of Science, grant 0062004.
We thank M. Wagner for help in the generation of the recombinant; S. Seelmeir, S. Boos, N. Röder, and D. Franzen for expert technical assistance; and T. Sacher (LMU) and J. Nikolich Zugich (OHSU) for helpful discussions.
The authors have no conflicts of interest.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Adler, S. P., S. H. Hempfling, S. E. Starr, S. A. Plotkin, and S. Riddell. 1998. Safety and immunogenicity of the Towne strain cytomegalovirus vaccine. Pediatr. Infect. Dis. J. 17:200-206. [DOI] [PubMed] [Google Scholar]
- 2.Adler, S. P., S. E. Starr, S. A. Plotkin, S. H. Hempfling, J. Buis, M. L. Manning, and A. M. Best. 1995. Immunity induced by primary human cytomegalovirus infection protects against secondary infection among women of childbearing age. J. Infect. Dis. 171:26-32. [DOI] [PubMed] [Google Scholar]
- 3.Arase, H., E. S. Mocarski, A. E. Campbell, A. B. Hill, and L. L. Lanier. 2002. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 296:1323-1326. [DOI] [PubMed] [Google Scholar]
- 4.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 6.Borst, E. M., and M. Messerle. 2003. Construction of a cytomegalovirus-based amplicon: a vector with a unique transfer capacity. Hum. Gene Ther. 14:959-970. [DOI] [PubMed] [Google Scholar]
- 7.Borst, E. M., and M. Messerle. 2005. Analysis of human cytomegalovirus oriLyt sequence requirements in the context of the viral genome. J. Virol. 79:3615-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 9.Bubić, I., M. Wagner, A. Krmpoti, T. Saulig, S. Kim, W. M. Yokoyama, S. Jonjić, and U. H. Koszinowski. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J. Virol. 78:7536-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunde, T., A. Kirchner, B. Hoffmeister, D. Habedank, R. Hetzer, G. Cherepnev, S. Proesch, P. Reinke, H. D. Volk, H. Lehmkuhl, and F. Kern. 2005. Protection from cytomegalovirus after transplantation is correlated with immediate early 1-specific CD8 T cells. J. Exp. Med. 201:1031-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 13.Cobbold, S. P., A. Jayasuriya, A. Nash, T. D. Prospero, and H. Waldmann. 1984. Therapy with monoclonal antibodies by elimination of T-cell subsets in vivo. Nature 312:548-551. [DOI] [PubMed] [Google Scholar]
- 14.Cox, G. W., B. J. Mathieson, L. Gandino, E. Blasi, D. Radzioch, and L. Varesio. 1989. Heterogeneity of hematopoietic cells immortalized by v-myc/v-raf recombinant retrovirus infection of bone marrow or fetal liver. J. Natl. Cancer Inst. 81:1492-1496. [DOI] [PubMed] [Google Scholar]
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Val, M., H. J. Schlicht, H. Volkmer, M. Messerle, M. J. Reddehase, and U. H. Koszinowski. 1991. Protection against lethal cytomegalovirus infection by a recombinant vaccine containing a single nonameric T-cell epitope. J. Virol. 65:3641-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Èièin-Šain, L., W. Brune, I. Bubić, S. Jonjić, and U. H. Koszinowski. 2003. Vaccination of mice with bacteria carrying a cloned herpesvirus genome reconstituted in vivo. J. Virol. 77:8249-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elek, S. D., and H. Stern. 1974. Development of a vaccine against mental retardation caused by cytomegalovirus infection in utero. Lancet i:1-5. [DOI] [PubMed] [Google Scholar]
- 20.Farrell, H. E., and G. R. Shellam. 1991. Protection against murine cytomegalovirus infection by passive transfer of neutralizing and non-neutralizing monoclonal antibodies. J Gen. Virol. 72:149-156. [DOI] [PubMed] [Google Scholar]
- 21.Farrell, H. E., H. Vally, D. M. Lynch, P. Fleming, G. R. Shellam, A. A. Scalzo, and N. J. Davis-Poynter. 1997. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature 386:510-514. [DOI] [PubMed] [Google Scholar]
- 22.Fowler, K. B., S. Stagno, and R. F. Pass. 2003. Maternal immunity and prevention of congenital cytomegalovirus infection. JAMA 289:1008-1011. [DOI] [PubMed] [Google Scholar]
- 23.Gill, T. A., P. J. Morley, and C. Sweet. 2000. Replication-defective mutants of mouse cytomegalovirus protect against wild-type virus challenge. J. Med. Virol. 62:127-139. [DOI] [PubMed] [Google Scholar]
- 24.González Armas, J. C., C. S. Morello, L. D. Cranmer, and D. H. Spector. 1996. DNA immunization confers protection against murine cytomegalovirus infection. J. Virol. 70:7921-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grzimek, N. K., D. Dreis, S. Schmalz, and M. J. Reddehase. 2001. Random, asynchronous, and asymmetric transcriptional activity of enhancer-flanking major immediate-early genes ie1/3 and ie2 during murine cytomegalovirus latency in the lungs. J. Virol. 75:2692-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahn, G., M. G. Revello, M. Patrone, E. Percivalle, G. Campanini, A. Sarasini, M. Wagner, A. Gallina, G. Milanesi, U. Koszinowski, F. Baldanti, and G. Gerna. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023-10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holtappels, R., M.-F. Pahl-Seibert, D. Thomas, and M. J. Reddehase. 2000. Enrichment of immediate-early 1 (m 123/pp89) peptide-specific CD8 T cells in a pulmonary CD62Llo memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J. Virol. 74:11495-11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobson, M. A., E. Sinclair, B. Bredt, L. Agrillo, D. Black, C. L. Epling, A. Carvidi, T. Ho, R. Bains, and S. P. Adler. 2006. Antigen-specific T cell responses induced by Towne cytomegalovirus (CMV) vaccine in CMV-seronegative vaccine recipients. J. Clin. Virol. 35:332-337. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson, M. A., E. Sinclair, B. Bredt, L. Agrillo, D. Black, C. L. Epling, A. Carvidi, T. Ho, R. Bains, V. Girling, and S. P. Adler. 2006. Safety and immunogenicity of Towne cytomegalovirus vaccine with or without adjuvant recombinant interleukin-12. Vaccine 24:5311-5319. [DOI] [PubMed] [Google Scholar]
- 31.Jonjić, S., M. Del Val, G. M. Keil, M. J. Reddehase, and U. H. Koszinowski. 1988. A nonstructural viral protein expressed by a recombinant vaccinia virus protects against lethal cytomegalovirus infection. J. Virol. 62:1653-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jonjic, S., W. Mutter, F. Weiland, M. J. Reddehase, and U. H. Koszinowski. 1989. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J. Exp. Med. 169:1199-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jonjic, S., I. Pavic, B. Polic, I. Crnkovic, P. Lucin, and U. H. Koszinowski. 1994. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J. Exp. Med. 179:1713-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karrer, U., S. Sierro, M. Wagner, A. Oxenius, H. Hengel, U. H. Koszinowski, R. E. Phillips, and P. Klenerman. 2003. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol. 170:2022-2029. (Erratum, 171:3895.) [DOI] [PubMed] [Google Scholar]
- 35.Karrer, U., M. Wagner, S. Sierro, A. Oxenius, H. Hengel, T. Dumrese, S. Freigang, U. H. Koszinowski, R. E. Phillips, and P. Klenerman. 2004. Expansion of protective CD8+ T-cell responses driven by recombinant cytomegaloviruses. J. Virol. 78:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kempkes, B., D. Pich, R. Zeidler, B. Sugden, and W. Hammerschmidt. 1995. Immortalization of human B lymphocytes by a plasmid containing 71 kilobase pairs of Epstein-Barr virus DNA. J. Virol. 69:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleijnen, M. F., J. B. Huppa, P. Lucin, S. Mukherjee, H. Farrell, A. E. Campbell, U. H. Koszinowski, A. B. Hill, and H. L. Ploegh. 1997. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 16:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi, H., M. Kobayashi, R. L. McCauley, D. N. Herndon, R. B. Pollard, and F. Suzuki. 1999. Cadaveric skin allograft-associated cytomegalovirus transmission in a mouse model of thermal injury. Clin. Immunol. 92:181-187. [DOI] [PubMed] [Google Scholar]
- 39.Koffron, A. J., M. Hummel, B. K. Patterson, S. Yan, D. B. Kaufman, J. P. Fryer, F. P. Stuart, and M. I. Abecassis. 1998. Cellular localization of latent murine cytomegalovirus. J. Virol. 72:95-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krmpotic, A., I. Bubic, B. Polic, P. Lucin, and S. Jonjic. 2003. Pathogenesis of murine cytomegalovirus infection. Microbes Infect. 5:1263-1277. [DOI] [PubMed] [Google Scholar]
- 41.Krmpotic, A., D. H. Busch, I. Bubic, F. Gebhardt, H. Hengel, M. Hasan, A. A. Scalzo, U. H. Koszinowski, and S. Jonjic. 2002. MCMV glycoprotein gp40 confers virus resistance to CD8+ T cells and NK cells in vivo. Nat. Immunol. 3:529-535. [DOI] [PubMed] [Google Scholar]
- 42.Krmpotic, A., M. Hasan, A. Loewendorf, T. Saulig, A. Halenius, T. Lenac, B. Polic, I. Bubic, A. Kriegeskorte, E. Pernjak-Pugel, M. Messerle, H. Hengel, D. H. Busch, U. H. Koszinowski, and S. Jonjic. 2005. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J. Exp. Med. 201:211-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krmpotic, A., M. Messerle, I. Crnkovic-Mertens, B. Polic, S. Jonjic, and U. H. Koszinowski. 1999. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J. Exp. Med. 190:1285-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, A., H. Xu, and W. Yan. 2007. Modulation of HLA expression in human cytomegalovirus immune evasion. Cell. Mol. Immunol. 4:91-98. [PubMed] [Google Scholar]
- 45.Lodoen, M. B., G. Abenes, S. Umamoto, J. P. Houchins, F. Liu, and L. L. Lanier. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J. Exp. Med. 200:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loewendorf, A., C. Kruger, E. M. Borst, M. Wagner, U. Just, and M. Messerle. 2004. Identification of a mouse cytomegalovirus gene selectively targeting CD86 expression on antigen-presenting cells. J. Virol. 78:13062-13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDonald, M. R., X. Y. Li, R. M. Stenberg, A. E. Campbell, and H. W. Virgin. 1998. Mucosal and parenteral vaccination against acute and latent murine cytomegalovirus (MCMV) infection by using an attenuated MCMV mutant. J. Virol. 72:442-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ménard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mocarski, E. S., and C. T. Courcelle. 2000. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 51.Morello, C. S., L. D. Cranmer, and D. H. Spector. 1999. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83). J. Virol. 73:7678-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morello, C. S., L. D. Cranmer, and D. H. Spector. 2000. Suppression of murine cytomegalovirus (MCMV) replication with a DNA vaccine encoding MCMV M84 (a homolog of human cytomegalovirus pp65). J. Virol. 74:3696-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morello, C. S., M. Ye, and D. H. Spector. 2002. Development of a vaccine against murine cytomegalovirus (MCMV), consisting of plasmid DNA and formalin-inactivated MCMV, that provides long-term, complete protection against viral replication. J. Virol. 76:4822-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morley, P. J., P. F. Ertl, and C. Sweet. 2003. High-frequency interferon-gamma-secreting splenocytes specific for murine cytomegalovirus immediate-early-1 (IE-1) peptide 168YPHFMPTNL176 are insufficient to provide complete protection from viral challenge. J. Med. Virol. 69:240-250. [DOI] [PubMed] [Google Scholar]
- 55.Oliveira, S. A., S. H. Park, P. Lee, A. Bendelac, and T. E. Shenk. 2002. Murine cytomegalovirus m02 gene family protects against natural killer cell-mediated immune surveillance. J. Virol. 76:885-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pass, R. F. 2000. Cytomegalovirus, p. 2675-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 57.Plachter, B., C. Sinzger, and G. Jahn. 1996. Cell types involved in replication and distribution of human cytomegalovirus. Adv. Virus Res. 46:195-261. [DOI] [PubMed] [Google Scholar]
- 58.Plotkin, S. A. 1994. Cytomegalovirus vaccines, p. 803-807. In S. A. Plotkin and E. A. Mortimer (ed.), Vaccines. WB Saunders, Philadelphia, PA.
- 59.Plotkin, S. A., T. Furukawa, N. Zygraich, and C. Huygelen. 1975. Candidate cytomegalovirus strain for human vaccination. Infect. Immun. 12:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plotkin, S. A., S. E. Starr, H. M. Friedman, E. Gonczol, and R. E. Weibel. 1989. Protective effects of Towne cytomegalovirus vaccine against low-passage cytomegalovirus administered as a challenge. J. Infect. Dis. 159:860-865. [DOI] [PubMed] [Google Scholar]
- 61.Podlech, J., R. Holtappels, N. K. A. Grzimek, and M. J. Reddehase. 2002. Animal models: murine cytomegalovirus, p. 493-525. In S. H. E. Kaufmann and D. Kabelitz (ed.), Methods in microbiology. Academic Press, San Diego, CA.
- 62.Presti, R. M., J. L. Pollock, A. J. Dal Canto, A. K. O'Guin, and H. W. Virgin. 1998. Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J. Exp. Med. 188:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reddehase, M. J., W. Mutter, K. Münch, H. J. Bühring, and U. H. Koszinowski. 1987. CD8-positive T lymphocytes specific for murine cytomegalovirus immediate-early antigens mediate protective immunity. J. Virol. 61:3102-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddehase, M. J., F. Weiland, K. Münch, S. Jonjić, A. Luske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reusch, U., W. Muranyi, P. Lucin, H. G. Burgert, H. Hengel, and U. H. Koszinowski. 1999. A cytomegalovirus glycoprotein re-routes MHC class I complexes to lysosomes for degradation. EMBO J. 18:1081-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schleiss, M. 2005. Progress in cytomegalovirus vaccine development. Herpes 12:66-75. [PubMed] [Google Scholar]
- 68.Sinzger, C., K. Schmidt, J. Knapp, M. Kahl, R. Beck, J. Waldman, H. Hebart, H. Einsele, and G. Jahn. 1999. Modification of human cytomegalovirus tropism through propagation in vitro is associated with changes in the viral genome. J. Gen. Virol. 80:2867-2877. [DOI] [PubMed] [Google Scholar]
- 69.Tolpin, M. D., S. E. Starr, A. M. Arbeter, and S. A. Plotkin. 1980. Inactivated mouse cytomegalovirus vaccine: preparation, immunogenicity, and protective effect. J. Infect. Dis. 142:569-574. [DOI] [PubMed] [Google Scholar]
- 70.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wagner, M., S. Jonjić, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 73.Wang, D., and T. Shenk. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. USA 102:18153-18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ziegler, H., R. Thale, P. Lucin, W. Muranyi, T. Flohr, H. Hengel, H. Farrell, W. Rawlinson, and U. H. Koszinowski. 1997. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity 6:57-66. [DOI] [PubMed] [Google Scholar]