Abstract
Variants of herpes simplex virus type 2 (HSV-2) generated by virus passage in GMK-AH1 cells in the presence of the sulfated oligosaccharide PI-88 were analyzed. Many of these variants were substantially resistant to PI-88 in their initial infection of cells and/or their cell-to-cell spread. The major alteration detected in all variants resistant to PI-88 in the initial infection of cells was a frameshift mutation(s) in the glycoprotein G (gG) gene that resulted in the lack of protein expression. Molecular transfer of the altered gG gene into the wild-type background confirmed that the gG-deficient recombinants were resistant to PI-88. In addition to PI-88, all gG-deficient variants of HSV-2 were resistant to the sulfated polysaccharide heparin. The gG-deficient virions were capable of attaching to cells, and this activity was relatively resistant to PI-88. In addition to having a drug-resistant phenotype, the gG-deficient variants were inefficiently released from infected cells. Purified gG bound to heparin and showed the cell-binding activity which was inhibited by PI-88. Many PI-88 variants produced syncytia in cultured cells and contained alterations in gB, including the syncytium-inducing L792P amino acid substitution. Although this phenotype can enhance the lateral spread of HSV in cells, it conferred no virus resistance to PI-88. Some PI-88 variants also contained occasional alterations in gC, gD, gE, gK, and UL24. In conclusion, we found that glycoprotein gG, a mucin-like component of the HSV-2 envelope, was targeted by sulfated oligo- and polysaccharides. This is a novel finding that suggests the involvement of HSV-2 gG in interactions with sulfated polysaccharides, including cell surface glycosaminoglycans.
It is well-established that cell surface heparan sulfate (HS) chains provide the binding sites for the initial interactions with cells of many viruses, including herpes simplex virus type 1 (HSV-1) and HSV-2 (38). The two types of HSV differ in their interactions with HS with respect to both the viral glycoproteins and the HS motifs involved. In particular, glycoprotein C (gC) of HSV-1 was identified as a component of the viral envelope that interacts with HS/heparin chains, thus mediating the attachment of the virus to cells (15). Although gC of HSV-2 can bind to HS/heparin chains and was found to be responsible for several HSV type-specific differences, such as polycation (28) and the hypertonic medium (36) resistance of HSV-2 infection of cells, this protein did not mediate HSV-2 attachment to cells (11). Instead, gB, another HS-binding component of the HSV envelope, was identified as the major virus attachment protein (5). In addition to gB and gC, gD of HSV-1, but not its HSV-2 homolog, can bind to HS chains modified by several isoforms of 3-O-sulfotransferase (31), an interaction that triggers HSV-1 entry into cells. Thus, interaction of HSV with HS seems to be a complex process that involves several kinds of viral proteins promoting virus attachment to and entry into cells.
Compounds such as sulfated oligo- and polysaccharides that mimic HS chains have long been known as inhibitors of HSV infectivity (37). Sulfated polysaccharides were shown to inhibit HSV attachment to cells (37, 38), suggesting that these compounds act via competition with HS chains for binding to the virus attachment proteins. It is not known whether sulfated polysaccharides can interfere with some postattachment steps, such as the initiation of virus entry by gD or virus-cell fusion, in the HSV invasion of cells. Although HSV-1 gD can trigger virus entry by interaction with 3-O-sulfated HS (31), it can also use two alternative receptors, herpesvirus entry mediator (25) and nectin (10), to perform this task.
We recently observed that the sulfated oligosaccharide PI-88 exhibits potent anti-HSV activity, including efficient reduction of the cell-to-cell transmission of the virus (27). PI-88 is a mixture of extensively sulfated mannose-containing di- to hexasaccharides that is currently undergoing clinical trial as an anticancer drug. PI-88 also shows anti-dengue and -encephalitic flavivirus (18) and anti-malarial (1) activities. One approach to further explore the interaction of HSV with HS chains has been the analysis of HSV-1 mutants resistant to sulfated oligo- and polysaccharides, such as heparin (12, 29) and carrageenan (4). Furthermore, our recent analysis of HSV-1 variants resistant to the sulfated oligosaccharide PI-88 revealed that this compound specifically targets the mucin-like region at the amino-terminal portion of viral gC (7). Because no similar studies were performed with HSV-2, in the present work we analyzed PI-88-resistant variants of this virus. Our studies revealed that PI-88 targeted HSV-2 gG, a mucin-like component of the virus envelope, which so far has not been reported to interact with or modulate virus binding to sulfated oligo- and polysaccharides or glycosaminoglycans.
MATERIALS AND METHODS
Cells, viruses, and PI-88 oligosaccharide.
African green monkey kidney (GMK AH1) cells (14) were cultivated in Eagle's minimum essential medium (EMEM) supplemented with 2% fetal calf serum, 0.05% Primaton RL (Kraft Inc., Norwich, CT), 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Human skin keratinocyte (HaCaT) cells (3) were propagated in Dulbecco′s modified EMEM supplemented with 10% fetal calf serum, 1% l-glutamine, and antibiotics. The HSV-2 strain 333 was used (6). The PI-88 oligosaccharide was prepared as described previously (39).
Preparation of PI-88 escape variants of HSV-2.
The HSV-2 strain 333 (uncloned stock) was subjected to 10 passages in GMK AH1 cells in the presence of 100 μg/ml of PI-88. Three procedures differing in the extent of virus exposure to PI-88 were employed. (i) The virus was incubated with PI-88 for 15 min prior to and during a 2-h period of infection of GMK AH1 cells at 37°C. The cells were then washed with EMEM and incubated in EMEM supplemented with PI-88 until the development of the complete cytopathic effect (CPE) (AC variants). (ii) The PI-88 oligosaccharide was incubated with the virus for 15 min prior to and during a 2-h period of virus infection of the cells. Subsequently, the cells were washed and incubated in EMEM without PI-88 until the development of CPE (A variants). (iii) The PI-88 oligosaccharide was added to the cells after a 2-h period of virus attachment to cells in the absence of an inhibitor. The cells were incubated with the inhibitor until the development of CPE (C variants). Following each passage, the infected cells and culture medium were harvested and subjected to one cycle of freezing and thawing. After being centrifugated for 10 min at 1,000 × g, the supernatant medium was diluted in EMEM and used for the next passage. After passage 10, several variants, AC, A, and C, were subjected to three rounds of plaque purification in the absence of PI-88.
Viral plaque assays.
Plaque number and size reduction assays were carried out as described previously (8, 27). Briefly, in the plaque number reduction assay, approximately 200 PFU of the virus was incubated with PI-88 (0.16 to 100 μg/ml) for 15 min prior to and during a 2-h period of virus infection of GMK AH1 cells. The cells were then washed and overlaid with EMEM supplemented with 1% methylcellulose and antibiotics. Following incubation of the cells for 1 to 3 days at 37°C, the viral plaques were visualized by staining with a 1% solution of crystal violet. In the plaque size reduction assay, the cells were infected with approximately 200 PFU of the virus in the absence of PI-88 for 2 h at 37°C. The cells were then washed and overlaid with EMEM supplemented with 1% methylcellulose and PI-88 (100 μg/ml). Following incubation of the cells for 1 to 3 days at 37°C, the viral plaques were stained with crystal violet solution. Images of 20 viral plaques were captured, using a DC300 digital camera (Leica, Heerbrugg, Switzerland) attached to a Diavert microscope (Leitz-Wetzlar, Germany). The area of the viral plaques was measured using IM500 software (Leica, Cambridge, United Kingdom).
Virus purification and binding of purified virions to cells.
GMK AH1 cells were infected with HSV-2 for 1 h at 37°C. Subsequently, the cells were washed and incubated in EMEM supplemented with [methyl-3H]thymidine (25 μCi/ml) for 40 h at 37°C. The infectious culture medium and infected cells were harvested and centrifuged at 1,000 × g for 10 min. The sedimented cells were frozen and thawed in a −70°C ethanol and 37°C water bath, respectively, and centrifuged again at 1,000 × g for 10 min. The supernatant fluid and infectious culture medium were combined and used for purification of HSV-2 virions by centrifugation through the three-step discontinuous sucrose gradient as previously described (36). To remove sucrose, purified virions were either pelleted by centrifugation at 22,000 × g for 2 h or centrifuged over a microcentrifugal concentrator filter with a 1,000-kDa cutoff (PallGelman, Lund, Sweden). For the cell-binding assay, confluent monolayers of GMK AH1 cells, precooled for 30 min at 4°C, were washed with cold phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 1 mM CaCl2, and 0.5 mM MgCl2) and blocked with PBS containing 1% bovine serum albumin for 1 h at 4°C. Purified virions of different HSV-2 preparations adjusted to contain the same number of the major virus capsid protein (VP5) units (38) were incubated with PI-88 (100 μg/ml) for 15 min at 4°C prior to the addition of the mixture to and incubation with GMK AH1 cells under moderate agitation for 1 h at 4°C. The cells were then extensively washed with PBS and lysed in a 5% solution of sodium dodecyl sulfate in PBS. Finally, the lysates were transferred to scintillation vials for the quantification of radioactivity.
Purification of viral glycoproteins and assays of their binding to cells and heparin.
gB, gC, and mature gG of HSV-2 were purified from pelleted HSV-2 virions and infected GMK AH1 cells by affinity chromatography (36) with the use of monoclonal antibodies B11D8 (anti-gB), E5F7 (anti-gC), and O1C5 (anti-gG) coupled to CNBr-Sepharose beads. To minimize the amount of detergent in the purified proteins, the immunosorbent beads with the attached viral glycoproteins were washed with detergent-free washing buffer just prior to their elution from the column. The eluted material was centrifuged to near dryness over a Microsep Omega concentrator with a 10-kDa cutoff (Pall Life Sciences, Lund, Sweden) to exchange the elution buffer with PBS (36). For the cell-binding assay, confluent monolayers of 3-day-old GMK AH1 cells or 6-day-old HaCaT cells in 96-well cluster plates were washed with EMEM (GMK AH1 cells) or Dulbecco's modified EMEM (HaCaT cells) and then precooled for 30 min at room temperature and for 30 min at 4°C. The purified proteins (4 μg) were incubated with specific concentrations of PI-88 for 15 min at room temperature prior to being added to the cells. Following incubation of the glycoprotein-PI88 mixture with the cells for 1 h at 4°C under moderate agitation, the medium was aspirated and the cells were washed once with 200 μl of PBS. The cells were then fixed with 0.25% glutaraldehyde in PBS for 15 min and triple washed with PBS. The bound glycoproteins were detected by an enzyme-linked immunosorbent assay-based procedure with the use of monoclonal antibodies B11D8, C2H12 (anti-gC), and O1C5. For the heparin-binding assay, purified gG (20 μg) in 0.5 ml of 0.065 M NaCl in phosphate buffer (see PBS buffer above) was applied to a disposable column containing 0.6 ml of Sephadex G10-heparin beads or Sephadex G10 beads alone. The column was washed with 10 ml of the same buffer and then stepwise eluted with increasing concentrations of NaCl in phosphate buffer.
Marker transfer assay.
HSV-2 DNA was extracted (24) from strain 333 that had been subjected to three rounds of plaque purification. Specific fragments of altered viral DNA were amplified by PCR, using Pfu Turbo polymerase (Stratagene, La Jolla, CA) and the flanking primers 222 to 240 and 2140 to 2119 for gG and −77 to −57, 1624 to 1605, or 1470 to 1488 and 2871 to 2850 for gB. One-microgram quantities of purified viral DNA and PCR-amplified fragments of the altered gG or gB gene were mixed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and incubated for 20 min at room temperature. Subsequently, the mixture was transfected into nearly confluent monolayers of 1-day-old cultures of GMK AH1 cells. After being incubated for 4 to 5 h at 37°C, the cells were washed and further incubated with fresh EMEM supplemented with 2% fetal calf serum and antibiotics until the development of CPE. For identification of the recombinant viruses altered in gB, several plaque variants of the virus exhibiting extensive, moderate/weak, or no syncytium-forming activity were selected for plaque purification and sequencing of the gB gene. For identification of the recombinant viruses possessing a frameshift mutation(s) in the gG gene, the viral plaques were immunostained with the monoclonal antibody O1C5 and those negative for gG expression were selected for plaque purification and nucleotide sequencing.
Primers used for amplification and sequencing.
The preparation of HSV-2 DNA and nucleotide sequencing of the coding regions of the gB, gC, gD, gE, gG, gK, and UL24 genes were performed as previously described (20). The primers used in the amplification and sequencing reactions were designed based on the complete genome sequence of strain HG52 of HSV-2 (GenBank accession number, NC 001798), and their positions were numbered relative to the first nucleotide of the initiation codon of the particular gene. The positions of the sense primers for the HSV-2 gB gene were −77 to −57, 401 to 420, 759 to 778, 1139 to 1158, 1470 to 1488, 1724 to 1744, 2133 to 2151, and 2523 to 2543, and of the antisense primers, 448 to 427, 866 to 845, 1198 to 1178, 1624 to 1605, 2003 to 1983, 2311 to 2290, 2599 to 2578, and 2871 to 2850. The positions of the sense primers for the HSV-2 gC gene were −122 to −102, 59 to 76, 211 to 230, 600 to 621, 866 to 885, and 1180 to 1197, and of the antisense primers, 133 to 116, 361 to 342, 705 to 688, 1062 to 1043, 1382 to 1364, and 1561 to 1542. The positions of the sense primers for the HSV-2 gD gene were −88 to −68, 345 to 365, 590 to 610, and 925 to 942, and of the antisense primers, 455 to 437, 780 to 763, 969 to 951, and 1350 to 1332. The positions of the sense primers for the HSV-2 gE gene were −47 to −27, 275 to 296, 350 to 368, 636 to 650, 775 to 793, and 1132 to 1150, and of the antisense primers, 61 to 44, 474 to 456, 816 to 796, 902 to 884, 1314 to 1297, and 1682 to 1662. The positions of the sense primers for the HSV-2 gG gene were −57 to −39, 222 to 240, 515 to 534, 847 to 866, 1305 to 1322, and 1761 to 1780, and of the antisense primers, 306 to 287, 600 to 582, 880 to 899, 1196 to 1177, 1620 to 1599, and 2140 to 2119. The positions of the sense primers for the HSV-2 gK gene were −105 to −86, 237 to 254, and 570 to 587, and of the antisense primers, 394 to 374, 704 to 686, and 1055 to 1038. The positions of the sense primers for the HSV-2 UL24 gene were −78 to −61, 261 to 280, and 568 to 588, and of the antisense primers, 321 to 304, 663 to 646, and 1006 to 988. To facilitate the sequencing of the DNA, the coding regions of the gB and gG genes were amplified as two and three overlapping fragments, respectively. All sequences were related to the original gene sequence of the HSV-2 333 strain from which the viral variants were selected.
Nucleotide sequence accession numbers.
The GenBank accession numbers are EU018080 for AC1 gB, EU018113 for AC2 gB, EU018081 for AC3 gB, EU018114 for AC4 gB, EU018082 for AC6 gB, EU018083 for AC8 gB, EU018120 for AC9 gB, EU018115 for AC10 gB, EU018116 for A2 gB, EU018121 for A4 gB, EU018117 for A6 gB, EU018084 for A9 gB, EU018118 for C1 gB, EU018085 for C3 gB, EU018086 for C5 gB, EU018119 for C6 gB, EU018088 for AC1 gC, EU018089 for AC8 gC, EU018122 for A2 gC, EU018123 for A6 gC, EU018124 for AC1 gD, EU018092 for AC2 gD, EU018125 for AC8 gD, EU018093 for A6 gD, EU018095 for AC4 gE, EU018096 for AC6 gE, EU018097 for A9 gE, EU018126 for AC1 gG, EU018099 for AC2 gG, EU018100 for AC3 gG, EU018101 for AC4 gG, EU018102 for AC6 gG, EU018127 for AC8 gG, EU018128 for AC9 gG, EU018103 for A4 gG, EU018104 for A9 gG, EU018106 for AC6 gK, EU018107 for AC8 gK, EU018109 for AC1 UL24, EU018110 for AC8 UL24, EU018111 for A4 UL24, and EU018112 for C6 UL24. The nucleotide sequences of the remaining AC, A, and C variants were identical with those of parental strain 333 gB (EU018079), 333 gC (EU018087), 333 gD (EU018091), 333 gE (EU018094), 333 gG (EU018098), 333 gK (EU018105), or 333 UL24 (EU018108).
RESULTS
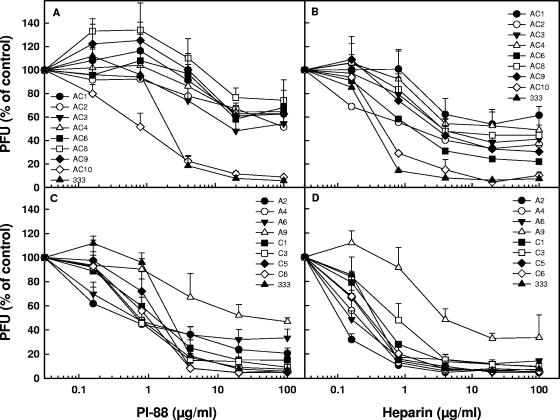
To expand our knowledge of initial events in the infection of cells by HSV-2, we sought to analyze variants of the HSV-2 333 strain resistant to the sulfated oligosaccharide PI-88, an inhibitor of HSV attachment to cells (27). To this end, the virus was subjected to 10 passages in GMK AH1 cells in the presence of PI-88. Because PI-88 can interfere with both the attachment of the virus to cells and the cell-to-cell spread of the virus (27), three different procedures were employed for selection of resistant variants. (i) PI-88 was present throughout virus propagation in the cells, i.e., during the initial virus attachment to and entry into the cells and the spread of progeny virus via released and cell-to-cell-transmitted virions (AC variants). (ii) HSV-2 was exposed to PI-88 only during attachment of the virus to and entry into the cells (A variants). (iii) Infection of the cells with HSV-2 took place in the absence of PI-88, followed by the addition of the drug and its coincubation with the cells during the spread of the progeny virus (C variants). For purposes of comparison, HSV-2 333 was also passaged 10 times in GMK AH1 cells in the absence of PI-88 (333-10p). Analysis of uncloned mixtures of the viral plaques revealed that while ∼2.7% of the plaques of strain 333 and ∼11.3% of the plaques of strain 333-10p exhibited syncytial or partly syncytial morphology (Table 1), >95% of the PI-88-selected variants AC, A, and C formed syncytial plaques. To verify the extent of the resistance of the PI-88 escape viruses, several variants from each group were plaque purified and then tested for sensitivity to PI-88 in plaque number and plaque size reduction assays (Table 1). The former assay assesses the sensitivity of the virus to PI-88 during its initial infection of cells, while the latter evaluates the effect of PI-88 on the cell-to-cell spread of the virus, measured as a reduction in plaque size (see Materials and Methods). All AC variants, except AC10, and variant A9 were significantly more resistant than strain 333 or 333-10p to the presence of PI-88 during the initial infection of the cells (Table 1). The remaining variants from group A and all the variants of group C were sensitive to the presence of PI-88 during the initial infection of the cells (Table 1). In contrast, the cell-to-cell-spread activity of all C and AC (except AC1 and AC4) variants, but not the A variants, was more resistant to PI-88 than the parent strain (Table 1). Note that the HSV-2 variants resistant to PI-88 in their initial infection of the cells were not necessarily those resistant to this compound in the cell-to-cell-spread activity (Table 1). This finding suggests that different viral components can be targeted by PI-88 during the initial infection of cells and the cell-to-cell spread of HSV-2.
TABLE 1.
Biological activities of PI-88-resistant variants of HSV-2 strain 333
| Virus varianta | Formed syncytia | Resistance to PI-88 as determined by indicated assay:
|
|
|---|---|---|---|
| Plaque no. reduction (IC50; μg/ml)b | Plaque size reductionc | ||
| 333 | −d | 2 | 16.3 (0.137) |
| 333-10pe | −f | 0.4 | 16.8 (0.179) |
| AC1 | + | >100 | 15.5 (1.218) |
| AC2 | + | >100 | 51.7 (2.753) |
| AC3 | + | >100 | 71.2 (0.589) |
| AC4 | + | >100 | 22.2 (0.644) |
| AC6 | + | >100 | 83.1 (1.296) |
| AC8 | + | >100 | 72.6 (0.314) |
| AC9 | +/− | >100 | 55.3 (0.527) |
| AC10 | + | 1.8 | 79.6 (0.397) |
| A2 | + | 0.5 | 2.7 (0.562) |
| A4 | − | 0.7 | 8.1 (0.307) |
| A6 | + | 0.7 | 1.8 (0.208) |
| A9 | + | 38 | 3.0 (0.749) |
| C1 | + | 1.2 | 60.7 (1.684) |
| C3 | + | 0.9 | 93.1 (4.067) |
| C5 | + | 1.5 | 49.9 (0.685) |
| C6 | + | 1.5 | 102.3 (1.868) |
| 333+AC2gBg | + | 0.3 | 22.8 (0.132) |
| 333+AC3gBg | +/− | 0.5 | 16.0 (0.168) |
| 333+AC6gGg | − | 35 | 14.9 (0.138) |
| 333+AC9gGg | − | 100 | 16.7 (0.129) |
Variants selected for by the presence of PI-88 were limited to either the initial infection of cells (A2 to A9), the spread of progeny virus (C1 to C6), or both (AC1 to AC10).
Serial PI-88 dilutions were incubated with ∼200 PFU of the virus for 10 min prior to and during a 2-h period of infection of GMK AH1 cells at 37°C. Results are expressed as the concentrations of PI-88 that reduced the number of viral plaques by 50% (IC50).
PI-88 (100 μg/ml) in an overlay methylcellulose medium was added to cells after they were infected with HSV-2 and was incubated with the cells throughout the development of viral plaques. Results are expressed as the percentages of the average areas of 20 viral plaques developed in the presence of PI-88 medium relative to that for mock-treated controls. The average areas (in mm2) of 20 viral plaques developed in the absence (control) of PI-88 are shown in parentheses.
Noncloned mixture of HSV-2 strain 333 plaques. Sporadic (2.7%) syncytial plaques were seen.
HSV-2 strain 333 that was passaged 10 times in GMK AH1 cells.
Noncloned mixture of HSV-2 strain 333-10p. Approximately 11.3% of plaques had the syncytial phenotype.
Recombinants prepared by molecular transfer of a specific gene fragment into strain 333.
To identify a specific alteration(s) responsible for the PI-88-resistant phenotype, the viral variants as well as the parent 333 strain were subjected to sequencing of their genes that code for gB, gC, gD, gE, gG, gK, and UL24 (Table 2). All PI-88-selected variants contained single or multiple mutations in gB. Eight different amino acid substitutions, i.e., N21K, N243T, A335V, Y353H, A524T, E662G, L792P, and R822Q, were found. The two most frequent amino acid substitutions (E662G and L792P) occurred simultaneously in 11 out of 16 variants examined. These included variants resistant to PI-88 in either the initial infection of cells (AC1, AC4, A9) or the cell-to-cell spread activity (AC10, C1, C3, C6) or both (AC2, AC6). However, these two alterations also occurred in variants sensitive to PI-88 in their initial infection and cell-to-cell spread (A2, A6), suggesting a lack of their contribution to the PI-88-resistant phenotype. To verify this assumption, a fragment of the gB gene of the AC2 variant which comprises these two mutations (Table 2) was transfected along with DNA of the 333 strain into GMK AH1 cells. The resulting recombinant (333+AC2gB) (Table 1) appeared to be sensitive to PI-88 both in its initial infection of cells and in its cell-to-cell-spread activity. This indicates that the E662G and L792P amino acid substitutions in gB conferred no resistance of HSV-2 to PI-88. Instead, the 333+AC2gB recombinant virus formed syncytia in cultured cells (Table 1). Because the E662G alteration occurs as the sole change in gB of a nonsyncytial variant, A4, the L792P amino acid substitution in the gB endodomain seems to be responsible for the syncytium-forming activity of AC2 and other variants containing this amino acid change. The remaining alterations in gB were either silent mutations or amino acid substitutions in the endodomain (R822Q) or ectodomain (N21K, N243T, A335W, Y353H, and A524T) of the protein. The recombinant virus containing the N243T amino acid substitution present in the gB ectodomain of the AC3 variant (333+AC3gB) was prepared by the marker transfer technique (Table 1). This virus produced little syncytia in cultured cells and appeared to be sensitive to PI-88 in its initial infection of cells and cell-to-cell-spread activity (Table 1). In conclusion, the most frequent alterations (E662G, L792P) and the N243T change found in the gB of PI-88-selected variants did not contribute to their drug-resistant phenotypes. In contrast to the numerous alterations detected in the gB gene, only sporadic mutations, which occurred in both PI-88-resistant and PI-88-sensitive variants (Table 2), were found in the gC, gD, gE, gK, and UL24 genes.
TABLE 2.
Nucleotide sequence analysis of the PI-88 escape variants of HSV-2 strain 333
| Virus variant | PI-88-resistant to indicated activity:
|
Formed syncytiaa | Alteration in indicated geneb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Initial infection | Cell-cell spread | gB | gC | gD | gE | gG | gK | UL24 | ||
| AC1 | + | − | + | c513t | g490c | a155g | None | ▴c1655 | None | c730t |
| Silent | A164P | Q27R | gG-neg. | P244S | ||||||
| a2051g | g957a | |||||||||
| E662G | Silent | |||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| AC2 | + | + | + | a2051g | None | c273c | None | ▴c1341 | ||
| E662G | Silent | ▴c1655 | ||||||||
| t2441c | gG-neg. | |||||||||
| L792P | None | None | ||||||||
| AC3 | + | + | + | a794c | None | None | None | ▾c624 | None | None |
| N243T | gG-neg. | |||||||||
| a2051g | ||||||||||
| E662G | ||||||||||
| AC4 | + | - | + | a2051g | None | None | g1473a | ▾g268 | None | None |
| E662G | Silent | ▴c1593 | ||||||||
| t2441c | gG-neg. | |||||||||
| L792P | ||||||||||
| AC6 | + | + | + | c798t | None | None | c648t | ▴c1857 | c181t | None |
| Silent | Silent | gG-neg. | Silent | |||||||
| a2051g | g840a | |||||||||
| E662G | Silent | |||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| AC8 | + | + | + | a2051g | c1383t | a155g | None | ▴c1655 | g235a | c173t |
| E662G | Silent | Q27R | gG-neg. | A79T | T58M | |||||
| g2531a | ||||||||||
| R822Q | ||||||||||
| AC9 | + | + | +/− | a2051g | None | None | None | ▴c1655 | None | None |
| E662G | gG-neg. | |||||||||
| AC10 | − | + | + | a2051g | None | None | None | None | None | None |
| E662G | ||||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| A2 | − | − | + | a2051g | t593c | None | None | None | None | None |
| E662G | L198P | |||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| A4 | − | − | − | a2051g | c63t | None | None | c1740t | None | c500t |
| E662G | Silent | Silent | T167M | |||||||
| A6 | − | − | + | a2051g | t593c | g534a | None | None | None | None |
| E662G | L198P | M148I | ||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| A9 | + | − | + | t1123c | None | None | g1255a | c555t | None | None |
| Y353H | G419R | Silent | ||||||||
| a2051g | ▴c783 | |||||||||
| E662G | gG-neg | |||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| C1 | − | + | + | a2051g | None | None | None | None | None | None |
| E662G | ||||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| C3 | − | + | + | c129a | None | None | None | None | None | None |
| N21K | ||||||||||
| g1636a | ||||||||||
| A524T | ||||||||||
| a2051g | ||||||||||
| E662G | ||||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
| C5 | − | + | + | c1070t | None | None | None | None | None | None |
| A335V | ||||||||||
| c1203t | ||||||||||
| Silent | ||||||||||
| C6 | − | + | + | a2051g | None | None | None | None | None | g88a |
| E662G | V30M | |||||||||
| t2441c | ||||||||||
| L792P | ||||||||||
+, positive for syncytium formation; −, negative for syncytium formation.
Nucleotide and predicted amino acid alterations are denoted with lowercase and uppercase letters, respectively. Nucleotide numbering begins from the first nucleotide of the initiation codon. Amino acid numbering in gC, gE, gG, gK, and UL24 begins from the initiation methionine, while that in gD and gB begins from the first amino acid of the mature protein. The symbols ▴ and ▾ denote deletion and insertion, respectively, of a single nucleotide. neg., negative.
Interestingly, all HSV-2 variants resistant to the presence of the drug during the initial infection of the cells (variants AC1 to AC9 and A9) (Table 1) carried one or two deletions and/or insertions of a single nucleotide in the gG gene (Table 2). These frameshift mutations resulted in the premature termination codon and thus in the absence of expression of mature gG at the surfaces of the infected cells. This was confirmed by the immunostaining of viral plaques with the monoclonal anti-gG antibody O1C5 (data not shown). Moreover, the presence of the premature stop codon may theoretically lead to the secretion into the culture medium of proteins truncated at amino acid residues 304 (A9), 306 (AC3 and AC4), 562 (AC2), or 641 (AC1, AC6, AC8, and AC9). To verify whether the frameshift mutations in gG can contribute to the PI-88-resistant phenotype of HSV-2, recombinant viruses comprising the altered gG genes of the AC6 and AC9 variants (333+AC6gG and 333+AC9gG) were prepared by the marker transfer technique (Table 1). In contrast to the parent 333 strain, both recombinant viruses were substantially resistant to PI-88 in their initial infections of the cells. These results indicate that the lack of gG expression provides the virus with a selective advantage to infect cells in the presence of PI-88.
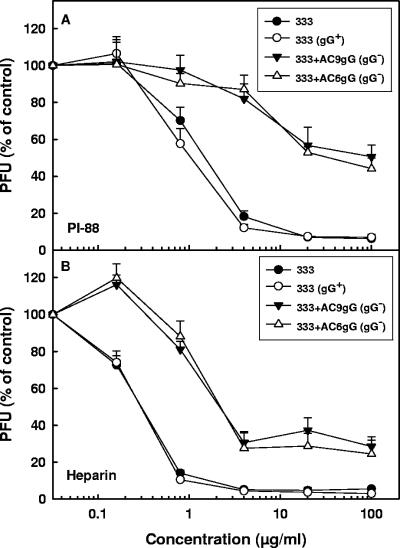
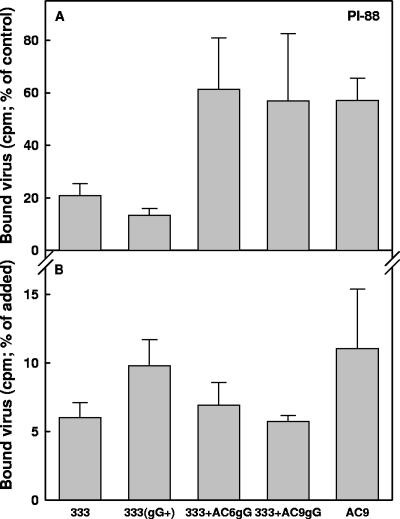
To investigate whether this finding could be extended to the sulfated polysaccharide heparin (a known inhibitor of HSV attachment to cells), all HSV-2 variants shown in Table 1 were compared for their sensitivity to PI-88 and heparin (Fig. 1). In comparison to heparin, PI-88 is composed of shorter but more extensively sulfated oligosaccharide chains. All AC variants resistant to PI-88 in their initial infections of the cells (AC1-9; gG deficient) (Fig. 1A and Table 1) were also resistant to heparin (Fig. 1B). In contrast, the PI-88-sensitive variant AC10 and the parental 333 strain (Fig. 1A and Table 1) were also sensitive to heparin (Fig. 1B). Likewise, of all the A and C variants, only A9 (gG deficient) was resistant to PI-88 (Fig. 1C and Table 1) and to heparin (Fig. 1D). We also tested the effects of PI-88 and heparin on the gG-deficient recombinant viruses (333+AC6gG and 333+AC9gG) and the gG-proficient 333 strain. Because noncloned preparations of HSV strains may contain very low levels of spontaneously generated variants deficient in expression of glycoproteins that are nonessential for replication in cultured cells (e.g., gC or gG), a plaque-purified 333 strain (333gG+) prepared based on its reactivity with the anti-gG O1C5 antibody was also included in this experiment. The gG-deficient recombinant viruses were substantially more resistant than the gG-proficient strains to both PI-88 (Fig. 2A) and heparin (Fig. 2B). These results indicate that HSV-2 gG is targeted by sulfated oligo- and polysaccharide inhibitors during the initial infection of cells by the virus.
FIG. 1.
Effect of PI-88 or heparin on infectivity of PI-88 escape variants of HSV-2. Approximately 100 to 200 PFU of a specific AC variant (A and B) or an A or C variant (C and D) were mixed with PI-88 (0.16 to 100 μg/ml) or heparin (0.16 to 100 μg/ml) and incubated for 15 min at room temperature prior to being added to and incubated with GMK AH1 cells for 2 h at 37°C. The cells were then washed and overlaid with methylcellulose solution without an inhibitor. The results are expressed as percentages of the number of viral plaques of the PI-88/heparin-treated virus relative to that of the mock-treated controls. The values shown are the means of duplicate determinations from two separate experiments.
FIG. 2.
Effect of PI-88 or heparin on infectivity of gG-deficient mutants of HSV-2. Approximately 200 PFU of gG-deficient recombinants 333+AC9gG and 333+AC6gG or gG-proficient strains 333 and 333(gG+) were mixed with 0.16 to 100 μg/ml of PI-88 (A) or heparin (B) and incubated for 15 min at room temperature prior to being added to and incubated with GMK AH1 cells for 2 h at 37°C. The cells were then washed and overlaid with methylcellulose solution without an inhibitor. The results are expressed as percentages of the number of viral plaques of the PI-88/heparin-treated virus relative to that of the mock-treated controls. The values shown are the means of duplicate determinations from two separate experiments.
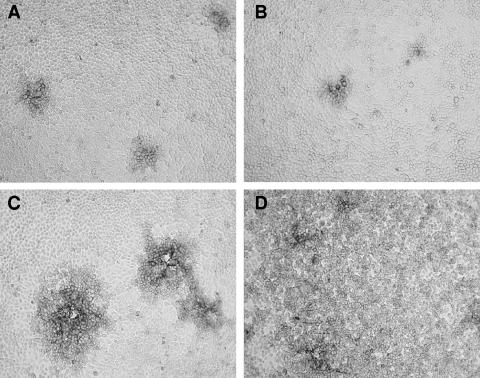
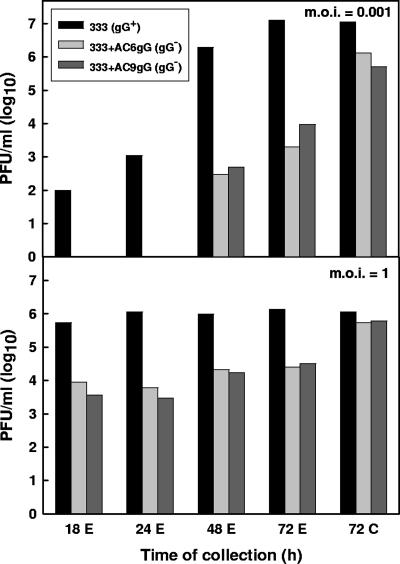
We sought to determine how the lack of gG expression could confer resistance of HSV-2 to PI-88. To this end, we investigated the biological activities of mutant gG-deficient viruses and purified gG components of HSV-2. During preparation of the HSV-2 stock viruses, we noticed that infections of cultured cells with gG-deficient variants or recombinant viruses progressed more slowly than infections with the gG-proficient strain 333. In particular, upon infection of GMK AH1 cells at a relatively low multiplicity of infection (MOI) and in the absence of methylcellulose or human gamma globulin in the overlay medium, several clusters of cells infected with gG-deficient or gG-proficient viruses were visible at 24 h after infection (Fig. 3A and B). However, at 48 h after infection, the gG-proficient 333 strain caused generalized infection of the cells (Fig. 3D), while infection with the gG-deficient virus progressed mainly via cell-to-cell spread, as deduced from the enlargement of the viral plaques (Fig. 3C). We assumed that the gG-deficient virions were either inefficiently released from the infected cells or were weakly infectious, due to, for example, a defect in their attachment to cells. To clarify this issue, the cells were infected with the gG-deficient recombinants 333+AC6gG and 333+AC9gG and with strain 333(gG+) at relatively low (0.001) and high (1.0) MOIs (Fig. 4). After infecting the cells at a low MOI, the gG-deficient viruses produced at least 100-times-less infectious virus in the extracellular medium than strain 333(gG+). However, at 72 h after infection, the amount of cell-associated gG-deficient virus was approximately 10 times lower than that of strain 333(gG+) (Fig. 4). After infection of the cells at a MOI of 1, extensive CPE was already observed at 18 h after infection with all the viruses tested. In spite of this, there were approximately 100-times-fewer infectious extracellular gG-deficient mutants than strain 333(gG+) viruses (Fig. 4). However, at 72 h after infection, the amount of cell-associated, gG-deficient virus was only approximately 2 to 3 times lower than that of strain 333 (Fig. 4). These results indicate that the gG-deficient viruses are released inefficiently from infected cells; however, when forcibly liberated from cells (by freezing and thawing), these viruses can infect cells efficiently.
FIG. 3.
Infection of GMK AH1 cells with the gG-deficient mutant of HSV-2. The cells were infected with the gG-deficient recombinant virus 333+AC6gG (A and C) or the gG-proficient 333(gG+) strain (B and D) at a multiplicity of infection of 0.001. The cells were washed and maintained in Eagle's medium without methylcellulose or human gamma globulin. The cells were immunostained with the monoclonal anti-HSV gB antibody B11D8 at 24 h (A and B) or 48 h (C and D) after infection.
FIG. 4.
Production of infectious virus in GMK AH1 cells infected with gG-deficient mutants of HSV-2. The cells were infected with the gG-deficient recombinant viruses 333+AC6gG and 333+AC9gG or the gG-proficient 333(gG+) strain at a MOI of 0.001 or 1. At specific times after infection, the production of extracellular (E) or cell-associated (C) virus was assessed by titration in GMK AH1 cells. Two separate experiments were carried out for each virus. The values shown were derived from the first experiment.
To identify the step in HSV-2 infection of cells that is affected by PI-88 and evaded by selection for the drug-resistant variants, purified radiolabeled virions of several gG-deficient viruses were tested for their binding to GMK AH1 cells in the presence of PI-88. For accurate comparison, prior to the experiments, all viruses specified in Fig. 5 were adjusted to contain the same number of relative VP5 units. Due to difficulties in preparing purified gG-deficient virions in sufficient quantities, their binding to cells was studied only in the presence of 100 μg/ml of PI-88. The cell-binding activities of gG-deficient variant AC9 and recombinants 333+AC6gG and 333+AC9gG were more resistant to PI-88 than similar activities of the gG-proficient 333 and 333(gG+) strains (Fig. 5A). These results clearly indicate that the lack of gG expression provides the virus with a selective advantage to attach to cells in the presence of PI-88. Furthermore, we compared the cell-binding activities of several gG-deficient and gG-proficient viruses in the absence of PI-88. The gG-deficient variant AC9 and recombinants 333+AC6gG and 333+AC9gG demonstrated no impairment in their attachment to GMK AH1 cells compared to that of the gG-proficient strains (Fig. 5B).
FIG. 5.
Effect of PI-88 on attachment of gG-negative mutants of HSV-2 to cells. (A) Purified radiolabeled virions of gG-deficient mutant viruses 333+AC6gG, 333+AC9gG, or AC9 and gG-proficient strains 333 and 333(gG+) were mixed with PI-88 (100 μg/ml) and incubated for 10 min at 4°C prior to being added to and incubated with GMK AH1 cells for 90 min at 4°C. The results are expressed as percentages of the counts per minute (cpm) of viruses attaching to cells in the presence of an inhibitor relative to that for the mock-treated controls. (B) Purified radiolabeled virions of gG-deficient and gG-proficient viruses were incubated with GMK AH1 cells for 90 min at 4°C. The results are expressed as percentages of the counts per minute of viruses binding to cells relative to that for the added virus. The values shown are the means of the results of two (333 and AC9) or five (333gG+, 333+AC6gG, and 333+AC9gG) separate experiments.
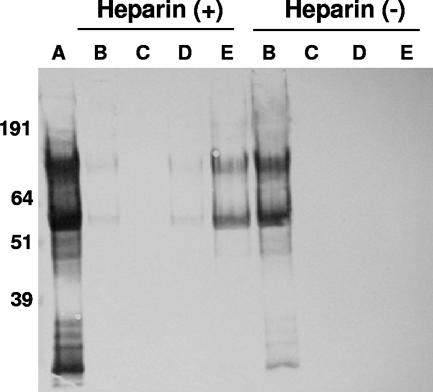
To determine whether gG of HSV-2 is capable of interacting with sulfated polysaccharides, purified protein suspended in a buffer containing 0.065 M NaCl was run through a column loaded with heparin covalently coupled to Sephadex G10 beads. Purified gG bound to heparin-Sephadex but not to uncoupled Sephadex beads (Fig. 6). Approximately 0.4 M NaCl was necessary to trigger elution of gG from the heparin column. The two major gG bands eluted from the heparin column represent the high-mannose intermediate (∼60 kDa) and the fully O-glycosylated mature (∼120 kDa) gG.
FIG. 6.
Binding of purified gG of HSV-2 to immobilized heparin. Purified gG was run through columns loaded with Sephadex G10-heparin or Sephadex G10 alone. The beads were eluted in a stepwise manner with increasing concentrations of NaCl. The input (A), flow-through (B), wash-out (not shown), and pooled fractions of protein eluted with 0.15 to 0.3 M (C), 0.4 to 0.6 M (D), or 0.7 to 2 M NaCl (E) were concentrated and electrophoresed under reducing conditions on a 10% acrylamide NuPAGE Bis-Tris precast gel. The electroblotted proteins were immunostained with the monoclonal anti-HSV-2 gG antibody O1C5. +, with heparin; −, without heparin.
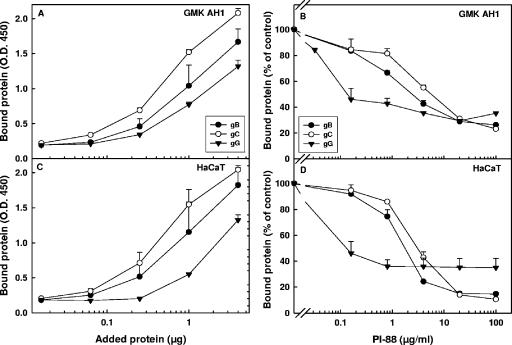
The gG of HSV-2 was also tested for its cell-binding activity. For purposes of comparison, purified gB and gC, the known cell-binding components of HSV-2 (34, 36), were also included. Purified gG bound to both GMK AH1 (Fig. 7A) and HaCaT (Fig. 7C) cells, although its cell-binding activity was somewhat less than that of purified gB and gC. PI-88 reduced the binding of gG to cells by approximately 60%; however, the concentration of PI-88 required to inhibit gG binding by 50% was approximately 30 times lower than those estimated for gB and gC (Fig. 7B and D). Altogether, our data obtained with purified gG indicate that this protein can bind to immobilized heparin and to surfaces of cultured cells and that PI-88 can inhibit the cell-binding activity of gG.
FIG. 7.
Binding of purified gG of HSV-2 to cells. Purified proteins at specific concentrations were incubated with GMK AH1 (A) or HaCaT (C) cells for 1 h at 4°C. Alternately, purified proteins (4 μg) were incubated with PI-88 for 15 min prior to being added to and during a 1-h period of protein adsorption to GMK AH1 (B) or HaCaT (D) cells at 4°C. The bound glycoproteins were detected by an enzyme-linked immunosorbent assay-based procedure. The amount of bound protein is expressed as an absorbance value (A and C) or as a percentage of cell-bound protein detected in the PI-88-treated sample relative to that in the mock-treated controls (B and D). The values shown are the means of duplicate determinations from two separate experiments. O.D.450, optical density at 450 nm.
DISCUSSION
Based on the analysis of HSV-2 variants resistant to PI-88, we provided genetic evidence that gG of the HSV-2 envelope is targeted by sulfated oligo- and polysaccharide inhibitors of attachment of the virus to cells. This is a novel biological function of HSV-2 gG, as this glycoprotein has not so far been reported to promote or modulate the initial interaction of HSV-2 with cells. Furthermore, our data demonstrate that glycoprotein gG of HSV-2, although nonessential for virus replication in cultured cells, appeared to be important for the egress of HSV-2 virions from cells.
gG of HSV-2 is known to elicit an HSV-2-specific antibody response and has therefore been used as an antigen for HSV type-discriminating serology (13, 19). The gG gene of HSV-2 codes for a 699-amino-acid-long precursor protein which is processed by proteolytic cleavage to generate a secreted amino-terminal fragment of gG (sgG) and a carboxy-terminal cell- and viral membrane-associated mature gG (mgG) (2, 23, 32, 33). The sgG is secreted into the extracellular medium (32) and can be purified from culture medium based on its affinity for heparin (21). The mgG is heavily O glycosylated, and the amino-terminal region occupied with clustered O-linked glycans constitutes more than half of the mgG amino acid sequence. Therefore, this region of mgG resembles a mucin-like structure. Interestingly, variants of HSV-1 resistant to PI-88 in their initial infection of cells all carried a large deletion of a mucin-like region at the amino-terminal part of the viral attachment protein gC (7). This region of HSV-1 gC was reported to promote attachment of the virus to cells (34). Because all HSV-2 variants resistant to PI-88 in their initial infection of cells were deficient in expression of a mucin-like protein mgG (this report), it is likely that sulfated oligosaccharide PI-88 specifically targeted the mucin-like structures on viral proteins. Recombinant gG-deficient HSV-2, prepared by molecular transfer of the altered gG gene into wild-type DNA, appeared to be resistant to PI-88, thus confirming that the absence of mgG expression provided HSV-2 virions with a selective advantage to infect the cells in the presence of this inhibitor. Furthermore, this observation was extended to the sulfated polysaccharide heparin, a known inhibitor of HSV attachment to cells. Our data showing that PI-88, an inhibitor of HSV attachment to cells (27), invariably selected for the mgG-deficient variants of HSV-2 suggest that mgG either is directly involved in the attachment of HSV-2 to cells or plays a role in the modulation of this activity. The first possibility is less probable, since purified virions of mutant gG-deficient HSV-2 demonstrated no clear impairment in their attachment to GMK AH1 cells.
Mucin-like regions are known to occur in different proteins, including those that function as adhesion components. Well-known examples of viral adhesion proteins that promote virus binding to cell surface HS and contain mucin-like structures are gC of HSV-1 (15) and gG of respiratory syncytial virus (RSV) (17). The mucin-like segment(s) may extend these proteins to be the outermost structure of the virus particle, a feature of importance for initial virus-cell contact. Furthermore, the HS-binding domains in these proteins comprise clusters of basic amino acid residues (9, 22, 30, 35) which can be unspecifically targeted by proteolytic enzymes and other proteins. The mucins located at the periphery of the protein ectodomain may selectively shield the HS-binding sites so that possible nonspecific interactions are limited but inherent biological activity is retained. Finally, the mucin-like structures may modulate the adhesion function of these proteins. It is not known whether this modulation includes the initial binding of mucin to the HS chain or preservation of the specificity of the protein-HS interaction. A wide variety of sulfated oligo- and polysaccharides, polysulfonated noncarbohydrate compounds, and other polyanions are known to inhibit infection of cells by HSV (37). Their antiviral effect is based on the reversible and nonvirucidal interaction with the HSV particle (26). One cannot exclude that relatively weak multiple interactions of these compounds with mucin components of viral proteins may explain their antiviral potency. It is noteworthy that the mgG of HSV-2 resembles attachment proteins of HSV-1 and RSV in that it contains the mucin-like region in addition to several basic amino acid residues sparsely distributed at the membrane proximal end of the mucin segment. These two domains of mgG may support its interaction with HS/heparin chains. Indeed, like HSV-1 gC (15) and RSV gG (17), purified mgG bound heparin. The presence of O-linked glycans may not be required for this activity, as both a high-mannose intermediate form of mgG, which contains no O-linked glycans, and a fully O-glycosylated mgG bound to heparin (Fig. 6). Because our preparation of mgG was derived from HSV-2 virions and the virus-infected cells, one cannot exclude the possibility that part of its binding activity was due to the presence of a noncleaved precursor protein composed of both mgG and sgG. Although sgG was not detected in preparations of mgG purified from infected cells based on its affinity to Helix pomatia lectin, this form of gG has an overall net positive charge and its heparin binding activity has been demonstrated (21). More importantly, we also observed that purified mgG bound to cultured cells, and the reduction of this activity by PI-88 suggested that mgG could interact with negatively charged molecules at the cell surface. PI-88 affected the cell-binding activity of purified mgG, gB, and gC in different ways. Although this activity of mgG was more sensitive to PI-88 than that of gB or gC, a fraction of purified mgG bound to HaCaT cells in a PI-88-resistant manner. Further studies are needed to determine whether this fraction is identical with the high-mannose intermediate or fully processed mgG. Finally, glycoprotein gG of HSV-2 seems to be a multifunctional component of the viral envelope which, in addition to serving as a target for sulfated oligo- and polysaccharides, promotes the egress of infectious virus particles from cells.
The presence of the mucin-like region in different glycoproteins of HSV-1 and HSV-2 is an example of the progressing structural and functional divergence of these viruses. HSV-2 gC, in contrast to its HSV-1 counterpart, does not contain a typical mucin-like domain and is not regarded as the virus attachment protein (11). Conversely, HSV-2 gG, unlike HSV-1 gG, possesses mucin-like structures and is targeted by inhibitors of virus attachment to cells.
The frameshift mutations in the gG gene, which as discussed above conferred virus resistance to PI-88 during the initial infection of cells, did not seem to contribute to the PI-88-resistant cell-to-cell spread of HSV-2, since this activity in some of the gG-deficient variants of the virus was sensitive to PI-88. These data suggest that different components of HSV-2 can be targeted by PI-88 during the initial infection of cells and the cell-to-cell spread of the virus. Although the same viral proteins that support HSV infection of the cell via the apical surface are thought to promote the cell-to-cell spread of the virus, the latter activity may not require gG, because the gG-deficient variants of HSV-2 exhibited efficient lateral spread in cultured cells (Fig. 3C). gE of HSV-1 is known to promote the cell-to-cell-spread activity by sorting nascent virions to cell junctions (16). However, the only amino acid substitution (G419R) detected in gE of PI-88-selected HSV-2 occurred in a variant that was sensitive to PI-88 in its cell-to-cell-spread activity. It is likely that specific mutations in several viral genes, including gB and/or gD, may in part contribute to the PI-88-resistant cell-to-cell-spread activity of HSV-2. However, knowing that all HSV-2 variants resistant to PI-88 in their initial infection of cells were altered in gG, one cannot exclude that an alteration(s) in the sole HSV-2 gene not sequenced in the present study may confer the drug-resistant phenotype in the cell-to-cell spread of HSV-2.
The majority of the PI-88-selected variants of HSV-2 exhibited syncytium-forming activity in cultured cells, and our results suggest that the L792P change was responsible for this phenotype, at least in several variants. Variants of HSV-1 resistant to heparin (12, 29) or carrageenan (4) produced syncytia in cultured cells due to mutations in gK (29) or the gB endodomain (12) or were predicted to contain an alteration(s) in gB (4). However, in line with our results obtained with PI-88-selected variants of HSV-2, the syncytium-inducing mutations in gB were not found to contribute to HSV-1 resistance to heparin (12) or carrageenan (4). The problem of frequent selection of syncytium-inducing mutants of HSV by sulfated polysaccharides and an unexpected lack of contribution of this phenotype to the drug resistance of the virus are difficult to explain. It is likely that the resistance of the virus to sulfated polysaccharides is coupled with its functional impairment and that the syncytium-inducing mutations may compensate for these alterations by enhancing lateral spread of HSV in cultured cells.
In conclusion, based on an analysis of PI-88-resistant variants of HSV-2, we report that glycoprotein mgG is the major target for sulfated oligo- and polysaccharide inhibitors of the attachment of HSV to cells. It is likely that this glycoprotein can facilitate the initial interaction of HSV-2 with cells and the egress of viral particles.
Acknowledgments
This work was supported by grants from the Mizutani Foundation for Glycoscience, the Swedish Research Council, the Sahlgren′s University Hospital Läkarutbildningsavtal, and the Scandinavian Society for Antimicrobial Chemotherapy.
Footnotes
Published ahead of print on 10 September 2007.
REFERENCES
- 1.Adams, Y., C. Freeman, R. Schwartz-Albiez, V. Ferro, C. R. Parish, and K. T. Andrews. 2006. Inhibition of Plasmodium falciparum growth in vitro and adhesion to chondroitin-4-sulfate by the heparan sulfate mimetic PI-88 and other sulfated oligosaccharides. Antimicrob. Agents Chemother. 50:2850-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balachandran, N., and L. M. Hutt-Fletcher. 1985. Synthesis and processing of glycoprotein gG of herpes simplex virus type 2. J. Virol. 54:825-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boukamp, P., R. T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N. E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlucci, M. J., L. A. Scolaro, and E. B. Damonte. 2002. Herpes simplex virus type 1 variants arising after selection with an antiviral carageenan: lack of correlation between drug susceptibility and syn phenotype. J. Med. Virol. 68:92-98. [DOI] [PubMed] [Google Scholar]
- 5.Cheshenko, N., and B. C. Herold. 2002. Glycoprotein B plays a predominant role in mediating herpes simplex virus type 2 attachment and is required for entry and cell-to-cell spread. J. Gen. Virol. 83:2247-2255. [DOI] [PubMed] [Google Scholar]
- 6.Duff, R., and F. Rapp. 1971. Oncogenic transformation of hamster cells after exposure to herpes simplex virus type 2. Nat. New Biol. 233:48-50. [DOI] [PubMed] [Google Scholar]
- 7.Ekblad, M., B. Adamiak, K. Bergefall, H. Nenonen, A. Roth, T. Bergström, V. Ferro, and E. Trybala. 2007. Molecular basis for resistance of herpes simplex virus type 1 mutants to the sulfated oligosaccharide inhibitor PI-88. Virology 367:244-252. [DOI] [PubMed] [Google Scholar]
- 8.Ekblad, M., T. Bergström, M. G. Banwell, M. Bonnet, J. Renner, V. Ferro, and E. Trybala. 2006. Anti-herpes simplex virus activities of two novel disulphated cyclitols. Antivir. Chem. Chemother. 17:97-106. [DOI] [PubMed] [Google Scholar]
- 9.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 11.Gerber, S. I., B. J. Belval, and B. C. Herold. 1995. Differences in the role of glycoprotein C of HSV-1 and HSV-2 in viral binding may contribute to serotype differences in cell tropism. Virology 214:29-39. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, J. L., and J. P. Engel. 1991. Altered pathogenesis in herpes simplex virus type 1 infection due to a syncytial mutation mapping to the carboxy terminus of glycoprotein B. J. Virol. 65:1770-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Görander, S., B. Svennerholm, and J.-Å. Liljeqvist. 2003. Secreted portion of glycoprotein G of herpes simplex virus type 2 is a novel antigen for type-discriminating serology. J. Clin. Microbiol. 41:3681-3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunalp, A. 1965. Growth and cytopathic effect of rubella virus in a line of green monkey kidney cells. Proc. Soc. Exp. Biol. Med. 118:185-190. [PubMed] [Google Scholar]
- 15.Herold, B. C., D. WuDunn, N. Soltys, and P. G. Spear. 1991. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J. Virol. 65:1090-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 18.Lee, E., M. Pavy, N. Young, C. Freeman, and M. Lobigs. 2006. Antiviral effect of the heparan sulfate mimetic, PI-88, against dengue and encephalitic flaviviruses. Antivir. Res. 69:31-38. [DOI] [PubMed] [Google Scholar]
- 19.Lee, F. K., R. M. Coleman, L. Pereira, P. D. Bailey, M. Tatsuno, and A. J. Nahmias. 1985. Detection of herpes simplex virus type 2-specific antibody with glycoprotein G. J. Clin. Microbiol. 22:641-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liljeqvist, J.-Å., B. Svennerholm, and T. Bergström. 1999. Herpes simplex virus type 2 glycoprotein G-negative clinical isolates are generated by single frameshift mutations. J. Virol. 73:9796-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liljeqvist, J.-Å., E. Trybala, J. Hoebeke, B. Svennerholm, and T. Bergström. 2002. Monoclonal antibodies and human sera directed to the secreted glycoprotein G of herpes simplex virus type 2 recognize type-specific antigenic determinants. J. Gen. Virol. 83:157-165. [DOI] [PubMed] [Google Scholar]
- 22.Mårdberg, K., E. Trybala, J. C. Glorioso, and T. Bergström. 2001. Mutational analysis of the major heparan sulfate-binding domain of herpes simplex virus type 1 glycoprotein C. J. Gen. Virol. 82:1941-1950. [DOI] [PubMed] [Google Scholar]
- 23.Marsden, H. S., A. Buckmaster, J. W. Palfreyman, R. G. Hope, and A. C. Minson. 1984. Characterization of the 92,000-dalton glycoprotein induced by herpes simplex virus type 2. J. Virol. 50:547-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mettenleiter, T. C., B. Lomniczi, N. Sugg, C. Schreurs, and T. Ben-Porat. 1988. Host cell-specific growth advantage of pseudorabies virus with a deletion in the genome sequences encoding a structural glycoprotein. J. Virol. 62:12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 26.Neyts, J., and E. De Clercq. 1995. Effect of polyanionic compounds on intracutaneous and intravaginal herpesvirus infection in mice: impact on the search for vaginal microbicides with anti-HIV activity. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 10:8-12. [PubMed] [Google Scholar]
- 27.Nyberg, K., M. Ekblad, T. Bergström, C. Freeman, C. R. Parish, V. Ferro, and E. Trybala. 2004. The low molecular weight heparan sulfate-mimetic, PI-88, inhibits cell-to-cell spread of herpes simplex virus. Antivir. Res. 63:15-24. [DOI] [PubMed] [Google Scholar]
- 28.Oyan, A. M., K. E. Dolter, N. Langeland, W. F. Goins, J. C. Glorioso, L. Haarr, and C. S. Crumpacker. 1993. Resistance of herpes simplex virus type 2 to neomycin maps to the N-terminal portion of glycoprotein C. J. Virol. 67:2434-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertel, P. E., and P. G. Spear. 1996. Modified entry and syncytium formation by herpes simplex virus type 1 mutants selected for resistance to heparin inhibition. Virology 226:22-33. [DOI] [PubMed] [Google Scholar]
- 30.Shields, B., J. Mills, R. Ghildyal, P. Gooley, and J. Meanger. 2003. Multiple heparin binding domains of respiratory syncytial virus G mediate binding to mammalian cells. Arch. Virol. 148:1987-2003. [DOI] [PubMed] [Google Scholar]
- 31.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 32.Su, H. K., R. Eberle, and R. J. Courtney. 1987. Processing of the herpes simplex virus type 2 glycoprotein gG-2 results in secretion of a 34,000-Mr cleavage product. J. Virol. 61:1735-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su, H. K., J. D. Fetherston, M. E. Smith, and R. J. Courtney. 1993. Orientation of the cleavage site of the herpes simplex virus glycoprotein G-2. J. Virol. 67:2954-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tal-Singer, R., C. Peng, M. Ponce de Leon, W. R. Abrams, B. W. Banfield, F. Tufaro, G. H. Cohen, and R. J. Eisenberg. 1995. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J. Virol. 69:4471-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trybala, E., T. Bergström, B. Svennerholm, S. Jeansson, J. C. Glorioso, and S. Olofsson. 1994. Localization of a functional site on herpes simplex virus type 1 glycoprotein C involved in binding to cell surface heparan sulphate. J. Gen. Virol. 75:743-752. [DOI] [PubMed] [Google Scholar]
- 36.Trybala, E., J.-Å. Liljeqvist, B. Svennerholm, and T. Bergström. 2000. Herpes simplex virus types 1 and 2 differ in their interaction with heparan sulfate. J. Virol. 74:9106-9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaheri, A. 1964. Heparin and related polyionic substances as virus inhibitors. Acta Pathol. Microbiol. Scand. Suppl. 171:1-98. [PubMed] [Google Scholar]
- 38.WuDunn, D., and P. G. Spear. 1989. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 63:52-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, G., N. S. Gunay, R. J. Linhardt, T. Toida, J. Fareed, D. A. Hoppensteadt, H. Shadid, V. Ferro, C. Li, K. Fewings, M. C. Palermo, and D. Podger. 2002. Preparation and anticoagulant activity of the phosphosulfomannan PI-88. Eur. J. Med. Chem. 37:783-791. [DOI] [PubMed] [Google Scholar]