Abstract
The mammalian APOBEC3 family of cytidine deaminases includes members that can act as potent inhibitors of retroviral infectivity and retrotransposon mobility. Here, we have examined whether the alpharetrovirus Rous sarcoma virus (RSV) is susceptible to inhibition by a range of human APOBEC3 proteins. We report that RSV is highly susceptible to inhibition by human APOBEC3G, APOBEC3F, and APOBEC3B and moderately susceptible to inhibition by human APOBEC3C and APOBEC3A. For all five proteins, inhibition of RSV infectivity was associated with selective virion incorporation and with C-to-T editing of the proviral DNA minus strand. In the case of APOBEC3G, editing appeared to be critical for effective inhibition. These data represent the first report of inhibition of retroviral infectivity and induction of proviral DNA editing by human APOBEC3A and reveal that alpharetroviruses, which do not normally encounter APOBEC3 proteins in their avian hosts, are susceptible to inhibition by all human APOBEC3 proteins tested. These data further suggest that the resistance of mammalian retroviruses to inhibition by the APOBEC3 proteins expressed in their normal host species is likely to have evolved subsequent to the appearance of this family of mammalian antiretroviral proteins some 35 million years ago; i.e., the base state of a naïve retrovirus is susceptibility to inhibition.
The antiretroviral activity of the APOBEC3 family of cytidine deaminases was first identified during research focusing on the activity of the human immunodeficiency virus type 1 (HIV-1) Vif protein, which was found to inhibit human APOBEC3G (A3G) function by binding A3G and targeting it for polyubiquitinylation and proteasomal degradation (10, 33, 35, 45, 46, 56). HIV-1 mutants lacking a functional vif gene (HIV-1ΔVif) selectively package A3G into progeny virions (1, 8, 43, 57). The antiretroviral activity of A3G results, at least in part, from its ability to induce hypermutation of the nascent proviral DNA minus strand by editing deoxycytidine residues to deoxyuridine during a subsequent infection (23, 31, 55, 58). In addition to introducing numerous deleterious point mutations, this editing activity may also compromise the stability of HIV-1 reverse transcripts and/or inhibit proviral integration (21, 30, 34, 48, 51).
In addition to A3G, the human genome encodes six other APOBEC3 proteins, including APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), and APOBEC3F (A3F) (12, 26). Analysis of other mammalian species has shown that the mouse genome contains a single APOBEC3 gene (the mA3 gene), while cats express at least three distinct APOBEC3 proteins (11, 37). While nonmammalian vertebrates, such as birds and fish, do not carry any genes encoding any APOBEC3 proteins, all vertebrates do carry genes encoding activation-induced deaminase (AID), a DNA-editing enzyme that plays a role in the diversification of antibody genes (11). While AID is thought to be the evolutionary precursor to the various APOBEC3 proteins (11), there is currently no evidence to suggest that it can repress retroviral replication (3).
In addition to inhibition of HIV-1ΔVif by human A3G, numerous other examples of the inhibition of retrovirus species by APOBEC3 family members have been reported (3, 4, 15-17, 27-29, 32, 44, 53, 54). The general pattern that has been observed is that retroviruses that replicate effectively in a given species are resistant to inhibition by APOBEC3 proteins expressed in the relevant tissues of that species (12). However, retroviruses are often strongly inhibited by APOBEC3 proteins derived from other species or from nontarget tissues in their normal host. Thus, wild-type HIV-1 is resistant to human A3G and A3F, both of which are normally expressed in human lymphocytes and macrophages, but is sensitive to human A3B (3, 16, 28, 53, 54), which is not expressed at significant levels in these cells, as well as to mA3 and simian A3G variants (3, 17, 27, 32, 44). HIV-1ΔVif, in contrast, is inhibited by all of these proteins. Conversely, murine leukemia virus (MLV) is resistant to mA3 but highly sensitive to human A3G and A3B, although it is resistant to human A3F (3, 17, 27). While the ability of wild-type HIV-1 to resist inhibition by a given APOBEC3 protein correlates with the ability of the HIV-1 Vif protein to induce degradation of that protein (4, 44), MLV and other simple retroviruses appear to have evolved mechanisms that permit the exclusion of the APOBEC3 proteins found in their normal host species from progeny virions, although they often continue to incorporate inhibitory noncognate APOBEC3 proteins (14, 15, 17, 27, 32).
The APOBEC3 protein family can be divided into two subgroups that differ based on the presence of only a single consensus cytidine deaminase active site (CDA) (e.g., A3A and A3C) or two CDAs (e.g., A3B, A3F, A3G, and mA3) (12, 26). The larger two CDA APOBEC3 proteins appear to be particularly effective inhibitors of retrovirus replication, with the more-N-terminal CDA playing a key role in the specific incorporation of the APOBEC3 protein into virion particles, while the more-C-terminal CDA is responsible for most, and sometimes all, of the DNA-editing activity of the protein (5, 22, 38). Single CDA APOBEC3 proteins are generally less effective inhibitors of retrovirus replication, although A3C has been shown to reduce the infectivity of simian immunodeficiency virus (3, 53, 54). However, A3A is an extremely potent inhibitor of the mobility of a wide range of retrotransposons, including intracisternal A particle, LINE-1, and Alu elements (6, 7, 9).
In this report, we have examined the ability of human APOBEC3 proteins to inhibit the infectivity of the alpharetrovirus Rous sarcoma virus (RSV) in avian cells. Because alpharetroviruses replicate exclusively in birds (20), which do not normally express any APOBEC3 protein members (11), they should differ from mammalian retroviruses in that they should not have evolved the capacity to block inhibition of their infectivity by these innate antiretroviral defense factors. Consistent with this hypothesis, we observed extremely potent (≥100-fold) inhibition of RSV infectivity by moderate levels of human A3B, A3G, and A3F and significant (∼10-fold) inhibition by human A3A and A3C. This inhibition was associated with the editing of RSV reverse transcripts by all five of these human APOBEC3 proteins, and, in the case of A3G, this editing was critical for effective inhibition. These data are consistent with the hypothesis that the resistance of certain mammalian retroviral species to inhibition by specific APOBEC3 proteins has coevolved with these cellular resistance factors and represent the first report of retroviral inhibition and proviral editing by human A3A in vivo.
MATERIALS AND METHODS
Cell culture and transfection.
The quail cell line QCl-3 was cultured as previously described (13) and was transfected using Fugene HD (Roche) by the protocol recommended by the manufacturer. Based on pilot studies using a green fluorescent protein expression plasmid, we estimate a transfection efficiency of ∼40% (data not shown).
Molecular clones.
We have previously described expression plasmids, based on pK, that encode full-length forms of human A3A, A3B, A3C, A3F, A3G, and β-arrestin (β-arr) bearing carboxy-terminal influenza hemagglutinin (HA) epitope tags (6). The vesicular stomatitis virus glycoprotein (G) expression plasmid pHIT/G has previously been described (53). An RSV-based retroviral indicator vector was derived by insertion of the Renilla luciferase (luc) gene into the unique ClaI site present in the RSV vector DANBP to give pRSV/luc. DANBP, a member of the RCAS series of alpharetroviral vectors developed by the Hughes laboratory (19, 25), was derived from a clone of the high-titer Bryan strain of RSV by substitution of a unique ClaI site in the place of the src gene. Therefore, pRSV/luc carries full-length RSV gag and pol genes in addition to the luc indicator gene. Infectious RSV particles can be generated by cotransfection of pRSV/luc and an envelope expression plasmid, in this case pHIT/G, into avian cells. Western analyses were performed as previously described (53), using a mouse monoclonal anti-HA antibody (Covance) or a rabbit polyclonal anti-RSV capsid antiserum (52).
Analysis of RSV proviral editing.
The level of C-to-T editing of RSV reverse transcripts was determined as previously described (16, 53), using virions derived from QCl-3 cells transfected with pRSV/luc, pHIT/G, and an APOBEC3 expression plasmid. Briefly, 24 h after retroviral infection of QCl-3 cells, we isolated total cellular DNA and subjected this to exhaustive digestion with DpnI to eliminate any plasmid DNA carried over from the transfected cells. We then used PCR (35 cycles) to isolate a 600-bp fragment of the Renilla luc gene present in pRSV/luc (this fragment contains four DpnI sites). After cloning and sequencing were performed, nucleotide differences from the wild-type luc sequence were identified and tabulated.
RESULTS
To determine whether human APOBEC3 proteins would be able to exert an inhibitory effect on the infectivity of the alpharetrovirus RSV (20), we cotransfected the quail cell line QCl-3 with an RSV-based retroviral vector bearing the Renilla luc gene (pRSV/luc) together with a vector encoding HA-tagged versions of human A3A, A3B, A3C, A3F, or A3G. A plasmid expressing an HA epitope-tagged form of the irrelevant cytoplasmic human protein β-arr was used as a control. The pRSV/luc vector expresses wild-type RSV Gag and Pol, and an envelope was provided in trans by cotransfection of the vesicular stomatitis virus G expression plasmid pHIT/G. At 48 h posttransfection, supernatant media were collected, passed through a 0.45-μm filter, and then used to infect a second, naïve QCl-3 cell culture. After a further 24-h incubation, these cells were lysed and used to quantify induced luciferase activities. In addition, at the time of supernatant harvest, the producer QCl-3 cells were also harvested and lysed, and the resultant extracts used to quantify the intracellular level of APOBEC3 protein expression.
As shown in Fig. 1A, all five human APOBEC3 proteins were found to exert a marked inhibitory effect on the infectivity of the alpharetrovirus RSV. Specifically, human A3B reduced RSV infectivity ∼200-fold, and A3G and A3F had an ∼100-fold inhibitory effect, while A3A and A3C, the two single CDA APOBEC3 proteins, exerted an ∼10-fold inhibitory effect (Fig. 1A). Analysis of the levels of intracellular expression of the various human APOBEC3 proteins in the transfected QCl-3 producer cells (Fig. 1B) revealed comparable levels of expression, with A3F consistently slightly underexpressed. Together these data demonstrate that all five of the human APOBEC3 proteins tested in this analysis are able to exert a strong inhibitory effect on the infectivity of the alpharetrovirus RSV.
FIG. 1.
RSV infectivity is inhibited by several human APOBEC3 proteins. (A) QCl-3 cells (35 mm culture) were transfected with 2 μg of pRSV/luc, 500 ng of pHIT/G, and 500 ng of pK-βarr (a control), pK-A3B, pK-A3F, or pK-A3G, or 250 ng of pK-A3A or pK-A3C plus 250 ng of the empty pK vector as filler. Lower levels of the A3A and A3C expression plasmids were used in an attempt to achieve equivalent levels of intracellular expression of A3A and A3C, compared to the levels of the three double-CDA APOBEC3 proteins. At 48 h posttransfection, supernatant media were harvested, filtered, and used to infect naïve QCl-3 cells (35 mm culture). A further 24 h later, the infected QCl-3 cells were harvested and lysed, and luciferase activities quantified. Average results of four independent experiments with standard deviations are indicated. The level of luciferase activity induced by RSV virion particles derived from the control culture was arbitrarily set at 100%. (B) The virus producer QCl-3 cultures depicted in panel A were lysed at the time of supernatant harvest and subjected to Western blot analysis using a monoclonal antibody specific for the HA epitope tags found at the carboxy-terminal end of all the APOBEC3 proteins, as well as the β-arr control. Results of a representative experiment are shown.
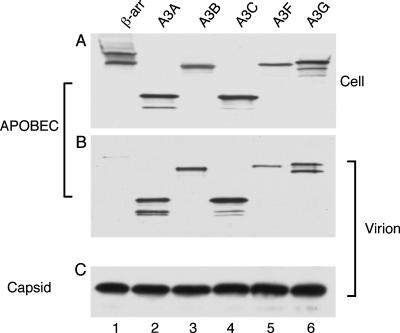
Analysis of the inhibition of other retroviruses by APOBEC3 family members has revealed that inhibition is dependent on the packaging of APOBEC3 proteins into virion particles (14, 15, 17, 32) and is frequently correlated with a significant level of C-to-T editing of the minus strand of retroviral reverse transcripts (23, 31, 55, 58). We therefore sought to determine whether these human APOBEC3 proteins would indeed be selectively packaged into RSV particles. For this purpose, we again transfected QCl-3 cells with pRSV/luc and an APOBEC3 expression plasmid and then harvested and filtered the supernatant media 48 h later. Released retroviral virions were then collected by ultracentrifugation through a sucrose cushion, as previously described (4, 17, 53). In Fig. 2, we present a Western analysis of the intracellular expression (Fig. 2A) and the intravirion packaging (Fig. 2B) of the various APOBEC3 proteins, using a monoclonal antibody specific for the HA epitope tags. The control used in this experiment was again an HA epitope-tagged version of β-arr, which is not selectively packaged into retroviral virions (53).
FIG. 2.
Human APOBEC3 proteins are selectively packaged into RSV virion particles. (A) Western analyses of the virion producer QCl-3 cell culture were performed as described in the legend for Fig. 1. (B) Virions released from the producer cell culture analyzed as described for panel A were pelleted, lysed, and then analyzed by Western blotting using the same HA epitope-specific antibody as that for panel A. (C) The virion pellets analyzed as described for panel B were also analyzed for the total level of released RSV virions by Western blotting using a rabbit polyclonal anti-RSV capsid antiserum.
As shown in Fig. 2B, all five of the APOBEC3 proteins analyzed were packaged into RSV virion particles in proportion to their expression levels in the producer QCl-3 cells (Fig. 2A). In contrast, we did not detect significant packaging of the β-arr protein (Fig. 2B, lane 1), even though this protein was expressed at a level in the producer cells comparable to those of the APOBEC3 proteins (Fig. 2A, lane 1). We therefore conclude that all five of the human APOBEC3 proteins analyzed here are indeed capable of being selectively packaged into RSV virions, a result which is consistent with the inhibition of RSV infectivity induced by these human antiretroviral defense factors (Fig. 1A).
In Fig. 2C, we also present a Western analysis quantifying the level of RSV Gag protein present in the pelleted virions analyzed in Fig. 2B. We did not observe any inhibitory effect of these APOBEC3 proteins on the release of RSV Gag, which suggests that the APOBEC3 proteins are not exerting a significant nonspecific toxic effect in QCl-3 cells. We note, however, that QCl-3 cells are constitutively infected with the replication-defective high-titer Bryan strain of RSV, i.e., the same virus used to make the pRSV/luc indicator vector (19), and constitutively release noninfectious RSV particles even before transfection with the pRSV/luc vector (13). Therefore, as our transfection efficiency in these cells was only ∼40%, we might not have been able to detect a modest inhibitory effect of the APOBEC3 proteins on RSV virion production. However, this phenotype has not previously been observed in studies addressing the effect of APOBEC3 proteins on a range of other retroviral species (3, 4, 15-17, 27-29, 32, 44, 53).
We next addressed the question of whether these human APOBEC3 proteins are, in fact, editing RSV reverse transcripts during retroviral infection of QCl-3 cells. For this purpose, we used PCR to recover an ∼600-bp-long stretch of the Renilla luc indicator gene present in pRSV/luc from infected cells at 24 h postinfection. As shown in Table 1, we observed significant levels of C-to-T editing of the luc gene during RSV reverse transcription for all five human APOBEC3 proteins analyzed. The level of C-to-T editing observed was ∼4.7% of all available C residues for A3F, ∼3.7% for A3G, ∼1.8% for A3B, ∼1.3% for A3C, and ∼0.7% for A3A. Analysis of the sequence context of the edited C residues showed that A3A, A3B, A3C, and A3F all preferred to edit C residues flanked by a 5′ T residue (79% of edited C residues were flanked by a 5′ T for A3A, 70% for A3B, 78% for A3C, and 88% for A3F). In contrast and as previously reported (3, 18, 22, 23, 53), A3G was found to preferentially edit C residues flanked by a 5′ C residue (70% of all edited C's), although C's flanked by a 5′ T were also edited by A3G, albeit at a lower frequency (25% of all edited C's). Importantly and as expected, sequencing of 4,804 bp of RSV reverse transcripts derived from virions produced in the absence of any exogenous APOBEC3 protein revealed only four mutations, none of which were C-to-T changes (data not shown).
TABLE 1.
Editing of RSV reverse transcripts in avian cells by human APOBEC3 proteinsa
RSV virions were produced in transfected QCl-3 cells as described in the legend for Fig. 1, except that the levels of pK-A3A and pK-A3C were increased to 500 ng per transfection (the same as pK-A3F and pK-A3G), while the level of pK-A3B was decreased to 250 ng. This was necessitated by the essentially complete block of the production of intact RSV proviruses seen when RSV was produced in the presence of 500 ng of pK-A3B (Fig. 1A, and data not shown). Twenty-four hours after infection, total QCl-3 DNA was harvested, exhaustively cleaved with DpnI, and then subjected to PCR using primers specific for the Renilla luc gene. This table compiles the observed point mutations observed after cloning and sequencing the luc DNA segments obtained. The expected nucleotide sequences are shown in the far left columns, while the observed sequences are listed in the top rows. The total number of bases sequenced for each APOBEC3 variant is given below the lower right corner of each box. No C-to-T editing was observed when 4,804 bp of RSV reverse transcripts derived from virions produced in the absence of any APOBEC3 protein were analyzed (data not shown).
There has been considerable controversy about the question of whether cytidine deaminase activity is important for the antiviral and antiretrotransposon activity of APOBEC3 proteins. Although one group has argued that this enzymatic activity is largely dispensable for the inhibition of HIV-1ΔVif infectivity by human A3G and A3F (2, 24, 39), others have reported a marked drop in the level of inhibition observed when stable, enzymatically inactive variants of A3G were analyzed (5, 31, 34, 38, 47). On the other hand, several groups have reported that inhibition of the infectivity of the hepadnavirus hepatitis B virus by A3G (40, 41, 50) and inhibition of retrotransposon mobility by APOBEC3 proteins (6, 7, 9, 36, 49) are largely or entirely independent of cytidine deaminase activity.
To address this question in the case of RSV, we analyzed two previously described A3G mutants called A3G-E2 and A3G-E1+E2 (5). Although A3G contains two consensus CDAs, only the more-carboxy-terminal CDA is enzymatically active (5, 22, 38, 39), while the more-amino-terminal CDA has been proposed to play a role in the specific packaging of A3G into retroviral virion particles (38). In A3G-E2, a key carboxy-terminal CDA glutamic acid residue at position 259 has been mutated to glutamine, which blocks entirely the cytidine deaminase activity of A3G (5, 39). In A3G-E1+E2, the analogous glutamic acid residue at position 67 in the amino-terminal CDA has also been changed to glutamine. As shown in Fig. 3, while all three of these A3G variants were expressed at comparable levels in the transfected QCl-3 producer cells (Fig. 3B), only the wild-type A3G protein and the A3G-E2 mutant were selectively packaged into RSV virion particles (Fig. 3C, lanes 2 and 3). Analyses of the effect of the A3G-E2 and A3G-E1+E2 mutants on RSV infectivity showed that both were severely compromised, with the observed inhibition dropping from the ∼100-fold seen with wild-type A3G to ∼2-fold for both A3G-E2 and A3G-E1+E2 (Fig. 1A). As predicted from earlier work (5, 39), we did not detect any evidence for C-to-T editing of RSV reverse transcripts produced by virions that had incorporated the A3G-E2 mutant (Table 1) or the A3G-E1+E2 mutant (data not shown). We therefore conclude that the amino-terminal CDA of A3G plays a critical role in the selective incorporation of A3G into RSV virions but that the editing activity of the carboxy-terminal CDA plays a key role in the subsequent inhibition of RSV infectivity.
FIG. 3.
The CDAs present in A3G are critical for inhibition of RSV infectivity. (A) This infection experiment was performed as described in the legend for Fig. 1A, except that the indicated wild-type or mutant forms of human A3G were analyzed. The pK/β-arr plasmid, which expresses HA epitope-tagged β-arr, was again used as a control (lane 1). (B) Western analysis of the intracellular expression levels of the β-arr and A3G protein variants described for panel A. (C) Western analysis of the intravirion expression levels of β-arr and the A3G protein variants described for panel A. This Western blotting, which was performed as described in the legend for Fig. 2, analyzed RSV virions released from the same QCl-3 cells described for panel B. (D) Western analysis of the levels of RSV capsid protein expression in the pelleted virions analyzed as described for panel C. The results of a representative experiment are shown.
DISCUSSION
All mammalian species examined so far express one or more APOBEC3 protein variants (11), and any mammalian retrovirus therefore should have evolved mechanisms that permit the retrovirus to at least attenuate the antiretroviral activity of the APOBEC3 proteins present in its normal host species (12). Consistent with this hypothesis, individual retroviruses such as HIV-1 and MLV have indeed been shown to be resistant to the APOBEC3 proteins expressed in the relevant target tissues of their normal host species, i.e., humans and mice, respectively. In contrast, HIV-1 is sensitive to mA3, while MLV is inhibited by human A3G (3, 17, 27, 32).
Analysis of the genomic sequence of the mammalian APOBEC3 locus in a range of species demonstrates that the ongoing evolution of the retrovirus-APOBEC3 interaction is observed not only in retroviruses. That is, mammals have clearly been under selective pressure to minimize their susceptibility to retroviral pathogens, and this pressure has resulted both in the rapid positive selection of sequence variants in mammalian APOBEC3 genes and in the amplification of the APOBEC3 locus present on chromosome 22 from the one APOBEC3 gene present in mice to the seven APOBEC3 genes now found in humans and other primates (11, 42, 59).
The fact that mammalian retroviruses are under selective pressure to evade inhibition by mammalian APOBEC3 proteins (42, 59) makes it difficult to assess the intrinsic antiretroviral activity of mammalian APOBEC3 proteins using mammalian retroviruses, as these are predicted to have evolved strong resistance to inhibition by cognate APOBEC3 proteins and, potentially at least, partial resistance to the noncognate APOBEC3 proteins expressed by heterologous mammalian species.
From this perspective, we were intrigued by the report that nonmammalian vertebrates, including birds, do not express any APOBEC3 homologs (11). If APOBEC3 proteins initially evolved as “universal” antiretroviral defense factors, then members of a retrovirus family that replicate exclusively in birds, i.e., alpharetroviruses, might not only be susceptible to inhibition by human APOBEC3 proteins but also, in fact, be hypersusceptible to inhibition by this family of, to them, entirely novel inhibitory proteins. Conversely, if the ability of APOBEC3 proteins to inhibit mammalian retroviruses represents a coevolved activity, then avian retroviruses might be relatively nonsusceptible to inhibition by these heterologous resistance factors. We note that both mammals and birds express the DNA-editing enzyme AID, which is believed to represent the evolutionary precursor of the various APOBEC3 proteins (11). However, there is currently no evidence to suggest that AID possesses any antiretroviral activity (3).
The data presented in this report are consistent with the first of the above-mentioned hypotheses, i.e., that retroviruses that are not normally exposed to APOBEC3 proteins are highly susceptible to inhibition by these proteins. Specifically, the relatively modest level of protein expression induced upon transfection of the QCl-3 quail cell line with nonreplicating vectors encoding human APOBEC3 proteins proved able to strongly inhibit the infectivity of the alpharetrovirus RSV (Fig. 1A). The double CDA human APOBEC3 proteins A3B, A3F, and A3G, all of which are effective inhibitors of HIV-1ΔVif infectivity (16, 28, 45, 46, 53), reduced RSV infectivity up to 200-fold. Moreover, the single CDA human APOBEC3 proteins A3A and A3C also proved able to inhibit RSV infectivity up to ∼10-fold. This result was particularly striking in the case of A3A, which has not previously been reported to be capable of inhibiting retrovirus infectivity (3, 6, 9). Together, these data therefore provide further support for the hypothesis that mammalian retroviruses and APOBEC3 genes are engaged in an ongoing process of rapid adversarial coevolution (42, 59) and suggest that retroviruses, in general, may well have been highly susceptible to inhibition by ancestral versions of APOBEC3 when they first appeared on the evolutionary scene some 35 million years ago.
Analysis of the ability of the various human APOBEC3 proteins to induce editing of RSV reverse transcripts revealed that all five indeed induced C-to-T hypermutation of the proviral minus strand (Table 1). This represents the first report of editing of retroviral reverse transcripts by A3A. Interestingly, while A3A has been reported to be a potent inhibitor of intracisternal A particle, LINE-1, and Alu retrotransposition, this inhibition does not appear to require DNA editing, and editing of retrotransposons by A3A has, in fact, not been reported (6, 7, 9).
Given the current controversy regarding the importance of DNA editing in the inhibition of retroviral infectivity by APOBEC3 proteins (12), we also asked whether inactivation of the carboxy-terminal CDA present in A3G, the only enzymatically active CDA (5, 38, 39), would reduce the level of inhibition of RSV infectivity. As shown in Fig. 3, an enzymatically inactive point mutant of A3G (A3G-E2) was in fact largely incapable of blocking RSV infectivity. Interestingly, while mutation of the carboxy-terminal CDA of A3G in A3G-E2 had no effect on the virion incorporation of A3G, the additional mutagenesis of the amino-terminal CDA in A3G-E1+E2 essentially abrogated A3G virion packaging. These data further substantiate the proposed division of labor for the two CDAs of A3G (38), with the enzymatically inactive amino-terminal CDA being primarily involved in virion packaging, while the enzymatically active carboxy-terminal CDA is critical for proviral editing.
Acknowledgments
We thank Stephen Hughes and John Wills for reagents used in this research. On the occasion of her retirement, we also thank Sharon Autry for assistance in the preparation of this report and many others over the last 20 years.
This research was sponsored by NIH grant AI065301 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S.-J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, and B. R. Cullen. 2007. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 364:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16:480-485. [DOI] [PubMed] [Google Scholar]
- 10.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 11.Conticello, S. G., C. J. F. Thomas, S. K. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22:367-377. [DOI] [PubMed] [Google Scholar]
- 12.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cullen, B. R., A. M. Skalka, and G. Ju. 1983. Endogenous avian retroviruses contain deficient promoter and leader sequences. Proc. Natl. Acad. Sci. USA 80:2946-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derse, D., S. A. Hill, G. Princler, P. Lloyd, and G. Heidecker. 2007. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl. Acad. Sci. USA 104:2915-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doehle, B. P., H. P. Bogerd, H. L. Wiegand, N. Jouvenet, P. D. Bieniasz, E. Hunter, and B. R. Cullen. 2006. The betaretrovirus Mason-Pfizer monkey virus selectively excludes simian APOBEC3G from virion particles. J. Virol. 80:12102-12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doehle, B. P., A. Schäfer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281-288. [DOI] [PubMed] [Google Scholar]
- 17.Doehle, B. P., A. Schäfer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannleux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 19.Federspiel, M. J., and S. H. Hughes. 1997. Retroviral gene delivery. Methods Cell Biol. 52:179-214. [PubMed] [Google Scholar]
- 20.Goff, S. P. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 21.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. Inhibition of tRNA3Lys-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haché, G., M. T. Liddament, and R. S. Harris. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920-10924. [DOI] [PubMed] [Google Scholar]
- 23.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, S. H., J. J. Greenhouse, C. J. Petropoulos, and P. Sutrave. 1987. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61:3004-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285-296. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi, M., A. Takaori-Kondo, K. Shindo, A. Abudu, K. Fukunaga, and T. Uchiyama. 2004. APOBEC3G targets specific virus species. J. Virol. 78:8238-8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liddament, M. T., W. L. Brown, A. J. Schumacher, and R. S. Harris. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:385-1391. [DOI] [PubMed] [Google Scholar]
- 29.Löchelt, M., F. Romen, P. Bastone, H. Muckenfuss, N. Kirchner, Y.-B. Kim, U. Truyen, U. Rösler, M. Battenberg, A. Saib, E. Flory, K. Cichutek, and C. Münk. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. USA 102:7982-7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, K., T. Wang, B. Liu, C. Tian, Z. Xiao, J. Kappes, and X.-F. Yu. 2007. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 81:7238-7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 32.Mariani, R., D. Chen, B. Schröfelbauer, F. Navarro, R. König, B. Bollman, C. Münk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 33.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 34.Mbisa, J. L., R. Barr, J. A. Thomas, N. Vandegraaff, I. J. Dorweiler, E. S. Svarovskaia, W. L. Brown, L. M. Mansky, R. J. Gorelick, R. S. Harris, A. Engelman, and V. K. Pathak. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. N. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 36.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Löwer, K. Cichutek, E. Flory, G. G. Schumann, and C. Münk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 37.Münk, C., J. Zielonka, H. Constabel, B.-P. Kloke, B. Rengstl, M. Battenberg, F. Bonci, M. Pistello, M. Löchelt, and K. Cichutek. 2007. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J. Virol. 81:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navarro, F., B. Bollman, H. Chen, R. König, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 39.Newman, E. N. C., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Anti-viral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen, D. H., S. Gummuluru, and J. Hu. 2007. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol. 81:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosler, C., J. Kock, M. Kann, M. H. Malim, H. E. Blum, T. F. Baumert, and F. Von Weizsacker. 2005. APOBEC-mediated interference with hepadnavirus production. Hepatology 42:301-309. [DOI] [PubMed] [Google Scholar]
- 42.Sawyer, S. L., M. Emerman, and H. S. Malik. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:1278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schäfer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the Gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 44.Schröfelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 46.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 47.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 48.Simon, J. H. M., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 50.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 51.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, Q., D. Chen, R. König, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379-53386. [DOI] [PubMed] [Google Scholar]
- 55.Yu, Q., R. König, S. Pillai, K. Chiles, M. Kearney, S. Palmer, D. Richman, J. M. Coffin, and N. R. Landau. 2004. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 11:435-442. [DOI] [PubMed] [Google Scholar]
- 56.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and S.-F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 57.Zennou, V., D. Perez-Caballero, H. Göttlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, J., and D. M. Webb. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 13:1785-1791. [DOI] [PubMed] [Google Scholar]