Abstract
Understanding the pathogenesis of dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS) requires the precise identification of dengue virus (DV)-permissive target cells. In a previous study using unfractionated human peripheral blood mononuclear cells, we found that monocytes, but not B or T cells, were the principal DV-permissive cells in the absence of DV-immune pooled human sera (PHS) and the major mediators of antibody-dependent enhancement in the presence of PHS. To further identify DV-permissive target cells in other tissues and organs, we isolated human splenic mononuclear cells (MNCs), inoculated them with DV type 2 (strain 16681) in the presence or absence of PHS, and assessed their infection either directly using flow cytometry and reverse transcription-PCR (RT-PCR) assays or indirectly by plaque assay. We found that in the absence of PHS, a small proportion of splenic macrophages appeared to be positive for DV antigens in comparison to staining controls by the flow cytometric assay (0.77% ± 1.00% versus 0.18% ± 0.12%; P = 0.07) and that viral RNA was detectable by the RT-PCR assay in MNCs exposed to DV. Additionally, supernatants from cultures of DV-exposed MNCs contained infectious virions that were readily detectable by plaque assay. The magnitude of infection was significantly enhanced in splenic macrophages in the presence of highly diluted PHS (5.41% ± 3.53% versus 0.77% ± 1.00%; P = 0.001). In contrast, primary T and B cells were not infected in either the presence or absence of PHS. These results provide evidence, for the first time, that human primary splenic macrophages, rather than B or T cells, are the principal DV-permissive cells in the spleen and that they may be uniquely important in the initial steps of immune enhancement that leads to DHF/DSS in some DV-infected individuals.
As one of the reemerging infectious diseases, dengue fever affects 50 to 100 million people annually in tropical and subtropical regions and causes severe disease that requires hospitalization in approximately 1% of infected individuals. Dengue fever is a mosquito-borne viral disease caused by infection with dengue virus (DV), a single-stranded RNA virus that belongs to the Flaviviridae family (5, 12, 27, 44). It exists as four serotypes, DV type 1 (DV1), DV2, DV3, and DV4, classified according to neutralization and complement-fixation assays (37, 40). Most primary DV infections present as a self-limiting, undifferentiated febrile illness, dengue fever. DV infection can cause more severe forms of disease, including dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS), in secondary infections and primary infections of infants 6 to 9 months of age. DHF/DSS is an acute vascular permeability syndrome that manifests clinically as hypotension, shock, and hemorrhage, carrying a high mortality rate of 10 to 40% without appropriate clinical management (44).
Antibody-dependent enhancement (ADE) is one of the central hypotheses for the pathogenesis of DHF/DSS that had been proposed 3 decades ago (5, 13-15, 39). The ADE hypothesis proposes that while neutralizing antibodies produced during primary infections confer life-long protection to a homologous virus, they may be paradoxically harmful in secondary infections with a heterotypic DV serotype. Preexisting antibodies to one serotype form infectious immune complexes with heterotypic DV serotypes and bind to Fc receptors (FcRs) on cells such as monocytes/macrophages. This facilitates dengue virus entry, replication, and spread and, hence, increased disease severity. Additionally, infected monocytes/macrophages may activate other effector mechanisms to produce vasoactive lymphokines that cause endothelial cell damage and the eventual DHF/DHS (8-11, 13-15, 19-21). In theory, cells bearing a putative DV receptor may be susceptible to DV infection, whereas cells bearing FcR can bind and internalize potentially infectious virus-antibody immune complexes independent of the putative DV receptor. In both instances, DV spread would be facilitated. In vitro experimental evidence suggests that both FcγRI and FcγRII mediate ADE of DV infection (22, 26, 38).
There are also alternative hypotheses of DHF/DSS that have proposed a major role for T or B cells, either as target cells for direct DV infection or as effector cells responding to DV antigens presented on infected antigen-presenting cells. In either case, T and B cells are thought to produce some cytokines that cause endothelium damage preceding the onset of DHF/DSS (5, 25, 39, 43). These alternative hypotheses are derived from several lines of experimental evidence. First, CD4+ and CD8+ T-cell clones, B-cell lines, or purified primary B cells were shown to be susceptible to infection by DV and capable of producing new virions in the absence of DV-immune serum (18, 25, 31, 41). Second, both DV-specific CD4+ and CD8+ T cells were detected in a proportion of subjects who acquired natural DV infection or received candidate DV vaccines (28, 29, 33, 34). Furthermore, these DV-specific T cells produced proinflammatory cytokines, including gamma interferon and tumor necrosis factor alpha, upon stimulation with DV antigens. Finally, diverse cell types including skin Langerhans cells, endothelial cells, and hepatocytes have been reported to be permissive to DV infection in the absence of enhancing antibodies (1, 3, 30, 42, 45). These DV-infected cells might present DV antigens that activate T or B cells to make undesirable cytokines that may cause microvasculature damage in DHF/DSS.
Central to the validation of these contrasting hypotheses is determining the identity of DV-permissive target cells and whether anti-DV antibody can direct DV infection to cells that are ordinarily poorly permissive. In a previous study using unfractionated human peripheral blood mononuclear cells (PBMCs), we found that monocytes, but not B or T cells, were the principal DV-permissive cells in the absence of DV-immune serum and the major mediators of antibody-dependent enhancement in the presence of DV-immune serum (23). In the current study, we extend our investigation to the use of mononuclear cells (MNCs) isolated from human spleens, an organ where dengue viral antigens were frequently detected in patients who succumbed to DHF/DSS (16) and in experimentally DV-infected mice and macaques (9, 24). Our results provide evidence for the first time that human primary splenic macrophages, rather than B or T cells, are most permissive to DV infection, and thus they may be uniquely important in the initial steps of immune enhancement that leads to DHF/DSS in some DV-infected individuals.
MATERIALS AND METHODS
Virus stocks.
Virulent strain DV2-16681 was provided by Richard Kinney (CDC, Fort Collins, CO). Virus stocks were prepared and titrated by plaque assay as previously described (38) and then aliquoted and stored at −80°C.
Source cells.
Extra splenic MNCs that were regularly purged from the freezer and discarded by the Histocompatibility Laboratory, University of Rochester, were saved and used in the current study, which has been approved by the institutional Research Subjects Review Board. Standard procedures were used to isolate MNCs from the spleen. Each spleen was obtained from a potential organ donor and then sectioned and distributed to tissue typing labs for use in tissue compatibility testing. Immediately after the splenic section was received, it was dissected into smaller pieces with a surgical scalpel. The pieces were kept in 3 to 5 ml of RPMI 1640 medium in a plastic bag, which was placed in a Stomacher 80 laboratory blender (Seward, Essex, United Kingdom) for 1 to 2 min. The suspension was filtered through a nitrocellulose membrane to remove larger clumps and diluted to 20 ml in phosphate-buffered saline (PBS). MNCs were isolated by standard Ficoll density gradient centrifugation methods, washed twice in RPMI 1640 medium, and cryopreserved at −150°C in RPMI medium with 20% fetal bovine serum and dimethyl sulfoxide at a concentration of 5 × 106 to 10 × 106 per ml until use in a cytotoxicity or flow cytometric cross-match to determine the human leukocyte antigen compatibility between the organ donor and organ recipient. Vero cells were obtained from the ATCC.
Staining antibodies.
A purified murine monoclonal antibody (7E1) specific for DV E antigen (anti-E) of all four DV serotypes was labeled with Alexa Fluor 647 (Molecular Probes, Invitrogen Corp.) and used for the enumeration of DV-infected cells as described previously (38). Other antibodies for cell surface markers were purchased from commercial sources, including CD3-phycoerythrin (PE)-Cy5, CD4-allophycocyanin, CD14-Alexa Fluor 700, CD16-PE-Cy7, CD19-PE, CD20-PE-Cy5, CD32-PE, CD56-PE-Cy5, and CD64-fluorescein isothiocyanate (BD Pharmingen, San Jose, CA) as well as CD3-Alexa Fluor 405 (Caltag, CA).
Phenotypic analysis of human splenic MNCs and PBMCs.
For multicolor phenotypic analysis, cryopreserved cells were thawed, washed twice with PBS, and resuspended in PBS at 2 × 107 cells per ml. A total of 100 μl of the cell suspension was stained for 30 min at 4°C with antibodies to various surface markers including CD3, CD4, CD14, CD20, CD64 (Fcγ RI), CD32 (Fcγ RII), and CD16 (Fcγ RIII). Finally, cells were washed twice with PBS and resuspended in 1% formaldehyde. Data were acquired on a BD LSR II machine (BD Immunocytometry System) and analyzed by using the FlowJo (Tristar) software.
Direct DV infection and ADE of infection in human splenic MNCs.
For direct infection, MNCs were inoculated with DV at various multiplicities of infection (MOIs) for 90 min at 37°C, washed twice with PBS, and then cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum, 100 IU/ml of penicillin, 100 μg/ml of streptomycin, and 2.0 mM of l-glutamine (RPMI 10 medium) until the day of assay. For the ADE experiment, DV-immune pooled human sera (PHS) were prepared by mixing equal amounts of serum obtained from three DV-immune subjects who had documented neutralizing reactivity to all four DV serotypes (J. J. Schlesinger, unpublished observation). The PHS was then filtered with a 0.2-μm-pore-size filter, aliquoted, and stored at −80°C. To prepare DV-immune complexes, known amounts of DV were mixed with serially diluted PHS in a volume of 500 μl and incubated for 30 min at 37°C before being added to a known number of splenic MNCs in suspension (at a final concentration of 2 × 106/ml). Infected cells were then washed twice, resuspended in RPMI 10 medium, and cultured at 37°C for two additional days. On the day of assay, cells were harvested and stained intracellularly for DV antigen in conjunction with a panel of surface markers to identify cell subsets. Results were expressed as the proportion of infected cells of a given phenotypically unique cell subset (such as monocytes that are CD14+ CD3− CD19−) under various conditions as detected by anti-E antibody.
FACS analysis of DV infection.
Standard as well as polychromatic fluorescence-activated cell sorting (FACS) analyses were performed to enumerate the number of DV-infected cells. DV-infected cells were harvested by gentle scraping, washed once with PBS, and then fixed and permeabilized with 500 μl of lysing buffer (BD Biosciences) for 10 min at room temperature. After being washed once with PBS, the cells were resuspended in 500 μl of PBS and incubated with an additional 500 μl of Perm 2 solution (BD Biosciences) for 10 min at room temperature. Following another wash with PBS, the cells were resuspended in PBS containing 50% human AB serum and stained with the anti-E antibody in conjunction with antibodies to cell subset markers for 30 min at 4°C. Labeled murine immunoglobulin G1 antibodies were used as staining controls in all assays. Finally, cells were washed twice with PBS and resuspended in 1% formaldehyde. FACS data acquisition was performed on a FACSCalibur or a BD LSR II machine (BD Immunocytometry Systems). For each sample, 20,000 to 100,000 total events were collected for analysis. The data acquired were analyzed using CellQuest (BD Biosciences) or FlowJo (TriStar) software.
Quantifying viral production by surrogate assays.
The amount of infectious virion produced in the infected splenic MNCs was quantified with the plaque assay as described previously (38). Briefly, 1.5 × 105 Vero cells were seeded to each well of a 24-well tissue culture plate and allowed to grow to 80% confluence overnight. A total of 200 μl of culture supernatant harvested from infected splenic MNCs was added to Vero cells and incubated for 90 min at 37°C. The cells were washed and then cultured in 2.0 ml of medium for 3 days before being assayed for DV production by an immunostaining assay as described previously (38). A parallel set of Vero cell cultures was set up for quantifying the number of infected Vero cells with anti-7E1 antibody staining and the FACS assay. Vero cells were seeded onto a six-well plate at a concentration of 5 × 105 cells per well and inoculated with 500 μl of supernatant harvested from each splenocyte infection condition for 90 min at 37°C. The cells were left in culture for 2 days and harvested, stained with our anti-E antibody, and then analyzed by FACS analysis.
RNA extraction and RT-PCR.
For reverse transcription-PCR (RT-PCR) analysis of DV infection, total RNA was extracted from 5 × 105 infected or control cells using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. Ample RNase Away (Invitrogen) was used on all tubes to limit RNA digestion. Each RT-PCR was performed using the Superscript III first-strand synthesis system (Invitrogen). First-strand cDNA was synthesized using oligo(dT)20 primers, and the reaction mixtures were incubated at 50°C for 50 min and then at 85°C for 5 min. Platinum PCR Supermix (Invitrogen) was used for the PCR at 33 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 80 s) in a GeneAmp PCR System 9700 model thermocycler (Applied Biosystems). Primers specific for the DV NS1 gene were used for detecting virus-infected cells [pNS1(BamHI)Forward, 5′-GCCGGATCCGATAGTGGTTGCGTTGTGAGCTGG-3′; pNS1(Xho1)Reverse, 5′-GGCCGAGCTCTTAAGCTGTGACCAAGGAGTTGACC-3′]. The PCR products were analyzed by electrophoresis on a 1% agarose gel.
Statistics.
The comparisons between data groups were performed with the Mann-Whitney test; for comparisons among data groups, one-way analysis of variance in GraphPad Prism, version 4, software (San Diego, CA) was used. A P value of less than 0.05 was considered significant.
RESULTS
Splenic MNCs are phenotypically different from blood PBMCs.
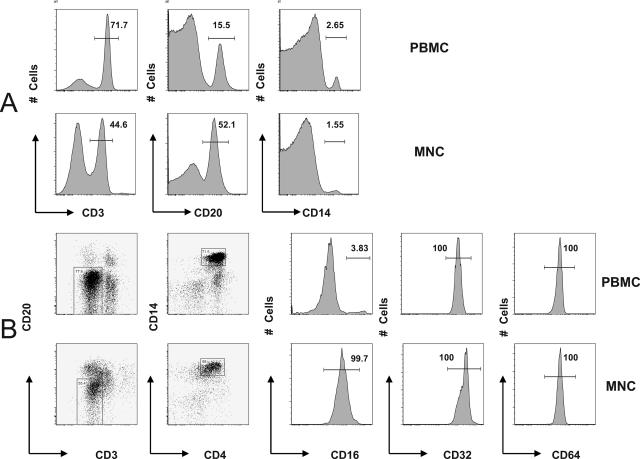
Other than being the primary filter of blood, the spleen is an important immunologic organ that plays a major role in processing and presenting antigens, stores one-third of the body's platelets, and functions as a major hematopoietic site (35). It contains approximately equal percentages of B and T cells (6) and a small proportion of other cells including macrophages, granulocytes, and NK cells. The spleen may play an important role in the development of DHF/DSS; however, the specific type of cells permissive to DV and capable of mediating ADE is largely unknown. We first compared the relative proportion of major subsets of immune cells of interest between splenic MNCs and PBMCs and then the phenotypic features of splenic macrophages versus blood monocytes. Because paired MNCs and PBMCs from the same donor were not available, we used PBMCs from one group of three individuals and MNCs from another group of three individuals. We first compared the overall phenotypic characteristic between blood PBMCs (sample 322-L007) and splenic MNCs (sample OL3020). The proportion of cells of interest differed between blood and spleen. PBMCs in blood were made up of mostly CD3+ T cells (71.7%), followed by CD19+ B cells (15.5%) and CD14+ monocytes (2.65%). In contrast, splenic MNCs consisted of a smaller proportion of T cells (44.6%) but more B cells (52.1%) and similar percentage of macrophages (1.55%) (Fig. 1A). The observation was reproducible in three experiments (in six subjects) (data not shown). There were statistically significant differences between PBMCs and MNCs in the proportions of T cells (76.1% ± 4.0% versus 35.2% ± 11.2%; P = 0.004) and B cells (10.0% ± 4.9% versus 55.6% ± 11.6%; P = 0.003) but not monocytes/macrophages (3.6% ± 2.6% versus 2.2% ± 1.2%; P = 0.420). In the same seven-color FACS analysis, we also examined the FcγRIs expression on blood monocytes (sample 322-L007) and splenic macrophages (sample OL3020). We first gated on CD14+ CD3− CD4+ CD20− monocytes/macrophages and then displayed the expression of FcγRI (CD64), FcγRII (CD32), and FcγRIII (CD16). While blood monocytes and splenic macrophages expressed similar levels of CD32 and CD64, a larger proportion of splenic macrophages than blood monocytes expressed CD16 (99.7% versus 3.83%) (Fig. 1B). Again, the contrast of expression levels among FcγR was reproducibly observed in three experiments (in six subjects) (data not shown).
FIG. 1.
Comparison of cell subsets between blood PBMCs and splenic MNCs. PBMCs and splenic MNCs were stained with cell subset makers and FcγR markers and compared using FACS analysis as described in Materials and Methods. (A) Distribution of T cells (CD3+), B cells (CD20+), and monocytes/macrophages (CD14+). (B) FcγRI (CD64), FcγRII (CD32), and FcγRIII (CD16) expression in blood PBMCs and splenic MNCs.
It is interesting that the mean fluorescent intensity (MFI) of CD16 expression in splenic macrophages was lower than that of the CD16+ blood monocytes but higher than the majority of CD16− monocytes. Although the explanation for this is not clear, it might be that subsets of blood monocytes have different fates after migrating to the spleen: the majority-CD16− monocytes acquire low levels of CD16 expression, and the minority-CD16+ monocytes differentiate into dendritic cells which become negative for CD16 surface expression (4, 36). The different compositions of T and B cells, as well as the contrasting patterns of expression among FcγR, highlighted the differences between blood monocytes and splenic macrophages. Additionally, because the adult spleen holds only 20 to 40 ml of blood (35), the likelihood that these splenic macrophages are contaminating blood monocytes is small.
Splenic macrophages, not T or B cells, appear to be permissive for DV infection.
Various hypotheses on the pathogenesis of DHF/DSS have placed emphasis on a pivotal role of monocytic phagocytes, T cells, or B cells. We believe that examining all these cell types under the same experimental conditions will provide new insight into DHF/DSS pathogenesis. In a previous study, we examined the relative permissiveness among different cell subsets in unfractionated human PBMCs by a FACS-based assay that allowed simultaneous assessment of multiple cell types in the same test tube (23). We now used the same approach to assess DV permissiveness among cell subsets in MNCs isolated from spleen.
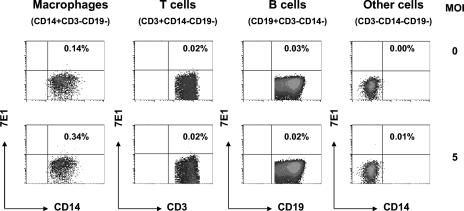
FACS analysis was applied to examine which cell types in human splenic MNCs are permissive to DV infection in the absence of “enhancing” antibodies. Figure 2 shows a representative result of this assay. MNCs were resurrected from the freezer and inoculated with DV2-16681 at an MOI of 5 or mock infected. Two days later, the cells were harvested and simultaneously stained with anti-E antibody (7E1) and antibodies to CD3, CD14, and CD19 as well as the corresponding isotype-matched immunoglobulin G control antibody. Results showed that among major cell types of interest, only resident macrophages (CD14+ CD3− CD19−) exhibited a slightly higher proportion of anti-E staining (0.34%) than the background control staining (0.14%). There was no evidence of DV infection in T cells (CD3+ CD14− CD19−), B cells (CD19+ CD3− CD14−), or other cells negative for T-cell, B-cell, and macrophage markers (CD3− CD14− CD19−).
FIG. 2.
Simultaneous assessment of DV permissiveness among cell subsets. Splenic MNCs were either uninfected (top panel) or infected with DV2-16681 at an MOI of 5 (bottom panel) and stained with anti-E antibody and antibodies to CD3, CD14, and CD19. Results show that only the resident macrophages (CD14+ CD3− CD19−) exhibited slightly higher anti-E staining (0.34%) than background staining (0.14%); T cells (CD3+ CD14− CD19−), B cells (CD19+ CD3− CD14−), and cells that were negative for T-cell, B-cell and monocyte/macrophage markers (CD3− CD14− CD19−) exhibited no significant increase in anti-E staining above background.
Splenic macrophages, not T or B cells, exhibit enhanced DV infection in the presence of highly diluted DV-immune human serum.
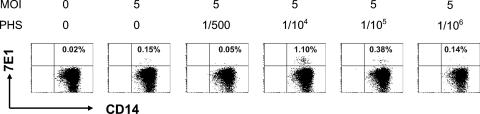
We showed previously that DV infection could be enhanced in primary monocytes, but not T and B cells, in the presence of DV-immune serum (23). To examine whether the same is true for splenic MNCs that consist of T and B cells, as well as resident macrophages, we inoculated MNCs with DV2-16681 at an MOI of 5 in the presence or absence of DV-immune PHS at a range of dilutions (1/500 to 1/106). The cells were incubated for 2 days and then assessed for infection, neutralization, and enhancement with labeled anti-E, as well as CD3, CD14, and CD19 antibodies that identify T cells, macrophages, and B cells, respectively. Since no infection of T and B cells was detectable, only the percentage of macrophages (CD14+ CD3− CD19−) infected by DV is presented. Results showed modest infection in the absence of PHS (0.15% versus 0.02%) but a sevenfold enhancement of infection in the presence of a 1/104 dilution of PHS (1.10% versus 0.15%) (Fig. 3). These results are in agreement with earlier findings that ADE of DV replication in monocytes/macrophages was optimal when subneutralizing antibody concentrations were used (14, 15, 20).
FIG. 3.
Antibody-dependent enhancement of DV infection in splenic macrophages, but not T or B cells. Splenic MNCs were infected by DV2-16681 at an MOI of 5 in the presence or absence of serially diluted PHS (dilutions of 1/500 to 1/106), incubated for 2 days, and then harvested and stained with labeled anti-E antibody as well as antibodies to T cells (CD3), B cells (CD19), or macrophages (CD14); cells were then analyzed using flow cytometry. No infection of T or B cells was detected under any condition. Results show percentages of anti-E antibody staining on macrophages (CD14+ CD3− CD19−) for each of the conditions.
Splenic macrophages support DV infection in the absence of DV-immune human serum.
Our previous work consistently demonstrated that blood monocytes were DV permissive even in the absence of PHS (23); it was less straightforward to show that splenic macrophage are DV permissive because the percentage of splenic macrophages positive for DV antigen staining was small (Fig. 3). To further examine whether splenic MNCs are DV permissive, we used a more sensitive RT-PCR method. Control experiments for the assay are shown in Fig. 4 and 5.
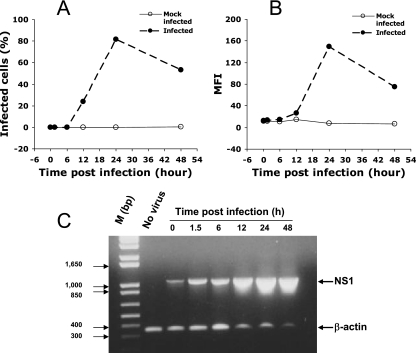
FIG. 4.
Association between detecting DV RNA and protein. Vero cells were either mock infected or infected by DV2-16681 (MOI of 5); samples were harvested at various time points. The proportion of infected cells (A) and the MFI of infected cells (B) were analyzed by intracellular anti-E antibody staining and FACS analysis. In parallel, viral RNA in infected cells was assessed by RT-PCR for the NS1 gene (C). Results show that infected cells were first detectable at 12 h and peaked at 24 h postinfection; in contrast, mock-infected cells changed little in staining over time (A and B). The relative amount of viral RNA in infected cells remained constant during the first 6 h of infection and increased dramatically from 12 to 48 h postinfection (C). As a control for input cellular RNA, β-actin levels were assessed in the same RT-PCR (C). M, molecular size markers.
FIG. 5.
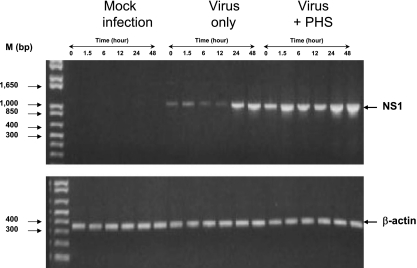
Kinetics of detecting DV gene expression in primary cells. (Top) PBMCs were either mock infected or infected by DV2-16681 (MOI of 5) alone or in the presence of a 1/104 dilution of PHS. Cells were washed and harvested at various times (0, 1.5, 6, 12, 24, and 48 h), and cellular RNAs were extracted for analysis. The presence of the DV NS1 gene product was detected using the RT-PCR assay. A 353-bp β-actin gene fragment was amplified in a separate reaction and used as a control for input cellular RNA. Results show a complete lack of NS1 signal in mock-infected cells and a progressive diminution of NS1 signal during the first 12 h after inoculation with DV alone and its augmentation after 24 to 48 h of infection. After infection by virus-PHS immune complexes, there is a persistence of NS1 during the first 12 h and a late increase NS1 signal at 24 to 48 h postinfection. (Bottom) The amounts of β-actin control are equal in all samples.
Vero cells were either mock infected or infected by DV2-16681 at an MOI of 5, and samples were harvested at various times. The proportion of infected cells (Fig. 4A) and the MFI of infected cells (Fig. 4B) were analyzed by intracellular anti-E antibody staining and FACS analysis. In parallel, the presence of viral RNA in infected cells was assessed by RT-PCR (Fig. 4C). Results show that infected cells were first detectable at 12 h and peaked at 24 h postinfection; mock-infected cells had little staining at all time points (Fig. 4A and B). The relative amount of viral NS1 gene product remained constant during the first 6 h of infection and increased dramatically from 12 to 48 h postinfection (Fig. 4C). A 353-bp β-actin gene fragment was also amplified in the same RT-PCR and was included as a control for input cellular RNA (Fig. 4C), the results of which indicate that the late increase in NS1 is not due to more input template RNA in RT-PCRs.
Next, the kinetics of DV infection was examined in PBMCs because of their easy availability and because their cellular composition is similar to that of splenic MNCs. PBMCs were either mock infected, infected by DV2-16681 (MOI of 5) only, or infected in the presence of a 1/104 dilution of PHS, which optimally enhanced DV infection in our previous studies (23) (Fig. 5). Cells were washed and harvested at various time points (0, 1.5, 6, 12, 24, and 48 h), and cellular RNA was extracted for analysis. The presence of the DV NS1 gene product was detected using the RT-PCR assay as described previously. The β-actin gene fragment was amplified in a separate RT-PCR and used as a control for input cellular RNA. Results show a complete lack of NS1 detection in mock-infected cells (Fig. 5, top), a progressive decline of NS1 signal during the first 12 h of infection with DV alone, and increased detection of NS1 signal after 24 to 48 h of infection (Fig. 5, top). The observation is consistent with published data showing that the amount of infectious virus produced into the culture supernatant of DV2-16681-infected human monocytes/macrophages peaks at 24 to 48 h postinfection (2). After infection by virus-PHS immune complexes, there were elevated levels of NS1 signal during the first 12 h of infection in comparison to the level of the virus-only group and a late increase in NS1 between 24 to 48 h postinfection (Fig. 5, top). The elevated NS1 level during the initial period of infection may be a reflection of increased attachment and uptake of virus-PHS immune complexes via FcRs expressed on monocytes/macrophages, while the late increase may be indicative of de novo viral replication. The amounts of β-actin control in these assays are equal in all samples (Fig. 5, bottom panel). Having determined the specificity and limitation of the RT-PCR assay for detecting DV infection using Vero cells and PBMCs, we next applied the method to examine the presence of viral RNA in splenic MNCs inoculated with DV.
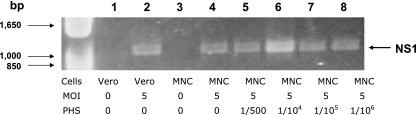
Splenic MNCs (sample PAQ028) were inoculated with DV2-16681 at an MOI of 5 in the presence or absence of various dilutions of PHS (1/500 and 1/106 dilutions). Two days later, cells were harvested, RNA was extracted, and RT-PCR was performed using primers for the NS1 gene to detect viral replication, as described in Materials and Methods. Results show that there were positive signals in all except the negative control samples in which no virus was added (Fig. 6, lanes 1 and 3). The results were reproduced using splenic MNCs from another subject. While the specific cell subset that produced the virus is uncertain from this experiment, some cells were clearly infected in the absence of PHS. It is likely that these cells are macrophages rather than T cells and/or B cells because the latter were nonpermissive even in the presence of PHS (see Table 2) (23).
FIG. 6.
Detection of DV in splenic MNCs in the absence of DV-immune human serum. Splenic MNCs were infected by DV2-16681 at an MOI of 5 in the presence or absence of serially diluted PHS for 2 days. Infection was examined using the RT-PCR assay as described in Materials and Methods. DV-infected or mock-infected Vero cells were used as positive and negative controls, respectively. Results show the detection of the DV NS1 gene in MNCs inoculated with DV alone or in the presence of a range of PHS dilutions (1/500 to 1/106).
TABLE 2.
Modification of DV permissiveness by PHS in splenic cells
| MOI | PHS (dilution) | na | Percentage of DV-positive cells in the indicated subset:
|
|||
|---|---|---|---|---|---|---|
| T cell | B cell | Macrophage | Other | |||
| 0 | 0 | 10 | 0.05 ± 0.09 | 0.08 ± 0.06 | 0.18 ± 0.12 | 0.10 ± 0.14 |
| 5 | 0 | 10 | 0.06 ± 0.13 | 0.09 ± 0.06 | 0.77 ± 1.00b | 0.11 ± 0.12 |
| 5 | 1/500 | 10 | 0.10 ± 0.22 | 0.10 ± 0.09 | 0.27 ± 0.31 | 0.11 ± 0.14 |
| 5 | 1/104 | 10 | 0.06 ± 0.08 | 0.15 ± 0.14 | 5.41 ± 3.53b | 0.18 ± 0.16 |
| 5 | 1/105 | 10 | 0.07 ± 0.14 | 0.11 ± 0.07 | 2.44 ± 1.85 | 0.11 ± 0.16 |
| 5 | 1/106 | 8 | 0.10 ± 0.18 | 0.11 ± 0.09 | 1.49 ± 1.43 | 0.13 ± 0.24 |
n, number of subjects.
For ADE versus viral infection only, P = 0.001. The peak infection value is shown in boldface.
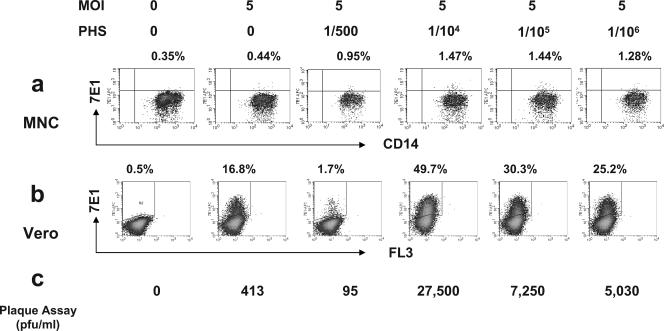
To further examine whether the small percentage of anti-E antibody-positive staining in splenic macrophages and positive NS1 signal in spleen cells inoculated with virus only are reflective of true infection or merely contamination of input viral RNA, we performed additional surrogate assays. Figure 7 shows representative results of three such experiments. MNCs were inoculated with DV2-16681 at an MOI of 5 in the presence or absence of PHS at different concentrations as described previously. Results show that 0.44% of cells were specific for E-antigen staining in the absence of PHS, but the infection was enhanced by threefold in the presence of a 1/104 dilution of PHS (Fig. 7a). Supernatant from each of these experimental conditions was harvested and used to infect Vero cells as described in Materials and Methods. The extent of Vero cell infection was assessed by FACS analysis and plaque assay. In the virus-only group, the percentage of Vero cells positive for anti-E staining was much higher than the control cells (16.8% versus 0.5%) (Fig. 7b). More importantly, we obtained 413 PFU/ml of infectious virus in the virus-only group, as well as a 67-fold enhancement of virus production (27,500 versus 413 PFU/ml) in the presence of a 1/104 dilution of PHS (Fig. 7c). Thus, we demonstrated a strong agreement in results obtained from all four types of assays: direct assessment of infection using FACS analysis or RT-PCR assays with splenic MNC or indirect measurement of infection by using a viral amplification assay in Vero cells and the traditional plaque assay in Vero cells with culture supernatants harvested from DV-infected splenic MNCs.
FIG. 7.
Verification of DV infection using surrogate assays. Splenic MNCs were infected by DV2-16681 at an MOI of 5 in the presence or absence of serially diluted PHS (dilutions of 1/500 to 1/106), incubated for 2 days, and then harvested and stained with labeled anti-E antibody as well as antibodies to T cells (CD3), B cells (CD19), or macrophages (CD14); cells were then analyzed using flow cytometry (a). Culture supernatants were harvested and used to infect Vero cells for three days, and viral infection was assessed using two assays: FACS assay with anti-E antibody staining (b) and plaque assay by immunostaining (c) as described in Materials and Methods. All three methods showed that splenic MNCs were permissive to DV even in the absence of PHS and that the infection was greatly enhanced in the presence of highly diluted PHS.
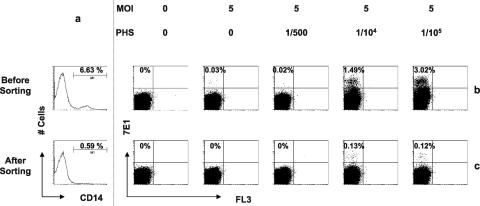
To more definitively establish that splenic macrophages were the principal infected cells that produced infectious virions detected by the surrogate plaque assay, we performed another type of experiment, the results of which are summarized in Fig. 8. Splenic MNCs (sample OL3026) depleted of CD14+ macrophages using magnetic beads were compared to unfractionated MNCs. The cells were inoculated with DV2-16681 at an MOI of 5 in the presence or absence of different dilutions of PHS (0, 1/500, 1/104, and 1/105). Supernatant from each of the experimental conditions was harvested 2 days later and inoculated onto Vero cells. After 3 days in culture, Vero cell infection was measured by staining with a labeled anti-E antibody and analyzed by FACS analysis. The CD14+ cell depletion procedure removed 90% of the macrophages from the MNCs (6.63% to 0.59%) (Fig. 8a) and enriched T-cell (21.1% to 24.8%) and B-cell (58.5% to 66.1%) populations (data not shown). The amount of virus produced from MNCs depleted of CD14+ cells was markedly reduced, and 90% fewer Vero cells were infected (1.49% to 0.13% at a 1/104 dilution of PHS and 3.02% to 0.12% at a 1/105 dilution of PHS) (Fig. 8b and c). Collectively, these data (Fig. 4, 5b and c, and 6) strongly suggest that splenic macrophages, not T and B cells, are permissive to DV and support DV replication.
FIG. 8.
Splenic macrophages are a major source of DV production. Splenic MNCs (sample OL3026) were either depleted of CD14+ macrophages or not as described in our previous study (23) and then inoculated with DV2-16681 at an MOI of 5 in the presence or absence of different dilutions of PHS (0, 1/500, 1/104, and 1/105). Supernatant from each of the experimental conditions was harvested 2 days postinfection and used to inoculate Vero cells. After 2 days, Vero cell infection was assessed by staining with labeled anti-E antibody and then analyzed by FACS analysis. Results show the efficiency of CD14 depletion (a) and Vero cell infection with supernatant harvested from unfractionated MNCs (b) or CD14-depleted MNCs (c) under various experimental conditions.
Only splenic macrophages, not B or T cells, were DV-permissive and capable of mediating ADE of DV infection.
To confirm that splenic macrophages were the most permissive target cells for direct DV infection, we repeated the DV infection experiment described in the legend of Fig. 3 in nine additional subjects. Results from all 10 subjects showed a fourfold increase in the average levels of anti-E staining in DV-infected macrophages compared to control mock-infected cells (0.77% ± 1.00% versus 0.18% ± 0.12%; P = 0.07, paired Student t test). The normal subject variability, however, rendered this difference statistically insignificant. In contrast, the levels of anti-E staining were similar with or without DV inoculation in T cells (0.10% ± 0.18% versus 0.06% ± 0.13%) and B cells (0.15% ± 0.14% versus 0.09% ± 0.06%), as well as cells that were negative for T-cell, B-cell, and monocyte markers (0.18% ± 0.16% versus 0.11% ± 0.12%) (Table 1).
TABLE 1.
Differential DV permissiveness in subsets of human spleen cells
| Treatment | Percentage of DV-positive cells in the indicated subset:
|
|||
|---|---|---|---|---|
| T cell | B cell | Macrophages | Other | |
| Mock infection | 0.05 ± 0.09 | 0.07 ± 0.05 | 0.18 ± 0.12a | 0.10 ± 0.14 |
| Virus infection | 0.06 ± 0.13 | 0.08 ± 0.06 | 0.77 ± 1.00a | 0.11 ± 0.12 |
For the values of splenic macrophages, P = 0.07.
To verify that only primary macrophages, not T and B cells, support enhanced DV infection in the presence of DV-immune serum, we assessed DV infection in splenic MNCs isolated from 10 subjects with DV2-16681 at an MOI of 5 in the presence or absence of a range of PHS dilutions (for 1/500 to 1/105 in two subjects and 1/500 to 1/106 in eight subjects). Results show that among the splenic macrophages, there was an apparent DV infection in the absence of PHS (0.77% ± 1.00% versus 0.18% ± 0.12%; P = 0.07), neutralization in the presence of PHS at a 1/500 dilution, and definitive enhancement of DV infection over a range of PHS dilutions (1/104 to 1/106), which peaked at the 1/104 PHS dilution (5.41% ± 3.53% versus 0.77% ± 1.00%; P = 0.001). In contrast, no infection of either T cells (0.10% ± 0.18% versus 0.06% ± 0.13%) or B cells (0.15% ± 0.14% versus 0.09% ± 0.06%) was detected under any of the experimental conditions. In addition, there was no infection in cell subsets that were negative for T-cell, B-cell, or macrophage markers (peak of 0.18% ± 0.16% versus 0.11% ± 0.12% of background staining) (Table 2).
DISCUSSION
DHF and DSS affect multiple organs, including liver, spleen, kidney, lung, and bone marrow (16). The precise identity of DV-permissive cells in tissues and organs, however, is still largely unknown. Halstead had speculated 25 years ago that “the only site of replication of dengue virus in man is cells of mononuclear phagocyte lineage” (7). Due to the lack of sophisticated experimental tools, though, this speculation had never been conclusively tested. We have previously established an assay that simultaneously identified DV-infected cells and their phenotypes among infected cells in unfractionated human PBMCs and found that only primary monocytes were permissive to DV (23). We now provide additional experimental support for the Halstead speculation. Our current study shows that, under identical experimental conditions in vitro, (i) splenic resident macrophages were modestly permissive to DV infection and exhibited significantly enhanced infection in the presence of highly diluted DV-immune human serum, and (ii) primary splenic T and B cells were not permissive to DV in either the presence or absence of DV-immune human serum.
To our knowledge, this is the first time that the permissiveness to DV infection and ability to mediate ADE were examined in unfractionated primary human splenic MNCs ex vivo. The experimental conditions mimic more closely the in vivo environment where these cell types may encounter DV and DV-immune complexes in a common milieu. Consequently, findings from this study may be more pertinent to the interpretation of DHF/DSS pathogenesis. In a recent study using a large number of blood and tissue samples obtained from DV-infected patients, DV antigens were detected with immunohistochemistry techniques in multiple tissues and organs including liver, spleen, kidney, lung, bone marrow, and blood clots. However, DV RNA, a surrogate of viral replication, was detected with an in situ hybridization technique only in splenic macrophages and monocytes of blood clots (16). Our current results demonstrating that splenic MNCs are permissive to DV infection and support DV replication offer a specific cellular basis to explain the observations made using biopsy and autopsy tissue specimens (16). Some interesting questions that warrant further study are (i) whether macrophages could be infected within the spleen and (ii) whether the infected splenic macrophages are merely an archive of monocytes that were infected in the blood, arrived to the spleen, and differentiated into splenic macrophages. Although there are no data on the precise route of DV spread in humans, in monkeys experimentally infected by DV, the virus was found in regional lymph nodes within a day and later in various tissue and organs, including the spleen (9).
Neither our previous study using PBMCs (23) nor the current study using splenic MNCs found any evidence of direct DV infection of T and B cells in vitro, suggesting that neither cell type contributes much to overall DV production in vivo. However, our results do not exclude a role for T and B cells as responder cells in the pathogenesis of DHF/DSS. In future studies, it would be important to examine whether cytokines produced by infected monocytes/macrophages and/or activated T and B cells are the culprits that induce endothelial damage prior to bleeding diathesis and plasma leakage during DHF/DSS.
We believe that our results, in addition to providing clarification on the relative permissiveness to DV infection and capacity for mediating ADE among cell subsets in human splenic MNCs, also allow for the formulation of new hypotheses. Splenic rupture and hepatosplanchnic circulatory dysfunction have occasionally been reported in patients who succumb to DHF/DSS (17, 32). Based on our presented data, it is tempting to postulate that this is caused by a sequence of events. First, preexisting anti-DV antibodies form infectious immune complexes with a heterotypic DV during a secondary infection. Such immune complexes are engulfed by resident splenic macrophages through FcγRs. Second, while attempting to destroy virus-antibody immune complexes, abundant viral antigens are presented on the surface of the macrophage, which activates T cells to produce a variety of cytokines. Lastly, some of the cytokines modulate endothelial linings to promote plasma leakage and bleeding, which cause the enlargement of the spleen. Whether such a hypothesis is true has yet to be tested.
In summary, results from the current study and our previous study (23) provide strong support to the Halstead speculation made 25 years ago that cells of mononuclear phagocyte lineage are the only site of DV replication in humans. The evidence provides additional experimental support to the ADE hypothesis of the pathogenesis of DHF/DSS and argues against the notion of T and B cells as major reservoirs for DV replication.
Acknowledgments
This study was supported in part by Pediatric Dengue Vaccine Initiative of the International Vaccine Institute, awards TR16 (X.J.) and TR03/04 (J.J.S).
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Avirutnan, P., P. Malasit, B. Seliger, S. Bhakdi, and M. Husmann. 1998. Dengue virus infection of human endothelial cells leads to chemokine production, complement activation, and apoptosis. J. Immunol. 161:6338-6346. [PubMed] [Google Scholar]
- 2.Chen, Y. C., and S. Y. Wang. 2002. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J. Virol. 76:9877-9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamond, M. S., D. Edgil, T. G. Roberts, B. Lu, and E. Harris. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon, S., and P. R. Taylor. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953-964. [DOI] [PubMed] [Google Scholar]
- 5.Green, S., and A. Rothman. 2006. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr. Opin. Infect. Dis. 19:429-436. [DOI] [PubMed] [Google Scholar]
- 6.Hahn, A. B., G. A. Land, and R. M. Strothman (ed.). 2000. American Society for Histocompatibility and Immunogenetics laboratory manual, 4th ed., vol. 1. American Society for Histocompatibility and Immunogenetics, New York, NY.
- 7.Halstead, S. B. 1981. The Alexander D. Langmuir Lecture. The pathogenesis of dengue. Molecular epidemiology in infectious disease. Am. J. Epidemiol. 114:632-648. [DOI] [PubMed] [Google Scholar]
- 8.Halstead, S. B. 1989. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev. Infect. Dis. 11(Suppl. 4):S830—S839. [DOI] [PubMed] [Google Scholar]
- 9.Halstead, S. B. 1980. Immunological parameters of togavirus disease syndromes, p. 107-173. In R. W. Schlesinger (ed.), The togaviruses: biology, structure, replication. Academic Press, New York, NY.
- 10.Halstead, S. B. 1970. Observations related to pathogenesis of dengue hemorrhagic fever. VI. Hypotheses and discussion. Yale J. Biol. Med. 42:350-362. [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead, S. B. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 12.Halstead, S. B., F. X. Heinz, A. D. Barrett, and J. T. Roehrig. 2005. Dengue virus: molecular basis of cell entry and pathogenesis, 25-27 June 2003, Vienna, Austria. Vaccine 23:849-856. [DOI] [PubMed] [Google Scholar]
- 13.Halstead, S. B., and E. J. O'Rourke. 1977. Antibody-enhanced dengue virus infection in primate leukocytes. Nature 265:739-741. [DOI] [PubMed] [Google Scholar]
- 14.Halstead, S. B., and E. J. O'Rourke. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 146:201-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halstead, S. B., E. J. O'Rourke, and A. C. Allison. 1977. Dengue viruses and mononuclear phagocytes. II. Identity of blood and tissue leukocytes supporting in vitro infection. J. Exp. Med. 146:218-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jessie, K., M. Y. Fong, S. Devi, S. K. Lam, and K. T. Wong. 2004. Localization of dengue virus in naturally infected human tissues, by immunohistochemistry and in situ hybridization. J. Infect. Dis. 189:1411-1418. [DOI] [PubMed] [Google Scholar]
- 17.Khongphatthanayothin, A., P. Lertsapcharoen, P. Supachokchaiwattana, P. Satupan, K. Thongchaiprasit, Y. Poovorawan, and C. Thisyakorn. 2005. Hepatosplanchnic circulatory dysfunction in acute hepatic infection: the case of dengue hemorrhagic fever. Shock 24:407-411. [DOI] [PubMed] [Google Scholar]
- 18.King, A. D., A. Nisalak, S. Kalayanrooj, K. S. Myint, K. Pattanapanyasat, S. Nimmannitya, and B. L. Innis. 1999. B cells are the principal circulating mononuclear cells infected by dengue virus. Southeast Asian J. Trop. Med. Public Health 30:718-728. [PubMed] [Google Scholar]
- 19.Kliks, S. 1990. Antibody-enhanced infection of monocytes as the pathogenetic mechanism for severe dengue illness. AIDS Res. Hum. Retrovir. 6:993-998. [DOI] [PubMed] [Google Scholar]
- 20.Kliks, S. C., S. Nimmanitya, A. Nisalak, and D. S. Burke. 1988. Evidence that maternal dengue antibodies are important in the development of dengue hemorrhagic fever in infants. Am. J. Trop. Med. Hyg. 38:411-419. [DOI] [PubMed] [Google Scholar]
- 21.Kliks, S. C., A. Nisalak, W. E. Brandt, L. Wahl, and D. S. Burke. 1989. Antibody-dependent enhancement of dengue virus growth in human monocytes as a risk factor for dengue hemorrhagic fever. Am. J. Trop. Med. Hyg. 40:444-451. [DOI] [PubMed] [Google Scholar]
- 22.Kontny, U., I. Kurane, and F. A. Ennis. 1988. Gamma interferon augments Fc gamma receptor-mediated dengue virus infection of human monocytic cells. J. Virol. 62:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kou, Z., M. Quinn, H. Chen, W. W. S. I. Rodrigo, R. C. Rose, J. J. Schlesinger, and X. Jin. Monocytes, but not T or B cells, are the principal target cells for dengue virus infection among human peripheral blood mononuclear cells. J. Med. Virol., in press. [DOI] [PubMed]
- 24.Kyle, J. L., P. R. Beatty, and E. Harris. 2007. Dengue virus infects macrophages and dendritic cells in a mouse model of infection. J. Infect. Dis. 195:1808-1817. [DOI] [PubMed] [Google Scholar]
- 25.Lin, Y. W., K. J. Wang, H. Y. Lei, Y. S. Lin, T. M. Yeh, H. S. Liu, C. C. Liu, and S. H. Chen. 2002. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J. Virol. 76:12242-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Littaua, R., I. Kurane, and F. A. Ennis. 1990. Human IgG Fc receptor II mediates antibody-dependent enhancement of dengue virus infection. J. Immunol. 144:3183-3186. [PubMed] [Google Scholar]
- 27.Mackenzie, J. S., D. J. Gubler, and L. R. Petersen. 2004. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat. Med. 10:S98-109. [DOI] [PubMed] [Google Scholar]
- 28.Mangada, M. M., T. P. Endy, A. Nisalak, S. Chunsuttiwat, D. W. Vaughn, D. H. Libraty, S. Green, F. A. Ennis, and A. L. Rothman. 2002. Dengue-specific T cell responses in peripheral blood mononuclear cells obtained prior to secondary dengue virus infections in Thai schoolchildren. J. Infect. Dis. 185:1697-1703. [DOI] [PubMed] [Google Scholar]
- 29.Mangada, M. M., and A. L. Rothman. 2005. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J. Immunol. 175:2676-2683. [DOI] [PubMed] [Google Scholar]
- 30.Marianneau, P., A. M. Steffan, C. Royer, M. T. Drouet, D. Jaeck, A. Kirn, and V. Deubel. 1999. Infection of primary cultures of human Kupffer cells by Dengue virus: no viral progeny synthesis, but cytokine production is evident. J. Virol. 73:5201-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mentor, N. A., and I. Kurane. 1997. Dengue virus infection of human T lymphocytes. Acta Virol. 41:175-176. [PubMed] [Google Scholar]
- 32.Miranda, L. E., S. J. Miranda, and M. Rolland. 2003. Case report: spontaneous rupture of the spleen due to dengue fever. Braz. J. Infect. Dis. 7:423-425. [DOI] [PubMed] [Google Scholar]
- 33.Mongkolsapaya, J., W. Dejnirattisai, X. N. Xu, S. Vasanawathana, N. Tangthawornchaikul, A. Chairunsri, S. Sawasdivorn, T. Duangchinda, T. Dong, S. Rowland-Jones, P. T. Yenchitsomanus, A. McMichael, P. Malasit, and G. Screaton. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 9:921-927. [DOI] [PubMed] [Google Scholar]
- 34.Mongkolsapaya, J., T. Duangchinda, W. Dejnirattisai, S. Vasanawathana, P. Avirutnan, A. Jairungsri, N. Khemnu, N. Tangthawornchaikul, P. Chotiyarnwong, K. Sae-Jang, M. Koch, Y. Jones, A. McMichael, X. Xu, P. Malasit, and G. Screaton. 2006. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J. Immunol. 176:3821-3829. [DOI] [PubMed] [Google Scholar]
- 35.Neiman, R. S., and A. Orazi. 1999. Disorders of the spleen, 2nd ed., vol. 38. W. B. Saunders Co., Philadelphia, PA.
- 36.Newman, K. C., and E. M. Riley. 2007. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat. Rev. Immunol. 7:279-291. [DOI] [PubMed] [Google Scholar]
- 37.Porterfield, J. S. 1980. Antigenic characteristics and classification of Togaviridiae, p. 13-42. In R. W. Schlesinger (ed.), The togaviruses: biology, structure, replication. Academic Press, New York, NY.
- 38.Rodrigo, W. W. S. I., X. Jin, S. D. Blackley, R. C. Rose, and J. J. Schlesinger. 2006. Differential enhancement of dengue virus immune complex infectivity mediated by signaling-competent and signaling-incompetent human FcγRIA (CD64) or FcγRIIA (CD32). J. Virol. 80:10128-10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rothman, A. L. 2004. Dengue: defining protective versus pathologic immunity. J. Clin. Investig. 113:946-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, P. K., and A. Nisalak. 1967. Dengue virus identification by the plaque reduction neutralization test. J. Immunol. 99:291-296. [PubMed] [Google Scholar]
- 41.Theofilopoulos, A. N., W. E. Brandt, P. K. Russell, and F. T. Dixon. 1976. Replication of dengue-2 virus in cultured human lymphoblastoid cells and subpopulations of human peripheral leukocytes. J. Immunol. 117:953-961. [PubMed] [Google Scholar]
- 42.Thepparit, C., and D. R. Smith. 2004. Serotype-specific entry of dengue virus into liver cells: identification of the 37-kilodalton/67-kilodalton high-affinity laminin receptor as a dengue virus serotype 1 receptor. J. Virol. 78:12647-12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welsh, R. M., and A. L. Rothman. 2003. Dengue immune response: low affinity, high febrility. Nat. Med. 9:820-822. [DOI] [PubMed] [Google Scholar]
- 44.Wilder-Smith, A., and E. Schwartz. 2005. Dengue in travelers. N. Engl. J. Med. 353:924-932. [DOI] [PubMed] [Google Scholar]
- 45.Wu, S. J., G. Grouard-Vogel, W. Sun, J. R. Mascola, E. Brachtel, R. Putvatana, M. K. Louder, L. Filgueira, M. A. Marovich, H. K. Wong, A. Blauvelt, G. S. Murphy, M. L. Robb, B. L. Innes, D. L. Birx, C. G. Hayes, and S. S. Frankel. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816-820. [DOI] [PubMed] [Google Scholar]