Abstract
UL9, a superfamily II helicase, is a multifunctional protein required for herpes simplex virus type 1 replication in vivo. Although the C-terminal 317-amino-acid DNA binding domain of UL9 exists as a monomer, the full-length protein behaves as a dimer in solution. Thus, it has been assumed that the N-terminal 534 residues contain a region necessary for efficient dimerization and that UL9 dimers are in a head-to-head configuration. We recently showed, however, that residues in the N terminus could modulate the inhibitory properties of UL9 by decreasing the DNA binding ability of the C terminus (S. Chattopadhyay and S. K. Weller, J. Virol. 80:4491-4500, 2006). We suggested that a direct interaction between the N- and C-terminal portions of UL9 might exist and serve to modulate the DNA binding activities of the C terminus. In this study, we used a coimmunoprecipitation assay to show that the N-terminal portion of UL9 can indeed directly interact with the C terminus. A series of truncation mutant proteins were used to show that a region in the N terminus between residues 293 and 321 is necessary for efficient interaction. Similarly, a region in the C terminus between residues 600 and 800 is required for this interaction. The simplest model to explain these data is that UL9 dimers are oriented in a head-to-tail arrangement in which the N terminus is in contact with the C terminus.
Herpes simplex virus type 1 (HSV-1) encodes seven proteins that are essential for the replication of its 152-kb double-stranded DNA genome. They include the origin binding protein (UL9), the single-stranded DNA binding protein (UL29/ICP8), the heterotrimeric helicase-primase complex (UL5/UL8/UL52), and the viral polymerase (UL30) and its processivity factor (UL42) (reviewed in references 8 and 29).
UL9, a member of superfamily II of helicases, is an 851-residue-long multifunctional protein that can bind specifically and cooperatively to the HSV-1 origins of replication; in addition, it exhibits ATPase and limited helicase activities (6, 11, 16). UL9 forms stable dimers in solution and interacts with several other viral and cellular proteins (reviewed in reference 29). Although UL9 is required for viral DNA replication in vivo (7) and it is assumed that it functions during initiation, many questions remain unanswered about its mechanism of action and the regulation of its activities.
The UL9 protein is organized into two distinct functional domains, the N-terminal two-thirds (amino acids [aa] 1 to 534) and the C-terminal one-third (aa 535 to 851). The N-terminal portion of UL9 contains seven conserved helicase motifs shared by all of the members of superfamily II of helicases, contains the interaction domain for two viral replication proteins (UL8 and UL42) (30, 31), and is responsible for dimerization and cooperative binding (12, 17). On the other hand, the C-terminal one-third contains the DNA binding domain, a nuclear localization signal, and residues required for interaction with the single-stranded DNA binding protein UL29 (ICP8) (1, 4, 5, 10, 25).
The C-terminal DNA binding domain forms a monomer in solution (12); however, the full-length protein exists as a dimer under these conditions (6, 16). It was thus assumed that the dimerization domain is located within the N-terminal portion. Over the last 10 years, several attempts to map residues important for dimerization have been initiated. Residues 147 to 171 in the N terminus of UL9 contain a putative leucine zipper similar to those associated with protein-protein interaction domains (19, 20). A mutant protein containing two leucine-to-valine substitutions was shown to sediment as a dimer by velocity sedimentation, indicating that the putative leucine zipper is not involved in dimerization (26). In addition, mutations in the conserved residues of UL9 helicase motifs I to VI were engineered and analyzed by gel filtration chromatography to examine their oligomeric status. All of the helicase motif mutants except IV (which aggregated) were found to be able to form dimers, as well as the wild-type UL9 protein (28). In another attempt, immunofluorescence (IF) microscopy was used to show that C-terminally truncated versions of UL9 can form heterodimers with full-length UL9 and thus be taken into the nucleus because of the nuclear localization signal located at the extreme C terminus of full-length UL9 (25). A fragment containing residues 1 to 321 could be transported to the nucleus by full-length UL9, suggesting that at least one region involved in dimerization lies within the first 321 aa (27).
Studies with temperature-sensitive mutant forms of UL9 indicate that it is required only in the first 6 h of lytic infection (3, 35). Several lines of evidence, in fact, indicate that UL9 can be inhibitory to HSV-1 infection. Overexpression of wild-type UL9 is inhibitory in a plaque reduction assay (24, 33, 37); cell lines containing a high copy number of the wild-type UL9 gene do not efficiently complement a null mutant and appear to inhibit wild-type infection (24); furthermore, UL9 can inhibit replication in insect cells expressing HSV replication proteins (36). On the basis of these observations, we have suggested that HSV replication occurs in two stages. The early stages of DNA replication are UL9 dependent; however, later in infection DNA replication switches to a UL9-independent mode involving recombination, rolling-circle replication, or both (29, 39). According to this model, it may be necessary for HSV-1 to regulate either UL9 levels or activities during infection to avoid inhibition. We have proposed that during the transition between the two stages, UL9 is removed from the origin of replication so that it will not be a physical barrier for the progressing replication fork. We speculate that if excess UL9 is present and bound to viral DNA, origin clearance may be delayed or inhibited, resulting in lower overall viral yields. In support of this model, we recently reported that residues in the N-terminal region of UL9 can regulate the inhibitory properties of UL9 by modulating the DNA binding ability of the C terminus (9). The ability of residues within the N terminus of UL9 to modulate the DNA binding domain in the C terminus may be a result of physical interaction between these two apparently independent functional domains of UL9. In this study, we constructed a series of N- and C-terminally truncated proteins and used them in coimmunoprecipitation assays to confirm that there is indeed a direct interaction between the N- and C-terminal portions of UL9. We also mapped two interaction domains, one in the N terminus and another in the C terminus. The simplest explanation for these results is that this interaction results in the formation of a head-to-tail dimer in which the N terminus of one monomeric unit is in contact with the C terminus of another. This is in contrast to the previous assumption that residues in the N terminus are in direct contact with each other to form UL9 dimers.
MATERIALS AND METHODS
Cells and antibodies.
African green monkey kidney fibroblasts (Vero cells) were purchased from the American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) with 5% fetal bovine serum, penicillin, and streptomycin (Invitrogen). Four UL9 antibodies were used in this study. Monoclonal antibody 17B is directed against the N-terminal 33 aa of UL9 (25), the polyclonal rabbit RH-7 antibody (a kind gift from D. Tenney, Bristol Myers Squib, Wallingford, CT) was raised against a fusion protein in which glutathione S-transferase was fused to the C-terminal domain of UL9 (residues 535 to 851), rabbit polyclonal antibody R249 is directed against the C-terminal decapeptide of UL9 (generously provided by Mark Challberg, NIH, Bethesda, MD), and KST-1 is a polyclonal antibody directed against the full-length UL9 protein (kindly provided by Deborah S. Parris, Ohio State University, Columbus). The six-His antibody was purchased from BD Biosciences (San Diego, CA).
Plasmids.
Plasmids pCDNA3-UL9-WT, UL9-1-450, UL9-1-354, UL9-1-321, and UL9-C were reported previously (9, 27). The UL9 mutant plasmids used in this study and the PCR primers used for generating these plasmids are listed in Tables 1 and 2, respectively. The pCDNA3-UL9-WT plasmid was used as a template in PCRs for the generation of plasmids UL9-1-292, UL9-1-236, UL9-236-534-His, UL9-His-600-851, and UL9-His-535-800. The fragments used to generate the UL9-1-292 and UL9-1-236 mutant forms were made by step-down PCR with UL9-start-BamHI as the downstream primer and oligonucleotide A or B as the upstream primer, respectively. Plasmid UL9-236-534-His was generated with oligonucleotide C as the downstream primer and oligonucleotide VII as the upstream primer (9). Plasmid UL9-His-600-851 was generated with oligonucleotide D as the downstream primer and UL9-end-EcoRI as the upstream primer (9). Plasmid UL9-His-535-800 was constructed with oligonucleotide III as the downstream primer and oligonucleotide E as the upstream primer (9). All of the plasmids described above were created by inserting the BamHI- and EcoRI-digested PCR fragments into the pCDNA3 vector. Plasmid UL9-N was generated with UL9-start-BamHI as the downstream primer and oligonucleotide F as the upstream primer. The baculovirus transfer vector of UL9-N was created by inserting a BamHI- and EcoRI-digested PCR fragment into the pFastBac vector. The UL9-C-His was subcloned into the pFastBac vector as a BamHI- and EcoRI-digested fragment to generate a baculovirus transfer vector.
TABLE 1.
UL9 plasmids used in this study
| Plasmid name | UL9 residues (amino acids) | Oligonucleotide used for construction |
|---|---|---|
| UL9-WT | 1-851 | |
| UL9-His-535-851(CT) | 535-851 | |
| UL9-1-450 | 1-450 | |
| UL9-1-354-MV | 1-354 | |
| UL9-1-321 | 1-321 | |
| UL9-1-292 | 1-292 | A |
| UL9-1-236 | 1-236 | B |
| UL9-236-534-His | 236-534 | C |
| UL9-His-600-851 | 600-851 | D |
| UL9-His-535-800 | 535-800 | E |
| UL9-N | 1-534 | F |
TABLE 2.
Oligonucleotides used in PCRs to generate UL9 mutant constructs
| Oligonucleotide name | Sequence (5′ to 3′) |
|---|---|
| A | GCCGAGGTGAATTCTTAGCGCGCTTCCAGCTC |
| B | GCCGAGGTGAATTCTTAGACCACCACATGCACGTT |
| C | GCCGAGGTGGATCCATGGCTGGCGAGTACGCC |
| D | GGTGGATCCATGCATCATCATCATCATCATCGCAGCAGCGCCATA |
| E | GGTGAATTCTTAGCGCGCGCCCGCAAACTCCCG |
| F | GGTGAATTCTTACCGGCACCGCAGCTC |
Transient transfection and Western blot analysis.
Transient transfection and Western blot analysis were performed as previously described (9). In brief, subconfluent monolayers of Vero cells in 60-mm2 dishes were transfected with 2 μg of plasmid DNA containing wild-type or mutant versions of the UL9 gene with the Lipofectamine Plus reagent (Invitrogen, CA) according to the manufacturer's instructions. After 20 h, concentrated cell lysates were prepared in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer, subjected to electrophoresis in 10% SDS-polyacrylamide gels, and transferred onto enhanced chemiluminescence membranes (Amersham Biosciences, Freiburg, Germany). Membranes were incubated with primary and alkaline phosphatase-conjugated secondary antibodies and developed with the color detection reagents nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate (BCIP) according to the manufacturer's (Promega, Madison, WI) instructions.
Immunoprecipitation.
Confluent Vero cells grown in 60-mm2 dishes were transfected with plasmids of interest and allowed to incubate at 37°C for 18 h. Cells were collected after centrifugation at 4,000 rpm and lysed on ice for 1 h in 500 μl RIPA buffer (300 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0], complete protease inhibitor [Roche]). The cleared supernatant was collected after whole-cell lysates were subjected to centrifugation at 10,000 rpm for 10 min at 4°C. Each cell lysate was precleared with normal rabbit immunoglobulin G. For each immunoprecipitation reaction, 500 μl (approximately 500 μg of total protein) of transfected cell lysate was incubated with 7 μl of primary antibody RH-7 for 2 h at 4°C. Forty microliters of a protein A-agarose slurry (Santa Cruz Biotech, Santa Barbara, CA) was added to each reaction mixture, which was then incubated overnight at 4°C. The beads were collected by centrifugation at 2,500 rpm for 5 min and washed four times with RIPA buffer. Finally, 45 μl of phosphate-buffered saline (PBS) and 15 μl of 4× SDS-polyacrylamide gel electrophoresis buffer were added and samples were boiled for 5 min. Immunoprecipitated proteins were subjected to Western blot analysis by probing with the 17B antibody.
IF microscopy.
Vero cells were grown on glass coverslips and transfected as described above. At 14 h posttransfection, coverslips were washed twice with cold 1× PBS. IF microscopy was performed as previously described (9). Fluorescence microscopic images were acquired with an Olympus BX-60 microscope with a 100× objective, and images were captured with a cooled charge-coupled device camera (Hammamatsu Corp., Boston, MA). Open Lab image software (Improvision, Inc., Lexington, MA) was used to analyze the images, and Adobe Photoshop 7.0 was used to generate the composite figures.
Protein expression by recombinant baculoviruses and pull-down assay with nickel magnetic beads.
Recombinant baculoviruses expressing the N terminus (1 to 534 aa) and the His-tagged C terminus (535 to 851 aa) of UL9 were generated with pFastBac (Life Technologies, Inc.) commercial systems according to the manufacturer's instructions. The baculoviruses were tested for optimal expression of recombinant proteins in SF9 insect cells.
SF9 cells at a density of 106/ml were coinfected with UL9-N and UL9-C-His recombinant baculoviruses and harvested at 48 h postinfection. The cell pellet was washed twice with PBS and resuspended in 500 μl of lysis buffer (20 mM Tris [pH 8.0], 5% glycerol, 400 mM NaCl, 0.1% NP-40, 1.5 mM MgCl2, protease inhibitor cocktail [Sigma]). The cells were placed on ice for 15 min, lysed with a Dounce homogenizer, and centrifuged at 10,000 rpm for 10 min. The supernatant was then mixed with 15 μl of Ni-nitrilotriacetic acid magnetic agarose beads (QIAGEN GmbH, Hilden, Germany) for 1 h at 4°C. The beads were washed three times with 500 μl of wash buffer (lysis buffer containing 30 mM imidazole), and proteins were eluted with 50 μl of elution buffer (lysis buffer containing 200 mM imidazole). The eluted proteins were analyzed by Western blotting with KST-1 antibody, which is directed against the full-length UL9 protein.
RESULTS
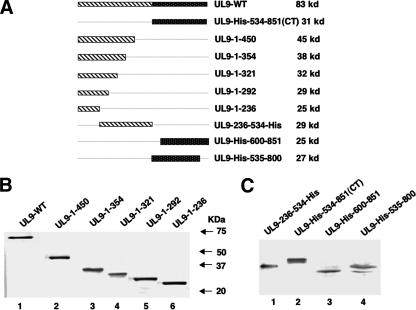
We recently reported that residues in the N terminus of UL9 could modulate the DNA binding ability of the C terminus in a context-dependent manner (9). To understand the mechanism of this regulation, we asked whether a direct interaction between these two domains could be detected. A series of N- and C-terminally truncated forms of UL9 were engineered into the pCDNA3 vector under the control of the cytomegalovirus promoter (Fig. 1A). To determine whether each of these truncated versions of the protein can be stably expressed, Vero cells were transfected with mutant plasmids and cell lysates were analyzed by Western blot analysis as described in Materials and Methods. The C-terminally truncated forms were detected by the 17B antibody, which recognizes the N-terminal 33 aa of UL9 (Fig. 1B); the N-terminally truncated forms and UL9-236-534-His were detected by the six-His monoclonal antibody (Fig. 1C). All of the N- and C-terminally truncated forms expressed mutant UL9 protein fragments of the expected sizes.
FIG. 1.
Expression of truncated UL9 proteins in Vero cells. (A) Schematic diagrams of the truncated forms of UL9 used in this study. The N-terminal domain of UL9 is depicted as a striped box, and the C-terminal of domain is depicted as a black dotted box. The size of the protein synthesized from each truncated form is indicated at the right. (B) The 17B antibody, directed against the N-terminal 33 aa of UL9, was used to detect C-terminally truncated UL9 proteins in concentrated cell lysates from Vero cells transfected with the plasmids as mentioned above. (C) The six-His monoclonal antibody was used to detect the N-terminally truncated UL9 proteins, as well as the UL9-236-534-His fragment, in concentrated Vero cell lysates.
Detection of heterodimer formation by coimmunoprecipitation in Vero cells.
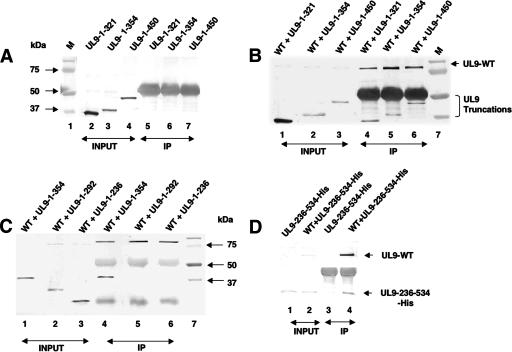
In order to determine whether the C-terminally truncated forms of UL9 were able to interact with full-length UL9, Vero cells were cotransfected with plasmids of interest and lysed in RIPA buffer 18 h after transfection. Coimmunoprecipitation was performed as described in Materials and Methods, with the RH-7 antibody raised against the C-terminal 317 aa residues of UL9, and the resulting precipitates were subjected to Western blot analysis. RH-7 would be expected to precipitate only the full-length UL9 protein and not the truncated versions lacking the C terminus. The 17B monoclonal antibody, which reacts with the N-terminal 33 aa of UL9, was used as a probe to detect the full-length sequence, as well as C-terminally truncated forms, in a Western blot analysis. The C-terminally truncated forms UL9-1-321, UL9-1-354, and UL9-1-450 were detected in cell lysates (Fig. 2A, lanes 2 to 4) but were not precipitated by the RH-7 antibody (Fig. 2A, lanes 5 to 7) or by protein A-agarose beads alone (data not shown). On the other hand, the C-terminally truncated forms UL9-1-321, UL9-1-354, and UL9-1-450 could be immunoprecipitated with the full-length UL9 protein (Fig. 2B, lanes 4 to 6). These results suggest that there is a domain within the first 321 aa which can interact with full-length UL9.
FIG. 2.
Detection of heterodimer formation by coimmunoprecipitation in Vero cells. Confluent Vero cells grown in 60-mm2 dishes were cotransfected with plasmids of interest and allowed to incubate at 37°C for 18 h. Cells were lysed in RIPA buffer and subjected to immunoprecipitation with the RH-7 antibody as described in Materials and Methods. In panels A, B, and C, the 17B antibody, which can recognize the N-terminal 33 aa of UL9, was used to detect the UL9 fragments. (A) Lanes 2, 3, and 4 represent the total cell lysates after transfection of plasmids UL9-1-321, UL9-1-354, and UL9-1-450. Lanes 5 to 7 represent proteins recovered by immunoprecipitation with the RH-7 antibody, which can recognize the C terminus of UL9. Prestained protein markers are shown in lane 1. In panels B and C, lanes 1 to 3 represent the input samples after cotransfection of plasmids containing the genes for full-length UL9 and the mutant forms, as indicated above the lanes. Lanes 4 to 6 represent the proteins recovered by immunoprecipitation with the RH-7 antibody. Lane 7 represents prestained molecular size markers (M). (D) The 17B antibody was used to probe the upper portion of the membrane to detect full-length UL9, and the six-His monoclonal antibody was used to probe the lower portion of the membrane to detect the truncated protein UL9-236-534-His. Lanes 1 and 2 represent the input samples after transfection of plasmids of interest. Lanes 3 and 4 represent proteins recovered by immunoprecipitation with the RH-7 antibody. WT, wild type; IP, immunoprecipitate.
In order to determine whether the interaction between full-length UL9 and the N-terminal fragments reflects direct protein-protein interactions or is mediated through DNA, the immunoprecipitated complex was treated with DNase I. No change in the coprecipitation patterns was observed, however, suggesting that coimmunoprecipitation of the C-terminally truncated forms and full-length UL9 is not dependent on the presence of DNA (data not shown). The interaction between the N-terminal fragments and full-length UL9, in theory, could reflect dimerization or higher-order interactions since UL9 is also known to cooperatively bind the origins of viral DNA (11). We believe that we have identified a domain involved in dimer formation because cooperativity is only observed in the presence of an origin plasmid containing multiple recognition sites for UL9 (11). The observation that the C-terminally truncated forms can be coprecipitated with full-length UL9 even after DNase I treatment thus suggests that we have identified a domain within the first 321 aa which is involved in dimer formation.
To map the N-terminal interaction domain more precisely, we constructed additional C-terminally truncated forms, i.e., UL9-1-292 and UL9-1-236, which were shown to express proteins of the expected sizes (Fig. 2C, lanes 2 and 3). The mutant protein UL9-1-354, described above, was included for comparison (Fig. 2C, lane 1). As expected, UL9-1-354 is efficiently precipitated by full-length UL9 (Fig. 2C, lane 4); however, the C-terminally truncated forms UL9-1-292 and UL9-1-236 were not (Fig. 2C, lanes 5 and 6, respectively). Unfortunately, the 1-to-236 fragment migrates at a position very close to the light-chain immunoglobulin G band, potentially obscuring whether or not it can be coprecipitated; however, despite this problem, it is clear that this band is not present at the same intensity as the 1-to-354 fragment (Fig. 2C, lane 4). In order to confirm that fragments smaller than aa 1 to 292 had indeed lost the ability to interact efficiently with full-length UL9, several other C-terminally truncated forms were constructed (aa 1 to 145, 1 to 253, 1 to 265, and 1 to 282), none of which was able to interact efficiently with full-length UL9 (data not shown). All six of the truncated forms ending at residues 145, 236, 253, 265, 282, and 292 exhibited a very weak coprecipitated band when the Western blot assays were developed for a longer time (data not shown). The weaker binding may reflect a low level of aggregation or alternatively may be a result of a secondary weaker interaction domain within the first 145 aa of UL9. Taken together, these results suggest that a region spanning residues 293 to 321 is important for efficient interaction with full-length UL9. Furthermore, it appears that fragments containing residues from the N terminus are not able to efficiently interact with full-length UL9 on their own. In order to determine whether a fragment lacking the N-terminal 236 residues is still capable of interacting efficiently with full-length UL9, a plasmid capable of expressing residues 236 to 534 was generated and analyzed as described above, in cells cotransfected with full-length UL9. The upper portion of the membrane was probed for full-length UL9 with the 17B antibody, and the lower portion was probed with the six-His monoclonal antibody to detect the UL9-236-534-His fragment. As shown in Fig. 2D (lane 4), the full-length UL9 protein was able to immunoprecipitate the UL9-236-534-His fragment efficiently, indicating that the region responsible for efficient heterodimer formation lies within residues 236 to 534.
N-terminal fragments can be coimmunoprecipitated with C-terminal fragments.
We previously showed that residues within the N terminus of UL9 could affect the DNA binding activity of the C-terminal domain, suggesting that an interaction between these two domains might exist (9). To test this hypothesis directly, cells were cotransfected with the N-terminal fragments described above and a clone containing the C-terminal one-third of UL9, UL9-535-851-His(CT). Immunoprecipitation was performed as described above. The 17B monoclonal antibody, which interacts with the N-terminal 33 aa, was used for Western blot analysis and would thus not be expected to react with UL9-CT. The presence of UL9-CT (His tagged) in the transfected cell lysates, as well as in the immunoprecipitated complex, was confirmed with an anti-His antibody by Western blot analysis (data not shown). The C terminus of UL9 was able to precipitate the N-terminal fragments (UL9-1-450 and UL9-1-354) as efficiently as full-length UL9 (compare Fig. 3A, lanes 5 and 6, with Fig. 2B, lanes 5 and 6). As shown in Fig. 3B (lane 2), the N-terminal UL9-1-321 fragment was also precipitated efficiently by the C terminus of UL9. On the other hand, a small amount of the N-terminal fragments UL9-1-292 and UL9-1-236 could be detected (Fig. 3A, lanes 7 and 8) but only after a very long exposure. The possibility that the interaction between the N and C termini might be mediated through DNA was ruled out by treating the immunoprecipitation complex with DNase I (data not shown). Thus, the data presented here suggest that indeed there is a direct interaction between the N- and C-terminal fragments of UL9. We observed strong interactions between the C-terminal fragment and the aa 1-to-450, 1-to-354, and 1-to-321 N-terminal fragments and relatively weak interactions between the C terminus and the smaller clones. These results suggest that the region between residues 293 and 321 contains important residues necessary for a strong interaction, whereas residues at the very N terminus of UL9 may play a role in weaker interactions.
FIG. 3.
The N-terminal fragments were coimmunoprecipitated with C-terminal fragments. A coimmunoprecipitation assay was performed with the RH-7 antibody as mentioned before after cotransfection of subconfluent Vero cells with plasmids as shown. In panels A and B, the 17B antibody, directed against the N-terminal 33 aa of UL9, was used to detect the N-terminal fragments in a Western blot analysis. The C-terminal fragments of UL9 are not expected to react with the 17B antibody in a Western blot analysis. Prestained protein molecular size markers are shown in lane M. (A) Lanes 1 to 4 indicate the N-terminal fragments present in the cell lysate. Lanes 5 to 8 indicate the N-terminal fragments immunoprecipitated along with the C terminus of UL9. (B) Lane 2 represents the proteins recovered by immunoprecipitation with the RH-7 antibody, and lane 3 represents the proteins present in the total cell lysate. (C) The six-His monoclonal antibody was used to detect the UL9 mutant protein fragments in a Western blot analysis. Lanes 2 to 4 represents the proteins present in total cell lysates. Lanes 5 to 7 represent the proteins recovered after immunoprecipitation with the RH7 antibody. (D) The 17B antibody was used to detect the N-terminal fragment UL9-1-354 in cell lysates, as well as after immunoprecipitation with the RH-7 antibody. The C-terminal fragments are not expected to react with the 17B antibody. IP, immunoprecipitate.
In order to confirm that the N-terminal 236 residues are not essential for efficient interaction with the C terminus, the N-terminal fragment UL9-236-534-His was subjected to coimmunoprecipitation with UL9-CT. The UL9-CT fragment immunoprecipitated the UL9-236-534-His fragment efficiently (Fig. 3C, lane 7), supporting our conclusion that the N-terminal 236 residues are not required for efficient interaction between the N- and C-terminal fragments.
To map the interaction domain in the C-terminal one-third (aa 535 to 851) of UL9 more precisely, two more truncated forms, UL9-600-851 and UL9-535-800 were constructed (shown in Fig. 1A). The ability of these truncated forms to interact with the N terminus was examined by cotransfecting Vero cells with the C-terminally truncated form UL9-1-354 and the new C-terminal fragments. Figure 3D shows that the N-terminal fragment UL9-1-354 could be coimmunoprecipitated by these two fragments of the C terminus as efficiently (Fig. 3D, lane 2 and 3) as by the entire C terminus (Fig. 3D, lane 1). Therefore, it appears that the minimal essential region required for the interaction of the N terminus with the C terminus maps within residues 600 to 800. The C-terminal fragments were not expected to react with the 17B antibody but could be detected with the six-His antibody (data not shown). Taken together, our data suggest that there are two domains involved in UL9-UL9 interaction, one located in the N terminus and another in the C terminus.
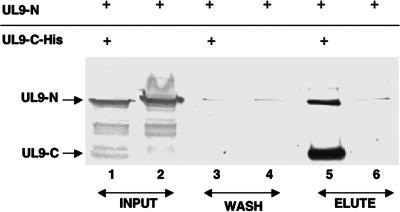
The observation that the N terminus can interact directly with the C terminus of UL9 is intriguing. To confirm this observation, the His-tagged whole C terminus (535 to 851) and an untagged whole N terminus (1 to 534) were expressed in insect cells infected with recombinant baculovirus and a pull-down assay was performed with nickel magnetic beads. The His-tagged C terminus was able to pull down the untagged N terminus efficiently (Fig. 4, lane 5). In order to rule out nonspecific interactions of the N terminus with nickel magnetic beads, control experiments were performed with cells infected with only the untagged N terminus. As expected, no significant binding of the untagged N terminus with nickel magnetic beads was observed (Fig. 4, lane 6). In order to rule out a nonspecific association of the untagged N terminus with the tagged C terminus, magnetic beads were washed thoroughly with wash buffer and samples from the third wash were analyzed by Western blotting. As shown in Fig. 4, no significant amount of the untagged N terminus was detected in the wash (Fig. 4, lane 3) but a significant amount of the untagged N terminus was eluted with the tagged C terminus (Fig. 4, lane 5), indicating that there is a specific interaction between the untagged N terminus and the tagged C terminus in the eluted sample. Taken together, this result indicates that the tagged C terminus is able to interact specifically and directly with the untagged N terminus and supports the conclusion that there are two interacting domains, one in the N terminus and another in the C terminus of UL9.
FIG. 4.
Coimmunoprecipitation of N- and C-terminal fragments expressed in insect cells from recombinant baculoviruses. The His-tagged C termini and the untagged N termini of UL9 proteins were expressed in insect cells by coinfecting insect cells with recombinant baculoviruses. The His-tagged C-terminal protein was pulled down by the addition of nickel magnetic beads as described in Materials and Methods. The KST-1 antibody, which is raised against the full-length UL9 protein, was used to detect the N and C termini of UL9 in a Western blot analysis.
The interaction between the N- and C-terminal fragments can be confirmed by IF microscopy.
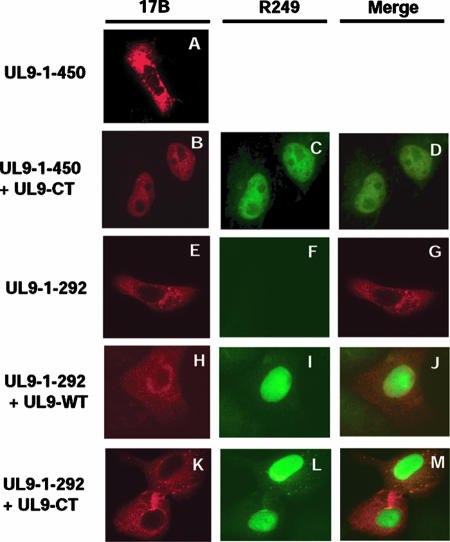
The previous report that the nuclear localization signal of UL9 is localized in the extreme C terminus between residues 744 and 851 (25) allowed us to confirm the interaction between the N- and C-terminal domains of UL9 by IF microscopy. Vero cells were cotransfected with various N- and C-terminally truncated forms, and IF microscopy was performed as described in Materials and Methods. N-terminal fragments were detected with the 17B antibody, and the C-terminal fragment (UL9-CT) was detected with the R249 antibody raised against the C-terminal decapeptide. Figure 5 shows that the N-terminal fragment UL9-1-450 remains in the cytoplasm when cells are transfected with this fragment alone (Fig. 5A) while full-length UL9 (Fig. 5I) and UL9-CT (Fig. 5C and L) localize to the nucleus in cotransfected cells, as previously reported (27). Interestingly, this UL9-1-450 N-terminal fragment was detected in the nucleus when expressed with either full-length UL9 (data not shown) or UL9-CT (Fig. 5B), indicating that the N-terminal fragment forms a heterodimer with either version of UL9. These results support the conclusion that the two domains of UL9 interact with each other. Similar results were obtained with the smaller N-terminal fragments UL9-1-354 and UL9-1-321 (data not shown).
FIG. 5.
Confirmation of the N- and C-terminal interaction by immunofluorescence microscopy analysis. Vero cells were transfected with UL9-1-450 (A), UL9-1-450 plus UL9-CT (B, C, and D), UL9-1-292 (E, F, and G), UL9-1-292 plus UL9-WT (H, I, and J), or UL9-1-292 plus UL9-CT (K, L, and M). Panels A, B, E, H, and K were stained with the 17B antibody (recognizes the N-terminal 33 aa of UL9). Panels C, F, I, and L were stained with the R249 antibody (directed against the C terminus of UL9). Panels A and E show that UL9-1-450 and UL9-1-292 stayed in the cytoplasm. Panels C, I, and L show that UL9-WT and UL9-CT went to the nucleus. Panel B shows that the N-terminal fragment UL9-1-450 went to the nucleus along with UL9-CT. Panels H and K show that UL9-1-292 stayed in the cytoplasm, even in the presence of UL9-WT or UL9-CT.
Next, we asked whether the N-terminal fragment UL9-1-292, which did not form strong heterodimers in the coimmunoprecipitation experiment, could localize to the nucleus along with UL9-WT or UL9-CT. In contrast to the previous experiment, the N-terminal fragment (UL9-1-292) remained in the cytoplasm (Fig. 5H and K) even when UL9-WT and UL9-CT both were detected in the nucleus (Fig. 5I and L). In less than 5% of the transfected cells, the mutant protein UL9-CT or the N-terminal fragments were detected in both the cytoplasm and nucleus (data not shown). In this case, the staining was very bright, which may indicate aggregation due to overexpression. This aggregation in a few cells may also explain the weak interactions observed in some of the coimmunoprecipitation experiments; however, as mentioned above, it is possible that another, weaker, interaction domain exists in the extreme N terminus of UL9. In any case, the IF data confirm that a region necessary for interaction with the C terminus maps within residues 293 to 321.
DISCUSSION
We previously reported that residues in the N terminus of UL9 can modulate its inhibitory properties by reducing the DNA binding ability of the C terminus (9). In this paper, we set out to demonstrate whether a direct physical interaction between these two domains of UL9 could be detected. A series of N- and C-terminally truncated forms of UL9 were constructed and tested for interactions with one another in a coimmunoprecipitation assay. We demonstrated that a direct interaction between the N- and C-terminal domains of UL9 can be detected. The N terminus may, in fact, contain two interaction domains: a region important for efficient interaction maps within residues 293 to 321, and another, weaker, interaction domain may exist in the very N terminus of UL9 (1 to 145). The N-terminal domain appears to interact with residues 600 to 800 in the C terminus. In theory, the interactions we have observed could reflect either intramolecular interactions of the N and C termini on one UL9 molecule or intermolecular interactions resulting in the formation of dimers or even higher-order structures. We favor the interpretation that the interactions reported here reflect the formation of UL9 dimers. The reasons for favoring the dimer model are as follows. (i) UL9 has been reported to be a stable homodimer in solution (6, 16); thus, intramolecular interactions are unlikely. (ii) The formation of higher-order structures requires the presence of origin DNA (11), and we showed that the interactions can be detected even after DNase I treatment.
The coimmunoprecipitation experiments described in this paper indicate that the N terminus may contain two domains which interact with the C terminus, one strong and the other much weaker. To provide additional support for the existence of two binding domains in the N terminus, we performed plaque reduction assays. We and others previously reported that the C-terminal DNA binding domain can drastically reduce plaque formation in a cotransfection assay with infectious DNA (26, 33, 37). More recently, we showed that residues in the N terminus could influence both DNA binding ability and inhibition of plaque formation exhibited by the C-terminal fragment (9). Preliminary data suggest that an N-terminal fragment containing both the putative weak and strong interaction domains was able to reduce plaque formation more efficiently than an N-terminal fragment containing only the weak interaction domain (Chattopadhyay and Weller, unpublished data). These data support the presence of two interaction domains within the N terminus. Additional support for the existence of an interaction domain at the very N terminus was provided by the observation that a UL9 construct lacking residues 1 to 35 was able to partially relieve inhibition that is exhibited by the overexpression of full-length UL9 in a plaque reduction assay (26). This may indicate that the deletion of residues 1 to 35 partially abrogates the formation of a heterodimer between the deletion-containing form and full-length UL9. Taken together, these data support the notion that residues in the N terminus influence the DNA binding and inhibition of plaque formation associated with the C terminus of UL9 and support the coimmunoprecipitation data presented in this paper.
UL9 dimer formation.
The results in this paper have profound implications regarding the orientation of the UL9 dimer. The previous observation that full-length UL9 forms stable dimers in solution while the C terminus is a monomer under the same conditions led to the suggestion that the N terminus of UL9 is important for dimer formation (12). The previous model of dimer formation was that dimers are formed by the interaction between residues in the N terminus of one subunit with similar residues in the N terminus of the second subunit; however, the results presented in this report suggest that the N terminus actually interacts directly with the C terminus of UL9. The simplest model of dimer formation consistent with all of the data is that the N terminus of one monomer interacts with the C terminus of a second monomer, resulting in a head-to-tail or antiparallel interaction. Alternatively, it is possible that the N- and C-terminal interaction domains come together to form one dimerization domain and a dimer is formed when two such noncontiguous domains come together. We favor the first model since it appears that the N- and the C-terminal domains of UL9 function separately. It is expected that a structural analysis such as cryoelectron microscopy is necessary to confirm that UL9 forms an antiparallel dimer.
The model in which UL9 forms a head-to-tail dimer in which the N terminus interacts directly with the C terminus is consistent with the way dimers are formed in several other proteins. For the dimeric APOBEC-1 protein, deletion of either seven residues from the N terminus or five residues from the C terminus disrupts dimer formation, suggesting that both the N and C termini are essential for dimerization (32). Similarly, the N and C termini of human androgen receptor have been shown to interact, resulting in an active antiparallel dimer conformation in the presence of ligand androgen; furthermore, dimer formation is essential for DNA binding and transcriptional activation (21).
Regulation of UL9.
Several questions remain about the regulation of UL9 during HSV-1 DNA replication. As mentioned in the introduction, it appears that HSV-1 DNA replication involves a UL9-dependent and a UL9-independent stage. In order to proceed to the UL9-independent second stage of infection, we suggest that it may be necessary to clear UL9 from the origin. This notion is supported by the observation that the inhibitory effects of UL9 are dependent in part upon its DNA binding ability (26, 37). Thus, the switch from a DNA binding to a non-DNA binding mode of UL9 may be important for the later stages of HSV-1 DNA replication. Several possible mechanisms of regulation can be envisioned. By analogy with the initiation protein of simian virus 40, large T antigen, whose activities are regulated through phosphorylation (15, 38), it is possible that UL9 activities are modulated by phosphorylation. Interestingly, Isler and Schaffer reported that UL9 is phosphorylated during HSV-1 infection by either viral or virus-induced cellular factors (18). We have also noted the presence of a sequence in the N terminus which resembles a PEST motif (29). These motifs have been shown in other proteins to play a role in targeting proteins for ubiquitination and degradation in a phosphorylation-dependent manner (34). Therefore, it will be of considerable interest to determine the role of the PEST region in the regulation of UL9 protein during infection. The suggestion that the levels of UL9 are regulated may be consistent with the report that UL9 can interact with the neuronal protein NFB42, an F-box protein which is involved in polyubiquitination (13, 14).
Another possible mechanism for the regulation of UL9 is suggested by the existence of the subgenomic fragment OBPC-1, a cathepsin B-mediated cleavage product of UL9 which comprises the C terminus of UL9 (2, 22, 23). As discussed above, we have shown that residues from the N terminus could influence the DNA binding ability of the C terminus (9). In fact, in that study we showed that fragments of the same approximate size as OBPC-1 could relieve inhibition of UL9 by reducing their origin binding ability. It is thus tempting to speculate that the presence of a UL9 fragment such as OBPC-1 may alter the conformation of full-length UL9 by forming a heterodimer which would bind the origins less efficiently. The presence of such a fragment may help clear UL9 from the origins and allow the switch from origin-dependent to origin-independent DNA replication. In summary, several levels of regulation of UL9 may exist; phosphorylation may regulate UL9 activity or protein levels through ubiquitinylation and proteolysis, and conformational changes exerted by subgenomic fragments of UL9 may regulate DNA binding activity.
In summary, we demonstrate in this paper that various N-terminal fragments can directly interact with the C terminus of UL9. Residues important for efficient interaction have been mapped between residues 293 and 321 in the N terminus and between residues 600 and 800 in the C terminus. The ability of the N terminus to interact directly with the C terminus suggests that UL9 dimers may exist in an antiparallel head-to-tail orientation. Furthermore, a weaker interaction domain was mapped between residues 1 and 145. These results have important implications for the function of UL9 not only in the initiation of DNA replication at the origins but also in the proposed regulation of UL9.
Acknowledgments
We gratefully acknowledge Mark Challberg for the R249 antibody, Daniel Tenney for the RH-7 antibody, and Deborah Parris for the KST-1 antibody. We thank Asis Das, Shlomo Eisenberg, and the members of our laboratory for helpful comments on the manuscript. We thank Ping Bai for the generation of recombinant baculoviruses and expression of recombinant proteins in SF9 insect cells and Boriana Marintcheva for generating the UL9-WT, UL9-1-450, UL9-1-354, and UL9-1-321 plasmids.
This investigation was supported by Public Health Service grant AI-21747 from the National Institutes of Health.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Arbuckle, M. I., and N. D. Stow. 1993. A mutational analysis of the DNA-binding domain of the herpes simplex virus type 1 UL9 protein. J. Gen. Virol. 74(Pt. 7):1349-1355. [DOI] [PubMed] [Google Scholar]
- 2.Baradaran, K., C. E. Dabrowski, and P. A. Schaffer. 1994. Transcriptional analysis of the region of the herpes simplex virus type 1 genome containing the UL8, UL9, and UL10 genes and identification of a novel delayed-early gene product, OBPC. J. Virol. 68:4251-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blümel, J., and B. Matz. 1995. Thermosensitive UL9 gene function is required for early stages of herpes simplex virus type 1 DNA synthesis. J. Gen. Virol. 76(Pt. 12):3119-3124. [DOI] [PubMed] [Google Scholar]
- 4.Boehmer, P. E. 1998. The herpes simplex virus type-1 single-strand DNA-binding protein, ICP8, increases the processivity of the UL9 protein DNA helicase. J. Biol. Chem. 273:2676-2683. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer, P. E., and I. R. Lehman. 1993. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc. Natl. Acad. Sci. USA 90:8444-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruckner, R. C., J. J. Crute, M. S. Dodson, and I. R. Lehman. 1991. The herpes simplex virus 1 origin binding protein: a DNA helicase. J. Biol. Chem. 266:2669-2674. [PubMed] [Google Scholar]
- 7.Carmichael, E. P., M. J. Kosovsky, and S. K. Weller. 1988. Isolation and characterization of herpes simplex virus type 1 host range mutants defective in viral DNA synthesis. J. Virol. 62:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challberg, M. 1996. Herpesvirus DNA replication. Cold Spring Harbor Press, Cold Spring Harbor, NY.
- 9.Chattopadhyay, S., and S. K. Weller. 2006. DNA binding activity of the herpes simplex virus type 1 origin binding protein, UL9, can be modulated by sequences in the N terminus: correlation between transdominance and DNA binding. J. Virol. 80:4491-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deb, S., and S. P. Deb. 1991. A 269-amino-acid segment with a pseudo-leucine zipper and a helix-turn-helix motif codes for the sequence-specific DNA-binding domain of herpes simplex virus type 1 origin-binding protein. J. Virol. 65:2829-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias, P., C. M. Gustafsson, and O. Hammarsten. 1990. The origin binding protein of herpes simplex virus 1 binds cooperatively to the viral origin of replication oriS. J. Biol. Chem. 265:17167-17173. [PubMed] [Google Scholar]
- 12.Elias, P., C. M. Gustafsson, O. Hammarsten, and N. D. Stow. 1992. Structural elements required for the cooperative binding of the herpes simplex virus origin binding protein to oriS reside in the N-terminal part of the protein. J. Biol. Chem. 267:17424-17429. [PubMed] [Google Scholar]
- 13.Eom, C. Y., W. D. Heo, M. L. Craske, T. Meyer, and I. R. Lehman. 2004. The neural F-box protein NFB42 mediates the nuclear export of the herpes simplex virus type 1 replication initiator protein (UL9 protein) after viral infection. Proc. Natl. Acad. Sci. USA 101:4036-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eom, C. Y., and I. R. Lehman. 2003. Replication-initiator protein (UL9) of the herpes simplex virus 1 binds NFB42 and is degraded via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 100:9803-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fanning, E. 1994. Control of SV40 DNA replication by protein phosphorylation: a model for cellular DNA replication? Trends Cell Biol. 4:250-255. [DOI] [PubMed] [Google Scholar]
- 16.Fierer, D. S., and M. D. Challberg. 1992. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J. Virol. 66:3986-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazuda, D. J., H. C. Perry, and W. L. McClements. 1992. Cooperative interactions between replication origin-bound molecules of herpes simplex virus origin-binding protein are mediated via the amino terminus of the protein. J. Biol. Chem. 267:14309-14315. [PubMed] [Google Scholar]
- 18.Isler, J. A., and P. A. Schaffer. 2001. Phosphorylation of the herpes simplex virus type 1 origin binding protein. J. Virol. 75:628-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouzarides, T., and E. Ziff. 1988. The role of the leucine zipper in the fos-jun interaction. Nature 336:646-651. [DOI] [PubMed] [Google Scholar]
- 20.Landschulz, W. H., P. F. Johnson, and S. L. McKnight. 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759-1764. [DOI] [PubMed] [Google Scholar]
- 21.Langley, E., Z. X. Zhou, and E. M. Wilson. 1995. Evidence for an anti-parallel orientation of the ligand-activated human androgen receptor dimer. J. Biol. Chem. 270:29983-29990. [DOI] [PubMed] [Google Scholar]
- 22.Link, M. A., and P. A. Schaffer. 2007. Herpes simplex virus type 1 C-terminal variants of the origin binding protein (OBP), OBPC-1 and OBPC-2, cooperatively regulate viral DNA levels in vitro, and OBPC-2 affects mortality in mice. J. Virol. 81:10699-10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Link, M. A., L. A. Silva, and P. A. Schaffer. 2007. Cathepsin B mediates cleavage of herpes simplex virus type 1 origin binding protein (OBP) to yield OBPC-1, and cleavage is dependent upon viral DNA replication. J. Virol. 81:9175-9182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malik, A. K., R. Martinez, L. Muncy, E. P. Carmichael, and S. K. Weller. 1992. Genetic analysis of the herpes simplex virus type 1 UL9 gene: isolation of a LacZ insertion mutant and expression in eukaryotic cells. Virology 190:702-715. [DOI] [PubMed] [Google Scholar]
- 25.Malik, A. K., L. Shao, J. D. Shanley, and S. K. Weller. 1996. Intracellular localization of the herpes simplex virus type-1 origin binding protein, UL9. Virology 224:380-389. [DOI] [PubMed] [Google Scholar]
- 26.Malik, A. K., and S. K. Weller. 1996. Use of transdominant mutants of the origin-binding protein (UL9) of herpes simplex virus type 1 to define functional domains. J. Virol. 70:7859-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marintcheva, B., and S. K. Weller. 2003. Existence of transdominant and potentiating mutants of UL9, the herpes simplex virus type 1 origin-binding protein, suggests that levels of UL9 protein may be regulated during infection. J. Virol. 77:9639-9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marintcheva, B., and S. K. Weller. 2001. Residues within the conserved helicase motifs of UL9, the origin-binding protein of herpes simplex virus-1, are essential for helicase activity but not for dimerization or origin binding activity. J. Biol. Chem. 276:6605-6615. [DOI] [PubMed] [Google Scholar]
- 29.Marintcheva, B., and S. K. Weller. 2001. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 70:77-118. [DOI] [PubMed] [Google Scholar]
- 30.McLean, G. W., A. P. Abbotts, M. E. Parry, H. S. Marsden, and N. D. Stow. 1994. The herpes simplex virus type 1 origin-binding protein interacts specifically with the viral UL8 protein. J. Gen. Virol. 75(Pt. 10):2699-2706. [DOI] [PubMed] [Google Scholar]
- 31.Monahan, S. J., L. A. Grinstead, W. Olivieri, and D. S. Parris. 1998. Interaction between the herpes simplex virus type 1 origin-binding and DNA polymerase accessory proteins. Virology 241:122-130. [DOI] [PubMed] [Google Scholar]
- 32.Navaratnam, N., T. Fujino, J. Bayliss, A. Jarmuz, A. How, N. Richardson, A. Somasekaram, S. Bhattacharya, C. Carter, and J. Scott. 1998. Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J. Mol. Biol. 275:695-714. [DOI] [PubMed] [Google Scholar]
- 33.Perry, H. C., D. J. Hazuda, and W. L. McClements. 1993. The DNA binding domain of herpes simplex virus type 1 origin binding protein is a transdominant inhibitor of virus replication. Virology 193:73-79. [DOI] [PubMed] [Google Scholar]
- 34.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 35.Schildgen, O., S. Graper, J. Blumel, and B. Matz. 2005. Genome replication and progeny virion production of herpes simplex virus type 1 mutants with temperature-sensitive lesions in the origin-binding protein. J. Virol. 79:7273-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skaliter, R., and I. R. Lehman. 1994. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc. Natl. Acad. Sci. USA 91:10665-10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stow, N. D., O. Hammarsten, M. I. Arbuckle, and P. Elias. 1993. Inhibition of herpes simplex virus type 1 DNA replication by mutant forms of the origin-binding protein. Virology 196:413-418. [DOI] [PubMed] [Google Scholar]
- 38.Weisshart, K., M. K. Bradley, B. M. Weiner, C. Schneider, I. Moarefi, E. Fanning, and A. K. Arthur. 1996. An N-terminal deletion mutant of simian virus 40 (SV40) large T antigen oligomerizes incorrectly on SV40 DNA but retains the ability to bind to DNA polymerase alpha and replicate SV40 DNA in vitro. J. Virol. 70:3509-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weller, S. K. 1995. Herpes Simplex virus DNA replication and genome maturation. American Society for Microbiology, Washington, D.C.