Abstract
Cellular cathepsins are required for Ebola virus infection and are believed to proteolytically process the Ebola virus glycoprotein (GP) during entry. However, the significance of cathepsin cleavage during infection remains unclear. Here we demonstrate a role for cathepsin L (CatL) cleavage of Ebola virus GP in the generation of a stable 18-kDa GP1 viral intermediate that exhibits increased binding to and infectivity for susceptible cell targets. Cell binding to a lymphocyte line was increased when CatL-proteolysed pseudovirions were used, but lymphocytes remained resistant to Ebola virus GP-mediated infection. Genetic removal of the highly glycosylated mucin domain in Ebola virus GP resulted in cell binding similar to that observed with CatL-treated full-length GP, and no overall enhancement of binding or infectivity was observed when mucin-deleted virions were treated with CatL. These results suggest that cathepsin cleavage of Ebola virus GP facilitates an interaction with a cellular receptor(s) and that removal of the mucin domain may facilitate receptor binding. The influence of CatL in Ebola virus GP receptor binding should be useful in future studies characterizing the mechanism of Ebola virus entry.
Zaire ebolavirus, a member of the family Filoviridae, is a negative-strand RNA virus that causes severe hemorrhagic fever in humans and nonhuman primates. The Ebola virus glycoprotein (GP) is the main determinant of viral tropism and mediates entry into target cells (20, 24). Ebola virus GP is a type I membrane protein that is comprised of a globular unit, GP1, and a transmembrane region, GP2, which also possesses the fusion machinery. GP1 is thought to contain the receptor-binding domain; however, a direct interaction of this region with a cellular receptor has not been documented (7). GP1 also possesses a highly N- and O-glycosylated region termed the mucin domain. This region in Ebola virus GP induces cell rounding and vascular cell cytopathogenicity (17, 25) and was shown to contribute to the binding of the C-type lectin, hMGL (human macrophage C-type lectin specific for galactose and N-acetylglucosamine), which enhanced infection of target cells (19). Despite the apparent benefits of the mucin domain during Ebola virus infection, genetic deletion of the mucin domain in Ebola virus GP renders pseudovirions more infectious than their wild-type Ebola virus GP counterparts (9, 11). This suggests that there may be costs associated with the mucin domain during viral entry. However, the basis for these consequences remains unclear.
Ebola virus enters cells via endocytosis and is sensitive to intracellular increases in pH (2, 5, 20, 24). However, Ebola virus entry is distinct from that of other pH-dependent viruses. Classic pH-dependent viruses, like influenza virus and Semliki Forest virus, display conformational changes in the viral GP following entry into the low-pH milieu of the cellular endosome. These structural rearrangements in GP trigger the viral fusion mechanism that drives mixing of the viral and cellular membranes (10). Ebola virus entry differs from this paradigm in that it appears that the prerequisite for low pH is an indirect effect of the requirement for cellular cathepsins (3, 15).
Recent studies suggest that Ebola virus GP, like the spike proteins of severe acute respiratory syndrome (SARS) coronavirus and mouse hepatitis virus 2 and the F proteins of Hendra and Nipah viruses, is proteolysed by endosomal cathepsins and that Ebola virus in particular requires the activity of cathepsin B (CatB) and cathepsin L (CatL) for entry into host cells (6, 12-14, 16). In a current model of SARS coronavirus entry, receptor binding and conformational changes in spike occur prior to CatL proteolysis, which is then believed to activate the fusion potential of the protein (16). This is supported by evidence that proteolysis of SARS coronavirus S protein before receptor binding renders virions noninfectious (16). In contrast, CatB and CatL pretreatment of Ebola virus GP on virions appears to produce a stable viral intermediate that retains infectivity (3, 15). Here, we further characterized the proteolysed Ebola virus GP viral intermediate and found that CatL-treated virions not only are more infectious but also exhibit greater cell binding activity than untreated virions. In contrast, Ebola virus GP virions lacking the mucin domain do not demonstrate an overall enhancement of cell binding or infectivity.
MATERIALS AND METHODS
Cell lines and plasmids.
pCAGGS-Ebola GP (strain Zaire '76 Mayinga), pCAGGS-Ebola GP-Δmuc, and pCB6-VSV(G) (vesicular stomatitis virus) are described in reference 17. pNL-Luc, pHit60, and pHit111 were also previously described (4, 18). Jurkat cells were maintained in RPMI with 10% fetal bovine serum and l-glutamine. VeroE6 and 293T cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (DMEM10).
Pseudovirion preparation.
Human immunodeficiency virus (HIV) pseudovirions were produced by standard calcium phosphate transfection of 293T cells with 10 μg of a luciferase-encoding HIV vector (pNL-luc) and 30 μg of pCAGGS-Ebola GP or 10 μg of pCB6-VSV(G). Viral supernatants were collected 48 h posttransfection and concentrated through a 20% sucrose cushion by ultracentrifugation at 40,000 rpm in an SW41 rotor for 1 h at 4°C. Pelleted virions were gently resuspended in phosphate-buffered saline (PBS) for 2 h at 4°C. Murine leukemia virus (MLV) pseudovirions were similarly generated by transfecting 293T cells with 10 μg of pHit60, 10 μg of pHit111, and 30 μg of pCAGGS-Ebola GP.
Single-round infection assay.
VeroE6 cells were plated at 1 × 104 cells per well in a 96-well plate 24 h before infection. Virus diluted in DMEM10 was added to cells and spin infected at 2,000 rpm for 2 h at 4°C. Cells were incubated at 37°C for 5 h. The medium was then replaced with fresh DMEM10, and the cells were incubated for 48 h at 37°C. The supernatant was removed, and cells were lysed in 0.5% Triton X-100. Virus infection was analyzed by measuring firefly luciferase enzyme activity in cell lysates using a luciferase assay system per the manufacturer's instructions (Promega).
Chemical entry inhibitors.
VeroE6 cells were pretreated for 1 h with 50 mM ammonium chloride, 10 μM leupeptin, 10 μM E64d (EMD Biosciences), 10 μM Z-Leu-Leu-Leu-fluoromethyl ketone (Z-LLL-FMK; EMD Biosciences), or 100 μM CA074 (EMD Biosciences). For infection, the medium was replaced with an equal concentration of fresh inhibitors diluted in DMEM10 containing the viral inoculum. After 6 h at 37°C, the inoculum was replaced with DMEM10 without inhibitors. Infected cells were cultured for an additional 40 h before luciferase activity was assessed.
Virion proteolysis.
HIV-Ebola virus GP was treated with 10 μg/ml recombinant CatL (Calbiochem) at pH 5.5 in 40 mM HEPES, 40 mM MES (morpholineethanesulfonic acid), 50 mM NaCl, and 4 mM dithiothreitol (DTT). Thermolysin (0.5 mg/ml)-treated virions were incubated in the aforementioned buffer at pH 7.5 and without DTT. All reactions, including those with mock-treated (no-enzyme) samples, were carried out at 37°C. Cathepsin proteolysis was terminated at various time points using 10 μg/ml leupeptin, while thermolysin activity was terminated with the addition of 0.1 mM EDTA. All samples were treated with the appropriate inhibitor at the designated time point and incubated at 37°C until completion of the time course. Mock-treated and proteolysed virions were added to cells for infection or binding assays. Alternatively, virions were analyzed by immunoblotting for HIV p24 (MAb 24-3; NIH AIDS Research and Reference Reagent Program) or Ebola virus GP using a polyclonal anti-GP1 rabbit antiserum raised against soluble GP.
Virion-cell binding assay.
VeroE6 cells, plated 24 h in advance, were chilled to 4°C before the addition of mock-treated or proteolysed virion preparations. Virions were diluted in chilled DMEM10 and spun onto cells at 2,000 rpm for 1 h at 4°C. Cells were washed five times in DMEM10 at 4°C and lysed in 0.5% Triton X-100 with protease inhibitors. HIV p24 levels were quantified by enzyme-linked immunosorbent assay (ELISA) as conducted by the University of Pennsylvania Center for AIDS Research Core. To strip bound virions from the cell surface, cells were washed twice in cold PBS after binding. Proteinase K (50 μg/ml) diluted in PBS was next added to cells and incubated at room temperature for 30 min. Cells were washed three times in cold PBS and lysed as described above. To permit internalization of bound virions before stripping, cells were incubated at 37°C for 30 min following binding. Cells were then washed twice in PBS before the addition of proteinase K and lysis as described above.
RESULTS
Infectivity of proteolysed Ebola virus GP pseudovirions.
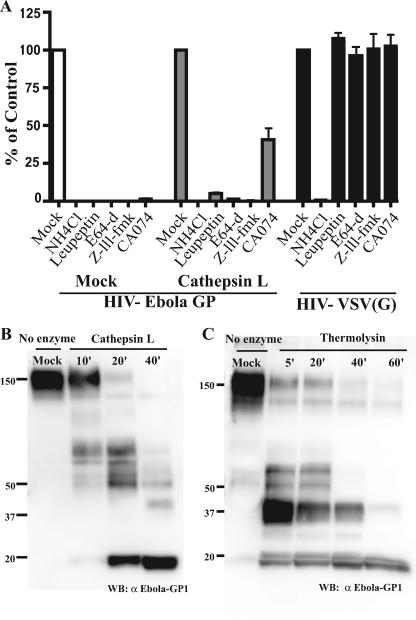
Ebola virus entry is sensitive to lysosomotropic agents and inhibition of the cellular cathepsins B and L (2, 3, 15). However, the relative contributions of CatB and CatL during entry are currently in question. Chandran et al. (3) support a model in which CatB is critical in mediating the final stages of Ebola virus entry. In contrast, data from Schornberg et al. (15) suggest that CatL plays a prominent role in the triggering of Ebola virus GP. In order to clarify the role of these cellular proteases during entry, we examined the effect of various protease inhibitors during infection of VeroE6 cells with HIV-Ebola virus GP pseudovirions (Fig. 1A). Virus entry was abolished when cells were pretreated with leupeptin, a broad-spectrum inhibitor of both serine and cysteine proteases, or E64-d, which specifically inhibits cysteine proteases. Furthermore, the potency of the narrow-spectrum inhibitors CA074, a CatB inhibitor, and Z-LLL-FMK, a CatL inhibitor, supports the previously demonstrated role for these cathepsins during HIV-Ebola virus GP entry.
FIG. 1.
Proteolysis of Ebola virus GP and inhibition of infection using cathepsin inhibitors. (A) VeroE6 cells were treated with NH4Cl (50 mM), leupeptin (10 μM), E64-d (10 μM), Z-LLL-FMK (10 μM), or CA074 (100 μM) for 1 h at 37°C prior to infection with mock- or CatL-treated HIV-Ebola virus GP pseudovirions encoding a luciferase reporter gene. HIV-VSV(G) pseudovirion infection was monitored as a control. Infectivity was quantified 48 h postinfection by measuring luciferase activity in cell lysates. Results are reported as the percent infection of mock-treated cells and are the means and standard deviations for samples run in triplicate. HIV-Ebola virus GP pseudovirions were mock treated or treated with 10 μg/ml of CatL at pH 5.5 with 4 mM DTT (B) or 0.5 mg/ml thermolysin at pH 7.5 (C) for the indicated times. CatL reactions were terminated with 10 μM leupeptin, and thermolysin activity was blocked with the addition of 0.1 mM EDTA. Samples were analyzed on a 4 to 15% sodium dodecyl sulfate-polyacrylamide gel and immunoblotted with polyclonal Ebola virus GP1 antisera.
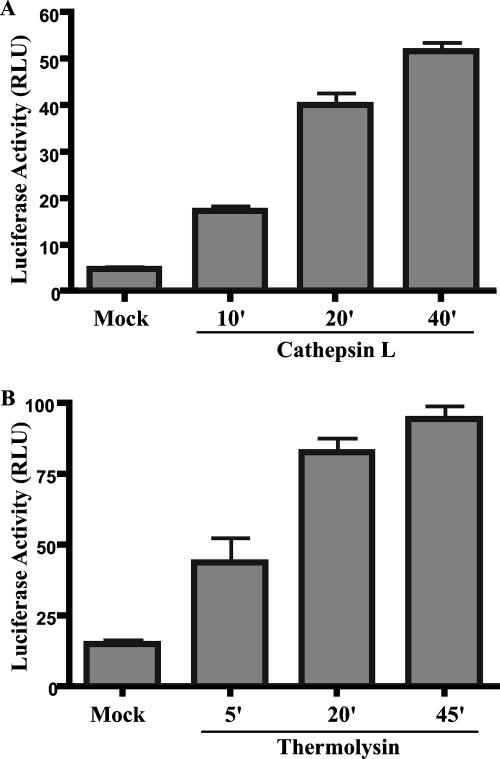
CatL treatment of virions resulted in truncation of GP1 to an 18-kDa fragment that accumulated during prolonged cleavage (Fig. 1B). CatL-treated virions remained infectious, and after 40 min of proteolysis, infectivity was enhanced approximately 10-fold relative to mock-treated virions (Fig. 2A), similar to results obtained by Schornberg et al. (15). CatL-mediated enhancement of infection mirrored the appearance of the 18-kDa GP1 fragment and the diminution of full-length GP1 and proteolysed intermediates. Similar results were obtained using the bacterial protease thermolysin (Fig. 1C and 2B).
FIG. 2.
Protease treatment enhances infectivity of Ebola virus GP pseudovirions. HIV-Ebola virus GP virions were treated with 10 μg/ml CatL (A) or 0.5 mg/ml thermolysin (B) at 37°C for the indicated times. Reactions were terminated as described for Fig. 1. Equal volumes of mock- or protease-treated virion preparations were used to infect VeroE6 cells. Luciferase activity was measured 48 h postinfection. Results are reported in relative light units (RLU) and are the means and standard deviations for samples run in triplicate. Similar results were obtained with 293T cells as infectious targets (data not shown).
Infectious CatL-treated virions remained sensitive to lysosomotropic agents and specific cathepsin inhibitors (Fig. 1A). Infection of VeroE6 cells by virions pretreated with CatL was potently inhibited by the CatL inhibitor, Z-LLL-FMK, but only weakly inhibited by the CatB inhibitor, CA074, suggesting that in vitro CatL-proteolysed HIV-Ebola virus GP entry requires the downstream activity of CatL, while the requirement for CatB is diminished. These data support the model proposed by Schornberg et al. (15) that CatL, but not CatB, is important in the downstream triggering of Ebola virus GP.
Proteolysis activates Ebola virus GP binding potential.
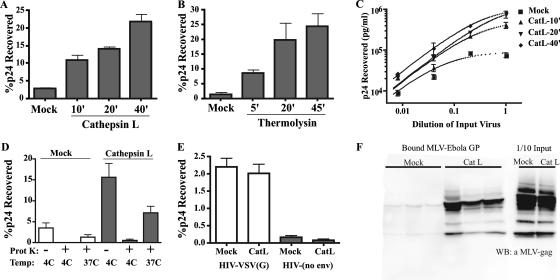
Proteolysed Ebola virus GP pseudovirions were more infectious than wild-type virions. As cell binding is the first step in virus entry, we hypothesized that an increase in binding could explain the enhanced infectivity of virions. To explore this possibility, we examined the efficiency of cell binding of virions treated with CatL or thermolysin over a time course. CatL- or mock-treated HIV-Ebola virus GP pseudovirions were then bound to VeroE6 cells at 4°C, and binding was quantified by HIV-p24 antigen ELISA. Interestingly, the degree of increase in binding at each time point (Fig. 3) closely resembled the increase in infectivity of the same sample, as shown in Fig. 2. Within 40 min of CatL proteolysis, virions exhibited a 10-fold increase in cell binding relative to mock-treated controls (Fig. 3A), which mirrored the 10-fold enhancement of infection observed at the same time point (Fig. 2A). Similar results were observed using thermolysin-treated HIV-Ebola virus GP (Fig. 3B). Proteolytic enhancement of binding was also independent of the level of virus that was added to cells during binding (Fig. 3C). Analogous results were obtained in both infection and binding experiments when virions were treated simultaneously with CatB and CatL or when assays were performed using HeLa or 293T cells (data not shown).
FIG. 3.
Proteolytic cleavage of Ebola virus GP enhances binding of virions to cells. HIV-Ebola virus GP pseudovirions were not treated (mock) or were treated with CatL (A) or thermolysin (B) at 37°C. Proteolytic reactions were terminated at the indicated times, and virions were added to VeroE6 cells chilled to 4°C. Virions were bound to cells by centrifugation. Unbound virus was removed by washing cells in medium at 4°C. Cell lysates were analyzed by p24 antigen ELISA. Results are the percentages of input p24 recovered after binding and are the means and standard deviations for samples run in triplicate. (C) HIV-Ebola virus GP virions were mock treated or treated with CatL for the indicated times, serially diluted fivefold, and bound to VeroE6 cells as described for panel A. Results are concentrations of p24 recovered in cell lysates and are the means and standard deviations for samples run in triplicate. (D) As a control for virus internalization during binding, mock- and CatL-treated virions were bound to cells at 4°C, washed, and lysed immediately or treated with 50 μg/ml proteinase K prior to lysis. Alternatively, cells with bound virions were incubated at 37°C for 30 min followed by treatment with proteinase K and cell lysis. Results are presented as in panel A. Similar results were obtained with 293T cells (data not shown). (E) HIV-VSV(G) pseudovirions and nonenveloped HIV core particles were mock treated or treated with CatL for 30 min. Virus preparations were bound to cells, washed, and lysed. The amount of cell-associated virus was quantified by p24 antigen ELISA. Results are percentages of input p24 recovered after binding and are the means and standard deviations for samples run in triplicate. (F) MLV-Ebola virus GP pseudovirions were mock treated or treated with CatL for 30 min. Virions were bound to cells, washed, and lysed. Samples were subjected to 4 to 15% sodium dodecyl sulfate-polyacrylamide gel and immunoblotted using polyclonal MLV Gag antisera. One-tenth of the virus input added to cells was directly analyzed by Western blotting to compare the level of virions present after proteolysis. Bound mock- and CatL-treated MLV-Ebola virus GP represent independent samples run in triplicate.
As a control for virus internalization during binding, virions were bound to VeroE6 cells at 4°C, and then virus remaining at the cell surface was stripped by proteinase K treatment. The amount of mock- or CatL-treated HIV-Ebola virus GP present on cells after proteinase K treatment was greatly diminished, suggesting that cell-associated virions remained at the cell surface and were not internalized during binding (Fig. 3D). Furthermore, bound virions were considerably protected from proteinase K digestion when cells were incubated at 37°C for 30 min after the virus-binding incubation (Fig. 3D), demonstrating that bound virions were capable of entering cells. Similar observations were obtained when mock- or CatL-treated virions were bound to cells in the presence of 0.02% sodium azide at 37°C to block endocytosis (data not shown).
To demonstrate that the effect of CatL proteolysis on binding was specific to Ebola virus GP pseudovirions, mock- or CatL-treated HIV-VSV(G) virions and HIV particles lacking a viral GP were examined in the cell binding assay. No difference between mock- and CatL-treated HIV-VSV(G) was observed (Fig. 3E). Similarly, HIV core particles lacking a viral envelope did not exhibit any change in binding to VeroE6 cells after CatL treatment (Fig. 3E). Analogous results were obtained when HIV-VSV(G) or nonenveloped HIV particles were pretreated with thermolysin (data not shown).
To confirm that the enhancement of cell binding was not limited to pseudovirions bearing an HIV core, we tested MLV-Ebola virus GP virions in our binding assay. Cell-associated virions were detected by immunoblotting using antibodies specific to MLV Gag proteins. As expected from the previous results, CatL-treated MLV-Ebola virus GP bound VeroE6 cells better than mock-treated virions (Fig. 3F). Collectively, these results suggest that protease treatment of Ebola virus GP pseudovirions increases cell binding, which results in enhanced infectivity of virions.
Effects of the mucin domain in Ebola virus GP-mediated infectivity and binding.
The cathepsin cleavage sites in Ebola virus GP have not yet been characterized, rendering the identity of the 18-kDa fragment associated with virions unknown. However, it is believed that the fragment represents an N-terminal product of GP that remains disulfide linked to GP2 by residue C53 in GP1 (3, 15). Thus, the highly glycosylated mucin domain of Ebola virus GP (amino acids 312 to 462) is likely removed from virions during CatL proteolysis in vitro. Using virions of Ebola virus GP-Δmuc, an Ebola virus GP mutant in which the mucin domain is genetically deleted, we sought to determine whether the increase in cell binding is due to accumulation of the 18-kDa fragment on virions or a consequence of removal of GP fragments, such as the mucin domain, that may inhibit binding.
CatL proteolysis of HIV-Ebola virus GP-Δmuc virions resulted in the accumulation of an 18-kDa band characteristic of the fragment observed during wild-type Ebola virus GP proteolysis (Fig. 4A). However, appearance of the 18-kDa fragment did not correlate with an increase in binding and infectivity (Fig. 4B and C). We instead observed that infectivity briefly decreased during the first 15 min of proteolysis and then recovered to near mock-treated levels after 60 min of proteolysis (Fig. 4B). When cell binding was examined, we similarly found that binding decreased during the early stages of proteolysis and returned to control levels after prolonged cleavage (Fig. 4C). When Ebola virus GP-Δmuc cell binding was directly compared to that of wild-type Ebola virus GP binding, we found that untreated Ebola virus GP-Δmuc virions bound cells more efficiently than untreated wild-type virions (Fig. 4C). However, after 60 min of CatL treatment, wild-type Ebola virus GP virions bound cells at nearly the same level as the Ebola virus GP-Δmuc virion counterparts, suggesting that removal of the mucin domain in wild-type Ebola virus GP virions by proteolysis contributes to enhanced cell binding and infectivity.
FIG. 4.
Cell binding and infectivity of mucin deletion Ebola virus GP pseudovirions. HIV p24 normalized HIV-Ebola virus GP-Δmuc or HIV-Ebola virus wild-type GP pseudovirions were mock treated or treated with CatL for 60 min, and the reactions were quenched at the indicated times. (A) Ebola virus GP-Δmuc samples were analyzed by immunoblotting using polyclonal GP1 antisera. (B) Samples were added to VeroE6 cells and incubated at 37°C for 48 h. Infection was determined by measuring luciferase activity in cell lysates. Results are reported as luciferase activity relative to the no-enzyme control. Mock-treated Ebola virus wild-type GP, 40 relative light units; mock-treated Ebola virus GP-Δmuc, 190 relative light units. (C) Proteolysed virions were bound to VeroE6 cells, and the amount of p24 recovered from cell lysates relative to the input was determined by p24 ELISA.
Infectivity and binding of protease-treated Ebola virus GP pseudovirions to Jurkat cells.
Lymphocytes represent a unique a cell type that are resistant to Ebola virus GP-mediated infection (20, 24). We hypothesized that CatL-treated Ebola virus GP pseudovirions would not exhibit an increase in binding and infectivity on Jurkat cells, a model lymphocyte cell line. Contrary to this hypothesis, HIV-Ebola virus GP virions treated with CatL (Fig. 5A) or thermolysin (data not shown) demonstrated a 10-fold enhancement of binding to Jurkat cells, concomitant with the level of enhancement observed with VeroE6 cells. However, Jurkat cells remained highly refractory to infection with CatL-treated virions (Fig. 5B). Similar results were obtained with K562 cells, a human leukemic cell line (data not shown).
FIG. 5.
Cathepsin proteolysis of Ebola virus GP enhances binding of virions to Jurkat cells, but resistance to infection is maintained. HIV-Ebola virus GP virions were mock treated or treated with CatL for 30 min and bound to Jurkat or VeroE6 cells at 4°C. A portion of the samples was immediately washed, lysed, and subjected to p24 ELISA (A). The remaining samples were incubated at 37°C for 48 h and analyzed for infection (B). Infection is reported as the percent GFP-positive cells, as determined by flow-cytometric analysis. Results are the means and standard deviations for samples run in triplicate.
DISCUSSION
The Ebola virus GP is a class I viral fusion protein that exists as a metastable complex of trimers on the virus surface (8, 21, 22). Membrane fusion catalyzed by viral fusion proteins is characterized by a triggered series of dramatic structural rearrangements that culminate in merger of the host and viral membranes. Recognized triggers for these structural changes include binding to cellular receptors, as typified by HIV, and exposure to a low-pH environment encountered as virions traffic through the endocytic pathway, as demonstrated by the influenza virus hemagglutinin protein. Variations of these triggering schemes exist. Notably, the avian sarcoma and leukosis virus Env fusion complex has evolved a unique multistep process in which receptor binding at neutral pH triggers initial structural changes that drive viral fusion peptide insertion into the host membrane, while low pH is subsequently required to initiate hemifusion and fusion pore formation (1). Another striking variation of these models was recently described in which an apparent pH trigger reflected the requirement for pH-dependent endosomal cathepsins for viral entry (3, 14-16).
The initial observation that CatL- and thermolysin-proteolysed Ebola virus GP pseudovirions were more infectious that untreated virions led us to question the role of GP proteolysis during entry (Fig. 2). We found that virions associated with the 18-kDa GP1 fragment bound cells more efficiently than virions with full-length GP, and this is likely the explanation for the observed increase in infectivity (Fig. 3). Our results are supported by work from Kuhn et al. (7) in which an approximately 16-kDa N-terminal fragment of Ebola virus GP1 (residues 54 to 201), and not full-length GP1, was capable of binding VeroE6 cells and blocking infection with Ebola virus (7). Taken together, these results suggest that truncation of Ebola virus GP1 activates the receptor binding potential of the protein, a process that likely occurs upon cathepsin proteolysis in vivo. Similar to Kuhn et al., we observed greatly enhanced cell surface binding of virions bearing a truncated GP1 fragment of nearly the same molecular weight. The idea that Ebola virus GP truncation enhances binding is further supported by studies of the SARS coronavirus S protein in which truncation of the S1 subunit yielded an independently folding receptor-binding domain that bound cells more efficiently than full-length S1 (23). It is possible that a key role for CatL during Ebola virus and SARS coronavirus entry is to proteolytically truncate extraneous regions of the GP, thereby exposing the receptor binding domain or allowing higher affinity receptor binding.
Ebola virus entry appears to be a multistep process in which CatB and CatL cleave GP1 to yield a stable 18-kDa intermediate in a manner that was recapitulated in vitro using CatL alone (Fig. 1B). It remains unclear what role CatL plays during the second step of viral entry, yet several explanations are possible. An additional cellular factor dependent on the activity of CatL may be required for triggering the fusion peptide, as proposed by Schornberg et al. (15). Alternatively, interaction with a cellular receptor within the endosome, or alteration of an existing receptor interaction after the initial proteolysis, may induce structural rearrangements in Ebola virus GP. Previously inaccessible regions in GP may then undergo further CatL proteolysis, thereby lowering the activation barrier and allowing the fusion machinery to trigger. The latter model assumes that our observations using in vitro proteolysed virions interacting with the cell surface recapitulate events that occur within the endosome. As many cell surface proteins are known to recycle through endosomal compartments, it seems reasonable to hypothesize that virions may encounter factors at the cell surface that are also present in CatL-enriched endosomes.
The mucin domain is involved in binding of Ebola virus GP to the C-type lectin hMGL (19). The significance of this interaction remains unclear, but it may contribute to trafficking of virions from the cell surface to endosomes, where cathepsin proteolysis proceeds. Our results analyzing cells that do not express hMGL suggest that the mucin domain may inhibit receptor binding and that removal of the mucin domain by genetic removal or by cathepsin proteolysis promotes virus-cell interactions. This is most obvious in the experiments using Ebola virus GP-Δmuc virions, which bound cells more effectively than wild-type Ebola virus GP virions (Fig. 4C). While the mucin domain may contribute to hMGL interactions, it appears that the costs associated with the mucin domain may include reduced affinity for a cellular factor. It is unclear why the mucin domain may inhibit virus entry, but is possible that the bulky glycosylated region may sterically prevent interactions of the Ebola virus GP N terminus with cellular factors. Removal of the mucin region upon cathepsin proteolysis in the endosome may promote receptor engagement and permit virus entry to proceed. Analysis of mutants in which the carbohydrate addition sites are altered will be required to discriminate whether loss of the glycan residues or deletion of the protein sequence is responsible for increased binding of the GP-Δmuc virions.
Unlike Kuhn et al. (7), who did not detect binding of a truncated GP1 construct to lymphocytes, we observed a significant increase in binding of CatL-proteolysed GP1 relative to virions possessing full-length GP (Fig. 5). We believe that the discrepancy in these findings is due to differences among the Ebola virus GP substrates used in our binding experiments. CatL-proteolysed Ebola virus GP in our experiments was a higher-molecular-weight species than the minimal fragment used by Kuhn et al. Moreover, in our experiments binding was examined in the context of virions rather than with purified nonoligomeric protein. Avidity-driven virion binding would likely reveal interactions that are not detected with purified protein. Furthermore, it remains unclear whether other proteolysed GP products remain noncovalently associated with virions in our studies, further contributing to cell binding. The observation that Jurkat cells remained largely resistant to infection with CatL-proteolysed virions despite enhancement of virus binding at the cell surface may suggest that the block to infection in Jurkat cells is downstream of virus binding. Identifying the block in lymphocytes may reveal additional factors or steps involved in Ebola virus entry.
Acknowledgments
We thank Bob Doms and members of the Bates lab their helpful discussions and thoughtful comments about the manuscript. We also thank the Penn CFAR core for performing the HIV p24 assays. An antibody to HIV p24 was kindly supplied by Bob Doms.
This work was funded by Public Health Service grants T32 GM07229 and T32 AI07324 (R.L.K.), U54 AI057168 (G.S.), and R01 AI43455 (P.B.).
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Barnard, R. J., D. Elleder, and J. A. Young. 2006. Avian sarcoma and leukosis virus-receptor interactions: from classical genetics to novel insights into virus-cell membrane fusion. Virology 344:25-29. [DOI] [PubMed] [Google Scholar]
- 2.Chan, S. Y., R. F. Speck, M. C. Ma, and M. A. Goldsmith. 2000. Distinct mechanisms of entry by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J. Virol. 74:4933-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandran, K., N. J. Sullivan, U. Felbor, S. P. Whelan, and J. M. Cunningham. 2005. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308:1643-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 5.Geisbert, T. W., and P. B. Jahrling. 1995. Differentiation of filoviruses by electron microscopy. Virus Res. 39:129-150. [DOI] [PubMed] [Google Scholar]
- 6.Huang, I. C., B. J. Bosch, F. Li, W. Li, K. H. Lee, S. Ghiran, N. Vasilieva, T. S. Dermody, S. C. Harrison, P. R. Dormitzer, M. Farzan, P. J. Rottier, and H. Choe. 2006. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 281:3198-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuhn, J. H., S. R. Radoshitzky, A. C. Guth, K. L. Warfield, W. Li, M. J. Vincent, J. S. Towner, S. T. Nichol, S. Bavari, H. Choe, M. J. Aman, and M. Farzan. 2006. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 281:15951-15958. [DOI] [PubMed] [Google Scholar]
- 8.Malashkevich, V. N., B. J. Schneider, M. L. McNally, M. A. Milhollen, J. X. Pang, and P. S. Kim. 1999. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Natl. Acad. Sci. USA 96:2662-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manicassamy, B., J. Wang, H. Jiang, and L. Rong. 2005. Comprehensive analysis of Ebola virus GP1 in viral entry. J. Virol. 79:4793-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marsh, M., and A. Helenius. 2006. Virus entry: open sesame. Cell 124:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina, M. F., G. P. Kobinger, J. Rux, M. Gasmi, D. J. Looney, P. Bates, and J. M. Wilson. 2003. Lentiviral vectors pseudotyped with minimal filovirus envelopes increased gene transfer in murine lung. Mol. Ther. 8:777-789. [DOI] [PubMed] [Google Scholar]
- 12.Pager, C. T., W. W. Craft, Jr., J. Patch, and R. E. Dutch. 2006. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pager, C. T., and R. E. Dutch. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12714-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu, Z., S. T. Hingley, G. Simmons, C. Yu, J. Das Sarma, P. Bates, and S. R. Weiss. 2006. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 80:5768-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schornberg, K., S. Matsuyama, K. Kabsch, S. Delos, A. Bouton, and J. White. 2006. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80:4174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 102:11876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simmons, G., R. J. Wool-Lewis, F. Baribaud, R. C. Netter, and P. Bates. 2002. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 76:2518-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soneoka, Y., P. M. Cannon, E. E. Ramsdale, J. C. Griffiths, G. Romano, S. M. Kingsman, and A. J. Kingsman. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takada, A., K. Fujioka, M. Tsuiji, A. Morikawa, N. Higashi, H. Ebihara, D. Kobasa, H. Feldmann, T. Irimura, and Y. Kawaoka. 2004. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 78:2943-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weissenhorn, W., L. J. Calder, S. A. Wharton, J. J. Skehel, and D. C. Wiley. 1998. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 95:6032-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 2:605-616. [DOI] [PubMed] [Google Scholar]
- 23.Wong, S. K., W. Li, M. J. Moore, H. Choe, and M. Farzan. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wool-Lewis, R. J., and P. Bates. 1998. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 72:3155-3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, Z. Y., H. J. Duckers, N. J. Sullivan, A. Sanchez, E. G. Nabel, and G. J. Nabel. 2000. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 6:886-889. [DOI] [PubMed] [Google Scholar]