Abstract
Recent advances in reverse genetics of hepatitis C virus (HCV) made it possible to determine the properties and biochemical compositions of HCV virions. Sedimentation analysis and characterization of HCV RNA-containing particles produced in the cultured cells revealed that HCV virions cover a large range of heterogeneous densities in sucrose gradient. The fractions of low densities are infectious, while the higher-density fractions containing the majority of HCV virion RNA are not. HCV core protein and apolipoprotein B and apolipoprotein E (apoE) were detected in the infectious HCV virions. The level of apoE correlates very well with HCV infectivity. Both apoE- and HCV E2-specific monoclonal antibodies precipitated HCV, demonstrating that HCV virions contain apoE and E2 proteins. apoE-specific monoclonal antibodies efficiently neutralized HCV infectivity in a dose-dependent manner, resulting in a reduction of infectious HCV by nearly 4 orders of magnitude. The knockdown of apoE expression by specific small interfering RNAs (siRNAs) remarkably reduced the levels of intracellular as well as secreted HCV virions. The apoE siRNA suppressed HCV production by more than 100-fold at 50 nM. These findings demonstrate that apoE is required for HCV virion infectivity and production, suggesting that HCV virions are assembled as apoE-enriched lipoprotein particles. Our findings also identified apoE as a novel target for discovery and development of antiviral drugs and monoclonal antibodies to suppress HCV virion formation and infection.
Hepatitis C virus (HCV) is a major cause of liver diseases, affecting approximately 170 million people worldwide (59). Most (∼85%) acutely HCV-infected individuals become chronic carriers that can develop cirrhosis and hepatocellular carcinoma (50). HCV is an enveloped RNA virus with a single-strand and positive-sense RNA genome and is classified as Hepacivirus in the Flaviviridae family (47). The genomic RNA consists of a long open reading frame and relatively short untranslated regions (UTR) at the 5′ and 3′ ends (11, 32, 36, 46, 53). The 5′ and 3′ UTR contain cis-acting RNA elements important for HCV polyprotein translation and RNA replication (16-18, 38, 39, 61, 62). The translation of HCV polyprotein is mediated by the internal ribosomal entry site within the 5′ UTR (46, 58). Upon translation, the HCV polyprotein is cleaved by cellular peptidases and viral proteases into different viral proteins in the order of C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B (36, 38). A number of studies demonstrated that the NS3 to NS5B proteins are sufficient for HCV RNA replication (4, 6, 37), which occurs in the membrane-bound replication complex consisting of HCV RNA and proteins as well as cellular proteins (13, 14, 42, 57). The core and NS5B coding regions also contain cis-acting RNA elements important for HCV RNA replication and regulation (40, 63). Last, the newly synthesized HCV proteins and genomic RNA are packaged to form progeny virus particles. However, the molecular aspects underlying HCV virion assembly, maturation, and egression have not been determined.
Accumulating evidence suggests that HCV virions possess unusual properties distinctive from other members of the Flaviviridae family (2). HCV particles isolated from chronic hepatitis C patients or produced in vitro display a striking density heterogeneity, covering a large range from 1.06 to 1.20 g/ml (2, 3, 9, 10, 26, 35, 43, 45, 54-56, 64). Biochemical and morphological studies by André et al. suggest that the low-density HCV virions are packaged as lipoviroparticles (LVPs) with densities similar to that of the very-low-density lipoprotein (VLDL) (2, 3). Purified LVPs were rich in triglycerides and contained at least apolipoprotein B (apoB), HCV RNA, and core protein (2, 3). Both apoB and apoE were detected in the low-density fractions of the HCV RNA-containing particles. HCV virions could also be precipitated by apoB- and apoE-specific antibodies (2, 43). However, the roles of lipoproteins in HCV replication and infection have not been defined due to the lack of a robust cell culture system for HCV propagation. Recent success in HCV reverse genetics in vitro made it possible to study the molecular aspects of HCV infection, replication, virion assembly, and egression (9, 21, 35, 56, 64). The characterization of HCV virions produced in vitro revealed a remarkable disparity between the abundance of HCV virion RNA (vRNA) and infectious titer (9, 35, 56, 64). The infectious HCV titer was at least 1,000-fold lower than the HCV vRNA copy number, suggesting that the majority of HCV RNA-containing particles were not infectious (9). However, the properties and biochemical compositions of HCV RNA-containing particles have not been determined. A recent report described that the intracellular infectious HCV virions exhibit higher buoyant densities than those secreted into the culture medium, suggesting an alteration of biochemical compositions conferring the low densities during HCV virion maturation and secretion (19). It was reported recently that inhibition of VLDL assembly by apoB-specific small interfering RNA (siRNA) or an inhibitor of microsomal triglyceride transfer protein (MTP) also suppressed HCV production, implying a possible coupling of HCV production with VLDL assembly (24). However, the roles of apolipoproteins in HCV infection and virion assembly have not been defined.
In the present study, we sought to determine the properties of HCV virions and the role of apoE in HCV infectivity and production. The HCV RNA-containing particles secreted into the media were separated by sucrose density gradient sedimentation. Characterization of the resulting HCV virions revealed that HCV virions display heterogeneous densities. The low-density HCV virions were rich in apoE protein, suggesting an important role of apoE in the HCV life cycle. Consistent with this finding, HCV virions were precipitated specifically by apoE- and HCV E2-specific monoclonal antibodies. Additionally, HCV infectivity was neutralized efficiently by apoE-specific monoclonal antibodies. Furthermore, HCV production was remarkably suppressed by siRNA-mediated knockdown of apoE expression. Collectively, these findings demonstrate that apoE is required for HCV infectivity and production, opening up a novel target for discovery and development of antiviral drugs and monoclonal antibodies against HCV infection.
MATERIALS AND METHODS
Cell lines and cell culture.
Human hepatoma cell lines that stably produce infectious HCV of genotype 2a (JFH1) were described previously (9). A Huh7.5 cell line was kindly provided by Charles M. Rice (7). Subgenomic JFH1 HCV replicon-containing Huh7.5 cells were made in the same way as reported previously (29). All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS), nonessential amino acids, penicillin, and streptomycin (Invitrogen). The JFH1 (2a) replicon cell lines were maintained in the presence of 0.5 mg/ml of G418, while the HCV-producing cells were maintained by the addition of 5 μg/ml of blasticidin (A.G. Scientific).
Production of apoE-specific monoclonal antibodies.
A hybridoma cell line producing an apoE (1506 A1.4) blocking monoclonal antibody was obtained from the American Type Culture Collection (Manassas, VA) (34), and these cells were maintained in RPMI 1640 (Invitrogen) medium supplemented with 10% FBS. Recombinant apoE proteins were purchased from PeproTech (Rocky Hill, NJ). Female RBF-DnJ mice (Jackson Laboratories) were immunized subcutaneously with 50 μg of purified recombinant apoE emulsified with Freund's complete adjuvant (Sigma). Three subsequent booster immunizations were given to each animal at 10-day intervals. At 3 days after the last booster immunization, animals were euthanized humanely and the spleens were removed with sterile scissors. Splenic lymphocytes were fused with P3X myeloma cells by use of polyethylene glycol 4000 (Sigma). Hybridomas were selected with RPMI 1640 medium supplemented with Hybrimax, controlled process serum replacement type 3 (Sigma), hypothanxine-aminopterin-thymidine, and hypothanxine-thymidine (Invitrogen). Hybridomas producing apoE monoclonal antibodies were identified by screening with enzyme-linked immunosorbent assay using recombinant apoE as an antigen. Positive clones were subcloned twice by limiting dilution using standard protocols (31). The apoE monoclonal antibodies were purified from hybridoma media by use of a protein A antibody purification kit (Sigma, St. Louis, MO). The concentration of purified monoclonal antibodies was determined by spectrophotometry.
HCV preparation and sucrose gradient sedimentation analysis.
The stable HCV-producing cells were grown in 175-cm2 cell culture flasks. A large quantity of cell culture media was prepared and cleared by being passed through a 0.22-μm filter unit (Corning). The HCV RNA-containing particles in the media were concentrated by ultracentrifugation at 27,000 rpm and 4°C for 6 h in a Beckman SW28 rotor. The concentrated HCV was subjected to 20 to 60% sucrose gradient sedimentation analysis, as previously described (9). A total of 12 fractions with 1 ml each were collected from the top to the bottom of the sucrose gradient. The sucrose in fractions was removed by dilution with 1× phosphate-buffered saline (PBS) at a 1:12 ratio and then ultracentrifugation at 40,000 rpm for 16 h at 4°C in a Beckman SW41 rotor. The resulting HCV pellets were dissolved in PBS and aliquoted for subsequent studies.
Immunoprecipitation (IP) of HCV RNA-containing particles.
A total of 300 μl twofold-diluted protein G-conjugated agarose beads (Invitrogen) were first incubated with various amounts (0.6, 3, and 15 μg) of normal mouse immunoglobulin G1 (mIgG1), HCV E2-specific monoclonal antibody (CBH5), or apoE-specific monoclonal antibody (mAb23) with shaking for 5 h at room temperature. The unbound antibodies were removed by washing with 1× PBS three times. The antibody-bound agarose beads were then mixed with 300 μl of infectious HCV at 4°C overnight. The unbound HCV virions were removed by washing with 1× PBS three times. The HCV-bound agarose beads were resuspended into 250 μl with 1× PBS and were subjected to vRNA extraction by the addition of 750 μl of Trizol LS reagent (Invitrogen). The resulted vRNA was quantified by RNase protection assay (RPA) using a radiolabeled RNA probe containing the negative strand of the HCV 3′ UTR (9).
HCV infection and antibody neutralization.
Huh7.5 cells in 12-well (for detection of HCV proteins) or 6-well (for quantification of HCV RNA) plates were infected with purified HCV or HCV-containing culture medium at 37°C for 2 to 3 h. The HCV-infected cells were washed twice with PBS and then incubated with 10% FBS-containing DMEM. For antibody neutralization of HCV infectivity, HCV was incubated with increasing concentrations (0, 0.4, 2.0, 10, and 50 μg/ml) of normal mIgG or apoE monoclonal antibodies, respectively, at room temperature for 1 h and then added onto Huh7.5 cells (9, 30). apoE monoclonal antibodies include 1506 A1.4 (34) and apoE mAb23 and mAb30, which were produced in our lab. At 2 h postinfection (p.i.), the antibody-HCV mixture was aspirated and cells were washed twice with 1× PBS and then incubated with DMEM containing 10% FBS for 3 to 4 days. The HCV titer in the medium was determined by immunofluorescence analysis (IFA). Cell lysate was used for detection of HCV NS3 protein by Western blotting, while total RNA was extracted with Trizol (Invitrogen) for determination of HCV RNA by RPA (9).
Knockdown of apoE expression by siRNA.
Initially, SmartPool siRNAs targeting apoE mRNA were obtained from Dharmacon. The most potent siRNAs identified from SmartPool siRNAs were chosen for subsequent experiments. The siRNA-targeting sequence for apoE mRNA is 5′-AGACAGAGCCGGAGCCCGA-3′. A nonspecific control (NSC) siRNA was purchased from Dharmacon. The apoE-specific siRNA was transfected into HCV-infected Huh7.5 cells by use of Lipofectamine RNAiMax according to the manufacturer's instruction (Invitrogen). The mixture of siRNA and RNAiMax was serially diluted with serum-free medium to siRNA concentrations as indicated. At 4 to 5 days posttransfection, cell culture medium was collected and cleared by centrifugation to remove any detached cells and cell debris. Cells attached to flasks were washed twice with 1× PBS and then scraped into 1× PBS. Cell pellets were resuspended in PBS and frozen and thawed three times to isolate the intracellular HCV virions (19). The infectivity of intracellular virions and HCV in the medium was determined by subsequent infection of Huh7.5 cells, in which the levels of HCV NS3 protein and positive-strand RNA were determined by Western blotting and RPA, respectively. The titers of infectious HCV in the cell and medium were determined by IFA. HCV RNA-containing particles in the medium were concentrated by ultracentrifugation at 27,000 rpm for 6 h in a Beckman SW28 rotor. The HCV pellet was dissolved in DMEM and used for vRNA extraction with Trizol LS reagent (Invitrogen).
Ectopic overexpression of human apoE proteins.
pcDNA3.1/hApoE3, which expresses human apoE3, was kindly provided by Theodore Mazzone (University of Illinois at Chicago). A human apoE4-expressing vector, pCMV/hApoE4, was purchased from OriGene (Rockville, MD). Increasing amounts (0, 0.25, 0.5, 1, and 2 μg) of apoE-expressing DNA were transfected into Huh7.5 cells in six-well cell culture plates. The amount of DNA transfected into each well was kept constant at 2 μg by adjustment with vector DNAs. The mixture of 2 μg DNA and 5 μl DMRIE-C lipids in Opti-MEM (Invitrogen) was incubated at room temperature for 15 min prior to being transferred onto Huh7.5 cells. At 6 h posttransfection, the DNA-lipid mixture was replaced with DMEM containing 10% FBS. At 24 h posttransfection, cells were infected with HCV. At 3 days p.i., the culture media were collected and cells were lysed. The culture media were used to infect naïve Huh7.5 cells to determine HCV infectivity in the media derived from Huh7.5 cells with apoE overexpression. The levels of apoE expression in the cell and apoE secreted into the medium were determined by Western blot analysis.
Western blot analysis.
The lysate of HCV-infected or subgenomic HCV replicon-containing cells was prepared with standard procedures (9). The protein concentration of cell lysate was determined using a protein assay reagent (Bio-Rad). Twenty-five micrograms of total protein was resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by transferring onto a nitrocellulose membrane. HCV NS3 was determined by Western blotting using an NS3-specific monoclonal antibody, as previously described (9). The β-actin protein was used as an internal control. For quantifying the levels of apoE secretion, 12 μl of culture medium was electrophoresed by 10% SDS-PAGE. apoE was determined by Western blotting using an apoE-specific monoclonal antibody (apoE mAb33).
RNA extraction and quantification by RPA.
Total RNAs were extracted with Trizol reagent (Invitrogen) from HCV-infected cells or subgenomic HCV replicon-harboring cells. HCV vRNA in the medium was extracted with Trizol LS reagent (Invitrogen). The levels of positive-strand HCV RNA in the cell or vRNA in the medium were determined by RPA. RPA was the same as described previously (9). After digestion with RNase A/T1, RNA products were analyzed on a 6% polyacrylamide-7.7 M urea gel and visualized by autoradiography (39).
IFA.
HCV in the media was serially diluted (10×) and then used to infect naïve Huh7.5 cells on coverslips in 24-well plates. At 3 to 4 days p.i., IFA was performed to determine the numbers of cell foci stained for HCV NS3 protein. The procedures of IFA were the same as those used in our previous studies (9, 13).
RESULTS
Correlation of apoE protein with HCV infectivity.
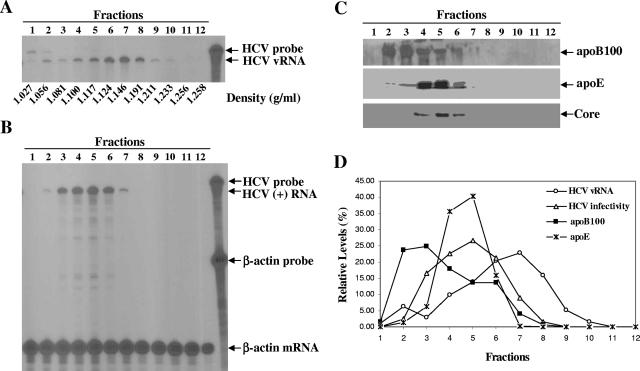
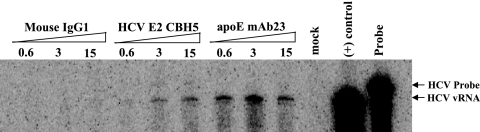
Previous studies revealed that the low-density HCV RNA-containing particles isolated from patients were assembled as LVPs. Findings derived from our recent studies as well as studies by others demonstrate that the HCV RNA-containing particles produced in cell culture display a broad range of buoyant densities, suggesting the existence of different forms of HCV virions (9, 35, 56, 64). To determine the properties of HCV virions and the role of apolipoproteins in HCV virion assembly and infection, the secreted HCV RNA-containing particles of various densities were separated by sucrose gradient sedimentation analysis. The levels of HCV vRNA in the fractions of sucrose gradient were quantified by RPA (Fig. 1A), while HCV infectivity in each fraction was determined by infection in cell culture (Fig. 1B). Also, HCV core, apoB, and apoE proteins in different fractions were detected by Western blotting (Fig. 1C). Similarly to previous findings (9), HCV RNA-containing particles cover a broad range of densities, varying from 1.03 to 1.23 g/ml, in sucrose gradient (Fig. 1A). The particles in the low-density fractions (fractions 3 to 6, with densities of 1.08 to 1.12 g/ml) were infectious, whereas those in the higher-density fractions (fractions 7 to 9, with densities of 1.15 to 1.21 g/ml), which accounted for the majority of HCV vRNA, were poorly or not infectious (Fig. 1B). Strikingly, fractions 4 to 6, with the highest HCV infectivity, also contained peak levels of apoE and HCV core protein (Fig. 1C). The levels of apoE in infectious fractions are consistent with the levels of HCV vRNA detected in the infectious fractions (Fig. 1D). In contrast, the highest levels of apoB were found in fractions 2 and 3, whose densities are lower than the fractions with the highest infectivity (Fig. 1C and D). To confirm the presence of apoE in infectious HCV virions, an IP experiment was carried out using apoE mAb23 and HCV E2-specific monoclonal antibody CBH5 (30). An isotype-matched normal mIgG1 was used as a control. Antibodies were bound to protein G-conjugated agarose beads and then incubated with infectious fractions (the mixture of fractions 3 to 6) (Fig. 1A). The levels of vRNA extracted from the precipitated HCV virions were determined by RPA. As shown in Fig. 2, both CBH5 and apoE mAb23 precipitated HCV RNA-containing particles in a dose-dependent manner. It appeared that the IP of HCV particles by apoE mAb23 reached a peak at 3 μg (Fig. 2). The lower IP at 15 μg was likely due to the presence of excess unbound mAb23 that prevented HCV from binding to mAb23-bound agarose beads (Fig. 2). In contrast, mIgG1 failed to precipitate HCV RNA-containing particles (Fig. 2). Taken together, these results demonstrate that infectious HCV virions are rich in apoE, suggesting a role of apoE in HCV assembly and infection.
FIG. 1.
Characterization of HCV RNA-containing particles. (A) Density gradient sedimentation analysis of HCV produced in cell culture. HCV virions secreted into the media were concentrated by ultracentrifugation and then fractionated through 20 to 60% sucrose density gradient centrifugation. Fractions (1 ml each) were collected from the top to the bottom of the sucrose gradient. The HCV vRNA was extracted with Trizol LS reagent and quantified by RPA, as described previously (9). (B) Determination of HCV infectivity in different fractions. Huh7.5 cells in six-well plates were infected with concentrated HCV of each fraction in DMEM at 37°C for 2 h. Cells were washed with PBS and incubated with 2 ml of DMEM containing 10% FBS. At 3 days p.i., total RNA was extracted with Trizol reagent and the levels of positive-strand (+) RNA were quantified by RPA. (C) Detection of apoB100 and apoE by Western blot analysis. apoB100 and apoE proteins in each fraction of the sucrose gradient were detected by Western blotting using apoB- and apoE-specific antibodies and visualized by enhanced chemiluminescence staining (9). (D) Comparison of HCV vRNA, infectivity, apoB100, and apoE levels in fractions of sucrose gradient. The data from panels A to C were quantified by densitometry as percentages. The relative levels of HCV vRNA, HCV infectivity, apoB100, and apoE in different fractions are plotted.
FIG. 2.
IP of infectious HCV virions. The procedures for IP of HCV virions by apoE mAb23, HCV E2-specific CBH5, and the isotype-matched mIgG1 are described in Materials and Methods. The vRNA of the immunoprecipitated HCV virions was determined by RPA, as described previously (9). Total RNAs extracted from mock- and HCV-infected Huh7.5 cells were used as negative and positive (+) controls, respectively. The HCV probe and vRNA product are highlighted, and the amounts (μg/ml) of antibodies used for IP are indicated.
Neutralization of HCV infectivity by apoE-specific monoclonal antibodies.
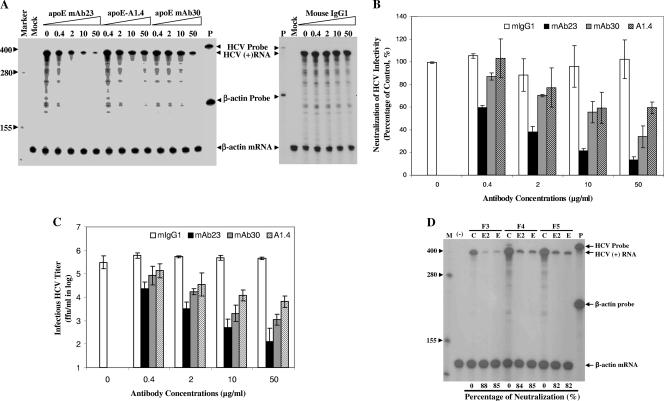
To determine whether apoE protein present in infectious HCV virions is required for HCV infectivity, HCV neutralization experiments were carried out using apoE-specific monoclonal antibodies. The commercially available apoE-blocking monoclonal antibody (1506 A1.4) was described previously (34). We also raised our own apoE-specific monoclonal antibodies by using purified recombinant apoE as an antigen. Upon selection and screening, a number of apoE-specific monoclonal antibodies were identified and purified. Five apoE monoclonal antibodies were able to neutralize HCV infectivity in cell culture (Fig. 3A and data not shown). Two of them, apoE mAb23 and mAb30, were titrated to determine their potency for neutralizing HCV infectivity. apoE mAb23 efficiently neutralized HCV infectivity in a dose-dependent manner (Fig. 3A and B). It lowered the HCV titers by 1, 2, 3, and nearly 4 orders of magnitude at 0.4, 2, 10, and 50 μg/ml, respectively (Fig. 3C). apoE mAb30 and 1506 A1.4 are much less potent than apoE mAb23 (Fig. 3A and B). As a control, the isotype-matched normal mIgG1 did not affect HCV infectivity (Fig. 3). The HCV-neutralizing activity of apoE mAb23 was further confirmed by using infectious HCV in the low-density fractions (F3 to F5) and compared in parallel with that of a human monoclonal antibody specific to HCV E2 protein, CBH5 (Fig. 3D). CBH5 was previously shown to be a very potent HCV-neutralizing monoclonal antibody (30). The HCV infectivities in fractions 3 to 5 (Fig. 1B) were neutralized equally by both apoE mAb23 and CBH5 at 10 μg/ml (Fig. 3D), consistent with the above-mentioned finding that HCV was immunoprecipitated specifically by both apoE mAb23 and CBH5 (Fig. 2). These results support the conclusion that infectious HCV virions contain both E2 and apoE proteins. Collectively, these findings demonstrate that apoE assembled in HCV virions is an important determinant of HCV infectivity and therein represents a novel target for antiviral intervention against HCV infection.
FIG. 3.
Neutralization of HCV infectivity by apoE-specific monoclonal antibodies. (A) Determination of apoE monoclonal antibody neutralization of HCV infectivity by RPA quantification of the levels of positive-strand HCV RNA. HCV (104 FFU/ml) was incubated with increasing concentrations (0, 0.4, 2, 10, and 50 μg/ml) of apoE mAb23, mAb30, 1506 A1.4 (ApoE-A1.4), or mIgG1 at room temperature for 1 h and was subsequently used to infect Huh7.5 cells in six-well cell culture plates. At 2 h p.i., the mixture of HCV and antibody was removed and cells were washed twice with PBS and then incubated with DMEM containing 10% FBS. At 3 days p.i., total RNA was extracted from the HCV-infected cells with Trizol reagent (Invitrogen). The levels of positive-strand (+) HCV RNA were determined by RPA. Antibody concentrations (μg/ml) are indicated. P, probe; values at left are RNA sizes in nucleotides. (B) Potency of HCV-neutralizing apoE monoclonal antibodies. The quantitative results derived from data shown in panel A were converted to percentages of the control. The levels of β-actin mRNA were used as controls to normalize the levels of HCV RNA. The levels of HCV RNA relative to that without apoE monoclonal antibody treatment (100%) were calculated. The relative levels of positive-strand HCV RNA on average are plotted against apoE monoclonal antibody concentrations. (C) Reduction of infectious HCV titer by apoE-specific monoclonal antibodies. Media were collected from cells infected with HCV in the presence of apoE-specific monoclonal antibodies, as described for panel A, at 3 days p.i. The titers of infectious HCV in the media were determined by IFA as FFU/ml. The average infectious HCV titers are plotted against antibody concentrations. (D) Neutralization of HCV infectivity in the low-density fractions (F3 to F5) by apoE mAb23 and HCV E2-specific human E2-specific monoclonal antibody CBH5 (30). An aliquot (20 μl) of concentrated HCV of fractions 3 to 5 diluted in DMEM was incubated with 10 μg/ml of either apoE mAb23 or CBH5 at room temperature for 1 h and then added onto Huh7.5 cells in six-well plates. The HCV-infected cells were washed twice with PBS and then incubated with DMEM containing 10% FBS for 3 days. Total RNA was extracted with Trizol reagent from the HCV-infected cells. The levels of positive-strand HCV RNA were determined by RPA and quantified by phosphorimager analysis. The levels of positive-strand HCV RNA relative to the control without antibody treatment were calculated as percentages of HCV neutralization. M, marker (values at left are RNA sizes in nucleotides); (−), mock infection; C, no antibody control; E, apoE mAb23; E2, HCV E2 monoclonal antibody CBH5.
Suppression of HCV virion production by siRNA-mediated knockdown of apoE expression.
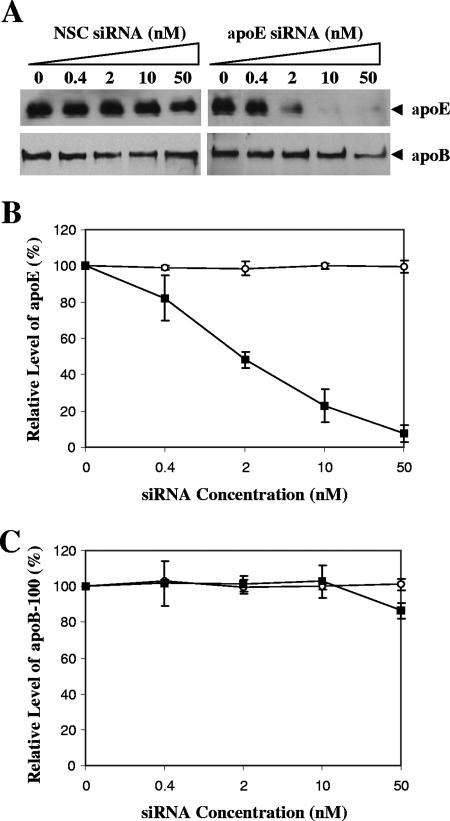
To determine the role of apoE in HCV virion assembly and egression, synthetic siRNA was used to specifically knock down apoE expression in the HCV-infected Huh7.5 cells. Huh7.5 cells were infected with HCV at a multiplicity of infection of approximately 0.01 at 37°C for 2 h and then transfected with apoE-specific or an NSC siRNA at concentrations of 0.4, 2, 10, and 50 nM. The levels of apoE secreted into the media were quantified by Western blotting using an apoE-specific monoclonal antibody (mAb33). The effect of apoE siRNAs on HCV production was subsequently determined by quantifying the levels of HCV RNA-containing particles and infectious HCV virions in the cell as well as in the media.
Most of the apoE secreted into the culture medium upon expression (data not shown). Therefore, the levels of apoE in the media were determined and compared. The apoE siRNA specifically reduced apoE secretion by 50%, 80%, and more than 90% at concentrations of 2, 10, and 50 nM, respectively (Fig. 4). However, apoE secretion was unaffected by the NSC siRNA, demonstrating the specificity of the siRNA-mediated knockdown of apoE expression. It was reported recently that apoB siRNA resulted in a reduction of HCV production (24). To examine whether apoE siRNA affected apoB secretion, we determined the levels of apoB secretion by Western blot analysis. Results show that apoE siRNA did not affect apoB secretion significantly (Fig. 4A and C). Taken together, these results demonstrate that the apoE siRNA efficiently and specifically knocked down apoE expression.
FIG. 4.
Knockdown of apoE expression by an apoE-specific siRNA. (A) Effects of apoE siRNA on apoE and apoB secretion. Huh7.5 cells were transfected with apoE-specific siRNA at various concentrations (nM) by use of Lipofectamine RNAiMax (Invitrogen) according to the manufacturer's instruction. An NSC siRNA was used as a negative control. At 4 days posttransfection, the levels of apoE and apoB100 secreted into the media were determined by Western blotting using polyclonal apoB antibodies (Calbiochem and Biodesign) and apoE mAb33 (raised in our lab), respectively. (B) Correlation of apoE secretion and siRNA concentrations. The levels of apoE relative to the control (without siRNA) were converted to percentages of the control. The relative levels of apoE secretion are plotted against siRNA concentrations. (C) Effect of apoE siRNA on apoB secretion. The quantitative data exemplified in panel A were converted into percentages of the control, considering the level of apoB100 in the absence of siRNA as 100%. The levels of apoB100 relative to controls are plotted against siRNA concentrations. The quantitative data in panels B and C represent means ± standard deviations obtained from three independent experiments. ○, NSC siRNA; ▪, apoE siRNA.
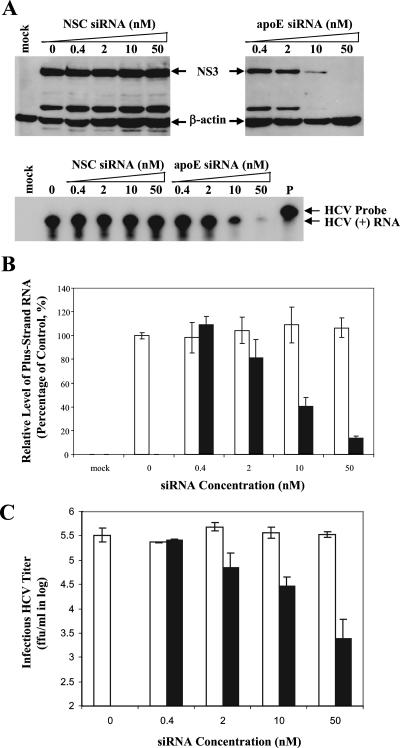
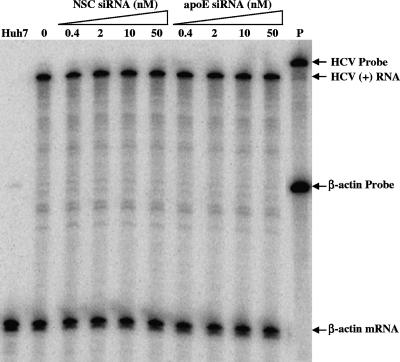
To determine the effect of apoE siRNA on HCV production, the levels of HCV RNA-containing particles in the media were quantified by several independent approaches. Initially, the infectious HCV virions secreted into the media were examined by infectivity. The media of the HCV-infected and siRNA-transfected cells were used to infect naïve Huh7.5 cells in 12-well (for detection of HCV NS3 protein) and 6-well (for quantification of positive-strand HCV RNA) cell culture plates. The levels of HCV NS3 protein and positive-strand HCV RNA in the infected naïve Huh7.5 cells were determined by Western blotting and RPA, respectively (Fig. 5A). Consistent with the reduction of apoE secretion, the levels of both NS3 and positive-strand RNA in Huh7.5 cells infected with the media correlated inversely with apoE siRNA concentrations (Fig. 5A and B). The apoE siRNA resulted in a decrease of NS3 protein by 80% at 10 nM and nearly 100% at 50 nM. The positive-strand HCV RNA was also reduced by 60% and 90% at 10 and 50 nM, respectively (Fig. 5A and B). The infectious HCV titers in the media of the HCV-infected and siRNA-transfected cells were then determined by IFA as focus-forming units (FFU)/ml. As shown in Fig. 5C, infectious titers of HCV virions secreted into the media were remarkably reduced by siRNA-mediated knockdown of apoE expression in a dose-dependent manner. The apoE siRNA reduced the infectious titers of secreted HCV virions by 100-fold at 50 nM (Fig. 5C). In contrast, the NSC siRNA did not affect HCV production, as determined by the levels of HCV protein and RNA in the infected cells as well as infectious HCV titers in the media of the siRNA-transfected cells (Fig. 5).
FIG. 5.
Suppression of HCV production by siRNA-mediated knockdown of apoE expression. Huh7.5 cells in 175-cm2 flasks were infected with HCV at a multiplicity of infection of approximately 0.01. At 2 h p.i., cells were transfected with various concentrations (nM) of apoE or NSC siRNA by use of Lipofectamine RNAiMax according to the manufacturer's instruction. After 4 days, the cell culture media and HCV-infected Huh7.5 cells were collected for determining the effects of siRNA-mediated knockdown of apoE expression on HCV production. (A) Effects of the siRNA-mediated knockdown of apoE expression on the production of infectious HCV. The media derived from the HCV-infected and siRNA-transfected Huh7.5 cells were used to infect naïve Huh7.5 cells in 12-well plates (for detection of NS3 by Western blotting) or 6-well plates (for determination of positive-strand RNA by RPA). At 3 days p.i., cell lysates of naïve Huh7.5 cells infected with HCV in 12-well plates were loaded into 10% SDS-PAGE gels and proteins were separated by electrophoresis and transferred onto a nitrocellulose membrane. HCV NS3 was detected by Western blotting using an NS3-specific monoclonal antibody (9). The total RNA was extracted with Trizol reagent from the HCV-infected Huh7.5 cells in six-well plates. The levels of positive-strand (+) HCV RNA were determined by RPA. The concentrations of apoE and NSC siRNAs are indicated. The NS3 and β-actin (as an internal control) proteins as well as positive-strand HCV RNA are highlighted. Mock, Huh7.5 cells without HCV infection; P, probe. (B) Dose-dependent reduction of positive-strand HCV RNA by apoE-specific siRNA. The levels of positive-strand HCV RNA exemplified in panel A were quantified by phosphorimager analysis and converted into percentages of the control, considering the HCV RNA level without siRNA as 100%. The relative levels of HCV RNA are plotted against siRNA concentrations. (C) Reduction of infectious HCV titers by knockdown of apoE expression. Huh7.5 cells on coverslips in 24-well plates were infected with 10× serially diluted HCV in the media. The titers of infectious HCV virions were determined by IFA as the numbers of cell foci that stained positive for NS3 (FFU/ml). The titers of infectious HCV virions are plotted against siRNA concentrations. Open bars, NSC siRNA; filled bars, apoE siRNA. The data shown in panels B and C are derived from at least two separate experiments.
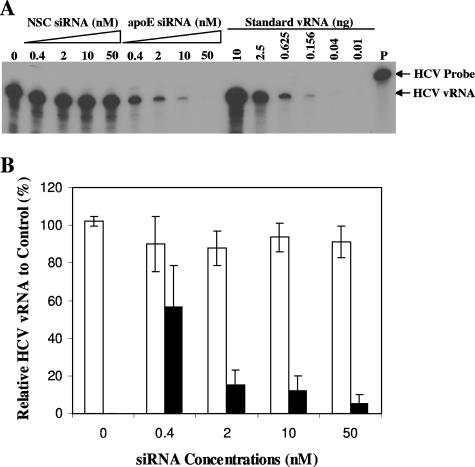
apoE was shown to be required for HCV infectivity (Fig. 3). Therefore, the reduction of HCV infectivity in the media of the apoE-specific siRNA-transfected cells could be due to either the absence of apoE in HCV virions or the blockage of HCV production. To determine whether the siRNA-mediated knockdown of apoE expression affected HCV virion production, HCV vRNA extracted from the media, which reflects the total level of HCV RNA-containing particles, was quantified by RPA. Similarly to the findings on HCV infectivity (Fig. 5), the apoE siRNA dramatically reduced the levels of HCV vRNA in proportion to the increase of siRNA concentrations, resulting in 50, 90, 95, and 99% reductions of HCV vRNA at 0.4, 2, 10, and 50 nM (Fig. 6). In contrast, the NSC siRNA did not cause any reduction of HCV vRNA in the media (Fig. 6). The degrees of HCV vRNA reduction were consistent with the levels of apoE and infectious HCV virions secreted into the media (Fig. 4 and 5). These findings demonstrate that apoE siRNA specifically knocked down apoE secretion and therein suppressed HCV virion production.
FIG. 6.
Suppression of the production of HCV RNA-containing particles by apoE-specific siRNA. The media of Huh7.5 cells infected with HCV and transfected with NSC or apoE siRNA were the same as those described in the legend for Fig. 5. The HCV RNA-containing particles in the media were concentrated by ultracentrifugation as described in Materials and Methods. HCV vRNA was extracted with Trizol LS reagent from concentrated HCV particles. (A) Determination of HCV vRNA levels by RPA. RPA was carried out using the same procedures as described previously (9). The in vitro T7 transcript of the JFH1 HCV RNA was used as a standard. The concentrations of NSC and apoE siRNAs are indicated. The radiolabeled RNA probe (P) and vRNA products are highlighted. (B) Correlation of HCV vRNA levels with siRNA concentrations. The levels of HCV vRNA shown in panel A were quantified by phosphorimager analysis and converted into percentages of the control, considering the vRNA level to be 100% in the medium derived from cells without siRNA treatment. Relative vRNA levels are plotted against siRNA concentrations. Open bars, NSC siRNA; filled bars, apoE siRNA.
Inhibition of HCV assembly by the siRNA-mediated knockdown of apoE expression.
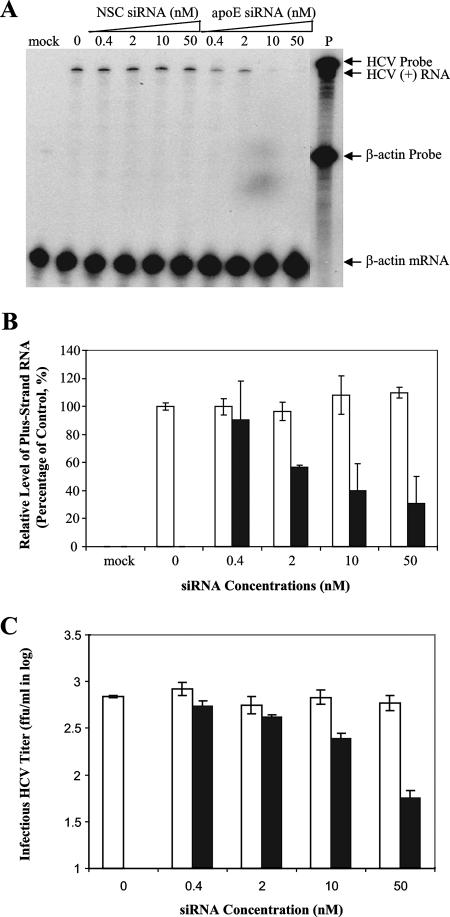
The reduction of HCV virions in the media could be caused by inhibition of HCV assembly and/or egression. To further determine whether apoE is required for HCV virion assembly, the levels of intracellular HCV virions were determined. The intracellular HCV virions were obtained by repeated freezing and thawing of the HCV-infected and siRNA-transfected Huh7.5 cells after extensive washing with PBS, followed by centrifugation to remove nuclei and cell debris. The resulting supernatant was used to determine the levels of intracellular HCV virions. Similarly to the reduction of infectious HCV in the media, the level of intracellular HCV virions was also decreased by apoE siRNA in a dose-dependent manner, as determined by the reduction of positive-strand HCV RNA (Fig. 7A and B). The infectious titers of intracellular HCV virions were more than 100-fold lower than those of secreted HCV virions. The apoE siRNA reduced infectious titers of intracellular HCV virions by at least 10-fold (Fig. 7C). These findings demonstrate that apoE is required for HCV virion assembly in the cell.
FIG. 7.
Inhibition of intracellular HCV assembly by knockdown of apoE expression. The HCV-infected and siRNA-transfected Huh7.5 cells were derived from the same experiments as described in the legend for Fig. 5. The intracellular HCV virions were prepared by the same methods reported by others (19). (A) Determination of intracellular HCV virions by the levels of positive-strand RNA in the infected cells. Huh7.5 cells were infected with intracellular HCV virions at 37°C for 2 h. At 3 days p.i., total RNA was extracted with Trizol reagent and used for the determination of positive-strand (+) HCV RNA by RPA. The concentrations of siRNA are indicated, and HCV RNA probe (P) and products are highlighted. (B) Correlation of the reduction of intracellular HCV virion production with siRNA concentrations. The levels of positive-strand HCV RNA exemplified in panel A were calculated as percentages of the control, considering the levels of positive-strand HCV RNA in infected Huh7.5 cells as 100%. The levels of HCV RNA relative to controls are plotted against siRNA concentrations. (C) Reduction of intracellular infectious HCV titers by apoE siRNA. The titers of intracellular infectious HCV were determined by IFA in the same way as described in the legend for Fig. 5. Open bars, NSC siRNA; filled bars, apoE siRNA.
Effect of ectopic apoE expression on HCV production.
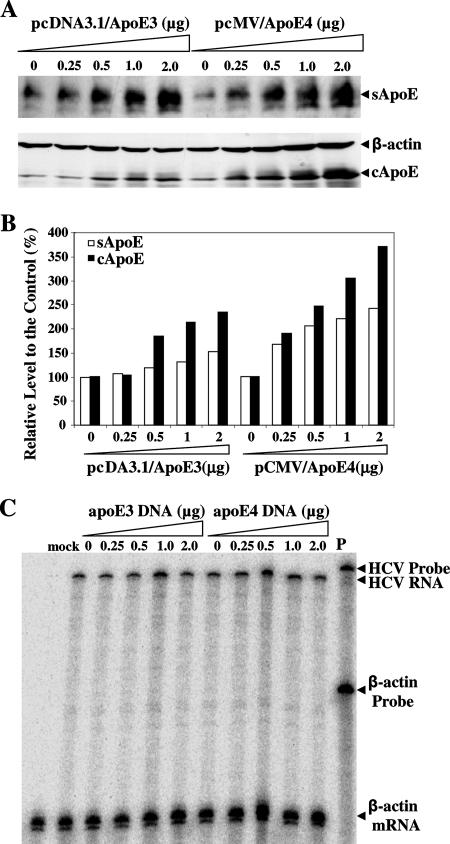
To examine whether ectopic apoE expression will affect HCV production, we transfected DNA vectors expressing human apoE3 and apoE4 into Huh7.5 cells prior to HCV infection. The levels of apoE expression in the cell and apoE secretion in the medium were determined by Western blotting. The levels of intracellular apoE3 and apoE4 were 2- to 3.5-fold higher in the apoE DNA-transfected cells than in normal cells (Fig. 8). Similarly, the levels of apoE3 and apoE4 secreted into the media were increased up to twofold (Fig. 8). However, the levels of HCV production were not significantly affected by ectopic overexpression of apoE3 and apoE4, as determined by HCV infectivity in the media derived from Huh7.5 cells with apoE overexpression. These results suggest that the level of apoE expression is not a limiting factor for HCV production, as apoE is expressed abundantly in Huh7.5 cells.
FIG. 8.
Effect of ectopic apoE expression on HCV production. Huh7.5 cells in six-well culture plates were transfected with increasing amounts (as indicated) of either pcDNA3.1/hApoE3 or pCMV-hApoE4 by use of DMRIE-C reagent (Invitrogen) following the manufacturer's instruction. At 6 h posttransfection, the medium was replaced with DMEM containing 10% FBS. At 24 h posttransfection, Huh7.5 cells were infected with HCV and then incubated for an additional 3 days. The media were collected, and cells were lysed as previously described (9). (A) Western blot analysis of apoE in the cell and media. The levels of apoE expression in the cell (cApoE) and secretion into the medium (sApoE) were determined by Western blotting in the same way as described in the legend for Fig. 4. (B) Quantification of apoE levels in the cell and medium. The intensities of apoE bands shown in panel A were quantified by densitometry. The levels of apoE were converted to percentages of the control, considering the level of apoE in normal Huh7.5 cells as 100%. The levels of apoE are plotted against the amounts of apoE-expressing DNA. (C) HCV infectivity, determined by the levels of positive-strand HCV RNA in infected Huh7.5 cells. The media derived from apoE DNA-transfected and HCV-infected cells were used to infect naïve Huh7.5 cells. At 3 days p.i., total RNA was extracted with Trizol reagent from HCV-infected cells. The levels of positive-strand HCV RNA were determined by RPA using 32P[UTP]-labeled RNA probe containing the negative-strand HCV 3′ UTR RNA. P, probe.
apoE siRNA did not affect HCV RNA replication.
The question arose whether apoE-specific siRNA affected HCV RNA replication, since apoE was found in the membrane-bound HCV replication complex (24; K.-S. Chang and G. Luo, unpublished data). The apoE and NSC siRNAs were transfected into Huh7.5 cells that harbored a subgenomic HCV replicon RNA of the same genotype (2a) (JFH1) as the infectious HCV used in this study (29). Similarly, apoE-specific siRNA but not NSC siRNA reduced apoE secretion in the HCV RNA replicon-containing cells (data not shown). However, the levels of positive-strand HCV RNA in the apoE siRNA-transfected cells were unaffected by apoE siRNA, demonstrating that apoE does not play a significant role in HCV RNA replication (Fig. 9). Therefore, the reduction of HCV virions by the siRNA-mediated knockdown of apoE expression in the cell was most likely due to the blockage of HCV virion assembly.
FIG. 9.
Effect of apoE siRNA on replication of a subgenomic HCV RNA replicon. Huh7.5 cells carrying a subgenomic JFH1 HCV replicon RNA were transfected with either apoE or NSC siRNA. After 3 days, the levels of positive-strand HCV RNA were determined by RPA. The levels of β-actin mRNA were used as internal controls. The positive-strand HCV RNA and β-actin mRNA products were analyzed in a 6% polyacrylamide-urea (7.7 M) gel. The concentrations of siRNA are shown, and the radiolabeled HCV RNA and β-actin probes (P) and positive-strand (+) HCV RNA and β-actin mRNA products are indicated.
DISCUSSION
Increasing evidence derived from a number of studies suggests that infectious HCV virions are packaged as LVPs (2, 3). Several previous studies found that the circulating HCV virions obtained from hepatitis C patients displayed a large range of low densities, as revealed by density gradient sedimentation analysis (10, 25, 41, 55). The HCV infectivity, when examined in chimpanzees, correlated inversely with the density of HCV particles (8, 23). These observations led to the speculation that HCV virions were associated with the plasma lipoproteins in vivo. However, it was not clear whether HCV virions were simply associated with circulating lipoproteins or assembled as integrated lipoprotein particles. To differentiate these possibilities, André et al. performed an elegant biochemical and morphological study of HCV particles isolated from the plasma of chronic hepatitis C patients. When viewed under an electron microscope, the HCV particles in the low-density fractions appeared as large spherical VLPs with a diameter of more than 100 nm. Delipidation of VLPs resulted in HCV capsid-like structures that reacted with core-specific antibodies. Purified LVPs were also shown to enter hepatocytes efficiently in vitro (2, 3). Supporting this concept, apoB- and apoE-specific polyclonal antibodies were able to precipitate more than 90% of the low-density HCV particles (43). Similarly, our findings described here demonstrate that both apoB and apoE were present in infectious HCV virions but that apoE is particularly enriched in infectious HCV virions (Fig. 1C). Furthermore, the infectivity of HCV in the low-density fractions was neutralized efficiently by HCV E2- and apoE-specific monoclonal antibodies, demonstrating that infectious HCV virions contain E2 and apoE proteins on the viral envelope (Fig. 3). Collectively, these findings suggest that infectious HCV virions are likely assembled as integrated apoE-enriched lipoprotein particles.
Several lines of evidence derived from our present study demonstrate that apoE is required for HCV virion production. This conclusion was suggested initially by the positive correlation between the level of apoE and HCV infectivity in the low-density fractions (Fig. 1) and the IP of HCV virions by apoE- and HCV E2-specific monoclonal antibodies (Fig. 2). More importantly, the siRNA-mediated knockdown of apoE expression resulted in a reduction of infectious HCV in the media by more than 2 orders of magnitude (Fig. 5). The level of HCV vRNA in the media was decreased to a level of underdetection by apoE-specific siRNA at 50 nM (Fig. 6), demonstrating that the knockdown of apoE expression efficiently suppressed HCV production. Likewise, the level of intracellular HCV virions was lowered considerably by apoE-specific siRNA (Fig. 7). The suppression of HCV production correlated closely with the siRNA-mediated knockdown of apoE expression. However, apoE expression and HCV production were unaffected by an NSC siRNA. Neither HCV RNA replication was affected by apoE-specific siRNA, as determined by HCV RNA replication in a subgenomic HCV RNA-harboring Huh7.5 cell line (Fig. 9). A recent study found that the siRNA-mediated knockdown of apoB expression and an MTP inhibitor blocking apoB secretion suppressed HCV production. However, the apoE-specific siRNA used in our studies did not affect apoB secretion significantly (Fig. 4). Collectively, these findings demonstrate that apoE is likely required for HCV assembly, although its role in virion maturation and egression cannot be excluded completely. Structural determination of the requirement of apoE for HCV virion assembly is warranted for future investigations. The blockage of apoE-containing lipoprotein assembly inhibits HCV production, opening up a novel target for discovery and development of antiviral drugs against HCV infection.
The underlying molecular mechanism for the assembly of HCV virions is not yet known. Previous studies demonstrated that HCV replication is closely related to lipid and lipoprotein metabolism. Many of the cholesterol and lipoprotein biosynthesis genes might play important roles in HCV RNA replication and virion assembly (27, 28, 51). The membrane fractions isolated from HCV replicon-bearing Huh7 cells, which contained the HCV replication complex, were also found to be enriched with proteins (apoB, apoE, and MTP) required for VLDL assembly (24). As with other positive-strand RNA viruses, HCV RNA replication takes place in the membrane-bound replication complex containing multiple viral and cellular proteins (12, 13, 38, 42). HCV RNA replication and/or HCV proteins were shown to induce an alteration of the endoplasmic reticulum (ER) membranes, resulting in distinct membrane structures designated “membranous webs” (33, 48). HCV RNA is thought to replicate within the modified ER membrane structures (12, 13, 38, 42). In HCV-infected cells, the E1/E2 glycoprotein heterodimer was also found to retain in the ER (48). Similarly to the HCV replication complex, the hepatic lipoproteins are assembled in the ER (15). Thus, the assembly of HCV virion and VLDL occurs in the same intracellular compartment, which provides a structural basis for the assembly of HCV LVPs. This model is supported by the findings that HCV core protein, E2, and apoE were all found in the same membrane fraction (Chang and Luo, unpublished), as determined by membrane flotation analysis (13), suggesting that the structural components necessary for formation of HCV virions are located in the same intracellular compartment.
In the present study, our findings demonstrate that apoE is required for HCV infectivity besides its important role in HCV virion assembly. The infectivity of HCV RNA-containing particles in cell culture correlated perfectly well with the levels of HCV vRNA and apoE protein in the low-density infectious fractions (Fig. 1). apoE-specific monoclonal antibody was also able to precipitate HCV virions in the infectious fractions (Fig. 2). These findings demonstrate that apoE is enriched in the infectious HCV virions. Most significantly, apoE-specific monoclonal antibodies were able to neutralize HCV infectivity in a dose-dependent manner. apoE mAb23 resulted in a reduction of infectious HCV titer by nearly 4 orders of magnitude at 50 μg/ml (Fig. 3). Both apoE mAb23 and HCV E2-specific CBH5 neutralized HCV infectivity in the infectious fractions equally, by more than 80% at 10 μg/ml (Fig. 3D). We have shown recently that the neutralization of HCV infection by CBH5 was not due to a physical hindrance, since some of the HCV E2-specific monoclonal antibodies did not neutralize HCV infectivity even though they had equal or higher HCV-binding affinity than CBH5 (30). The mechanism of action of apoE-specific monoclonal antibodies for HCV neutralization is under investigation. Nevertheless, our findings demonstrate that human apoE is required for HCV infection and thereby represents a novel target for antiviral intervention against HCV infection. One immediate application of the HCV-neutralizing apoE monoclonal antibodies is to be pursued as prophylactic agents for preventing HCV infection in liver transplant recipients. The humanized apoE monoclonal antibodies may also provide clinical benefits to the treatment of chronic hepatitis C in combination with other HCV inhibitors (52).
The requirement of apoE for HCV infectivity suggests that it may also play an important role in the development of persistent HCV infection. The most characteristic feature of HCV is its ability to establish a chronic infection in the majority of acutely HCV-exposed individuals. The advantage for HCV to use a host protein for its cell entry is to mask the host immune response so that it can establish a persistent infection. This concept is supported by recent findings derived from genetic studies that the outcome of HCV infection correlated with apoE polymorphism (44, 60). The apoE3 allele was found to correlate with persistent HCV infection (44), whereas the apoE2 and apoE4 alleles were associated with a reduced likelihood of chronic HCV infection (44, 60). Therefore, functional apoE gene polymorphisms may represent an important determinant of outcome in HCV infection. Whether apoE isoforms (E2, E3, and E4) will have different effects on HCV virion assembly and/or infectivity remains to be determined by future studies.
How apoE mediates the binding of HCV to cells remains an interesting question that waits to be addressed in future studies. It is known that apoE can interact with a variety of cell surface receptors (22), including the low-density-lipoprotein receptor (LDLR), VLDL receptor, scavenger receptor class B type I, LDLR-related protein, and apoE receptor. Both LDLR and scavenger receptor class B type I have already been shown to play important roles in HCV entry (1, 5, 20, 27, 49). It is not yet clear, however, whether other apoE-binding receptors play any role in mediating HCV cell entry. Future investigations are warranted to determine the receptor for apoE and the underlying molecular mechanism of apoE-mediated HCV entry into target cells.
Acknowledgments
We thank Charles M. Rice (Rockefeller University) for kindly providing the Huh7.5 cell line, Takaji Wakita (National Institute of Health, Japan) for the JFH1 replicon cDNA pSGR-JFH1, Theodore Mazzone (University of Illinois at Chicago) for pcDNA3.1/hApoE3, and Lei Cai and Deneys R. van der Westhuyzen (University of Kentucky) for apoB polyclonal antibodies. We also thank Fei Liu (visiting Ph.D. student from Central South University in China) for technical assistance and Deneys R. van der Westhuyzen for critical reading of the manuscript.
This work was supported by NIH grants AI051592 and AI070769.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.André, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.André, P., G. Perlemuter, A. Budkowska, C. Brechot, and V. Lotteau. 2005. Hepatitis C virus particles and lipoprotein metabolism. Semin. Liver Dis. 25:93-104. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., M. Frese, and T. Pietschmann. 2004. Novel insights into hepatitis C virus replication and persistence. Adv. Virus Res. 63:71-180. [DOI] [PubMed] [Google Scholar]
- 5.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley, D., K. McCaustland, K. Krawczynski, J. Spelbring, C. Humphrey, and E. H. Cook. 1991. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J. Med. Virol. 34:206-208. [DOI] [PubMed] [Google Scholar]
- 9.Cai, Z., C. Zhang, K. S. Chang, J. Jiang, B. C. Ahn, T. Wakita, T. J. Liang, and G. Luo. 2005. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 79:13963-13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrick, R. J., G. G. Schlauder, D. A. Peterson, and I. K. Mushahwar. 1992. Examination of the buoyant density of hepatitis C virus by the polymerase chain reaction. J. Virol. Methods 39:279-289. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Hage, N., and G. Luo. 2003. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J. Gen. Virol. 84:2761-2769. [DOI] [PubMed] [Google Scholar]
- 14.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. USA 101:13038-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher, E. A., and H. N. Ginsberg. 2002. Complexity in the secretory pathway: the assembly and secretion of apolipoprotein B-containing lipoproteins. J. Biol. Chem. 277:17377-17380. [DOI] [PubMed] [Google Scholar]
- 16.Friebe, P., and R. Bartenschlager. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76:5326-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friebe, P., J. Boudet, J. P. Simorre, and R. Bartenschlager. 2005. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79:380-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gastaminza, P., S. B. Kapadia, and F. V. Chisari. 2006. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 80:11074-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grove, J., T. Huby, Z. Stamataki, T. Vanwolleghem, P. Meuleman, M. Farquhar, A. Schwarz, M. Moreau, J. S. Owen, G. Leroux-Roels, P. Balfe, and J. A. McKeating. 2007. Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity. J. Virol. 81:3162-3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heller, T., S. Saito, J. Auerbach, T. Williams, T. R. Moreen, A. Jazwinski, B. Cruz, N. Jeurkar, R. Sapp, G. Luo, and T. J. Liang. 2005. An in vitro model of hepatitis C virion production. Proc. Natl. Acad. Sci. USA 102:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herz, J., and Y. Chen. 2006. Reelin, lipoprotein receptors and synaptic plasticity. Nat. Rev. Neurosci. 7:850-859. [DOI] [PubMed] [Google Scholar]
- 23.Hijikata, M., Y. K. Shimizu, H. Kato, A. Iwamoto, J. W. Shih, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1993. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J. Virol. 67:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, H., F. Sun, D. M. Owen, W. Li, Y. Chen, M. Gale, Jr., and J. Ye. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 104:5848-5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1994. Buoyant density of hepatitis C virus recovered from infected hosts: two different features in sucrose equilibrium density-gradient centrifugation related to degree of liver inflammation. Hepatology 19:296-302. [PubMed] [Google Scholar]
- 26.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Density analysis of hepatitis C virus particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J. Hepatol. 22:440-448. [DOI] [PubMed] [Google Scholar]
- 27.Kapadia, S. B., H. Barth, T. Baumert, J. A. McKeating, and F. V. Chisari. 2007. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81:374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kapadia, S. B., and F. V. Chisari. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl. Acad. Sci. USA 102:2561-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 30.Keck, Z. Y., J. Xia, Z. Cai, T. K. Li, A. M. Owsianka, A. H. Patel, G. Luo, and S. K. Foung. 2007. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J. Virol. 81:1043-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohler, G., and C. Milstein. 1975. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 256:495-497. [DOI] [PubMed] [Google Scholar]
- 32.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konan, K. V., T. H. Giddings, Jr., M. Ikeda, K. Li, S. M. Lemon, and K. Kirkegaard. 2003. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J. Virol. 77:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krul, E. S., M. J. Tikkanen, T. G. Cole, J. M. Davie, and G. Schonfeld. 1985. Roles of apolipoproteins B and E in the cellular binding of very low density lipoproteins. J. Clin. Investig. 75:361-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 36.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 37.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 38.Luo, G. 2004. Molecular virology of hepatitis C virus. Birkhauser, Basel, Switzerland.
- 39.Luo, G., S. Xin, and Z. Cai. 2003. Role of the 5′-proximal stem-loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 77:3312-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMullan, L. K., A. Grakoui, M. J. Evans, K. Mihalik, M. Puig, A. D. Branch, S. M. Feinstone, and C. M. Rice. 2007. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl. Acad. Sci. USA 104:2879-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyamoto, H., H. Okamoto, K. Sato, T. Tanaka, and S. Mishiro. 1992. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J. Gen. Virol. 73:715-718. [DOI] [PubMed] [Google Scholar]
- 42.Moradpour, D., R. Gosert, D. Egger, F. Penin, H. E. Blum, and K. Bienz. 2003. Membrane association of hepatitis C virus nonstructural proteins and identification of the membrane alteration that harbors the viral replication complex. Antivir. Res. 60:103-109. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen, S. U., M. F. Bassendine, A. D. Burt, C. Martin, W. Pumeechockchai, and G. L. Toms. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price, D. A., M. F. Bassendine, S. M. Norris, C. Golding, G. L. Toms, M. L. Schmid, C. M. Morris, A. D. Burt, and P. T. Donaldson. 2006. Apolipoprotein epsilon3 allele is associated with persistent hepatitis C virus infection. Gut 55:715-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prince, A. M., T. Huima-Byron, T. S. Parker, and D. M. Levine. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J. Viral Hepat. 3:11-17. [DOI] [PubMed] [Google Scholar]
- 46.Rijnbrand, R. C., and S. M. Lemon. 2000. Internal ribosome entry site-mediated translation in hepatitis C virus replication. Curr. Top. Microbiol. Immunol. 242:85-116. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shini, P. Simmonds, D. Smith, L. Stuyver, A. Weiner, et al. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 48.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seeff, L. B., and J. H. Hoofnagle. 2002. National Institutes of Health Consensus Development Conference: management of hepatitis C: 2002. Hepatology 36:S1-S2. [DOI] [PubMed] [Google Scholar]
- 51.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan, S. L., A. Pause, Y. Shi, and N. Sonenberg. 2002. Hepatitis C therapeutics: current status and emerging strategies. Nat. Rev. Drug Discov. 1:867-881. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomssen, R., S. Bonk, C. Propfe, K. H. Heermann, H. G. Kochel, and A. Uy. 1992. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 181:293-300. [DOI] [PubMed] [Google Scholar]
- 55.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182:329-334. [DOI] [PubMed] [Google Scholar]
- 56.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, C., M. Gale, Jr., B. C. Keller, H. Huang, M. S. Brown, J. L. Goldstein, and J. Ye. 2005. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18:425-434. [DOI] [PubMed] [Google Scholar]
- 58.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.WHO. 1998. WHO concerns hepatitis C. Lancet 351:1415. [Google Scholar]
- 60.Wozniak, M. A., R. F. Itzhaki, E. B. Faragher, M. W. James, S. D. Ryder, and W. L. Irving. 2002. Apolipoprotein E-epsilon 4 protects against severe liver disease caused by hepatitis C virus. Hepatology 36:456-463. [DOI] [PubMed] [Google Scholar]
- 61.Yi, M., and S. M. Lemon. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77:3557-3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi, M., and S. M. Lemon. 2003. Structure-function analysis of the 3′ stem-loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA 9:331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.You, S., D. D. Stump, A. D. Branch, and C. M. Rice. 2004. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78:1352-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]