Abstract
Recognition of polypyrimidine (Py) tracts typically present between the branch point and the 3′ splice site by the large subunit of the essential splicing factor U2AF is a key early step in pre-mRNA splicing. Diverse intronic sequence arrangements exist, however, including 3′ splice sites lacking recognizable Py tracts, which raises the question of how general the requirement for U2AF is for various intron architectures. Our analysis of fission yeast introns in vivo has unexpectedly revealed that whereas introns lacking Py tracts altogether remain dependent on both subunits of U2AF, introns with long Py tracts, unconventionally positioned upstream of branch points, are unaffected by U2AF inactivation. Nevertheless, mutation of these Py tracts causes strong dependence on the large subunit U2AF59. We also find that Py tract diversity influences the requirement for the conserved C-terminal domain of U2AF59 (RNA recognition motif 3), which has been implicated in protein-protein interactions with other splicing factors. Together, these results suggest that in addition to Py tract binding by U2AF, supplementary mechanisms of U2AF recruitment and 3′ splice site identification exist to accommodate diverse intron architectures, which have gone unappreciated in biochemical studies of model pre-mRNAs.
In animals, removal of introns from the majority (>80 to 90%) of nascent transcripts via pre-mRNA splicing is an important step for gene regulation (19). Alternative splicing serves important biological roles in diverse developmental contexts and provides an important mechanism to generate molecular diversity (7). The 5′ splice site (SS), the branch point sequence (BPS), and the polypyrimidine (Py) tract 3′ SS in the pre-mRNA are important splicing signals; they are recognized by the U1 snRNP, the U2 snRNP, and the U2 snRNP auxiliary factor U2AF, respectively, resulting in the formation of a dynamic RNA-protein complex called the spliceosome (23).
In humans, the essential splicing factor U2AF is a heterodimer of a large protein subunit (U2AF65) and a small protein subunit (U2AF35). U2AF65 binds to the Py tract (62), and U2AF35 recognizes the 3′ SS (33, 61, 65). Both subunits of U2AF are essential for viability in many model organisms, such as the zebrafish, the fruit fly, the nematode worm, and fission yeast (U2AF59) (14, 24, 36, 43, 56, 66). However, in budding yeast the large subunit is dispensable (1) and the small subunit is absent. U2AF65 interacts with other splicing factors such as BBP/SF1, UAP56, SAP155 (or SF3b155), and SRp54 (1, 10, 15, 39, 64). The branch point binding protein BBP/SF1 binds to the BPS and cooperates with U2AF65 for RNA binding (2, 6).
Detailed in vitro biochemical analyses using model splicing substrates in metazoans have significantly contributed to our mechanistic view of the role of U2AF65 function in splicing. The N terminus of U2AF65 harbors an arginine-serine-rich (RS) activation domain, and its C terminus contains three RNA recognition motifs (RRMs), each with a four-stranded antiparallel β-sheet and two α-helices (62). In metazoans, binding to the Py tract serves as the primary determinant for U2AF recruitment onto pre-mRNA. This interaction positions the RS domain to engage in a series of interactions with pre-RNA during spliceosome assembly, including stabilization of the base pairing between the BPS and the U2 snRNA (46, 53). U2AF65 is also an important target for splicing regulation, where splicing regulators such as SXL, PTB, hnRNP A1, ASF/SF2, SC35, and TRA can facilitate or antagonize its activity (7, 48).
Whereas U2AF65 is highly conserved from fission yeast to humans, its C-terminal RRM3 domain is the only recognizable portion in the budding yeast protein Mud2p (1). The human RRM3 interacts with BBP/SF1 and SAP155 (or Schizosaccharomyces pombe prp10) (1, 15), and RRM3-related domains are present in several splicing factors (27). Deletion of the conserved RRM3 domain of the large subunit of U2AF (U2AF59) is lethal in S. pombe (4). Intriguingly, RRM3 shows no detectable RNA binding and is not required for the splicing of model substrates in a HeLa cell nuclear extract. RRM1 and RRM2 domains of the human U2AF65 are sufficient for Py tract recognition and in vitro splicing (4). Previously, we proposed that the RRM3 domain might be important for the splicing of only a subset of introns in vivo.
Relative to Saccharomyces cerevisiae, S. pombe shares with mammals many more features of pre-mRNA splicing, including the presence of degenerate splicing signals, similarity of splicing factors (snRNAs and proteins), and a requirement for both subunits of U2AF (U2AF59 and U2AF23) (57, 59). Thus, by combining the power of genomics, molecular genetics, and biochemical analysis, S. pombe represents an excellent model system for analysis of the role of U2AF and Py tract in vivo.
Our study of splicing in S. pombe offers important new information on RRM3 and U2AF functions and Py tract requirements in vivo. There is a large diversity in the arrangement of intronic sequences relevant for 3′ splice site recognition, and in the requirements for U2AF subunits and domains, beyond what is known from detailed in vitro biochemical analysis of model pre-mRNAs in metazoans. These findings also help explain why deletion of RRM3 is lethal in S. pombe whereas deletion of the human RRM3 has no effect on RNA binding and on the splicing of model substrates in vitro. Finally, whereas splicing of introns that lack a Py tract remains dependent on U2AF59, upstream Py tracts, located between the 5′ splice site and the BPS, are required for splicing in vivo only under conditions of U2AF59 inactivation.
MATERIALS AND METHODS
Hosts and plasmids.
The heterozygous SpCR1 diploid (prp2::ura4/prp2+, ade6-M210/ade6-M216, ra4d18/ura4d18, leu1-32/leu1-32) and cdc2.2 and the Py tract variants (Y-long) (41), the prp2.1 mutant and the U2AF59 genomic fragment plasmid pIRT3-prp2+ (36), the U2AF23-ts mutant, (55), and the prp10-YH03 mutant (18) were previously described.
Computational search.
The intronic sequence database was downloaded from the S. pombe site (http://www.sanger.ac.uk/Projects/S_pombe/intron.shtml). Nucleotide strings containing 4 to 15 pyrimidines were searched using the regular expression [tc] {4,15} (for details, see reference 13) both upstream and downstream of the annotated BPS (URAY consensus) in the above-named database. Values for the frequency of introns, reflected by the sizes of the bubbles, with respect to the length of the Py tract(s) and its location relative to the annotated BPS were generated for various categories and plotted as shown. The search algorithm was written in Python and is available upon request.
Generation of haploid U2AF59 genomic knockout cells.
S. pombe cells were grown in EMM2 containing appropriate amino acids supplements (see the text in the supplemental material) as described in the Fission Yeast Handbook (http://www.sanger.ac.uk/PostGenomics/S_pombe/docs/nurse_lab_manual.pdf). For mutagenesis and genomic integration details, see the text in the supplemental material.
RNA preparation and analysis.
The protocols for RNA preparation and semiquantitative reverse transcription-PCR (RT-PCR) (56) were as previously described and used gene-specific primers (primers 17 through 36 [see Table S1 in the supplemental material]). For the quantitative RT-PCR assay, PCR products were stained with SYBR green dye and quantitated using a phosphorimager (Blue and 850V).
RESULTS
Diversity of introns with respect to the location and length of Py tracts.
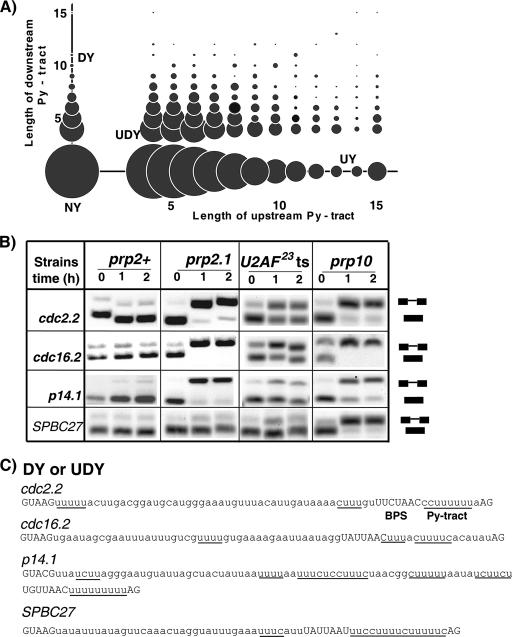
We performed a genome-wide S. pombe analysis to determine the distribution of Py tracts in the annotated database of introns; approximately 46% of S. pombe genes have introns (http://www.sanger.ac.uk/Projects/S_pombe/intron.shtml). The consensus splicing signals for S. pombe introns are degenerate (5′SS, G/GURWGU; BPS, YURAY; 3′ SS, YAG; A/U tract between the BPS and 3′ SS [R is purine, Y is pyrimidine, and W is A or U]) (28). We define Py tracts as four or more pyrimidines, because a typical RNA recognition motif recognizes four to seven nucleotides (48). The analysis revealed several intron architectures with respect to the length and location of Py tracts in relation to the BPS. We found that ∼5.4% of the 4,613 introns analyzed had Py tracts between the BPS and the 3′ SS, which is the conventional location of the Py tract (Fig. 1A) (category DY). A significant fraction (51.2%) of introns had Py tracts located upstream of the BPS (between the 5′ SS and the BPS; category UY), and 32.8% had Py tracts both upstream and downstream of the BPS (category UDY). Surprisingly, 10.6% had no Py tracts at all (category NY). Our searches for either uridine tracts (a more stringent requirement) or Py tracts with up to two purines (a less stringent sequence requirement) revealed all four categories (data not shown). The S. pombe BPS used for annotation is highly degenerate (YURAY) and is a predicted rather than an experimentally determined element. Thus, if a potential BPS located upstream of the annotated site were used instead, some of the introns labeled as UY or UDY could be DY. While this could result in underestimation of certain categories of introns, the effect, if any, would not be significantly large because, based on computational analysis, it has been argued that the distance between the 5′ SS and the BPS provides the major contribution to variability in intron length (28). Moreover, a scanning mechanism similar to that proposed for the recognition of the 3′ SS downstream of the BPS in mammals may favor a nearby BPS rather than an additional BPS(s) located further upstream (50). These results indicate that several intronic architectures (or four broad categories) with respect to the length and positions of Py tracts exist in the S. pombe genome. These observations guided the systematic analysis described below with respect to U2AF and RRM3 requirement.
FIG. 1.
Diversity of intronic sequence architectures and requirements for U2AF subunits in splicing. (A) Four categories of S. pombe introns with respect to the length and positions of Py tracts with respect to the BPS: upstream (UY), downstream (DY), both up and downstream (UDY), and none (NY). The circle size corresponds to the relative frequency of each fraction. (B) Introns with variable-length Py tracts between the BPS and the 3′ SS show diverse requirements for U2AF subunits. The splicing patterns were determined using semiquantitative RT-PCR for endogenous introns (indicated on the left) in wild-type (prp2+), prp2.1, U2AF23-ts, and prp10 mutant strains incubated for 0, 1, and 2 h at 37°C. Positions of spliced and unspliced RNAs are indicated on the right. (C) Sequences of the introns used above are shown. The 5′ and 3′ splice sites and the predicted BPS are shown in uppercase letters. The Py tracts in this and subsequent figures are underlined. Intron SPBC27B12. 07_b is abbreviated as SPBC27.
Conventional Py tracts show various U2AF requirements.
The Py tract typically present between the BPS and the 3′ SS and the large subunit of U2AF that interacts with it are important for the splicing of model pre-mRNAs in metazoans. Given the diversity of intronic sequence architectures (Fig. 1A), we asked how general the requirement for U2AF is for the splicing of introns with various sequence arrangements in vivo. We analyzed several pre-mRNAs by use of a semiquantitative RT-PCR. The first category of introns that we tested has conventional Py tracts located between the BPS and the 3′ SS (Fig. 1C) and resembles model mammalian introns routinely used for in vitro biochemical studies. The wild-type prp2+ (or U2AF59) allele showed no difference in the splicing patterns at 25°C and 37°C (Fig. 1B). However, a previously identified temperature-sensitive (ts) U2AF59 mutant (prp2.1; C387Y substitution) (36), as expected, rapidly accumulated precursors for cdc2.2, cdc16.2, and p14.1 introns within 1 h following a temperature shift to 37°C (Fig. 1B); the C387Y substitution is located at the very end of the RRM2 domain (residues 307 to 393). This is consistent with the idea that splicing patterns of these transcripts are insensitive to temperature shift alone and that impaired splicing is a direct consequence of inactivation of U2AF59. A temperature-sensitive mutant (Y108A, F111A) of the U2AF small subunit (U2AF23-ts) (55) caused only a partial splicing defect for cdc2.2 and cdc16.2 introns and a barely detectable defect for p14.1 at the restricted temperature.
Surprisingly, the prp2.1 and U2AF23-ts mutants supported efficient splicing under nonpermissive conditions for the SPBC27 substrate, which has a long natural Py tract between the BPS and the 3′ SS (Fig. 1B and C); we considered Py tracts longer than eight consecutive pyrimidines to be long. This contrasts with the conclusion from previous studies using the prp2.1 mutant that the length of the Py tract does not determine responsiveness to U2AF59 in the context of the cdc2.2 intron (41). We found that the prp10 (also known as SAP155) ts mutant (18), however, failed to splice this intron (Fig. 1B), suggesting that a lack of pre-mRNA accumulation in U2AF mutants may not be due simply to preferential pre-mRNA turnover. The spliceosomal factor SAP155 is a core component of the U2 snRNP that interacts with the RRM3 domain of the large subunit of U2AF (15). We conclude that under identical conditions U2AF59 inactivation causes splicing inhibition of the majority of introns but has no detectable effect on an intron with a long Py tract.
Introns lacking a Py tract remain U2AF dependent.
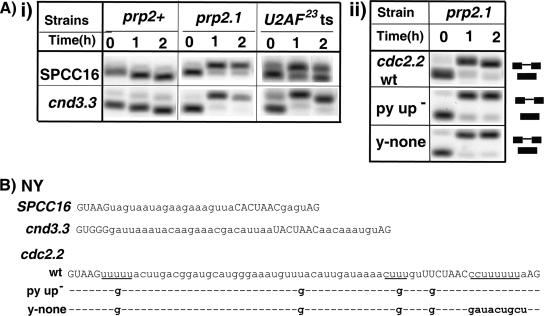
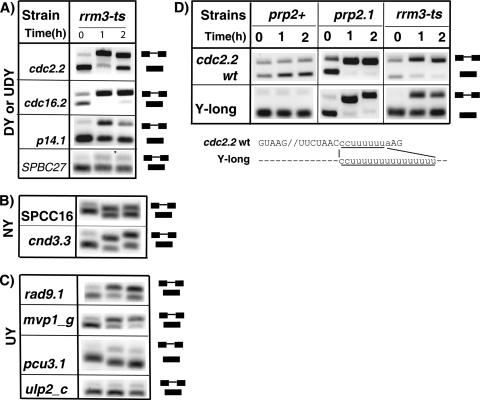
It was previously proposed that the presence of a Py tract between the BPS and the 3′ SS is not necessary for the involvement of U2AF59 in splicing (41). However, a careful inspection of the four introns used in those studies (cdc2.2, nda3.3, cam1.1, and cgs2.1) revealed that all RNAs have contiguous pyrimidines downstream, upstream, or on both sides of the BPS (Fig. 2B; also see Fig. S1 in the supplemental material), and mutation of only the downstream Py tract of the cdc2.2 intron was previously tested. Therefore, to rigorously test whether introns lacking a Py tract (Fig. 1A, NY) require U2AF activity for splicing in vivo, we chose to test the SPCC16 and cnd3.3 introns because they unambiguously lack Py tracts in the entire intron, with no more than one contiguous pyrimidine downstream of the BPS and no more than two contiguous pyrimidines upstream (Fig. 2B). Based on studies of the large subunit of the metazoan U2AF (21, 42, 49, 61), it is unlikely that U2AF59 would bind directly to these introns with reasonable affinity, although we cannot exclude the possibility that association with BBP/SF1 (22) alters the RNA binding properties of U2AF59. Under nonpermissive conditions, the U2AF59 mutant prp2.1 showed little to no splicing for both introns (Fig. 2Ai), and the U2AF23-ts mutant strain showed no splicing for the cnd3.3 intron. The U2AF23-ts mutation also significantly reduced the splicing of SPCC16 (Fig. 2Ai). In addition, we mutated all potential Py tracts (upstream and downstream of the BPS) in the cdc2.2 intron (Fig. 2B). We found that in the prp2.1 mutant neither the Py tract-less cdc2.2 intron (with mutations of all Py tracts of three or more contiguous pyrimidines; y-none) nor the py-up− cdc2.2 intron (with mutation of upstream three or more contiguous pyrimidines) was spliced (Fig. 2Aii). We chose guanosine substitutions because they represent the most disruptive residue for U2AF65 binding (47, 49). These results indicate that introns lacking Py tracts remain dependent on U2AF activity.
FIG. 2.
Introns lacking Py tracts show a U2AF requirement in vivo. (Ai) Splicing patterns of the endogenous SPCC16 and cnd3.3 introns in various strains incubated for 0, 1, and 2 h at 37°C. (Aii) Splicing patterns of the wild-type cdc2.2 and the Py tract-less cdc2.2 mutant introns (expressed from plasmid) in prp2.1. (B) Sequences of the wild-type (wt) and the Py tract mutant (py up− and y-none) introns. Dashes indicate identical nucleotides. Intron SPCC1688.11C_c is abbreviated as SPCC16. Positions of spliced and unspliced RNAs and splicing signals are as described for Fig. 1B.
No detectable U2AF requirement for certain introns with long unconventional Py tracts.
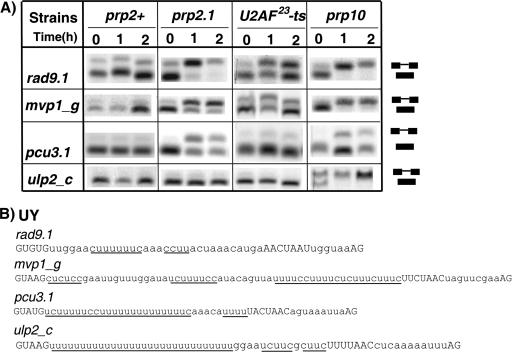
The U2AF large subunit is considered essential for splicing. A significant fraction of S. pombe introns have unusually long Py tracts located upstream of the BPS (Fig. 1A, category UY). Therefore, we tested the requirement of U2AF for the splicing of introns that lacked a Py tract between the BPS and the 3′ SS but had Py tracts of various lengths upstream of the BPS (Fig. 3B). Upstream Py tracts have not previously been linked to splicing or U2AF function in fission yeast. Analysis of these introns in the U2AF59 prp2.1 mutant at nonpermissive temperature showed that the rad9.1 intron (with the shortest upstream Py tract) was not spliced and that mvp1_g and pcu3.1 introns (with longer Py tracts) were partially spliced (Fig. 3A). In this mutant, the ulp2_c intron (with the longest upstream Py tract), however, showed efficient splicing (Fig. 3A), recapitulating the result seen with the SPBC27 intron with a long downstream Py tract (Fig. 1B). In the prp10 mutant, the ulp2_c intron, in similarity to the SPBC27 intron, showed no splicing (Fig. 3A). In the U2AF23-ts mutant, the rad9.1 and mvp1_g introns showed partial splicing, whereas pcu3.1 and ulp2_c introns showed no detectable splicing defects. Thus, we conclude that introns with very long upstream (or downstream) Py tracts are efficiently spliced in vivo under conditions that specifically inactivate U2AF (large and small subunit) function, sufficiently to abolish splicing of introns with shorter Py tracts.
FIG. 3.
Introns with unconventionally located Py tracts show a range of splicing responses to U2AF inactivation in vivo. (A) Splicing patterns of the endogenous introns, indicated on the left, for various strains incubated for 0, 1, and 2 h at 37°C. (B) Positions of the spliced and unspliced RNAs and splicing signals are as described for Fig. 1B.
Conditional requirement for upstream Py tracts in splicing.
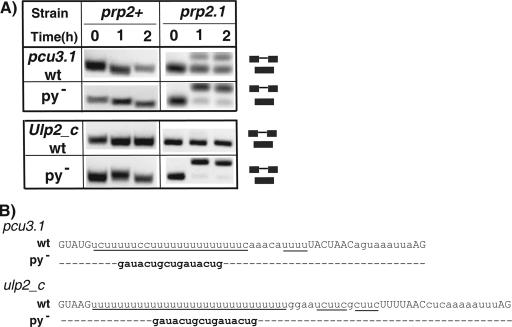
The conventional Py tract or U2AF65 binding site is present between the BPS and the 3′ SS. Given that introns that lack Py tracts also showed a U2AF requirement, we hypothesized that the upstream Py tracts might be irrelevant for splicing. To determine whether the unusually long upstream Py tracts were important for splicing, we mutated these sequences (pcu3.1 py− and ulp2_c py−) as shown in Fig. 4B and analyzed their effect on splicing. We chose the same sequence for mutagenesis that was used previously to disrupt the downstream Py tract in the cdc2.2 intron, where it inhibited splicing (41). We found that mutation of these Py tracts had no effect on splicing under permissive conditions (Fig. 4A, 0 h, prp2+). Surprisingly, however, under nonpermissive conditions in the prp2.1 mutant these mutations almost completely abolished splicing (Fig. 4A, 1 and 2 h). Our simplest interpretation for these results is that the upstream Py tracts are functional and important for splicing, although the effect of the Py tract mutations is conditional, requiring U2AF59 inactivation as well. Thus, disruption of the upstream Py tract likely provides a sensitive background to reveal a U2AF requirement for these introns.
FIG. 4.
Mutations of upstream Py tracts cause strong dependence on U2AF59. (A) Splicing patterns of the wild-type pcu3.1 and Ulp2_c introns and the mutant (py−) introns (expressed from plasmid) in prp2.1 are shown. (B) Sequences of the wild-type (wt) and mutant introns. Dashes indicate identical nucleotides. Positions of the spliced and unspliced RNAs and splicing signals are as described for Fig. 1B.
A genetic system allowed isolation of conditional mutants in U2AF59 RRM3.
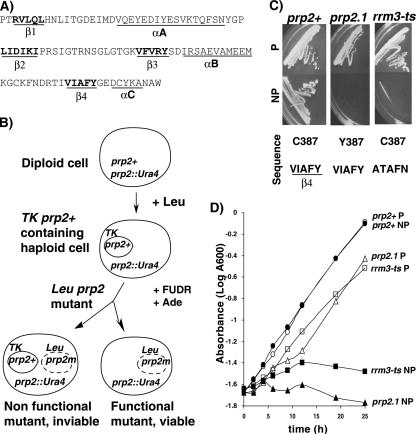
As noted above, the role of the RRM3 domain in splicing had been a mystery. We reasoned that isolation of a conditional mutation in RRM3 might help in several ways. First, it could help us link the RRM3 domain to splicing. Second, it could provide a more sophisticated tool to rule out the possibility that the differences in U2AF dependence were due simply to decreasing levels or activity of the factor under nonpermissive conditions. Third, it could help uncover aspects of U2AF function, in relation to various intron architectures, that escaped analysis using the original prp2.1 allele. Therefore, we developed a genetic system for oligonucleotide-directed mutagenesis of each of the four β-strands of RRM3 and isolation of conditional mutants. Amino acid sequences of RRM3 targeted for mutagenesis are indicated (Fig. 5A), and a diagram representing the screening of prp2 mutants is schematically shown (Fig. 5B). Approximately 2,500 viable (fluorodeoxyuridine-resistant) colonies from each β-strand mutagenesis library were replica plated to screen for ts growth phenotypes by incubating them at a permissive (25°C) or nonpermissive (37°C) temperature. We obtained eight ts mutants but no cold-sensitive mutants. As expected, whereas the wild-type prp2+ showed growth at both temperatures, the original prp2.1 mutant showed no growth at 37°C (Fig. 5C). Each of the selected β-1-, β-3-, and β-4-strand mutants showed ts growth (Fig. 5C; also see Fig. S2A in the supplemental material). The 3.9 allele displayed the weakest phenotype, the 1.46 allele was intermediate (see Fig. S2A in the supplemental material), and the 4.103 allele (hereafter referred to as rrm3-ts) displayed the strongest phenotype (Fig. 5C); the 4.99 allele also had a strong phenotype (see Fig. S2 in the supplemental material), but analysis was not pursued further. The growth phenotype of relevant strains was also confirmed using liquid culture (Fig. 5D). (Amino acid sequences for the RRM3 ts mutants used for this study are shown in Fig. 5C and in Fig. S2C in the supplemental material.) Thus, we have isolated conditional mutants within the RRM3 domain of U2AF59.
FIG. 5.
Isolation and characterization of temperature-sensitive mutants in RRM3. (A) Amino acid sequence of the RRM3 domain of U2AF59. The β strands (β1 to β4) and α helices (αA to αC) are indicated. Amino acids targeted for mutagenesis are shown in bold. (B) Schematics for the generation and screening of RRM3 mutants. Heterozygous diploid cells used to obtain a haploid U2AF59 genomic knockout strain carrying prp2+ in the TK plasmid were transformed with RRM3 mutant libraries (prp2m). The transformants were screened for viable mutants followed by replica plating for growth at permissive and nonpermissive temperatures as described in Materials and Methods. (C) Characterization of U2AF59 RRM3 ts mutants. The wild-type (prp2+), prp2.1, and rrm3-ts strains were incubated at permissive (P; 25°C) or nonpermissive (NP; 37°C) temperature. Amino acid sequences of the wild type and substitutions in prp2.1 and the RRM3 mutant are shown. (D) Growth curve. The strains shown in panel C were grown in liquid cultures at permissive and nonpermissive temperatures, and the cell density was measured using absorbance at 600 nm at various time intervals.
Introns show varied requirements for RRM3.
Deletion of RRM3 is lethal in S. pombe but causes no splicing defect in vitro for model introns in a HeLa nuclear extract (4). To determine whether the newly isolated RRM3 mutants showed any splicing defect and whether various fission yeast introns differed in their requirement for RRM3 in vivo, we analyzed the array of pre-mRNA substrates shown in Fig. 1 to 3. The RRM3 mutants showed a range of splicing defects, depending on the RRM3 mutant and the intron used. First, in rrm3-ts (Fig. 6A) and another RRM3 mutant, 1.46 (see Fig. S2 in the supplemental material), there was almost complete splicing inhibition of the cdc16.2 intron, which has a conventional Py tract between the BPS and the 3′ SS. However, in the 1.46 mutant there was little defect in the splicing of the p14.1 intron (see Fig. S2B in the supplemental material), and in the rrm3-ts mutant there was no splicing defect for SPBC27 (Fig. 6A). Thus, all three mutations in U2AF subunits under conditions that abolish the splicing of many other introns tested supported the splicing of the SPBC27 intron. This intron contains an unusually long Py tract at the conventional location. Other combinations showed partial or complete splicing defects (Fig. 6A; also see Fig. S2B in the supplemental material). Integration of the rrm3-ts allele in the genome recapitulated the results obtained with the plasmid-borne allele (see Fig. S3 in the supplemental material). Second, the rrm3-ts mutant under nonpermissive conditions showed partial splicing defects for introns lacking a Py tract (SPCC16 and cnd3.3) (Fig. 6B). Third, analysis of the introns that had different lengths of upstream Py tracts (Fig. 6C) showed that rad9.1 and mvp1_g introns were significantly affected in rrm3-ts but by less than was seen with prp2.1. However, pcu3.1 and ulp2_c introns, which were either partially or efficiently spliced in prp2.1 (Fig. 3A), were fully spliced in rrm3-ts (Fig. 6C). The ulp2_c intron thus behaved like the SPBC27 intron in showing no detectable splicing defect in all three U2AF mutants. These results indicate that under identical conditions certain introns show a stringent RRM3 requirement whereas others (with longer Py tracts) can be spliced efficiently in the RRM3 ts mutant.
FIG. 6.
The RRM3 domain is important for splicing in vivo. (A to C) Diverse response (partial or complete splicing inhibition or efficient splicing) of the RRM3 ts mutant in the splicing of the endogenous transcripts as described for Fig. 1 to 3. (D) Increasing the Py tract length reduces RRM3 dependence in vivo. Splicing patterns of cdc2.2 and the cdc2.2 Py tract variant introns (expressed from plasmids) in various strains incubated for 0, 1, and 2 h at 37°C are shown. The Py tract region of the cdc2.2 wt intron was replaced by a uridine tract (Y-long). Dashes indicate identical nucleotides.
We conclude that introns show differing requirements for RRM3 in vivo. These studies directly link the function of the RRM3 domain to splicing in vivo, explaining why deletion of RRM3 is lethal in S. pombe. Furthermore, identification of RRM3-insensitive introns in vivo such as SPBC27 and ulp2_c also lends credence to our previous observation that RRM3 is dispensable for in vitro splicing of model pre-mRNA substrates in a HeLa nuclear extract (4), implying that it is not an in vitro artifact.
Length of the Py tract in part influences dependence on RRM3.
The above-described experiments suggested that the length of the Py tract could influence RRM3 requirement. To address this issue directly, we used previously generated Py tract mutants of cdc2.2 (41). Consistent with previous results (41), substitution of the conventional Py tract with a long uridine stretch (Y-Long) allowed for efficient splicing in the wild-type background but not in the prp2.1 mutant under nonpermissive conditions (Fig. 6D). Most importantly, whereas the wild-type intron was not spliced, the Y-long mutant was spliced (>50%) in the rrm3-ts mutant. This finding is consistent with our observation that the SPBC27 intron, with a long natural Py tract, was efficiently (100%) spliced in the rrm3-ts mutant. However, the splicing behavior of the Y-long mutant differs in the prp2.1 and rrm3-ts mutants, perhaps because the Py tract is either not as long as those of SPBC27 and ulp2_c introns or is present in a different sequence context. We conclude that certain introns with long Py tracts are efficiently spliced in the rrm3-ts mutant, that our analysis with the rrm3-ts mutant has revealed that U2AF59 is indeed responsive to the length of the Py tract, its sequence composition, and/or its sequence context, and that the requirements for U2AF and RRM3 are not identical. It is tempting to speculate that analysis of additional introns might reveal introns that require U2AF but not RRM3.
Since each of the U2AF mutants (prp2.1, U2AF23-ts, and rrm3-ts) abolished splicing of some introns but had either no detectable effect or only a partial effect on the splicing of others, we interpret these outcomes as reflecting intron-specific differences in the requirements of U2AF subunits or the RRM3 domain rather than merely the strength of a particular allele. Our combined results have important implications for the model of U2AF function.
DISCUSSION
The most important finding of this study is that there is an unexpected diversity in 3′ splice site recognition with respect to intronic sequence arrangements and requirements for U2AF subunits and domains in S. pombe beyond what is known from detailed biochemical analysis of model pre-mRNAs. Our results suggest that splicing of introns that lack a Py tract remains dependent on U2AF59, whereas under identical conditions, splicing of certain introns with long upstream or downstream Py tracts is insensitive to U2AF59 inactivation. Furthermore, upstream Py tracts are required for splicing in vivo only under conditions of U2AF59 inactivation. We have also shown that Py tract length variations in part confer differential RRM3 sensitivities. We provide here a model of U2AF function that accounts for these unexpected findings (Fig. 7).
FIG. 7.
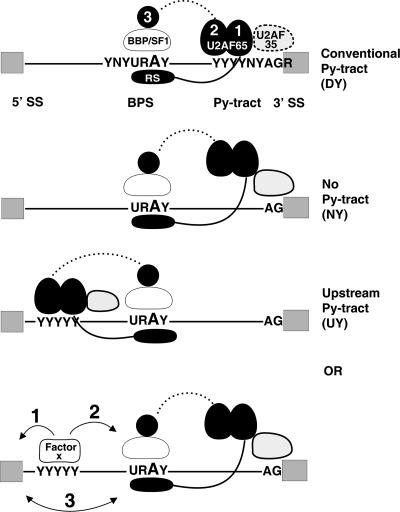
Diversity of intronic sequence architectures and U2AF requirements. The U2AF large subunit (U2AF65 RRM1, RRM2, and RRM3 and the RS domain; black fill), the small subunit (U2AF35; light-gray fill), BBP/SF1 (white fill), splicing signals (5′ SS, BPS, and 3′ SS), intron (line), and exons (gray boxes) are shown. The large subunit binds to the conventional Py tract (the DY panel) typically present between the BPS and the 3′ SS and recruits other components of the splicing machinery, including the U2 snRNP (metazoans; DY panel). For clarity, interactions between components of the 5′ SS machinery and the BPS or the 3′ SS machinery are not shown. S. pombe introns that lack a downstream Py tract depend on the large subunit, which presumably involves other interactions for its recruitment (the NY panel). Introns with long, unconventional Py tracts located upstream of the BPS may bind the large subunit (the UY panel) or an unknown factor (factor x) which directly collaborates with U2AF bound at the normal location to support splicing similar to that represented by the NY panel. Factor x could also facilitate recognition of the 5′ SS by the U1 snRNP (1) and/or the BPS by the BBP/SF1 (2), leading to interactions between U1 snRNP and BBP/SF1 and U2AF recruitment. The dashed line connecting RRM3 or the dashed oval for the small subunit U2AF35 reflects the fact that these domains are dispensable for some introns but required for others.
Species-specific differences in mechanisms of 3′ splice site recognition.
Species-specific preference for or dependence on certain sequence features, intron architectures, or splicing factors has been noted for 3′ splice site recognition (34, 40). In mammals, the BPS is degenerate, the large subunit of U2AF is required for the splicing of AG-independent introns (introns with long Py tracts support early spliceosomal assembly without the need for the 3′ SS AG dinucleotide), both U2AF subunits are required for the splicing of AG-dependent introns (with short Py tracts), and the RRM3 domain of U2AF is dispensable for splicing of model substrates from both categories in vitro (Fig. 7, conventional Py tracts, DY). In Caenorhabditis elegans, the BPS consensus is lacking, but U4CAGR is the predominant 3′ splice site sequence, and both subunits are important for 3′ splice site recognition (21, 65). In S. cerevisiae, interaction between the invariant BPS (UACUAAC) and SF1/BBP plays an important role in commitment complex formation, the U2AF small subunit is absent, the large subunit (Mud2p) is dispensable for viability, and the Py tract is generally dispensable for splicing (1, 35). In S. pombe, U2AF and BBP/SF1 form a stable complex (22). Unlike the mammalian introns, S. pombe introns show generally parallel requirements for both U2AF subunits (55). Moreover, various introns show differential RRM3 requirements (this study). Taken together, our findings with S. pombe indicate that an unexpected diversity of intronic sequence arrangements and of requirements for U2AF subunits, domains, and binding sites exists within the same organism (Fig. 7). We propose that a similar situation (U2AF recruitment via Py tract and supplementary mechanisms) is likely to pertain in mammals as well as other metazoans.
Role of U2AF in the splicing of introns without a Py tract.
The existing model of U2AF function in 3′ splice site definition cannot account for why the Py tract-less introns require U2AF59. It has been thought that binding of U2AF65 to the Py tract is the primary determinant for its association with the pre-mRNA, leading to U2 snRNP recruitment. However, introns without Py tracts remain dependent on U2AF activity in S. pombe, implying that U2AF is needed for splicing but must be recruited to the pre-mRNA via a mechanism that does not require Py tract binding. Based on previous studies of metazoans (21, 42, 49, 61), we expect that these intron sequences should exhibit no detectable binding for the RNA binding domain of the large subunit. Although it is possible that a tight association with BBP/SF1 (22) could alter the RNA binding activity of U2AF59 or that the S. pombe U2AF59 and human U2AF65 orthologs differ in sequence preferences, we have been unable to discern any consensus sequence between the BPS and the 3′ SS that may serve as unconventional U2AF59 binding site in such introns, including the five introns that were experimentally tested in this study. This scenario of a lack of the Py tract requirement is analogous to the situation seen with S. cerevisiae, where a more efficient interaction of BBP/SF1 with the invariant BPS (UACUAAC), unlike the degenerate consensus (YNCURAY) in mammals, could possibly compensate for a lack of the Py tract or of Mud2p or both (6). Contrary to our expectation, we found that only 8 of 55 randomly selected Py tract-less introns had a perfect BPS (UACUAAC), implying that a stronger BPS may not compensate for a lack of Py tract for most of such introns. Our preliminary experiments show that the recombinant RNA binding domain (RRM1-3) of the S. pombe U2AF59 protein has little or no detectable RNA binding activity (>100-fold lower affinity) in vitro relative to the RRM1-3 domain of the human U2AF65 (see Fig. S4 in the supplemental material). Although a trivial possibility is that the recombinant protein is inactive, it is tempting to speculate that the presence of the RS domain and association of the large subunit with the small subunit (42) and/or the SF1 (22) could influence the RNA binding property of the S. pombe large subunit. Furthermore, requirements for the two U2AF subunits are generally parallel in S. pombe, implying that the two subunits collaborate in vivo (55) (this study). In this respect, the situation seen with S. pombe is different from that in humans, where the large subunit recognizes the 3′ SS with long Py tracts (AG-independent introns) without the need for the small subunit, which is important for the AG-dependent introns. Another mechanism may involve bridging interactions across introns involving the small subunit of U2AF and the U1 snRNP 70K protein (60), BPS and U1 snRNP (45), or other factors such as SRp54 (25). We favor the idea that for certain introns these interactions could indeed provide a major contribution, as an alternative to the widely held view centering on the U2AF large subunit-Py tract, to early events during 3′ splice site definition. We propose that a variety of early interactions contribute to 3′ splice site recognition for different intron architectures (Fig. 7).
How do upstream Py tracts function?
The current model does not explain how upstream Py tracts function in constitutive splicing and why they become important only upon U2AF59 inactivation. There are two possible explanations for why some introns are unaffected upon U2AF inactivation. First, certain introns in S. pombe (and possibly in metazoans) are U2AF independent and involve a U2AF-Py tract-independent splicing mechanism for 3′ splice site definition in vivo. Although this may be regarded as a radical view, considering an essential requirement of U2AF65 in splicing, this proposal is indeed similar to that addressing the exceptional situation in S. cerevisiae, where both Mud2p and the Py tract are generally dispensable for splicing (1, 35). It is also known that certain introns can be spliced in U2AF-depleted extracts under unusual in vitro conditions such as an excess of SR proteins or in extracts prepared from adenovirus-infected cells (31, 32), and an E-like splicing complex (E′) has been observed in the absence of U2AF (26). Moreover, the U12-type (minor) introns lack a Py tract, and thus U2AF is unlikely to facilitate association of U12 snRNP with the BPS (58). Second, such introns could be U2AF dependent, because under nonpermissive conditions the prp2.1 mutant may retain residual U2AF activity (a limitation also applicable to biochemical depletion of U2AF in vitro) and both introns that are unaffected by U2AF mutations have long Py tracts. This residual activity could have supported the splicing of these introns, presumably because unusually long Py tracts offer multiple registers for binding and thus increased affinity to compensate for reduced U2AF activity (3). Splicing inhibition in this scenario requires, akin to a synthetic phenotype, a combination of Py tract mutation and U2AF inactivation. The experiments described here cannot distinguish whether the upstream Py tract binds U2AF directly or binds another factor such as SRp54, TIA-1-like, or Nam-8p-like proteins (Fig. 7, UY panel, factor x) to facilitate 5′ splice site and/or BPS recognition (11, 12, 25, 38), after which U1 snRNP can then support SF1/U2AF recruitment (2, 8, 30, 63). We note that the introns with upstream Py tracts tested here have optimal 5′ SS and are constitutively spliced, which differs from findings of the presence of suboptimal 5′ splice sites in examples of TIA-1- and Nam8p-sensitive regulated introns. The upstream Py tract could also function by providing a favorable sequence context. If U2AF functions from these upstream Py tracts, it could easily facilitate a series of sequential interactions between the RS domain and splicing signals during spliceosome assembly (Fig. 7, UY panel) as recently proposed by Shen and Green (46). Although the Py tract in the proposed model is located on the other side of the BPS, it could nonetheless serve to bind RRM1 and RRM2 and bring the RS and/or RRM3 domain in proximity to the BPS. This is feasible because the RNA chain is flexible and the RRM3 and RS domains are tethered via extended flexible linker sequences, which could permit U2AF function in a location-independent manner, akin to how activators work from either upstream or downstream enhancers to promote relevant steps during the assembly of multiprotein splicing (or transcription) complexes (20, 37).
Role of RRM3 in splicing.
We have shown that whereas the majority of fission yeast introns require RRM3, certain introns are insensitive to RRM3 inactivation in vivo. Whereas the human RRM3 is dispensable for model introns with short and long Py tracts in vitro (4), a requirement for RRM3 is sensitive to Py tract length (and possibly sequence composition and sequence context) in S. pombe. Although interactions of RRM3 with BBP/SF1 and SAP155 could explain the sequence conservation of RRM3 as well as synthetic lethality of S. cerevisiae mud2 with other components of the splicing machinery (1, 2, 6, 15, 62), our favored explanation is that during spliceosome assembly RRM3 is required for a step(s) that may be rate limiting for only a subset of introns and that RRM3 (or its interactions with other splicing factors) likely plays a kinetic rather than an essential role. This assertion is supported by several observations. First, a USx chimera (of U2AF65 and SXL) lacking RRM3 supports splicing of a 3′ splice site associated with a 17-nucleotide-long Py tract in a HeLa extract and in transgenic flies (16, 54). This is most likely because a higher affinity of USx for a rather long Py tract allows it to overcome a rate-limiting step that requires RRM3. Second, the S. cerevisiae ortholog Mud2p containing RRM3 as the only recognizable portion of U2AF65 is dispensable (1). Third, the RRM3 interacting partner SF1/BBP has a kinetic rather than an essential role in splicing and U2 snRNP binding in S. cerevisiae and humans (17, 44). In addition to the kinetic role of the RRM3-SF1 interaction, the potential RRM3-dependent rate-limiting process could contribute to subsequent steps during spliceosome assembly, such as recognition of the BPS by the U2 snRNP or any function prior to U2AF release from the spliceosome (5).
Biological importance—spliceosome assembly pathways beyond model substrates.
Development of an in vitro splicing system and characterization of model pre-mRNA substrates contributed enormously to the identification of splicing signals, factors, and a spliceosome assembly pathway(s). However, findings obtained with model substrates may not explain every instance of splicing. Numerous intronic sequence arrangements for 3′ splice site recognition exist; these may differ with respect to the length, strength, spacing, and sequence contexts of cis-acting splicing elements. Similarly, there are cell- and tissue-specific differences with respect to the concentration, activity, localization, and interacting partners for splicing factors and the presence or absence of splicing activators and repressors. Whereas these features obviously generate a highly complex landscape of assembly pathways with respect to requirements for cis- and trans-acting elements, studies with model substrates have captured only a limited fraction of the entire spectrum that actually exists. The presence of a large array of intron architectures and of requirements for trans-acting factors increases the potential for the generation of enormous diversity for molecular functions and/or regulation via combinatorial control (29, 48), which has played an important role during metazoan evolution. At the same time, these features increase the degree of freedom in regard to associations of cis- and trans-acting elements, thereby complicating intron-exon identification in eukaryotic genomes and contributing to peculiar splicing behaviors of introns.
These studies reinforce the notion that the spliceosome assembly pathway is flexible, that different pre-mRNAs likely have distinct rate-limiting steps, and that alternative (U1-, U2AF-, and RRM3-insensitive) spliceosome assembly pathways exist (4, 9, 32, 51, 52). Whether a splicing factor, a domain, or a particular assembly pathway is essential, dispensable, or partially required for splicing may not be absolute for each and every intron. Moreover, strengthening or weakening of one or more of the numerous potential interactions in different contexts could modify the need for other interactions. In conclusion, versatility of initial interactions early during spliceosomal assembly within the same species exists to accommodate a variety of intron architectures. This phenomenon is relevant not only for splicing but also for many steps along the gene expression pathway (transcription, RNA processing, translation, and signaling).
Supplementary Material
Acknowledgments
We thank Dick McIntosh and DeWight Williams for help with the yeast work, Erin Peden for help with computational analysis, Chris Webb, Jo Ann Wise, Judith Potashkin, and Tokio Tani for plasmids, strains, and reagents, and Tom Blumenthal, Dick McIntosh, Juan Valcárcel, Joaquin Espinosa, Greg Odorizzi, Jens Lykke-Andersen, Mark Robida, and Rajesh Gaur for helpful discussions and critical reading of the manuscript.
This work was supported in part by grants from the American Cancer Society, the Keck Foundation, and the National Institutes of Health to R.S.
Footnotes
Published ahead of print on 20 August 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abovich, N., X. C. Liao, and M. Rosbash. 1994. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 8:843-854. [DOI] [PubMed] [Google Scholar]
- 2.Abovich, N., and M. Rosbash. 1997. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403-412. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, H., A. Rahn, W. Davis, and R. Singh. 2003. Sex lethal and U2 small nuclear ribonucleoprotein auxiliary factor (U2AF65) recognize polypyrimidine tracts using multiple modes of binding. RNA 9:88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, H., A. Rahn, B. Gawande, S. Guth, J. Valcárcel, and R. Singh. 2004. The conserved RNA Recognition Motif 3 of U2 snRNA Auxiliary Factor (U2AF65) is essential in vivo but dispensable for activity in vitro. RNA 10:240-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, M., S. Michaud, J. Kingston, and R. Reed. 1992. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 6:1986-2000. [DOI] [PubMed] [Google Scholar]
- 6.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 8.Côté, J., J. Beaudoin, R. Tacke, and B. Chabot. 1995. The U1 small nuclear ribonucleoprotein/5′ splice site interaction affects U2AF65 binding to the downstream 3′ splice site. J. Biol. Chem. 270:4031-4036. [DOI] [PubMed] [Google Scholar]
- 9.Crispino, J. D., B. J. Blencowe, and P. A. Sharp. 1994. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science 265:1866-1869. [DOI] [PubMed] [Google Scholar]
- 10.Fleckner, J., M. Zhang, J. Valcárcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 11.Förch, P., L. Merendino, C. Martinez, and J. Valcárcel. 2003. U2 small nuclear ribonucleoprotein particle (snRNP) auxiliary factor of 65 kDa, U2AF65, can promote U1 snRNP recruitment to 5′ splice sites. Biochem. J. 372:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Förch, P., O. Puig, N. Kedersha, C. Martinez, S. Granneman, B. Seraphin, P. Anderson, and J. Valcárcel. 2000. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre- mRNA splicing. Mol. Cell 6:1089-1098. [DOI] [PubMed] [Google Scholar]
- 13.Friedl, J. E. F. 1997. Mastering regular expressions. O'Reilly and Associates, Sebastopol, CA.
- 14.Golling, G., A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S. Y. Lin, R. M. Nissen, and N. Hopkins. 2002. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31:135-140. [DOI] [PubMed] [Google Scholar]
- 15.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granadino, B., L. O. F. Penalva, M. R. Green, J. Valcárcel, and L. Sanchez. 1997. Distinct mechanisms of splicing regulation in vivo by the Drosophila protein Sex-lethal. Proc. Natl. Acad. Sci. USA 94:7343-7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guth, S., and J. Valcárcel. 2000. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J. Biol. Chem. 275:38059-38066. [DOI] [PubMed] [Google Scholar]
- 18.Habara, Y., S. Urushiyama, T. Tani, and Y. Ohshima. 1998. The fission yeast prp10(+) gene involved in pre-mRNA splicing encodes a homologue of highly conserved splicing factor, SAP155. Nucleic Acids Res. 26:5662-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 20.Hertel, K. J., K. W. Lynch, and T. Maniatis. 1997. Common themes in the function of transcription and splicing enhancers. Curr. Opin. Cell Biol. 9:350-357. [DOI] [PubMed] [Google Scholar]
- 21.Hollins, C., D. A. Zorio, M. MacMorris, and T. Blumenthal. 2005. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA 11:248-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, T., J. Vilardell, and C. C. Query. 2002. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23). EMBO J. 21:5516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jurica, M. S., and M. J. Moore. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5-14. [DOI] [PubMed] [Google Scholar]
- 24.Kanaar, R., S. E. Roche, E. L. Beall, M. R. Green, and D. C. Rio. 1993. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science 262:569-573. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy, C. F., A. Kramer, and S. M. Berget. 1998. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol. Cell. Biol. 18:5425-5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent, O. A., D. B. Ritchie, and A. M. Macmillan. 2005. Characterization of a U2AF-independent commitment complex (E′) in the mammalian spliceosome assembly pathway. Mol. Cell. Biol. 25:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kielkopf, C. L., S. Lucke, and M. R. Green. 2004. U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 18:1513-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupfer, D. M., S. D. Drabenstot, K. L. Buchanan, H. Lai, H. Zhu, D. W. Dyer, B. A. Roe, and J. W. Murphy. 2004. Introns and splicing elements of five diverse fungi. Eukaryot. Cell 3:1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine, M., and R. Tjian. 2003. Transcription regulation and animal diversity. Nature 424:147-151. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., and B. J. Blencowe. 1999. Distinct factor requirements for exonic splicing enhancer function and binding of U2AF to the polypyrimidine tract. J. Biol. Chem. 274:35074-35079. [DOI] [PubMed] [Google Scholar]
- 31.Lützelberger, M., E. Backstrom, and G. Akusjarvi. 2005. Substrate-dependent differences in U2AF requirement for splicing in adenovirus-infected cell extracts. J. Biol. Chem. 280:25478-25484. [DOI] [PubMed] [Google Scholar]
- 32.MacMillan, A. M., P. S. McCaw, J. D. Crispino, and P. A. Sharp. 1997. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc. Natl. Acad. Sci. USA 94:133-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcárcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 34.Moore, M. J. 2000. Intron recognition comes of AGe. Nat. Struct. Biol. 7:14-16. [DOI] [PubMed] [Google Scholar]
- 35.Patterson, B., and C. Guthrie. 1991. A U-rich tract enhances usage of an alternative 3′ splice site in yeast. Cell 64:181-187. [DOI] [PubMed] [Google Scholar]
- 36.Potashkin, J., K. Naik, and K. Wentz-Hunter. 1993. U2AF homolog required for splicing in vivo. Science 262:573-575. [DOI] [PubMed] [Google Scholar]
- 37.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 38.Puig, O., A. Gottschalk, P. Fabrizio, and B. Seraphin. 1999. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev. 13:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rain, J. C., Z. Rafi, Z. Rhani, P. Legrain, and A. Kramer. 1998. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA 4:551-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340-345. [DOI] [PubMed] [Google Scholar]
- 41.Romfo, C. M., S. Lakhe-Reddy, and J. A. Wise. 1999. Molecular genetic analysis of U2AF59 in Schizosaccharomyces pombe: differential sensitivity of introns to mutational inactivation. RNA 5:49-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudner, D. Z., K. S. Breger, R. Kanaar, M. D. Adams, and D. C. Rio. 1998. RNA binding activity of heterodimeric splicing factor U2AF: at least one RS domain is required for high-affinity binding. Mol. Cell. Biol. 18:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudner, D. Z., R. Kanaar, K. S. Breger, and D. C. Rio. 1996. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. USA 93:10333-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rutz, B., and B. Seraphin. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19:1873-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seraphin, B., and M. Rosbash. 1989. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell 59:349-358. [DOI] [PubMed] [Google Scholar]
- 46.Shen, H., and M. R. Green. 2004. A pathway of sequential arginine-serine-rich domain-splicing signal interactions during mammalian spliceosome assembly. Mol. Cell 16:363-373. [DOI] [PubMed] [Google Scholar]
- 47.Singh, R., H. Banerjee, and M. R. Green. 2000. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA 6:901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh, R., and J. Valcárcel. 2005. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 12:645-653. [DOI] [PubMed] [Google Scholar]
- 49.Singh, R., J. Valcárcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 50.Smith, C. W., T. T. Chu, and B. Nadal-Ginard. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 13:4939-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcárcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 20:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tarn, W. Y., and J. A. Steitz. 1994. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 8:2704-2717. [DOI] [PubMed] [Google Scholar]
- 53.Valcárcel, J., R. K. Gaur, R. Singh, and M. R. Green. 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273:1706-1709. [DOI] [PubMed] [Google Scholar]
- 54.Valcárcel, J., R. Singh, P. D. Zamore, and M. R. Green. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362:171-175. [DOI] [PubMed] [Google Scholar]
- 55.Webb, C. J., S. Lakhe-Reddy, C. M. Romfo, and J. A. Wise. 2005. Analysis of mutant phenotypes and splicing defects demonstrates functional collaboration between the large and small subunits of the essential splicing factor U2AF in vivo. Mol. Biol. Cell 16:584-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb, C. J., and J. A. Wise. 2004. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol. Cell. Biol. 24:4229-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wentz-Hunter, K., and J. Potashkin. 1995. The evolutionary conservation of the splicing apparatus between fission yeast and man. Nucleic Acids Symp. Ser. 33:226-228. [PubMed] [Google Scholar]
- 58.Will, C. L., C. Schneider, R. Reed, and R. Luhrmann. 1999. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284:2003-2005. [DOI] [PubMed] [Google Scholar]
- 59.Wood, V., et al. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871-880. [DOI] [PubMed] [Google Scholar]
- 60.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 61.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 62.Zamore, P. D., J. G. Patton, and M. R. Green. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355:609-614. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, D., and M. Rosbash. 1999. Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev. 13:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, W. J., and J. Y. Wu. 1996. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol. Cell. Biol. 16:5400-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zorio, D. A., and T. Blumenthal. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835-838. [DOI] [PubMed] [Google Scholar]
- 66.Zorio, D. A., and T. Blumenthal. 1999. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5:487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.