Abstract
The positive transcription elongation factor P-TEFb controls the elongation of transcription by RNA polymerase II. P-TEFb is inactivated upon binding to HEXIM1 or HEXIM2 proteins associated with a noncoding RNA, 7SK. In response to the inhibition of transcription, 7SK RNA, as well as HEXIM proteins, is released by an unknown mechanism and P-TEFb is activated. New partners of 7SK RNA were searched for as potential players in this feedback process. A subset of heterogeneous ribonuclear proteins, hnRNPs Q and R and hnRNPs A1 and A2, were thus identified as major 7SK RNA-associated proteins. The degree of association of 7SK RNA with these hnRNPs increased when P-TEFb-HEXIM1-7SK was dissociated following the inhibition of transcription or HEXIM1 knockdown. This finding suggested that 7SK RNA shuttles from HEXIM1-P-TEFb complexes to hnRNPs. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK complexes was attenuated when both hnRNPs A1 and A2 were knocked down by small interfering RNA. As hnRNPs are known to interact transiently with RNA while it is synthesized, hnRNPs released from nascent transcripts may trap 7SK RNA and thereby contribute to the activation of P-TEFb.
The positive transcription elongation factor P-TEFb is required to activate the transcription of most class II genes (48). P-TEFb comprises two subunits, CDK9, a cyclin-dependent protein kinase, and its corresponding cyclin T1 or cyclin T2. The activity of P-TEFb is regulated. It increases in response to the inhibition of transcription (45, 63) or cardiac cell hypertrophic stimulation (54). Previous studies have indicated that 7SK RNA associates with an inactive form of P-TEFb. 7SK RNA is an abundant (2 × 105-molecule-per-cell) noncoding nuclear RNA of 331 nucleotides (60, 68). The inhibition of P-TEFb activity relies upon the binding of HEXIM1 or HEXIM2 proteins to cyclin T1 or T2 (5, 13, 41, 64). This process requires the association of 7SK RNA with HEXIM1 or HEXIM2 (40, 65). Two hairpins in the 7SK RNA structure are involved in this association (14). 7SK binding to HEXIM proteins promotes a major conformational change allowing the C-terminal domains of the proteins to interact with the N-terminal domains of cyclin T's (41, 55). This transcription-dependent regulation may constitute a feedback loop fine-tuning the efficiency of the elongation step in class II gene transcription.
The molecular regulatory mechanism remains largely unknown. 7SK RNA is very stable even when it dissociates from HEXIM proteins; its degradation is unlikely to contribute to the dissociation. Posttranslational modifications of protein subunits in P-TEFb-HEXIM-7SK may determine their association. For instance, the phosphorylation of CDK9 on threonine residue T186 is required, but the in vivo regulation of this step has not been established (10, 35, 47). HEXIM proteins and 7SK RNA may be released when P-TEFb binds to components of the transcriptional machinery, such as transcription factors like NF-κB (2), retinoblastoma protein (56), androgen receptor (34), aryl hydrocarbon receptor (58), Tat (41, 55), STAT3 (17), Brd4 (27, 62), and the capping enzymes (47).
Alternatively, P-TEFb regulation may be driven by the competitive interaction of 7SK RNA with partners other than HEXIM proteins. The present study addresses this hypothesis. The heterogeneous nuclear ribonucleoproteins hnRNP Q, hnRNP R, hnRNP A1, and hnRNP A2 were thus identified as major 7SK-binding proteins.
Four related genes code for A0, A1, A2, and A3 proteins of the hnRNP A subgroup, of which hnRNP A1 is by far the most abundant. hnRNPs A1 and A2 are capable of high-affinity binding to sequences to modulate mRNA turnover and translation (4, 21, 29, 53). More relevant to our findings, hnRNPs have also been found to be involved in numerous nuclear processes. hnRNPs A1 and A2 associate with telomere ends and stimulate telomerase activity (16, 32, 46, 67). hnRNP A1 binds to pre-mRNA in the nucleus (49), and it antagonizes the alternative splicing activity of splicing factors such as splicing factor 2/alternative splicing factor and SC35 (7, 15, 36). High-affinity binding sites for the hnRNP A1 protein stimulate the use of a distal 5′ splice site in mammalian pre-mRNAs (25). Two distinct but related genes code for the hnRNP Q and hnRNP R proteins, which have also been designated synaptotagmin-binding cytoplasmic RNA-interacting proteins (SYNCRIP); NS1-associated protein 1 (NSAP1); and glycine, arginine, and tyrosine RNA-binding protein (GRY-RBP). This diversity reflects the large number of processes in which they have been found to be involved. Cytoplasmic and nuclear functions have been reported. hnRNP Q proteins are associated with polysomes (24) and prevent the deadenylation of unstable mRNAs (9, 20, 31). hnRNP Q and R proteins are components of mRNA granules transported in neuronal dendrites and growth cones (1, 52). hnRNPs Q and R interact with the apolipoprotein B RNA-binding protein 1 complementation factor to inhibit C-to-U editing by apolipoprotein B RNA-binding protein (3). hnRNP Q proteins are involved in mouse hepatitis virus cytoplasmic RNA synthesis (11). Both hnRNP Q and hnRNP R associate with the phosphorylated RNA polymerase II C-terminal domain in a far-Western assay (8) and are required for efficient pre-mRNA splicing in an in vitro assay (44).
When transcription was arrested or HEXIM1 was knocked down by RNA interference (RNAi), 7SK RNA was released from its P-TEFb complex and recovered mostly with hnRNP A1 and A2 and hnRNP Q and R proteins. Furthermore, the transcription-dependent dissociation of P-TEFb-HEXIM1-7SK complexes was attenuated when both hnRNPs A1 and A2 were knocked down by small interfering RNA. As both A and Q/R types of hnRNPs are involved in a great variety of posttranscriptional RNA processes, our findings further illustrate the coordinated regulation of transcription and posttranscriptional events (50, 70).
MATERIALS AND METHODS
Plasmids.
pCDNA3-HA plasmids (52) were used to express tagged wild-type hnRNP R and truncated forms ΔRRM (with amino acids [aa] 166 to 331 deleted) and ΔSMN (with aa 522 to 556 deleted). For tandem affinity purification (TAP)-tagged proteins, hnRNP A1 cDNA was cloned into the BamH1 and EcoR1 sites in pNTAP-B (Stratagene). The mutations F57D and F59D in hnRNP A1 RNA recognition motif 1 (RRM1) and F148D and F150D in RRM2 were generated using a QuikChange site-directed mutagenesis kit (Stratagene). p-GEX and p-GEX-hnRNPA1 (a gift from Joëlle Marie) were used to express glutathione S-transferase (GST) fusion proteins.
Antibodies and TAP tag purifications.
Antibodies to hnRNP A1 (monoclonal antibody [MAb] 4B10; Santa Cruz), cyclin T1 (H-245; Santa Cruz), hnRNP C1/C2 (MAb 4F4; AbCAM), Rpb1 (MAb 4H8; Euromedex), polypyrimidine tract-binding protein (PTB) (51), hnRNP A2 (21), hnRNP Q/R (MAb 18E4) (44), HEXIM1 (41), and HEXIM2 (5) were used. Secondary antibodies were coupled with horseradish peroxidase (Promega) and detected by enhanced chemiluminescence with an ECL kit (Pierce). Antihemagglutinin (anti-HA)-agarose beads (Sigma) were used for immunoprecipitations. Biotin beads from the TAP tag purification kit (Stratagene) were used to purify TAP-tagged proteins.
Cells, transfections, RNAi, and lysis.
HeLa cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal calf serum. Log-phase cells were treated for 1 h with actinomycin D (1 μg ml−1) or 5,6-dichloro-1-d-ribofuranosylbenzimidazole (DRB; 100 μM) or were not treated. Cells arrested in mitosis were detached by shaking after exposure to nocodazole (50 ng ml−1) alone or in combination with DRB (100 μM) for 6 h and pelleted by centrifugation at 200 × g. For RNAi knockdowns, log-phase cells were transfected both on day 1 and on day 2 after plating and lysed on day 3. For transfection, small interfering RNA (MWG-Biotech AG) targeting hnRNP A1 (UGGGGAACGCUCACGGACU-dT-dT), hnRNP A2 (CGUUUGAAACCACAGAAGA-dT-dT), HEXIM1 (GGAUCCGAGCCGAGAUGUU-dT-dT), or vimentin (CUACAUCGACAAGGUGCGC-dT-dT) was mixed with Opti-MEM medium and Oligofectamine (Invitrogen). To prepare extracts, cells were washed in buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 200 mM NaCl, 0.2 mM EDTA) and lysed in buffer B (buffer A supplemented with 1 mM dithiothreitol, 40 U of RNasin (Promega) ml−1, protease inhibitor cocktail [P-8340; Sigma], 1 mM phenylmethylsulfonyl fluoride, and 0.5% Nonidet P-40). The calcium phosphate method was used for transfection, adding pSP64 as a noncoding carrier plasmid at up to 20 μg of DNA per dish (25 cm2), and cells were lysed 48 h after transfection. Lysates were clarified by centrifugations for 5 min at 500 × g and 5 min at 9,000 × g at 4°C.
7SK snRNP affinity purification.
Cell lysates were fractionated on glycerol gradients. Streptavidin beads (Amersham) were preincubated with a solution of 100 μg of glycogen ml−1, 1 mg of bovine serum albumin ml−1, and 100 μg of tRNA ml−1 in buffer C (20 mM Tris-HCl [pH 7.6], 0.1% NP-40, 0.1% NaN3, and 50 mM NaCl) for 20 min at 4°C. Gradient fractions containing 7SK snRNA were dialyzed against buffer D (35 mM Tris-HCl [pH 7.5], 200 mM NaCl, 0.075% NP-40, 0.05% NaN3) and left for 30 min at 30°C with a mixture of 3 nmol of 7SK antisense (BBBBCCUUIAIAICUUIUUUIIAII, where B represents biotinylated dT and I represents 2′-O-methyl-inosine) or sense (BBBBIIAIIUUUIUUCIAIAIUUCC) 2′-O-methyl oligoribonucleotides (Xeragon), 3.2 mM MgCl2, 30 μg of tRNA (Sigma) ml−1, and 0.5 mM ATP. Preincubated streptavidin beads were added next for a further 90 min at 4°C, and the fractions were centrifuged. This procedure was performed first with sense 2′-O-methyl oligoribonucleotides, and then the beads were discarded and the procedure was repeated with the resulting supernatant and either sense or antisense oligoribonucleotides. The second batch of beads was washed in buffer E (20 mM Tris-HCl, 0.1% NaN3, and 250 mM NaCl) and next in ETO buffer (5% glycerol, 1 mM HEPES buffer [pH 7.9], 0.25 mM EDTA, 0.5 mM dithiothreitol, 0.02% Tween 20). 7SK snRNP was eluted at 30°C in ETO buffer with 12 nmol of an oligodeoxyribonucleotide complementary to the biotinylated 2′-O-methyl oligoribonucleotide. Following sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, Coomassie blue-stained bands were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry as described previously (41).
Northern blot assays and protein-RNA cross-linking.
RNAs were electrophoresed in polyacrylamide gels, transferred onto Hybond N+ (Amersham), and hybridized to nick end-translated 32P-labeled DNA probes. The membranes were washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 65°C. Radioactivity was quantified with a Fuji FLA 3000 phosphorimager using the Image Gauge software. For protein-RNA cross-linking, formaldehyde (1% final concentration) was added to the culture medium for 10 min and then neutralized with glycine (125 mM, pH 7) for 5 min. Cells were lysed in RIPA buffer (50 mM Tris [pH 7.9], 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% SDS, 1 mM EDTA, 150 mM NaCl) and sonicated. The lysate was clarified by centrifugation. Protein-RNA complexes were immunoprecipitated, and the cross-link was reversed by incubation for 2 h at 65°C in a mixture of 50 mM Tris [pH 7.9], 5 mM EDTA, 10 mM dithiothreitol, and 1% SDS. RNAs were isolated with the RNeasy micro kit (QIAGEN). cDNAs transcribed with the Superscript III reverse transcriptase kit (Invitrogen) were amplified by real-time PCR using a Roche LightCycler 480. cDNAs were amplified by real-time PCR using oligodeoxynucleotides CCTGTAGTCCCAGCTACTCG and CTGCTCCGTTTCCGACCTGG and ATCTGTCACCCCATTGATCG and GCGCAGCTACTCGTATACCC as primer pairs for 7SL and 7SK RNA. To compare target nucleic acid amounts in different samples, we determined crossing points as a function of sample dilution by using the second derivative maximum method (http://www.idahotec.com/lightcycler_u/lectures/quantification_on_lc.htm).
Electrophoretic mobility shift assay.
7SK RNA transcribed in vitro with the T7 RiboMAX large-scale production system (Promega) was denatured at 75°C in the presence of poly(dI-dC) (50 μg) and renatured at room temperature for 10 min. Reactions were carried out in a 50-μl final volume containing 25 mM HEPES (pH 7.9), 15% glycerol, 60 mM KCl, 0.1 mM EDTA, 0.01% NP-40, 1.5 mM MgCl2, 1 mM dithiothreitol, 1 U of RNasin/μl, and 85 μg of bovine serum albumin/ml with 10 ng of 7SK RNA and with GST or GST-hnRNP A1 fusion protein eluted from glutathione-agarose beads (Amersham) in the amounts indicated in Fig. 3. Reaction mixtures were incubated for 20 min at 23°C. A 5% polyacrylamide gel in Tris (41 mM)-glycine (192 mM) was prerun for 10 min at 4°C. Samples were loaded and run for 2 h at 4°C, and the gel was transferred and analyzed by Northern blotting.
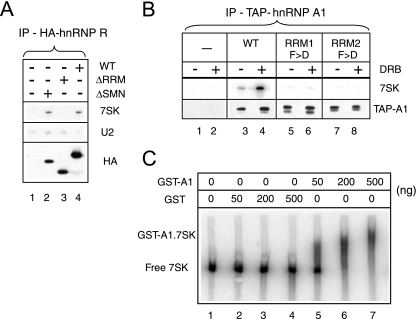
FIG. 3.
7SK RNA binding involves RRMs. Asynchronously growing cells were transiently transfected to express tagged hnRNPs or empty vector plasmids (-). Transfected cells were exposed to DRB (100 μM; +) or not (−) for 1 h prior to lysis. (A) RNAs immunoprecipitated with full-length (wild-type [WT]) or truncated (ΔRRM and ΔSMN) HA-tagged hnRNP R proteins. IP, immunoprecipitate. (B) RNAs were retained on biotin beads with wild-type TAP-tagged hnRNP A1 proteins or mutant forms carrying F→D (F>D) point mutations in either RRM1 or RRM2. HA-tagged hnRNP R or TAP-tagged hnRNP A1 on beads was detected by Western blotting with anti-HA or anti-hnRNP A1, respectively. RNAs were probed by Northern blotting using the same samples. Samples in all lanes were prepared from the same number of transfected cells. (C) The mobility of synthetic 7SK RNA decreased upon the addition of increasing amounts of GST-hnRNP A1 (lanes 5 to 7). The addition of genuine GST had no effect (lanes 2 to 4).
RESULTS
Affinity purification of 7SK RNA-associated proteins.
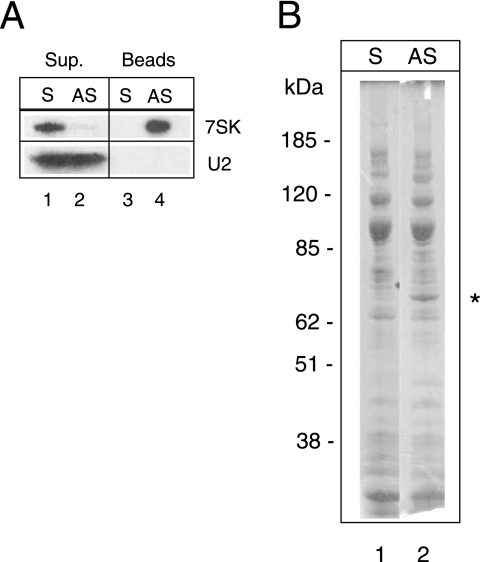
A pioneering work used antisense 2′-O-methyl RNA oligonucleotides to affinity select 7SK snRNPs and found eight unidentified proteins associated with 7SK RNA (60). Here, a similar strategy was followed. To eliminate the previously characterized P-TEFb-HEXIM-7SK complexes, lysates from cells in which transcription was arrested were fractionated on a glycerol gradient. Fractions containing 7SK RNA were pooled and incubated with a biotinylated 2′-O-methyl oligoribonucleotide corresponding either to the 7SK sequence from nucleotides 221 to 241 or to the complementary antisense sequence. The antisense oligonucleotides efficiently retained 7SK RNA on streptavidin beads (Fig. 1A, lane 4) and depleted it from the starting fraction (lane 2). In contrast, when a sense oligonucleotide was used, the 7SK RNA remained in the solution (lanes 1 and 3). Other RNAs, such as U2 snRNA, were not retained by either oligonucleotide. Despite the selectivity of the procedure, numerous proteins were retained on both beads and eluted by a competing oligodeoxyribonucleotide (Fig. 1B). The major band specifically retained on antisense beads had a molecular mass of close to 65 kDa (lane 2). It was digested with trypsin and analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry. The measured masses for 25 major peaks matched within 0.1 Da to the computed masses of tryptic peptides of hnRNP Q protein isoform Q1 (accession no. AY034483), Q2 (accession no. AY034482), or Q3 (accession no. AY034481). Most peptides were common to all three isoforms, giving an average protein sequence coverage of 39%, but two peptides were specific to Q1 and Q3 and one was specific to Q2 and Q3. hnRNP Q1, Q2, and Q3 are splicing variants encoded by the same gene and migrate as 55-, 60-, and 70-kDa proteins, respectively (44). This finding suggested that hnRNP Q proteins associate with 7SK RNA.
FIG. 1.
Purification of 7SK-associated proteins. Glycerol gradient fractions containing 7SK RNA were incubated with 7SK sense (S) or antisense (AS) biotinylated 2′-O-methylated oligoribonucleotides and streptavidin beads. Beads and supernatants (Sup.) were separated by centrifugation. (A) 7SK or U2 RNAs were detected by Northern blotting. (B) Proteins bound to streptavidin beads were stained by Coomassie blue after elution and SDS-polyacrylamide gel electrophoresis. The star indicates the protein identified as an hnRNP Q form by mass spectrometry.
hnRNPs Q and R and hnRNP A1 are 7SK-associated proteins.
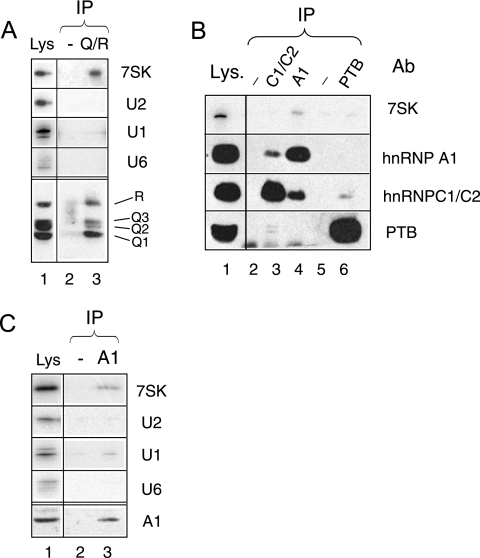
To validate the interaction of hnRNP Q proteins with 7SK RNA, coimmunoprecipitations using a specific antibody were performed. The 18E4 MAb recognizes all three members of the hnRNP Q protein family, as well as the hnRNP R protein encoded by a cognate gene (44). Indeed, it immunoprecipitated 7SK from a HeLa cell lysate (Fig. 2A). hnRNP Q and R proteins had been shown to be components of hnRNP complexes containing several hnRNPs, such as hnRNP C1/C2 (44). However, hnRNP C1/C2 antibodies immunoprecipitated hardly any 7SK RNA (Fig. 2B, lane 3). The possible interaction of 7SK with other hnRNPs was investigated next. PTB did not coimmunoprecipitate significantly with 7SK RNA (lane 6). In contrast, 7SK RNA immunoprecipitated with hnRNP A1 (lane 4). It is noteworthy that partial hnRNP A1 and hnRNP C1/C2 coimmunprecipitation (see lanes 3 and 4), as well as weak PTB and hnRNP C1/C2 coimmunoprecipitation (see lanes 3 and 6), was detected as reported previously (42, 44). Neither U2 nor U6 snRNAs immunoprecipitated as significantly with either hnRNP Q/R or hnRNP A1 (Fig. 2A and C). A small amount of U1 snRNA was reproducibly detected with hnRNP A1. Thus, A- and Q/R-type hnRNPs have been identified as new specific 7SK RNA-binding proteins.
FIG. 2.
Immunoprecipitation of 7SK RNA with hnRNP Q and R and hnRNP A1 proteins. Proteins and RNAs were immunoprecipitated with anti-hnRNP Q/R (A); anti-hnRNP C1/C2, anti-hnRNP A1, and anti-PTB antibodies (Ab) (B); and anti-hnRNP A1 antibody (C). Mock antibodies (-) were used for lanes 2 in panels A and C and lanes 2 and 5 in panel B. Proteins in input lysates (Lys) and protein A beads were detected by Western blotting. RNAs were probed by Northern blotting using the same samples. IP, immunoprecipitate.
Association of hnRNPs with 7SK requires intact RRMs.
To further evaluate the specificity of the interactions, hnRNP cDNAs were fused to epitope tags and transiently expressed in HeLa cells. Indeed, 7SK RNA immunoprecipitated efficiently with HA-tagged hnRNP R (Fig. 3A, lane 4). The RRMs, which are consensus RNA-binding domains of approximately 90 aa, are thought to be involved in the interactions of the hnRNP proteins with RNA. RRMs contain two sequences (RNP1 and RNP2) of eight conserved residues and have been maintained in a large number of ribonucleoproteins throughout evolution (37). Three such RRMs (designated RRM1, RRM2, and RRM3) are present in the hnRNP R sequence (52). The deletion of the first two RRMs (aa 166 to 331) suppressed binding (Fig. 3A, lane 3). In contrast, the deletion of a domain (the SMN-binding domain from aa 522 to 556) distinct from the RRMs did not affect 7SK binding (lane 2). Importantly, wild-type and mutant proteins showed the same nuclear localization patterns (not shown). The hnRNP A1 cDNA was next fused to a sequence encoding the TAP tag. When this fusion was expressed in HeLa cells, 7SK RNA was retained on biotin beads (Fig. 3B, lane 3). As discussed below, a greater amount of 7SK RNA was retained when the cells had been treated with DRB, an inhibitor of transcription, prior to lysis (Fig. 3B, lane 4) than when the cells had not been treated. Two RRMs (RRM1 and RRM2) are present in the hnRNP A1 sequence. The replacement of two conserved phenylalanines in the RNP2 sequence by aspartate residues in both RRMs had been shown previously to decrease the affinity of hnRNP A1 for pre-mRNA (38). Wild-type and mutant proteins all showed the same nuclear localization patterns. Introducing these mutations into either RRM independently had a dramatic effect and brought 7SK binding to background levels (Fig. 3B, lanes 5 to 8). The electrophoretic mobility of 7SK RNA was shifted by the recombinant GST-hnRNP A1 fusion protein (Fig. 3C, lanes 5 to 7). Taken together, these data suggested the direct binding of hnRNP A1 to 7SK RNA.
Increased association of Q/R- and A-type hnRNPs with 7SK RNA following P-TEFb-HEXIM-7SK complex dissociation.
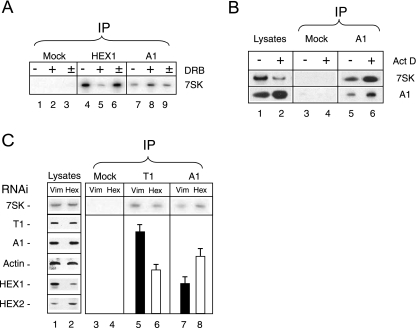
The arrest of transcription in growing asynchronous cells results in the disruption of the P-TEFb-HEXIM-7SK complex (41, 64). Yet the 7SK RNA is not degraded. Its transfer to hnRNP-containing particles was therefore investigated thoroughly. An increase in 7SK binding to epitope-tagged hnRNP A1 was observed when cells expressing TAP-tagged hnRNP A1 were treated with DRB to inhibit transcription (Fig. 3B, compare lanes 3 and 4). Exposure to DRB markedly decreased the amount of 7SK immunoprecipitated with HEXIM1 (Fig. 4A, lanes 4 and 5) but increased the amount of 7SK immunoprecipitated with endogenous hnRNP A1 (lanes 7 and 8). DRB is a reversible inhibitor of transcription. Indeed, 7SK binding to HEXIM1 was restored in less than 1 h when DRB was washed away (Fig. 4A, lane 6). Meanwhile, 7SK binding to hnRNP A1 decreased back to initial levels (Fig. 4A, lane 9). This experiment was performed in the presence of cycloheximide to block protein synthesis. Thus, the exchange of 7SK between HEXIM1 and hnRNP A1 was not linked to the assembly of newly synthesized proteins.
FIG. 4.
Association of 7SK with hnRNP A1 increases upon P-TEFb-HEXIM1-7SK dissociation. Cell lysates were subjected to immunoprecipitation (IP) with antibodies against HEXIM1 (HEX1), cyclin T1 (T1), hnRNP A1 (A1), or preimmune serum (mock). Histograms correspond to the quantification of immunoprecipitated (bound) 7SK by Northern blotting. Lysates were probed by Western blotting for cyclin T1, hnRNP A1, HEXIM1 (HEX1), HEXIM2 (HEX2), and actin (Act). (A) Cycloheximide-treated cells were treated with DRB (100 μM; + and ±) or not (−) for 1 h. DRB-treated cells were (±) or were not (+) allowed to recover in normal medium for 1 h prior to lysis. Cycloheximide (50 μg ml−1) was present throughout the entire experiment. (B) Cells were treated with actinomycin D (Act D) at 1 μg ml−1 (+) or not (−) for 1 h. (C) Cells were treated for 48 h with interfering RNA (RNAi) targeting vimentin (Vim) or HEXIM1 (Hex).
Reassortment between RNA and RNA-binding proteins has been shown to occur after cell disruption (43). Therefore, to test for true in vivo associations, 7SK RNA was quantified by real-time PCR analysis of reverse-transcribed RNAs immunoprecipitated from lysates of cells that had been cross-linked with formaldehyde prior to lysis in a dissociating buffer. A very large proportion of 7SK RNA immunoprecipitated with HEXIM1 (Table 1). A smaller amount immunoprecipitated with hnRNP Q/R or hnRNP A1 antibodies. However, this amount was more than 100-fold above the background levels observed with mock antibodies or with un-cross-linked lysates or an unrelated RNA such as 7SL RNA. Following the arrest of transcription promoted by DRB, the amount of 7SK cross-linked to HEXIM1 dropped from 25 to 0.6%; meanwhile, there was a strong increase in the level of 7SK RNA cross-linked to hnRNPs (Table 1).
TABLE 1.
Quantification of RNAs cross-linked to proteinsa
| Antibody | % of 7SK RNA immunoprecipitated from lysate of:
|
% of 7SL RNA immunoprecipitated from lysate of CL cells | ||
|---|---|---|---|---|
| CL cells | UL cells | DRB-treated CL cells | ||
| Mock | 0.01 ± 0.005 | 0.02 | 0.01 ± 0.005 | 0.01 |
| Anti-HEXIM1 | 26.00 ± 4.0 | 0.01 | 0.6 ± 0.1 | 0.04 |
| Anti-hnRNP Q/R | 0.75 ± 0.07 | 0.004 | 1.4 ± 0.1 | 0.02 |
| Anti-hnRNP A1 | 5.76 ± 0.40 | 0.009 | 10.3 ± 0.3 | 0.06 |
Immunoprecipitations from lysates of cells cross-linked (CL) or not (UL) with formaldehyde were performed. When indicated, cells were treated with DRB (100 μM) before cross-linking. 7SK and 7SL RNAs were quantified by real-time PCR analysis of cDNAs. Results are provided as the percentages of RNAs immunoprecipitated from the starting lysates. Numbers are the means ± the standard deviations (where indicated) of results from three independent experiments.
DRB impairs transcription by inhibiting the kinase activity of Cdk9. Hence, we assayed the immunoprecipitation of 7SK following the exposure of cells to actinomycin D, which is thought to inhibit transcription through a distinct mechanism, presumably intercalation into DNA. Like treatment with any inhibitor of transcription, actinomycin D treatment resulted in an increase in the extractability of hnRNP A1 that will be discussed later. Although there was decreased extractability of 7SK RNA, a larger amount immunoprecipitated with hnRNP A1 from extracts of actinomycin D-treated cells (Fig. 4B, lanes 5 and 6).
Next, the possible transfer of 7SK RNA from HEXIM1 to hnRNP A1 when HEXIM1 was knocked down by RNAi was investigated. This procedure did not modify the amount of 7SK RNA in lysates (Fig. 4C, lanes 1 and 2), but the amount of 7SK immunoprecipitated with hnRNP A1 increased twofold (lanes 7 and 8). However, the level of 7SK immunoprecipitated with the cyclin T1 subunit of P-TEFb decreased (lanes 5 and 6) only twofold. Indeed, HEXIM2 partially compensates for a HEXIM1 knockdown (5, 66) and an increase in HEXIM2 levels was observed in lysates from cells with knocked-down HEXIM1 (lane 2). This observation may explain why the consequences of HEXIM1 knockdown were less dramatic than the consequences of transcription inhibition. Despite several attempts, HEXIM2 knockdown was not successful, and hence, a complete HEXIM knockdown could not be achieved. To summarize, distinct procedures that lead to the dissociation of 7SK RNA from P-TEFb-HEXIM1 complexes resulted in a marked increase in the amount of 7SK RNA immunoprecipitated with hnRNPs.
Quantification of 7SK RNA transfer to Q/R- and A-type hnRNPs following P-TEFb-HEXIM-7SK complex dissociation.
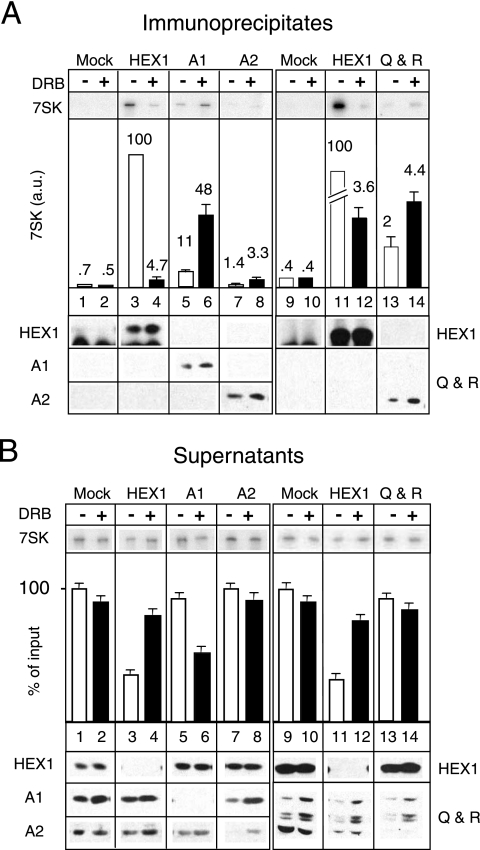
In order to evaluate a possible transfer of 7SK from P-TEFb-HEXIM to hnRNP complexes, the RNAs immunoprecipitated with the various proteins were quantified under conditions of immunodepletion. In contrast to HEXIM1, hnRNP A1 could not be depleted from the cross-linked cell lysates used to obtain the data in Table 1 (data not shown). The amount of immunoprecipitated antigen was rather small and did not increase with the use of 20 or 100 μl of MAb per assay. The cross-linking reaction likely damages the epitope in some hnRNP A1 molecules. However, hnRNP A1 was depleted from nondenatured cell lysates (Fig. 5B, lower panel). Therefore, to investigate a possible transfer of 7SK RNA from HEXIM to hnRNPs, we decided to perform an extensive quantitative study using nondenaturing lysates. A large amount of 7SK RNA immunoprecipitated with HEXIM1 (Fig. 5A, lanes 3 and 11), and although HEXIM1 had been depleted (Fig. 5B, lanes 3 and 11), roughly 50% of 7SK RNA remained free in postimmunoprecipitation supernatants. When cells were treated for 1 h with DRB to inhibit transcription, the amount of 7SK immunoprecipitated with HEXIM1 decreased markedly (Fig. 5A, compare lanes 3, 4, 11, and 12) and the level of 7SK in the postimmunoprecipitation supernatants increased (Fig. 5B, lanes 4 and 12). In contrast, the amount of 7SK immunoprecipitated with hnRNP A1 (Fig. 5A, lanes 5 and 6) from DRB-treated cells increased and less 7SK remained in hnRNP A1 postimmunoprecipitation supernatants from DRB-treated cells (Fig. 5B, lanes 5 and 6). A small amount of 7SK, which increased following DRB treatment, was also recovered with hnRNP A2 (Fig. 5A, lanes 7 and 8). Similarly, the small amount of 7SK recovered with hnRNP Q/R increased following DRB treatment (lanes 13 and 14).
FIG. 5.
Association of 7SK with hnRNPs A1 and A2 and hnRNPs Q and R increases in asynchronously growing cells exposed to DRB. HeLa cells were treated with DRB (100 μM; +) or not (−) for 1 h. Lysates were incubated with either mock serum or anti-HEXIM1, anti-hnRNP A1, anti-hnRNP A2, or anti-hnRNP Q/R. Immunoprecipitates (A) and postimmunoprecipitation supernatants (B) were probed for 7SK RNA and proteins by Northern or Western blotting. Histograms correspond to 7SK quantification by Northern blotting. In panel A, the amount of 7SK RNA immunoprecipitated with HEXIM1 (HEX1) from untreated cell lysates was set at 100 a.u.
The amount of 7SK immunoprecipitated with HEXIM1 from untreated cell lysates was set at 100 arbitrary units (a.u.) for the following discussion. In cells treated with DRB, the level of 7SK bound to HEXIM1 decreased from 100 to 4 a.u. In contrast, the amount of 7SK bound to hnRNP A1 increased from 11 to 48 a.u. This increase (37 a.u.) corresponded to 40% of the 7SK released from HEXIM1 (96 a.u.). Taken together, these data suggested that 7SK RNA shuttles from P-TEFb-HEXIM1 complexes to hnRNP complexes.
Increased solubility of hnRNP A1 following transcriptional arrest.
Efforts to quantify the results of the above-described experiments encountered a limitation linked to the use of nondenaturing conditions. Indeed, buffers are limited in ionic strength to avoid the disruption of RNA-protein complexes. Under these conditions, a large amount of hnRNPs Q and R and hnRNP A1 remained associated with nuclear pellets after centrifugation at 10,000 × g (Fig. 6A). Less hnRNP A1 was found in supernatants (lane 1) than in pellets (lane 5). In contrast, following the arrest of transcription, more hnRNP A1 was present in cytosolic supernatants (Fig. 6A, lane 3) than in nuclear pellets (lane 7), as previously reported (29, 30). The solubility of hnRNPs Q and R was much less sensitive to the inhibitors. The amount of 7SK RNA in supernatants decreased in response to DRB. This decrease was more pronounced with actinomycin D treatment (Fig. 4B, lanes 1 and 2).
FIG. 6.
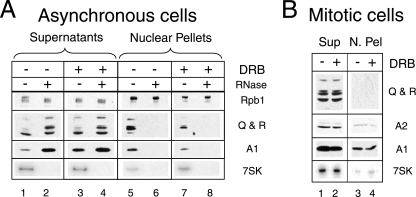
Complete extraction of hnRNPs A1 and A2 and hnRNPs Q and R from mitotic cells. Lysates from asynchronous (A) or mitotic (B) cells treated with DRB (100 μM; +) or not (−) and treated with RNase (10 μg ml−1; +) or not (−) were fractionated by centrifugation at 10,000 × g into supernatants (Sup) and nuclear pellets (N. Pel). RNA polymerase II (Rpb1) and hnRNPs were detected by Western blotting. 7SK RNA was detected by Northern blotting.
Neither hnRNP A1 nor hnRNPs Q and R remained in nuclear pellets when RNase was added to the lysis buffer (Fig. 6A, lanes 6 and 8). In contrast, RNA polymerase II extraction was insensitive to RNase. hnRNPs are thought to associate with nascent RNAs (49). The magnitude of the increase in solubilization following transcription inhibition suggests that most hnRNP A1 molecules are fractionated with nuclear pellets and are indeed associated with nascent RNAs.
To eliminate the possibility that an increase in hnRNP in the extracts from cells with arrested transcription was responsible for the increase in 7SK coimmunoprecipitations, we turned to mitotic cells, where the disruption of the nuclear membrane favors the solubilization of most nuclear proteins. Mitotic cells accumulate in the presence of nocodazole and detach from culture dishes (69). Mitotic HeLa cells that have been exposed to nocodazole for 6 h still efficiently reenter a normal cell cycle when the drug is removed. The extraction of 7SK RNA and hnRNPs A1, A2, Q, and R from such mitotic cells was found to be greatly enhanced; very small amounts of these macromolecules remained associated with pellets after centrifugation at 10,000 × g (Fig. 6B). The corresponding clarified lysates contained similar levels of 7SK RNA and hnRNPs whether the cells had been treated with DRB or not.
Quantification of 7SK transfer from HEXIM1 to hnRNPs in mitotic-cell extracts.
The mitotic cells permit the quantification of the DRB effect under conditions of immunodepletion as well as full solubility (and hence constant levels) of hnRNPs, P-TEFb, and 7SK RNA. The levels of 7SK RNA immunoprecipitated by anti-HEXIM1 antibodies from mitotic cell extracts were comparable to those obtained from asynchronous cell extracts (Fig. 7, lanes 3 and 11). This finding was unexpected because mitotic cells are transcriptionally inactive (19, 28). Furthermore, the addition of DRB to arrested cells that had entered mitosis and already detached from the culture plate had no influence on 7SK immunoprecipitation with HEXIM1 (data not shown). As the inhibition of transcription did not interfere with mitotic-cell accumulation, lysates from mitotic cells that had accumulated in the presence of both nocodazole and DRB were prepared. This procedure ensured that transcription was arrested before entry into mitosis.
FIG. 7.
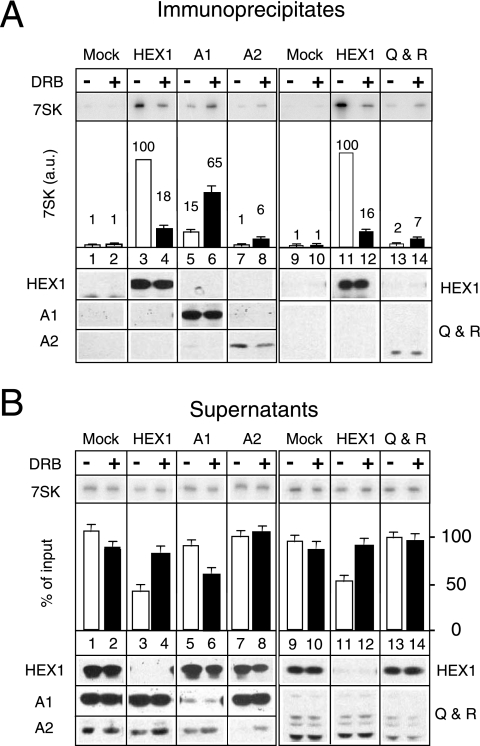
7SK RNA transfer from HEXIM1 to hnRNPs Q and R or to hnRNPs A1 and A2 in DRB-treated mitotic cells. HeLa cells arrested in mitosis were collected after being treated for 4 h with nocodazole in the absence (−) or in the presence (+) of DRB (100 μM). Lysates were incubated with either mock serum or anti-HEXIM1, anti-hnRNP A1, anti-hnRNP A2, or anti-hnRNP Q/R. Immunoprecipitates (A) and postimmunoprecipitation supernatants (B) were probed for 7SK RNA and proteins by Northern or Western blotting. Histograms correspond to 7SK quantification by Northern blotting. In panel A, the amount of 7SK RNA immunoprecipitated with HEXIM1 (HEX1) from untreated cell lysates was set at 100 a.u.
The amount of 7SK RNA immunoprecipitated by anti-HEXIM1 from DRB- and nocodazole-treated cells was much smaller than that immunoprecipitated from cells treated with just nocodazole, as previously observed with attached asynchronously growing cells (Fig. 7A, compare lanes 3 and 4). In contrast, the amount of 7SK RNA immunoprecipitated with hnRNP A1 (Fig. 7A, lanes 5 and 6) increased markedly. The amount of 7SK RNA immunoprecipitated with either anti-hnRNP A2 (Fig. 7A, lanes 7 and 8) or anti-hnRNP Q and R (lanes 13 and 14) antibodies was much smaller than that immunoprecipitated with hnRNP A1. However, it also increased when cells were treated with DRB in the presence of nocodazole.
HEXIM1, hnRNP A1, and hnRNP A2 were depleted through immunoprecipitation (Fig. 7B). In cells treated with DRB, the sum of the increases in the levels of 7SK bound to hnRNP A1 (50 a.u., from 15 to 65 a.u.) and hnRNP A2 (5 a.u., from 1 to 6 a.u.) roughly corresponded to 70% of the amount released from HEXIM1 (82 a.u., from 100 to 18 a.u.). Smaller amounts of 7SK RNA were immunoprecipitated with hnRNPs Q and R (an increase of 5 a.u., from 2 to 7 a.u.). However, despite several attempts using large amounts of antibodies, the depletion of hnRNPs Q and R was not successful; half of these proteins remained free (Fig. 7B, lanes 13 and 14). Hence, the amount of 7SK bound to hnRNPs Q and R was underestimated. Taken together, these data indicate that most (about 80%) of the 7SK RNA released from HEXIM1 complexes is transferred to complexes with hnRNP A1, hnRNP A2, and hnRNPs Q and R.
Knockdown of both hnRNP A1 and hnRNP A2 results in limited transcription-induced dissociation of P-TEFb-HEXIM1-7SK complexes.
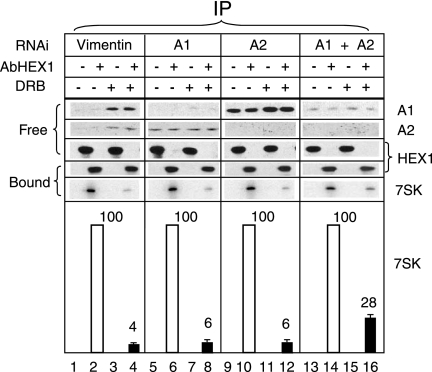
An RNAi strategy was used next to investigate whether 7SK RNA release from HEXIM1 upon the inhibition of transcription involves hnRNPs. Single RNAi knockdown of either hnRNP A1 or hnRNP A2 had little effect on the amount of 7SK RNA immunoprecipitated with HEXIM1 (Fig. 8). Following DRB treatment, the 7SK signal decreased to 4% ± 1% of that from untreated cells in which vimentin had been knocked down (Fig. 8, lanes 2 and 4). The 7SK signal from cells with the single knockdown of hnRNP A1 (lanes 6 and 8) or hnRNP A2 (lanes 10 and 12) decreased to 6% ± 1% of that from cells expressing both hnRNPs. In contrast, when both hnRNP A1 and A2 proteins were knocked down, the proportion of 7SK remaining with HEXIM1 following DRB treatment reached 28% ± 3% (lanes 14 and 16). Of interest, an hnRNP A1 single knockdown resulted in increased levels of hnRNP A2 (lanes 5 to 8). Conversely, an hnRNP A2 single knockdown resulted in increased levels of hnRNP A1 (lanes 9 to 12). A higher level of expression of one protein may compensate for the knockdown of the other one. This compensation may account for the insignificant consequences of single knockdowns for 7SK RNA binding to HEXIM1. It cannot be ruled out that the remaining hnRNP A1 and/or an increase in the highly similar but minor hnRNP A0 and A3 proteins also partly compensates for the double knockdown of hnRNPs A1 and A2. Nevertheless, the diminished dissociation of P-TEFb-HEXIM1-7SK complexes following transcription inhibition in cells with hnRNP A1-hnRNP A2 double knockdowns suggested that these proteins contribute to the dissociation process.
FIG. 8.
Decreased DRB-induced dissociation of P-TEFb-HEXIM1-7SK RNA following RNAi. HeLa cells were treated with interfering RNA targeting vimentin or hnRNPs A1 and A2 and exposed to DRB (+) or not (−) for 1 h prior to lysis. Lysates were subjected to immunoprecipitation (IP) with anti-HEXIM1 (AbHEX1; +) or mock (−) antibodies. The presence of hnRNPs A1 and A2 and HEXIM1 (HEX1) in supernatants (free) or on protein A beads (bound) was tested by Western blotting. 7SK RNA immunoprecipitated with HEXIM1 was detected by Northern blotting. Signal quantification (the average of results from three independent experiments) is shown in the lower panel. Histograms correspond to the quantification of immunoprecipitated (bound) 7SK by Northern blotting. The level of 7SK RNA immunoprecipitated with HEXIM1 from cells that had not been exposed to DRB was set at 100 a.u.
DISCUSSION
hnRNP A and hnRNP Q and R proteins are related.
In this study and in reports published after this work was submitted (23, 59), hnRNP K, hnRNPs Q and R, and hnRNPs A1 and A2 were shown to be major 7SK RNA-associated proteins. Heterogeneous nuclear ribonucleoproteins form a large group of very diverse RNA-binding proteins (12). Among them, hnRNP A and hnRNP Q and R proteins share related primary structures. They comprise related RRMs followed by a C-terminal arginine-, glycine-, and tyrosine-rich domain. The association of 7SK RNA with hnRNP R and hnRNP A1 involves the RRMs. hnRNP A1 possesses two RRMs. The inactivation of a single RRM by two point mutations is sufficient to abolish 7SK binding. It is noteworthy that the inactivation of both RRMs was previously shown to be required to abolish hnRNP A1 binding to human β-globin pre-mRNA in vitro (38). The same study reported that despite the homology of hnRNPs A1 and A2, hnRNP A2 binds more weakly to pre-mRNA than hnRNP A1. Less 7SK RNA was found associated with hnRNP A2 than with hnRNP A1; however, hnRNP A2 is less abundant than hnRNP A1. RRMs in hnRNPs A1 and A2 are almost identical and closely related to those in hnRNPs Q and R (33) (Fig. 3A). More distantly related RRM-containing proteins, such as hnRNP C1/C2 and PTB, did not immunoprecipitate with 7SK RNA. Thus, the 7SK-binding characteristic of RRMs in A- and Q-type hnRNPs supports a functional similarity between these proteins beyond their amino acid sequences.
Transcription-dependent interaction of hnRNPs with RNAs.
The binding of 7SK RNA to hnRNP K, hnRNPs Q and R, and hnRNPs A1 and A2 (references 23 and 59 and this work) increases when transcription is inhibited. hnRNPs A1, A2, and K belong to a subclass of hnRNPs which shuttle between the nucleus and the cytoplasm (6, 39, 49). Their intracellular localization is affected by drugs that inhibit transcription through distinct mechanisms. DRB inhibits P-TEFb activity, whereas actinomycin D intercalates into DNA. hnRNP A1 is mostly nuclear in asynchronously growing cells but does not return to the nucleus after mitosis in cells exposed to the inhibitors for several hours. The transcription-dependent nuclear import of hnRNP A1 relies on a small peptide domain (M9) distinct from the RRMs (26, 57, 61). Importantly, the conditions used in this work (1 h of exposure of asynchronous cells to 100 μM DRB) did not significantly alter hnRNP A1 and 7SK RNA localization patterns, as both hnRNP A1 and 7SK remained spread out in the nucleus but excluded from nucleoli (data not shown).
Most of the total hnRNP A1 remains associated with nuclear pellets of asynchronously growing cells. In contrast, following the exposure of the cells to either DRB or actinomycin, most hnRNP A1 is solubilized. The presence of hnRNP A1 in the nuclear material relies on RNAs, and the protein is suppressed when RNase is added to the buffers. This observation suggests that a majority of hnRNP A1 molecules remain trapped with chromatin as they bind nascent transcripts. HeLa cells have been estimated to contain 6 × 107 molecules of hnRNP A1 per cell (22) and 2 × 105 molecules of 7SK RNA per cell (60). When transcription is arrested, hnRNP A1 is released from mature transcripts but is not captured back by new nascent transcripts, and it thus becomes available in a very large molar excess (more than 100-fold) to bind 7SK RNA.
hnRNP cooperates in the P-TEFb-HEXIM-7SK dissociation process.
7SK RNA had previously been shown to be associated with HEXIM1 or HEXIM2 proteins and P-TEFb (Cdk9 and cyclin T1/T2) (41, 64). This complex is disrupted when transcription is inhibited, yet 7SK RNA is not degraded. In growing asynchronous cells, the disruption is seen within 15 min and is reversible. This observation suggests a permanent dynamic exchange between active free P-TEFb and inactive P-TEFb bound to HEXIM1-7SK or HEXIM2-7SK. In this study, we showed that a major proportion of 7SK RNA released from P-TEFb-HEXIM1-7SK complexes was recovered with Q- and A-type hnRNPs following DRB or actinomycin D treatment. Thus, 7SK RNA shuttles from P-TEFb to hnRNPs. In asynchronous cells, DRB and actinomycin D inhibit transcription at the elongation step following the initiation step. These drugs would not prevent P-TEFb-HEXIM-7SK dissociation when P-TEFb was loaded onto genes through an interaction with transcription factors during transcription initiation. The accumulation of free P-TEFb and hnRNP-7SK when transcription is arrested by a drug may indicate that both the assembly of P-TEFb-HEXIM-7SK complexes and the disassembly of hnRNP-7SK complexes require active transcription. The attenuated release of 7SK RNA from P-TEFb-HEXIM-7SK complexes upon the knockdown of hnRNPs A1 and A2 (this study) suggests that these proteins cooperate in the P-TEFb complex dissociation process. This cooperation is supported by the recent finding that a truncated 7SK RNA molecule unable to bind hnRNPs is resistant to inhibitor-induced disassembly from P-TEFb (59).
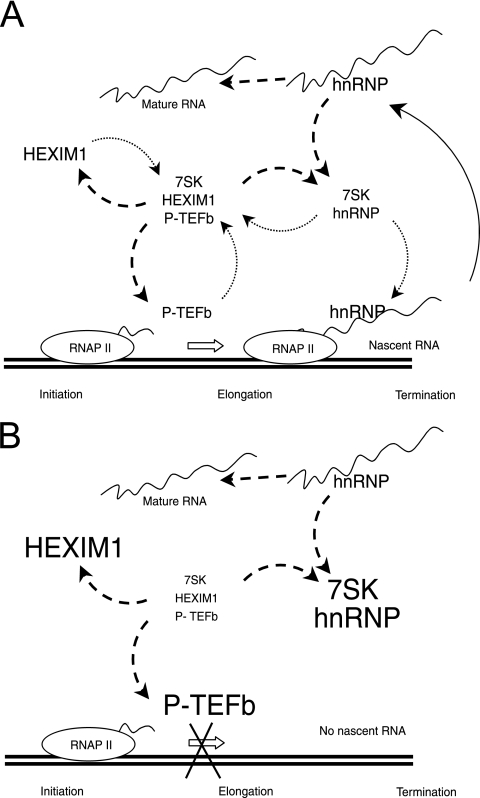
Our present knowledge may be summarized with a simple model (Fig. 9A). As the nascent transcripts are extruded out of RNA polymerase, they recruit hnRNPs that may release their 7SK RNA companion, which would in turn become liable to reassembly with HEXIM1 and P-TEFb on the transcription unit. Shortly after initiation, when P-TEFb is loaded onto the transcription machinery, 7SK would dissociate from P-TEFb and be trapped onto hnRNP molecules that had just been released from a mature transcript. Conversely, P-TEFb-HEXIM-7SK would reassemble upon the elongation-termination step as 7SK RNA was outcompeted by nascent transcripts. When transcription is inhibited, P-TEFb might be blocked on genes (18) and the pool of hnRNP would increase (Fig. 9B). As a result, hnRNP-7SK complexes as well as core P-TEFb would accumulate. RNA polymerase II inhibition has been proposed previously to increase the binding activity of hnRNP A1 (29). Such increased binding may also contribute to the trapping of 7SK RNA. The active dissociation of the P-TEFb-HEXIM-7SK complexes by free hnRNP is unlikely. It has not been observed in vitro following the addition of recombinant hnRNP A1 to such complexes (data not shown), and more significantly, the P-TEFb/P-TEFb-HEXIM-7SK ratio remains frozen in cells blocked in mitosis, although transcription is arrested. However, a major distinction can be made between drug-induced and mitotic inhibition of transcription. Both DRB and actinomycin D inhibit transcription at the elongation step. In contrast, both initiation and elongation are repressed during mitosis (19, 28). As P-TEFb is no longer recruited to transcriptional machinery during mitosis, it would not dissociate from HEXIM and 7SK RNA. The flow of RNA from nascent transcripts bound to hnRNPs to mature transcripts devoid of most hnRNPs may drive the balance between active and inactive forms of P-TEFb. In asynchronous cells, 7SK RNA trapping by hnRNPs may act as a sensor of transcription, thereby controlling P-TEFb activity and fine-tuning the overall class II gene transcription efficiency.
FIG. 9.
Model of coupled hnRNP-7SK and P-TEFb-HEXIM1-7SK RNA complex formation. (A) At the initiation of transcription, P-TEFb is loaded onto the transcription machinery and 7SK RNA is released from P-TEFb (and HEXIM1) and transferred onto an hnRNP that has been released from a mature RNA molecule (dashed-line arrows). Upon the elongation or termination of transcription, as hnRNPs are recruited onto the nascent transcripts, they release their 7SK RNA companion, which reassembles onto P-TEFb (dotted-line arrows). RNAP II, RNA polymerase II. (B) There are no nascent chains when transcription is inhibited. Core P-TEFb accumulates at the expense of P-TEFb-HEXIM-7SK. hnRNPs are released and trap 7SK RNA.
Acknowledgments
We are indebted to Anne Catherine Dock-Bregeon (Strasbourg, France), Xavier Darzacq, Gaëlle Diribarne, Annemieke Michels (Lausanne, Switzerland), and Béatrice Spiluttini for fruitful discussions and help and to Gideon Dreyfuss (Philadelphia, PA), Joëlle Marie (Gif sur Yvette, France), David Price (Iowa City, IA), William F. Rigby (Lebanon, NH), Michael Sendtner (Wuerzburg, Germany), and Chris Smith (Cambridge, United Kingdom) for reagents.
This work was supported by grants from Association pour la Recherche sur le Cancer, Agence Nationale de Recherche sur le SIDA, Agence Nationale pour la Recherche, Fondation pour la Recherche Médicale.
Footnotes
Published ahead of print on 20 August 2007.
REFERENCES
- 1.Bannai, H., K. Fukatsu, A. Mizutani, T. Natsume, S. Iemura, T. Ikegami, T. Inoue, and K. Mikoshiba. 2004. An RNA-interacting protein, SYNCRIP (heterogeneous nuclear ribonuclear protein Q1/NSAP1) is a component of mRNA granule transported with inositol 1,4,5-trisphosphate receptor type 1 mRNA in neuronal dendrites. J. Biol. Chem. 279:53427-53434. [DOI] [PubMed] [Google Scholar]
- 2.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327-337. [DOI] [PubMed] [Google Scholar]
- 3.Blanc, V., J. O. Henderson, E. P. Newberry, S. Kennedy, J. Luo, and N. O. Davidson. 2005. Targeted deletion of the murine apobec-1 complementation factor (acf) gene results in embryonic lethality. Mol. Cell. Biol. 25:7260-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnal, S., F. Pileur, C. Orsini, F. Parker, F. Pujol, A. C. Prats, and S. Vagner. 2005. Heterogeneous nuclear ribonucleoprotein A1 is a novel internal ribosome entry site trans-acting factor that modulates alternative initiation of translation of the fibroblast growth factor 2 mRNA. J. Biol. Chem. 280:4144-4153. [DOI] [PubMed] [Google Scholar]
- 5.Byers, S. A., J. P. Price, J. J. Cooper, Q. Li, and D. H. Price. 2005. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 280:16360-16367. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 8.Carty, S. M., and A. L. Greenleaf. 2002. Hyperphosphorylated C-terminal repeat domain-associating proteins in the nuclear proteome link transcription to DNA/chromatin modification and RNA processing. Mol. Cell Proteomics 1:598-610. [DOI] [PubMed] [Google Scholar]
- 9.Chen, C. Y., and A. B. Shyu. 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 23:4805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, R., Z. Yang, and Q. Zhou. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279:4153-4160. [DOI] [PubMed] [Google Scholar]
- 11.Choi, K. S., A. Mizutani, and M. M. Lai. 2004. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 78:13153-13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreyfuss, G., V. N. Kim, and N. Kataoka. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195-205. [DOI] [PubMed] [Google Scholar]
- 13.Dulac, C., A. A. Michels, A. Fraldi, F. Bonnet, V. T. Nguyen, G. Napolitano, L. Lania, and O. Bensaude. 2005. Transcription-dependent association of multiple positive transcription elongation factor units to a HEXIM multimer. J. Biol. Chem. 280:30619-30629. [DOI] [PubMed] [Google Scholar]
- 14.Egloff, S., E. Van Herreweghe, and T. Kiss. 2006. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol. Cell. Biol. 26:630-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Expert-Bezancon, A., A. Sureau, P. Durosay, R. Salesse, H. Groeneveld, J. P. Lecaer, and J. Marie. 2004. hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of beta-tropomyosin exon 6B. J. Biol. Chem. 279:38249-38259. [DOI] [PubMed] [Google Scholar]
- 16.Ford, L. P., W. E. Wright, and J. W. Shay. 2002. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene 21:580-583. [DOI] [PubMed] [Google Scholar]
- 17.Giraud, S., A. Hurlstone, S. Avril, and O. Coqueret. 2004. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene 23:7391-7398. [DOI] [PubMed] [Google Scholar]
- 18.Gomes, N. P., G. Bjerke, B. Llorente, S. A. Szostek, B. M. Emerson, and J. M. Espinosa. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197-202. [DOI] [PubMed] [Google Scholar]
- 20.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29-40. [DOI] [PubMed] [Google Scholar]
- 21.Hamilton, B. J., R. C. Nichols, H. Tsukamoto, R. J. Boado, W. M. Pardridge, and W. F. Rigby. 1999. hnRNP A2 and hnRNP L bind the 3′UTR of glucose transporter 1 mRNA and exist as a complex in vivo. Biochem. Biophys. Res. Commun. 261:646-651. [DOI] [PubMed] [Google Scholar]
- 22.Hanamura, A., J. F. Caceres, A. Mayeda, B. R. J. Franza, and A. R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 23.Hogg, J. R., and K. Collins. 2007. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 13:868-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hresko, R. C., and M. Mueckler. 2002. Identification of pp68 as the tyrosine-phosphorylated form of SYNCRIP/NSAP1. A cytoplasmic RNA-binding protein. J. Biol. Chem. 277:25233-25238. [DOI] [PubMed] [Google Scholar]
- 25.Hutchison, S., C. LeBel, M. Blanchette, and B. Chabot. 2002. Distinct sets of adjacent heterogeneous nuclear ribonucleoprotein (hnRNP) A1/A2 binding sites control 5′ splice site selection in the hnRNP A1 mRNA precursor. J. Biol. Chem. 277:29745-29752. [DOI] [PubMed] [Google Scholar]
- 26.Iijima, M., M. Suzuki, A. Tanabe, A. Nishimura, and M. Yamada. 2006. Two motifs essential for nuclear import of the hnRNP A1 nucleocytoplasmic shuttling sequence M9 core. FEBS Lett. 580:1365-1370. [DOI] [PubMed] [Google Scholar]
- 27.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, Y., M. Liu, C. A. Spencer, and D. H. Price. 2004. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol. Cell 14:375-385. [DOI] [PubMed] [Google Scholar]
- 29.JoNell Hamilton, B., C. M. Burns, R. C. Nichols, and W. F. C. Rigby. 1997. Modulation of AUUUA response element binding by heterogeneous nuclear ribonucleoprotein A1 in human T lymphocytes. J. Biol. Chem. 272:28732-28741. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura, H., Y. Tomozoe, T. Akagi, D. Daisuke, M. Ochiai, and M. Yamada. 2002. Identification of the nucleocytoplasmic shuttling sequence of heterogeneous nuclear ribonucleoprotein D-like protein JKTBP and its interaction with RNA. J. Biol. Chem. 277:2732-2739. [DOI] [PubMed] [Google Scholar]
- 31.Kim, T.-D., J.-S. Kim, J. H. Kim, J. Myung, H.-D. Chae, K.-C. Woo, S. K. Jang, D.-S. Koh, and K.-T. Kim. 2005. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 25:3232-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaBranche, H., S. Dupuis, Y. Ben-David, M. R. Bani, R. J. Wellinger, and B. Chabot. 1998. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet. 19:199-202. [DOI] [PubMed] [Google Scholar]
- 33.Lau, P. P., B. H. Chang, and L. Chan. 2001. Two-hybrid cloning identifies an RNA-binding protein, GRY-RBP, as a component of apobec-1 editosome. Biochem. Biophys. Res. Commun. 282:977-983. [DOI] [PubMed] [Google Scholar]
- 34.Lee, D. K., H. O. Duan, and C. Chang. 2001. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276:9978-9984. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., J. P. Price, S. A. Byers, D. Cheng, J. Peng, and D. H. Price. 2005. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 280:28819-28826. [DOI] [PubMed] [Google Scholar]
- 36.Marchand, V., A. Mereau, S. Jacquenet, D. Thomas, A. Mougin, R. Gattoni, J. Stevenin, and C. Branlant. 2002. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 323:629-652. [DOI] [PubMed] [Google Scholar]
- 37.Maris, C., C. Dominguez, and F. H. Allain. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272:2118-2131. [DOI] [PubMed] [Google Scholar]
- 38.Mayeda, A., S. H. Munroe, J. F. Caceres, and A. R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michels, A. A., A. Fraldi, Q. Li, T. E. Adamson, F. Bonnet, V. T. Nguyen, S. C. Sedore, J. P. Price, D. H. Price, L. Lania, and O. Bensaude. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23:2608-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mili, S., H. J. Shu, Y. Zhao, and S. Pinol-Roma. 2001. Distinct RNP complexes of shuttling hnRNP proteins with pre-mRNA and mRNA: candidate intermediates in formation and export of mRNA. Mol. Cell. Biol. 21:7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mili, S., and J. A. Steitz. 2004. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10:1692-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mourelatos, Z., L. Abel, J. Yong, N. Kataoka, and G. Dreyfuss. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK snRNA binds to and inhibits the activity of Cdk9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 46.Patry, C., B. Lemieux, R. J. Wellinger, and B. Chabot. 2004. Targeting heterogeneous nuclear ribonucleoparticule A1 and A2 proteins by RNA interference promotes cell death in transformed but not in normal mouse cell lines. Mol. Cancer Ther. 3:1193-1199. [PubMed] [Google Scholar]
- 47.Pei, Y., H. Du, J. Singer, C. Stamour, S. Granitto, S. Shuman, and R. P. Fisher. 2006. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol. Cell. Biol. 26:777-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterlin, B. M., and D. H. Price. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 23:297-305. [DOI] [PubMed] [Google Scholar]
- 49.Piñol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 50.Proudfoot, N. 2004. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell Biol. 16:272-278. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, G. C., C. Gooding, and C. W. Smith. 1996. Smooth muscle alternative splicing induced in fibroblasts by heterologous expression of a regulatory gene. EMBO J. 15:6301-6310. [PMC free article] [PubMed] [Google Scholar]
- 52.Rossoll, W., S. Jablonka, C. Andreassi, A. K. Kroning, K. Karle, U. R. Monani, and M. Sendtner. 2003. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 163:801-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rousseau, S., N. Morrice, M. Peggie, D. G. Campbell, M. Gaestel, and P. Cohen. 2002. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 21:6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8:1310-1317. [DOI] [PubMed] [Google Scholar]
- 55.Schulte, A., N. Czudnochowski, M. Barboric, A. Schonichen, D. Blazek, B. M. Peterlin, and M. Geyer. 2005. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 280:24968-24977. [DOI] [PubMed] [Google Scholar]
- 56.Simone, C., P. Stiegler, L. Bagella, B. Pucci, C. Bellan, G. De Falco, A. De Luca, G. Guanti, P. L. Puri, and A. Giordano. 2002. Activation of MyoD-dependent transcription by cdk9/cyclin T2. Oncogene 21:4137-4148. [DOI] [PubMed] [Google Scholar]
- 57.Siomi, M. C., P. S. Eder, N. Kataoka, L. Wan, Q. Liu, and G. Dreyfuss. 1997. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 138:1181-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian, Y., S. Ke, M. Chen, and T. Sheng. 2003. Interactions between the Ah receptor and P-TEFb: sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter. J. Biol. Chem. 278:44041-44048. [DOI] [PubMed] [Google Scholar]
- 59.Van Herreweghe, E., S. Egloff, I. Goiffon, B. E. Jady, C. Froment, B. Monsarrat, and T. Kiss. 2007. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 26:3570-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wassarman, D. A., and J. A. Steitz. 1991. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 11:3432-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weighardt, F., G. Biamonti, and S. Riva. 1995. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J. Cell Sci. 108:545-555. [DOI] [PubMed] [Google Scholar]
- 62.Yang, Z., J. H. Yik, R. Chen, N. He, M. K. Jang, K. Ozato, and Q. Zhou. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19:535-545. [DOI] [PubMed] [Google Scholar]
- 63.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 64.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 65.Yik, J. H., R. Chen, A. C. Pezda, C. S. Samford, and Q. Zhou. 2004. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 24:5094-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yik, J. H., R. Chen, A. C. Pezda, and Q. Zhou. 2005. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive P-TEFb complexes for control of transcription. J. Biol. Chem. 280:16368-16376. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Q. S., L. Manche, R. M. Xu, and A. R. Krainer. 2006. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA 12:1116-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zieve, G., and S. Penman. 1976. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell 8:19-31. [DOI] [PubMed] [Google Scholar]
- 69.Zieve, G. W., D. Turnbull, J. M. Mullins, and J. R. McIntosh. 1980. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole. Nocodazole accumulated mitotic cells. Exp. Cell Res. 126:397-405. [DOI] [PubMed] [Google Scholar]
- 70.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296:91-97. [DOI] [PubMed] [Google Scholar]