Abstract
The PLU-1/JARID1B nuclear protein, which is upregulated in breast cancers, belongs to the ARID family of DNA binding proteins and has strong transcriptional repression activity. To identify the target genes regulated by PLU-1/JARID1B, we overexpressed or silenced the human PLU-1/JARID1B gene in human mammary epithelial cells by using adenovirus and RNA interference systems, respectively, and then applied microarray analysis to identify candidate genes. A total of 100 genes showed inversely correlated differential expression in the two systems. Most of the candidate genes were downregulated by the overexpression of PLU-1/JARID1B, including the MT genes, the tumor suppressor gene BRCA1, and genes involved in the regulation of the M phase of the mitotic cell cycle. Chromatin immunoprecipitation assays confirmed that the metallothionein 1H (MT1H), -1F, and -1X genes are direct transcriptional targets of PLU-1/JARID1B in vivo. Furthermore, the level of trimethyl H3K4 of the MT1H promoter was increased following silencing of PLU-1/JARID1B. Both the PLU-1/JARID1B protein and the ARID domain selectively bound CG-rich DNA. The GCACA/C motif, which is abundant in metallothionein promoters, was identified as a consensus binding sequence of the PLU-1/JARID1B ARID domain. As expected from the microarray data, cells overexpressing PLU-1/JARID1B have an impaired G2/M checkpoint. Our study provides insight into the molecular function of the breast cancer-associated transcriptional repressor PLU-1/JARID1B.
The PLU-1 nuclear protein is expressed in most primary breast cancers and breast cancer cell lines (4, 46), while its expression in normal adult tissues is largely restricted to testes (46), where it is expressed in spermatogonia and in specific stages of meiosis (48). However, significant expression is also seen in the murine pregnant mammary gland (4) and in the embryonic mammary bud (4, 47). Thus, in addition to being elevated in breast cancer, PLU-1 is involved in the development and differentiation of the mammary gland.
The PLU-1 gene encodes a 1,544-amino-acid multidomain protein that is exclusively localized to the nucleus. Sequence homology analysis shows that it contains several conserved domains, including the ARID DNA binding domain (AT-rich-interacting domain), plant homeodomain/leukemia-associated protein domains (PHD domains), Jumonji domains, and putative nuclear localization signals (46). The ARID domain, the N-terminal and C-terminal Jumonji domains (JmJN and JmJC), two of the plant homeodomain domains, and a novel Trp/Tyr/Phe/Cys domain (the PLU domain), which overlaps the JmJC domain (62), are conserved in the four members of the JARID1 subfamily of the larger family of ARID proteins (15 proteins to date) (86). Although their sequence homology is high, these proteins appear to have diverse functions, with JARID1A (RBP2) being involved in activating transcription from nuclear receptors (14), PLU-1 (now referred to as PLU-1/JARID1B) being a strong transcriptional repressor (75, 88), and JARID1C (the SMCX gene) found mutated in some humans with X-linked mental retardation (36, 37, 73). Genes showing high homology with the JARID1 subfamily of ARID proteins are found across species, including the lower eukaryotes, and these genes are also involved in transcriptional regulation and chromatin modifications (62, 85, 86).
The highly conserved JmJC domain belongs to the cupid superfamily of metalloenzymes, and proteins containing this domain have been shown to have demethylase activity, targeting specific methylated lysines on histones (16, 43, 78, 83, 89). Euchromatic histone methylation can contribute either to transcriptional activation or to repression. Overall, di- and trimethylation at H3K4 and H3K36 at the promoter level have been associated with transcriptional activation, while H3K9 and H3K27 methylation is linked to the repression of transcription (49, 57, 60, 63, 79, 91). Recently, it has been shown that PLU-1/JARID1B specifically removes methyl groups from H3K4me3 and that the demethylation activity is required for transcriptional repression (36, 70, 88).
The earliest known member of the ARID family, the dead ringer protein (dri) in Drosophila, was identified through specific binding to AT-rich sequences in DNA to the ARID domain (28). While other family members, such as mouse Bright, MRF-1, and MRF-2, have shown a similar preference for binding AT-rich sequences, most have been reported to be less specific in binding to DNA (32, 35, 84). Here we show that sequences preferentially bound by both the PLU-1/JARIDB ARID domain and the full-length protein contain a GCACA/C motif and are relatively GC rich.
Although we have shown that PLU-1/JARID1B is a strong transcriptional repressor, binding class I and class IIa histone deacetylases (5), the specific genes regulated by this protein in breast cancer have not been identified. To identify these genes, we overexpressed and downregulated the PLU-1/JARID1B gene in human mammary epithelial cells by using a recombinant adenovirus and RNA interference (RNAi) technology, respectively. Microarray analysis showed that a total of 100 genes, including genes involved in the M phase of the mitotic cell cycle, in signal transduction, in primary metabolism, and in development, showed inversely correlated differential expressions in the two systems. As expected, most of the regulated genes (81%) were found to be downregulated by PLU-1/JARID1B overexpression; these included several members of the metallothionein (MT) family as well as the tumor suppressor gene BRCA1 and genes required for a functional G2/M and/or spindle checkpoint. Chromatin immunoprecipitation (ChIP) using a polyclonal antiserum to the carboxy terminus of PLU-1/JARID1B showed that the protein bound to the promoters of the MT genes and that the level of H3K4me3 associated with the promoter of the metallothionein gene MT1H is increased in cells where PLU-1/JARID1B was silenced. Since some of the genes downregulated by PLU-1/JARID1B are involved in cell cycle checkpoints, we also tested the effects of this protein on cell cycle progression after induction of a G2/M block by spindle-destabilizing agents.
MATERIALS AND METHODS
Cell culture.
HB2 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 0.3 μg/ml l-glutamine, 5 μg/ml hydrocortisone, and 10 μg/ml insulin in 5% CO2. MCF7 cells were maintained in 3.7% RPMI supplemented with 10% fetal bovine serum and 0.3 μg/ml glutamine at 37°C in 10% CO2. The MCF7 short-hairpin RNAi (shRNAi) stable transfectants were grown in the same 3.7% RPMI medium with 500 μg/ml G418 (Gibco). HeLa T-REx cells expressing the inducible PLU-1/JARID1B were grown in the 3.7% RPMI medium with 5 μg/ml blasticidin (Invitrogen) and 500 μg/ml Zeocin (Invitrogen).
Plasmid construction and transfection. (i) MCF7 shRNA clone and mix.
An shRNA synthetic oligodeoxynucletide cassette (against PLU-1/JARID1B) was cloned into the IMG800 expression vector (Imgenex Corporation) according to the instructions of the user manual. The sequences of the annealed oligonucleotides cloned into the IMG800 vector were 5′tcgaGGCATTGAAGCTTGCACCTgagtactgAGGTGCAAGCTTCAATGCCttttt-3′ and 5′ctagaaaaaGGCATTGAAGCTTGCACCTcagtactcAGGTGCAAGCTTCAATGCC-3′.
The PLU-1/JARID1B target sequences (positions 5035 to 5053 of the PLU-1/JARID1B cDNA sequence, NCBI RefSeq entry NM_006618) are in capitals; lowercase letters indicate flanking and linking sequences. The correct cloning and orientation of the insert were confirmed by sequencing. Five to 10 μg of PLU-1/JARID1B-shRNA expression vector was transfected into semiconfluent MCF7 cells plated into a 150-mm petri dish by using Lipofectamine (Invitrogen). After 3 days of incubation, cells were split and replated in the presence of 500 μg/ml G418 (Gibco). G418-resistant colonies were picked and expanded in culture to give the shRNA clone and shRNA mix. Control cell lines were developed by transfection of the empty vector.
(ii) Inducible HeLa clone.
A BamHI-XhoI fragment containing full-length PLU-1/JARID1B cDNA was excised from PLU-1pBS-SK(−) (46) and ligated into the pcDNA4/TO vector from the Invitrogen T-REx system. The resultant construct was linearized with SapI prior to transfection into HeLa T-REx cells (Invitrogen) by using SAINT reagent according to the manufacturer's protocol. Clones were isolated and grown in selective medium containing 5 μg/ml blasticidin and 500 μg/ml Zeocin. The control cell line was made in the same manner by using the empty pcDNA4/TO vector.
Preparation of the recombinant adenovirus and adenoviral transduction of HB2 cells.
The constructs pShuttleCMV, pAdeasy-1, and BJ5183 were gifts from Georges Vassaux (Cancer Research UK), and 293A cells were purchased from Quantum Biotechnologies. The Ad5-GFP vector (where Ad5 is adenovirus type 5 and GFP is green fluorescent protein) was made as described previously (69). To obtain the PLU-1/JARID1B coding sequence joined in frame with the GFP coding sequence and cloned into the pShuttleCMV vector, the following approach was used. The full-length untagged PLU-1/JARID1B cDNA sequence was excised from the pBS-SK(−)/PLU-1 construct (46) by using the BamHI and AccI restriction enzymes. A linker containing a BamHI site was then ligated to the AccI end at the 3′ end of this fragment. The resulting fragment was cloned into the BamHI site of the pShuttleCMV vector (Quantum Biotechnologies).
To tag the PLU-1/JARID1B coding sequence to the GFP coding sequence, a fragment containing the PLU-1/JARID1B cDNA was excised from the expression vector PLU-1-ORF/MYC-His (46) by using the NheI and XhoI restriction enzymes and cloned into the same restriction sites of the pEGFP-N1 vector (Clontech). This construct (PLU-1/JARID1B-pEGFP-N1) has a 4.781-kb fragment containing the full-length 5′ untranslated region (UTR) and the full-length coding sequence of the PLU-1/JARID1B gene, but not the 3′ UTR, ligated in frame to the GFP coding sequence.
Finally, to obtain a GFP-tagged PLU-1/JARID1B cDNA recombinant pShuttleCMV vector (PLU-1/JARID1B-GFP-pShuttleCMV), a NotI fragment from the untagged-PLU-1/JARID1B-pShuttleCMV construct was replaced with a NotI insert from the PLU-1/JARID1B-pEGFP-N1 construct. This fragment includes the sequence from the NotI site in position 309 of the PLU-1/JARID1B cDNA (NCBI RefSeq entry NM_006618) to the NotI site just downstream of the stop codon of the GFP gene. The final construct, PLU-1/JARID1B-GFP-pShuttleCMV, was linearized with PmeI and cotransfected with pAdeasy-1 into electrocompetent BJ5183 (Quantum Biotechnologies) for in vivo recombination. Recombinant, kanamycin-resistant clones were verified by restriction fragment analysis and PCR, amplified in DH5a, and then purified using a QIAGEN Maxi column. Purified plasmid was linearized with PacI and introduced by calcium phosphate transfection into 293A cells for virus propagation. The preparation and titration of Ad5-GFP and Ad5-PLU-1-GFP to determine the 50% tissue culture infective dose were performed as instructed in the manual for the AdEasy vector system (Quantum Biotechnologies). A total of 4 × 106 HB2 cells were seeded in a 175-cm2 flask and either infected or mock infected with Ad5-GFP or Ad5-PLU-1-GFP at a multiplicity of infection (MOI) of 0.5 ×103 to 1 × 104 as described previously (69).
High-density oligonucleotide microarrays.
Affymetrix (Santa Clara, CA) DNA microarray analysis was carried out on the HG-U133A chip according to the manufacturer's instructions and as described previously (69). Affymetrix Microarray Suite 5.0 was used for the quantification of the target gene expression levels. Global scaling was applied to the data to adjust the average recorded to a target intensity of 100. Data were exported from Affymetrix Microarray Suite 5.0 into GeneSpring 6 (Silicon Genetics, Redwood City, CA) for further analysis.
Data normalization (per-chip normalizations to the 50% percentile and per-gene normalization to specific samples) was performed using the protocol recommended by Silicon Genetics for Affymetrix data. When necessary, the P value was calculated by using a one-sample Student t test.
Western blot analysis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis were performed as described previously (4). The α-PLU-1-C antibody to the C-terminal end of PLU-1/JARID1B has been described previously (4). The antibody directed against HSC70 was purchased from Santa Cruz Biotechnology (catalog no. sc7298). The horseradish peroxidase-conjugated secondary antibodies were purchased from DakoCytomation.
RNA preparation and quantitative real-time reverse transcription-PCR.
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Two micrograms of total RNA was reverse transcribed using the reaction ready, first-strand cDNA synthesis kit (SuperArray Bioscience). Quantitative real-time reverse transcription-PCR (q-RT-PCR) was carried out using SYBR green JumpStart ReadyMix (Sigma) according to the manufacturer's instructions using a DNA Engine Opticon 2 (MJ Research, Inc.). The PLU-1/JARID1B (QPH12457A) and the BRCA1 (QPH00322A) primers were purchased from SuperArray Bioscience, while the beta-actin primers were described previously (69). Primer sequences and the expected product sizes of the MT1H, MT1F, BUB1B, BUB3, and STK6 genes were designed using the Primer3 Web interface (66) and are detailed in Table 1.
TABLE 1.
Primers for q-RT-PCR and ChIP analysis
| Gene or promoter | Forward primer | Reverse primer | Size (bp) |
|---|---|---|---|
| Genes | |||
| ACTB | CACCACACCTTCTACAATGAGCTGC | ACAGCCTGGATAGCAACGTACATGG | 158 |
| MT1H | GCTCCTGCAAGTGCAAAAAG | CAGCAGCTGCACTTCTCTGA | 130 |
| MT1F | TCCTGCAAGTGCAAAGAGTG | AAAGGTTGTCCTGGCATCAG | 151 |
| BUB1B | GGGATTCAACAGAAGGCTGA | TGCATCTGTTGAGGAAATGG | 268 |
| BUB3 | TACCAGACTCGCTGCATACG | AATTGGCACAGTCGCTTTTT | 278 |
| STK6 | GGCAAATGCCCTGTCTTACT | AGGCTCCAGAGATCCACCTT | 221 |
| Promoters for ChIP | |||
| MTIH | GCGGGAGTAGCAGGTAATCT | CAGTTGGCAGCTCCTTTTCT | 279 |
| MT1F | GCTCACCCAGGCACAAAG | GCTGTGTGCAGCAGAAGG | 114 |
| MT1X | ACTGCTCCCTCGCTATGCT | CCGGCGGCTCTCTTTATAGT | 387 |
| BRCA1 | TAAGCCGCAACTGGAAGAGT | CAGAAAGAGCCAAGCGTCTC | 152 |
| IGFBP2 | ACCCGCGAGTTATCCGTATT | TTTTGGAGACCACTCCCTTC | 187 |
Analysis of q-RT-PCR data was carried out using the comparative ΔΔCT method (87). For each sample, the intensity of the amplimer was normalized against that of the internal control human β-actin and then the data were each depicted as severalfold changes (2-ΔΔCT) with respect to the transcription level observed in the control experiment.
Flow cytometry analysis.
Flow cytometry analysis for detecting GFP expression was performed as described previously (69). For cell cycle analysis, asynchronous HB2 cells were noninfected or infected with Ad5-GFP or Ad5-PLU-1-GFP at an MOI of 500. Twenty-four hours after infection, the medium was replaced with fresh complete medium containing 0.4 μg/ml nocodazole or 20 μM CdCl2 and incubated at 37°C, 5% CO2, for a further 24 h before harvesting using trypsin. The asynchronously dividing MCF7 clone and mix cell lines were harvested with trypsin before proceeding with the staining. Cells were pulsed with 10 μM bromodeoxyuridine (BrdU) for 30 min before harvesting and then fixed with 1% paraformaldehyde on ice for at least 10 min. After washing with phosphate-buffered saline, cells were permeabilized using 0.5% Triton X-100 for 15 min at 25°C and then treated with 25 to 50 U of DNase (Promega; catalog no. M610A) for 45 min at 37°C. After three washes with 0.2% Tween 20-phosphate-buffered saline solution, cells were incubated with mouse anti-BrdU antibody (Becton Dickinson; catalog no. 347580), followed by staining with Alexa Fluor 647-labeled goat anti-mouse antibody (Invitrogen/Molecular Probes; catalog no. A21235). Finally, cells were resuspended in an appropriate volume of Hoechst 33342 solution (0.1% Triton X-100, 20 μg/ml Hoechst 33342). Stained cells were then analyzed by using an LSRII (Becton Dickinson, San Jose, CA).
ChIP assay.
Subconfluent MCF7 cells were cross-linked by adding formaldehyde directly to the culture medium at a final concentration of 1% for 10 min before quenching with 0.125 M glycine for 5 min, and after seven pulses of sonication at 70% amplitude with a Branson digital sonifier 250, the chromatin was subjected to ChIP assay using the ChIP-IT kit (Active Motif) according to the instruction manual. Two micrograms of antisera to H3K4me3 (Abcam; catalog no. 8585), H3K4me2 (Abcam; catalog no. 7766), H3K4me1 (Abcam; catalog no. 8898), or control immunoglobulin G (Santa Cruz Biotechnology; catalog no. sc2027) or 5 μl of α-PLU-1-C antiserum or prebleed antiserum was used for the ChIP experiment.
The PCR was performed using primer pairs designed using the Primer3 Web interface (66), and primers are listed in Table 1. The PCR was carried out in a final volume of 50 μl containing 5 μl of purified ChIP DNA, 2.5 μl of specific forward primer (20 ng/μl), 2.5 μl of specific reverse primer (20 ng/μl), 5 μl deoxynucleoside triphosphates (2 mM), 1× PCR buffer II (Applied Biosystems), 5 μl MgCl2 (25 mM), 2.5 U of AmpliTaq Gold (Applied Biosystems), and water to a 50-μl final volume. The reaction mixture was incubated at 95°C for 5 min and then amplified with a variable number of cycles (between 27 and 40 cycles, depending on the promoter analyzed) at 95°C for 30 s, 50 to 57°C for 30 s, and 72°C for 45 s. Twenty microliters of each PCR was subjected to electrophoresis on a 1.8% Tris-borate-EDTA agarose gel, and DNA bands were detected by ethidium bromide staining and by using a Typhoon PhosphorImager (Molecular Dynamics).
Protein expression.
The glutathione S-transferase (GST)/PLU-1/JARID1B-ARID domain fusion protein (GST-PLU-1/ARID) was obtained by cloning a 489-bp PCR product (covering the region from positions 303 to 792 of the human PLU-1/JARID1B cDNA [NCBI RefSeq entry NM_006618]) into the cloning BamHI and SmaI restriction sites of the pGEX2TK expression vector (Pharmacia). The GST-Fly/ARID expression vector was a generous gift from Robert Saint (28). These protein expression vectors were transformed into Escherichia coli BL21 cells, and protein expression was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Cells were sonicated, and the GST fusion proteins in the cell extract were bound to glutathione-Sepharose beads (Sigma) at room temperature for 30 min before washing. The GST fusion protein was eluted from the beads by using 20 mM glutathione, pH 8.25, quantitated, and stored at 4°C.
Full-length, His-tagged, human PLU-1/JARID1B protein was produced using the baculovirus system as described previously (5).
DNA binding activity to native DNA.
The GST-PLU-1/ARID fusion protein was passed over a native DNA cellulose column (Amersham), which was then washed with loading buffer containing 50 mM NaCl, and collected in a series of increasing NaCl salt concentration fractions. Equal aliquots of the flowthrough, wash, and eluted fractions were analyzed by SDS-PAGE.
PCR-assisted DNA binding selection from random oligonucleotides.
A mixture of 58-base oligonucleotides (oligoN2), in which the middle 20 bases consisted of random nucleotides between the primers F-AGACACGACACCTTTCTAAC and R-ACCCCTTATTTGCCTGCG, was converted to double-stranded oligonucleotides by Taq polymerase as follows. In the presence of [α-32P]dCTP (3,000 Ci/mmol) and 0.05 mM dATP, dGTP, and dTTP, 100 ng of the single-stranded oligoN2 was denatured at 95°C for 2 min, annealed to 100 ng of the primer R-ACCCCTTATTTGCCTGCG at 52°C for 3 min, and then made double stranded at 72°C for 9 min. The resulting radiolabeled, double-stranded oligonucleotide mixture was electrophoresed through a 10% acrylamide gel, and the radioactive 58-bp band was excised. The radioactivity was used to ensure that only double-stranded oligonucleotides were used in the first selection pool. The gel fragment was crushed and soaked in elution buffer (10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM EDTA) at 37°C overnight, before being precipitated and resuspended in Tris-EDTA buffer.
The oligonucleotides (approximately 10 ng) were allowed to bind to GST-PLU-1/ARID (approximately 100 μg) or full-length PLU-1/JARID1B (approximately 50 μg) on beads for 1 to 2 h at 4°C in the presence of 4 mg bovine serum albumin, 20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 10% glycerol, and 1 mM dithiothreitol as well as 2 μg poly (dA-dT) and 2 μg poly (dG-dC). Beads were washed three times under the conditions described above and treated with 20 μg of proteinase K at 50°C overnight. The DNA was extracted with phenol and precipitated before being amplified by PCR using both primers; the resulting band was excised from an acrylamide gel and eluted and precipitated as before. This binding selection process was repeated eight times.
oligoN2 was cloned into pCR2.1 using the TOPO TA cloning kit (Invitrogen) after zero, three, and eight rounds of binding and selection so that the oligonucleotides could be sequenced using BigDye (Applied Biosystems, Cheshire, United Kingdom).
Electrophoretic mobility shift assay (EMSA).
Oligonucleotides were end labeled with 32P and were used in binding reaction mixtures that contained between 0.5 and 2 μg GST-PLU-1/ARID fusion protein, 50 ng poly(dA-dT) (Sigma), 20 mM Tris, pH 8.0, 12.5% glycerol, 5 mM β-mercaptoethanol (Sigma), 50 μg/ml bovine serum albumin, 100 mM KCl, and 1 mM EDTA. Unlabeled competitor oligonucleotides, where required, were added at 5, 10, 20, 25, and 45 times molar excess. Binding reactions were allowed to proceed at room temperature for 30 min before being loaded onto a 6% acrylamide gel made with 0.5× Tris-borate-EDTA. Reactions were electrophoresed for 1 to 2 h at 4°C. Gels were dried onto Whatman paper before being scanned by using a Typhoon PhosphorImager (Molecular Dynamics).
RESULTS
Overexpression and silencing of PLU-1/JARID1B in mammary epithelial cells.
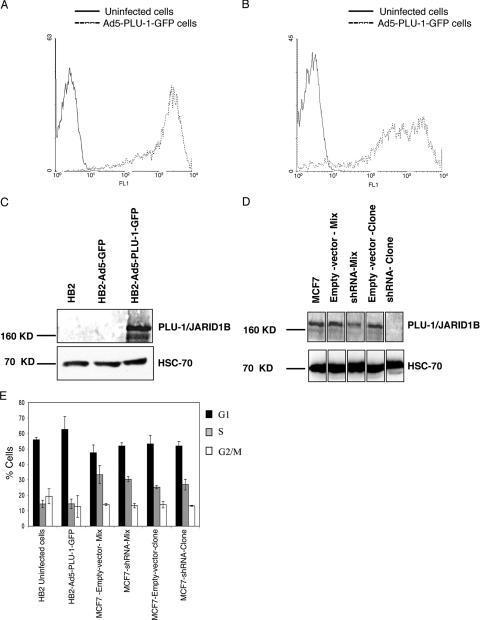
The immortalized nonmalignant human epithelial cell line HB2 (11) is a clonal derivative of the MTSV1-7 cell line (7), which was originally derived from human milk, and shows many features of normal luminal mammary epithelial cells. HB2 cells, which do not express endogenous PLU-1/JARID1B protein, were infected with a high MOI of a recombinant E1−/E3− adenovirus type 5 vector expressing a GFP-tagged PLU-1 protein (Ad5-PLU-1-GFP) or expressing GFP alone (Ad5-GFP). By using flow analysis to detect GFP expression, the effective transduction of more than 90% of the cells was demonstrated at 24 and 72 h after infection (Fig. 1 A and B), but a cytopathic effect was not seen, even 72 h after infection. The correct molecular weight of the overexpressed recombinant protein was verified by Western blot analysis using the α-PLU-1-C antibody specifically reactive with the PLU-1/JARID1B protein (Fig. 1C) (4).
FIG. 1.
Regulation of PLU-1/JARID1B expression in human mammary epithelial cells using a recombinant adenovirus and shRNA. (A and B) Uninfected or Ad5-PLU-1-GFP-infected HB2 cells were incubated for 24 (A) or 72 (B) hours and analyzed by flow cytometry. The y axis depicts cell number, and the x axis depicts log relative fluorescence intensity (FL1) for GFP expression. The graph of HB2 cells infected with the Ad5-GFP control vector is not shown because the reading was extremely bright and partially off the scale. (C) Whole-cell extracts prepared from HB2 cells infected for 24 h with Ad5-GFP or Ad5-PLU-1-GFP or uninfected were separated by SDS-PAGE and immunoblotted using α-PLU-1-C antiserum (upper panel) or anti-HSC70 antibody (lower panel) to check for protein load. (D) Western blot analysis of whole-cell protein lysate of shRNA-transfected MCF7 cell lines or the corresponding empty vector transfectants. The upper panel shows the immunoblot analysis using α-PLU-1-C antiserum, while the lower panel shows the immunoblot analysis using anti-HSC70 antibody. (E). Changing the level of expression of the PLU-1/JARID1B gene does not alter the cell cycle profile of HB2 and MCF7 cells. HB2 cells that were noninfected or infected for 48 h with Ad5-PLU-1-GFP and MCF7 cells stably transfected with an empty vector (MCF7-empty-vector-clone and MCF7-empty-vector-mix) or with a PLU-1-specific shRNA expression vector (MCF7-shRNA-clone and MCF7-shRNA-mix) were subjected to cell cycle analysis (BrdU incorporation and Hoechst staining) as described in Materials and Methods. Bars represent the means ± standard errors of the means of three independent experiments.
The MCF7 breast cancer cell line, which also shows features of luminal epithelial cells (8, 9), expresses high levels of PLU-1/JARID1B. To confirm and complement the data obtained when PLU-1/JARID1B is overexpressed, the expression of the endogenous PLU-1/JARID1B gene was stably silenced in MCF7 cells by using shRNA.
The sense and antisense sequences of one set of the RNAi oligonucleotides tested by transient transfection were linked together by an 8-nucleotide loop and cloned into an shRNA expression vector downstream of the of U6 promoter (see Materials and Methods). An MCF7-shRNA clone that showed consistent PLU-1/JARID1B silencing in long-term culture was isolated, as was the mixed transfected cell line (MCF7-shRNA-mix). PLU-1/JARID1B expression was almost completely silenced in the clone and was reduced by 50% in the mixed culture (Fig. 1D). One clone and a mixed population of the empty vector transfectants (MCF7-empty-vector-clone and MCF7-empty-vector-mix) were also isolated.
Identification of genes that are differentially expressed after manipulation of PLU-1/JARID1B expression.
Flow cytometric analysis showed no differences in the cell cycle profiles before and after the overexpressing or silencing of the PLU-1/JARID1B genes in HB2 and in MCF7 cells, respectively, thus excluding the possibility that any expression changes of target genes would be due to an effect on the cell cycle progression (Fig. 1E). We therefore proceeded to use microarray analysis to identify the specific genes regulated by PLU-1/JARID1B. Total RNA was extracted from uninfected control HB2 cells, Ad5-GFP-infected cells, or Ad5-PLU-1-GFP-infected cells 24 or 72 h after infection and subjected to microarray analysis using the Affymetrix HG-U133A gene chips. Each chip contains a total of 22,283 probe sets or probe features, representing approximately 18,400 human transcripts and expressed sequence tags. The expression value for each gene was obtained as the signal ratio between either the Ad5-GFP mock-infected or the Ad5-PLU-1-GFP-infected cell channel and the control channel of the uninfected cells. Four and three biologically independent replicates were used at 24 and 72 h, respectively. Genes were scored as upregulated or downregulated if they showed a difference of at least 1.5-fold (P < 0.05) in signal between the uninfected control cells and the Ad5-GFP- or Ad5-PLU-1-GFP-infected cells. To identify genes that were affected only by the PLU-1/JARID1B transgene and not by the adenovirus vector per se (19, 27, 69, 92), the genes that showed differential expression in the Ad5-GFP-infected cells (compared to uninfected cells) were excluded from further analysis (data not shown). A total of 780 and 294 probe sets showed a signal ratio of at least a 1.5-fold difference (P < 0.05) at 24 h and 72 h, respectively, in the Ad5-PLU-1-GFP-infected cells but not in the Ad5-GFP-infected cells (data not shown).
To identify consistent candidate genes regulated by PLU-1/JARID1B in mammary epithelial cells, the microarray results obtained with the recombinant adenovirus system were compared with the microarray results obtained from the PLU-1/JARID1B-silenced MCF7 cells (MCF7-shRNA-clone and MCF7-shRNA-mix cell lines).
Gene expression values from the shRNA-transfected MCF7 cell channels were normalized against the empty vector transfectants. Two biological microarray replicates were carried out for each of the shRNA MCF7 cell lines. The probe sets that were either upregulated or downregulated following infection with Ad5-PLU-1-GFP were filtered for their differential expression in the silenced MCF7 cell lines, with the expectation that specific genes that were upregulated by the overexpression of the PLU-1/JARID1B protein would be downregulated following the shRNA silencing of the PLU-1/JARID1B gene and vice versa.
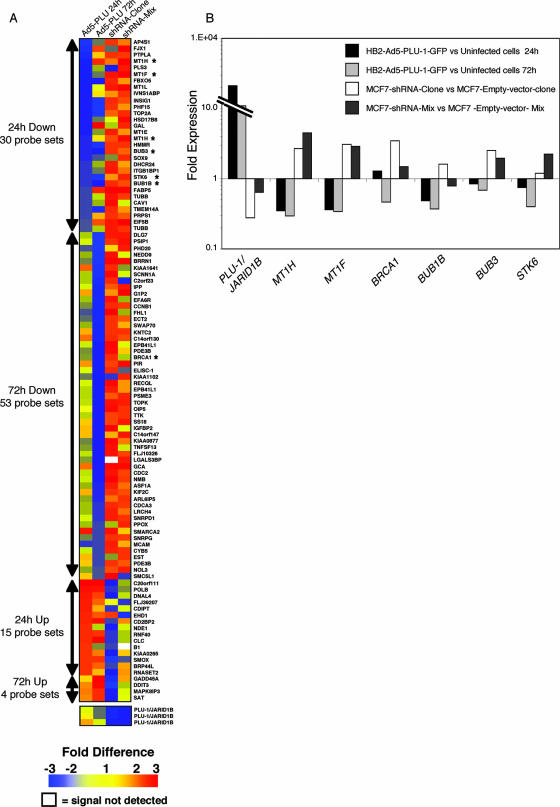
Figure 2 shows a TreeView diagram with color coding demonstrating the inverse correlation between the expression changes in the adenovirus and shRNA systems. Twenty-eight and 53 genes that were significantly downregulated (P < 0.05) by PLU-1/JARIDB1 overexpression in HB2 cells at 24 or 72 h, respectively, showed at least a 1.5-fold increase in expression in the MCF7-shRNA-clone or in the MCF7-shRNA-mix. Of the 28 genes showing downregulation at 24 h, 11 were also downregulated significantly at 72 h. However, only 15 and 4 genes, which were upregulated at 24 and 72 h, respectively, following the infection of HB2 cells with Ad5-PLU-1-GFP, were downregulated 1.5-fold or more in at least one of the two silenced MCF7 cell lines (Fig. 2; see Tables S1 and S2 in the supplemental material). The fact that most of the genes affected by PLU-1/JARID1B overexpression are downregulated confirms that PLU-1/JARID1B acts mainly as a transcriptional repressor.
FIG. 2.
Increased or decreased expression of PLU-1/JARID1B in mammary epithelial cells results in changes in the profile of gene expression. (A) TreeView diagram depicting the genes that were significantly deregulated following the manipulation of PLU-1/JARID1B expression. Red represents upregulated genes, blue represents downregulated genes, and yellow represents unchanged expressions. The numbers on the color scheme represent the changes in gene expression. White coloring represents a signal not detected. The columns designated Ad5-PLU 24 h and Ad5-PLU 72 h represent changes in gene expression in Ad5-PLU-1-GFP-infected cells normalized to uninfected cells at 24 and 72 h, respectively. The columns designated shRNA-clone and shRNA-mix represent changes in gene expression in the MCF7-shRNA-clone normalized to the MCF7-empty-vector-clone and in the MCF7-shRNA-mix cells normalized to MCF7-empty-vector-mix cells, respectively. The color values represent the averages between biological replicates. Four biological replicates were used for Ad5-PLU 24 h, three replicates were used for Ad5-PLU 72 h, and two replicates each were used for the shRNA-clone and shRNA-mix. The numbers of the probe sets significantly (P < 0.05, Student's t test) up- or downregulated at 24 and 72 h after infection with Ad5-PLU-1-GFP are shown on the left of the tree. Asterisks denote genes that were further analyzed for their expression with q-RT-PCR. Note that the PLU-1/JARID1B transgene in the adenovirus does not contain the 3′ UTR that is detected by the PLU-1/JARID1B-specific microarray probe set. Therefore, changes in the expression of PLU-1/JARID1B mRNA could be detected only in the RNAi cell lines where endogenous mRNA is silenced. (B) Confirmation by q-RT-PCR of expression changes shown by microarray data. RNA was isolated from HB2 cells 24 and 72 h after infection with the recombinant adenovirus and from the shRNA-MCF7 cells before reverse transcription and q-RT-PCR analysis. Data for each gene are normalized against primers for β-actin and are represented as the change (2-ΔΔCT) compared to the negative control (uninfected HB2 cells or the empty vector transfected MCF7 cells). One representative experiment out of at least two experiments is shown.
By using the gene ontology mining tool from Affymetrix, we identified genes by microarray analysis and grouped them into 21 different categories according to their biological functions (see Tables S1 and S2 in the supplemental material). Of the genes showing decreased expression levels in HB2 cells infected with Ad5-PLU-1-GFP and increased levels in the silenced MCF7 cells, several relate to the M phase of the mitotic cell cycle, to the spindle checkpoint (including BUB1B, BUB3, STK6, TTK, cyclin B1, and CDC2), to signal transduction pathways, and to primary metabolism. The downregulation of expression of the tumor suppressor BRCA1 by PLU-1/JARID1B is of great interest and could relate to PLU-1/JARID1B function in breast cancer. Moreover, the silencing of BRCA1 is known to result the in the downregulation of expression of genes related to the spindle checkpoint (2).
Four members of the MT family (1H, 1X, 1F, and 1E) were significantly downregulated by PLU-1/JARID1B overexpression at 24 h and upregulated following silencing with the specific shRNA (see Table S1 in the supplemental material), suggesting that the PLU-1/JARID1B protein regulates the expression of these small metal-binding proteins.
To validate the microarray data, the expression of some of the genes which were downregulated by overexpression of PLU-1/JARID1B and upregulated upon PLU-1/JARID1B silencing was confirmed by q-RT-PCR. Figure 2B shows the results for six of these genes: MTIH, MT1F, BRCA1, BUB1B, BUB3, and STK6. The PLU-1/JARID1B mRNA level was also measured by q-RT-PCR, and as expected from the Western blot analysis (Fig. 1D), the PLU-1/JARID1B silencing was more pronounced in the MCF7-shRNA-clone.
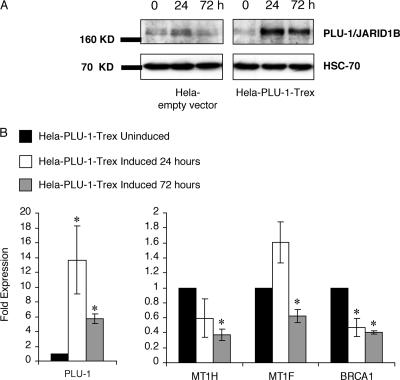
To investigate effects of changes in PLU-1/JARID1B expression on selected genes in another cell line, the repression of expression of BRCA1 and metallothionein genes was documented by q-RT-PCR in HeLa cells that had been stably transfected with a tetracycline-inducible PLU-1/JARID1B expression vector. After the addition of tetracycline, the expression of the PLU-1/JARID1B protein increases substantially at 24 and 72 h compared with that in uninduced cells (Fig. 3A). With increased expression of PLU-1/JARID1B, the expression of BRCA1, MT1H, and MT1F is reduced, with the effect being most significant 72 h after induction. Changes in the expression of these genes were not seen in cell lines derived by transfection of the empty vector (data not shown).
FIG. 3.
MT1H, MT1F, and BRCA1 expression is downregulated in HeLa cells following induction of a PLU-1/JARID1B transgene. (A) Western blot showing PLU-1/JARID1B expression from an inducible promoter. Lysates from HeLa cells transfected with the PLU-1/JARID1B-inducible vector (HeLa-PLU-1-Trex) or from cells transfected with the empty vector (HeLa empty vector) were prepared after inducing the expression of the transgene for 0, 24, and 72 h. The extracts were separated on a denaturing gel, and Western blot analysis was performed with α-PLU-1-C antiserum or anti-HSC70 antibody for loading control. (B) q-RT-PCR analysis of expression of PLU-1/JARID1B, MT1H, MT1F, and BRCA1 in the PLU-1/JARID1B transfected HeLa cells induced for 24 or 72 h. Data for each gene at 24- or 72-h time points are normalized against the internal expression of β-actin and are represented as the change (2-ΔΔCT) compared to the noninduced cells. Bars represent the means ± standard errors of the means of three independent experiments. The asterisks denote a significant difference (P < 0.05) between induced and uninduced cells by using Student's t test.
PLU-1/JARID1B binds directly to the promoter of metallothionein genes.
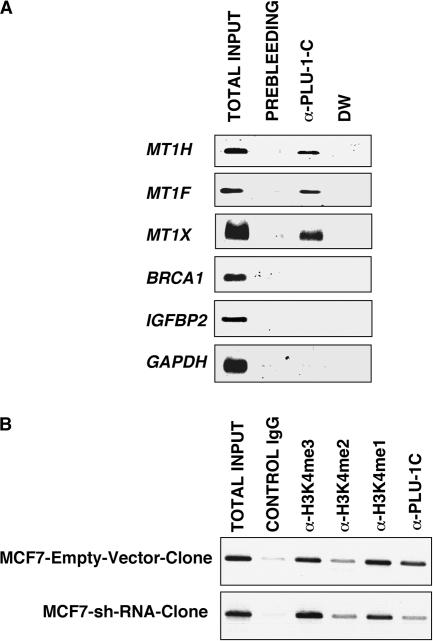
To determine whether the PLU-1/JARID1B protein can bind directly to the promoter of the target genes identified by microarray, we performed ChIP (see Materials and Methods) by using the α-PLU-1-C rabbit antiserum or preimmune serum. After purification of the DNA in the immunoprecipitate, the abundance of genomic DNA containing a promoter was determined by PCR amplification using sequence-specific primer pairs.
Since PLU-1/JARID1B acts mainly as a negative regulator of transcription, the ChIP assays focused on downregulated genes, with the binding of PLU-1/JARID1B to the promoters of the MT1H, MT1X, MT1F, and BRCA1 genes as well as the randomly selected IGFBP2 gene being examined. Representative ethidium bromide-stained agarose gels of the PCR products of the immunoprecipitated chromatin are shown in Fig. 4. Immunoprecipitation with PLU-1/JARID1B antiserum resulted in the recovery of considerably more PCR products in the MT1H, MT1F, and MT1X promoters than did the preimmune rabbit serum (prebleed) immunoprecipitation. However, there were no detectable amplifications within the BRCA1 and IGFBP2 promoters with the primers used. As a negative control, we used primers specific to the promoter region of the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene, which is unaffected by PLU-1/JARID1B. As shown in Fig. 4A, there was no enrichment in the GAPDH promoter PCR product from the chromatin immunoprecipitated with α-PLU-1-C compared with the chromatin immunoprecipitated with the preimmune serum. These data indicate that PLU-1/JARID1B can interact with the promoters of the metallothionein genes.
FIG. 4.
PLU-1/JARID1B binds to the promoter of MT genes and changes the methylation status of H3K4 in the promoter of MT1H. (A) ChIP analysis was performed with chromatin from MCF7 cells, which express high levels of PLU-1/JARID1B. Cross-linked protein-DNA complexes were immunoprecipitated using the α-PLU-1-C antiserum or the corresponding prebleed serum as a control. The immunoprecipitated chromatin was analyzed with PCR using primers specific to the promoters of the indicated genes. Input chromatin represents a portion of the sonicated chromatin before immunoprecipitation diluted 1:20. DW, a no-template control. One representative experiment out of at least three is shown. (B) ChIP assay was performed on chromatin extracted from the MCF7-empty-vector-clone and the MCF7-shRNA-clone by using the indicated antibodies.
Since PLU-1/JARID1B has been found to demethylate lysine 4 on histone 3 (36, 70, 88), we examined the level of methylation of H3K4 on the MT1H promoter after silencing PLU-1/JARID1B by RNAi in MCF7 cells. The knockdown of PLU-1/JARID1B strongly reduced PLU-1/JARID1B binding, indicating that the ChIP signal is specific, and also resulted in an increase in the trimethylated form of histone H3K4 (Fig. 4B). Changes in the levels of H3K4me2 and H3K4me1 were less obvious, but a decrease in all forms of methylated lysine 4 on histone 3 could be seen by immunohistochemistry when PLU-1/JARID1B was overexpressed in HB2 cells by the transduction of the PLU-1/JARID1B recombinant adenovirus (data not shown).
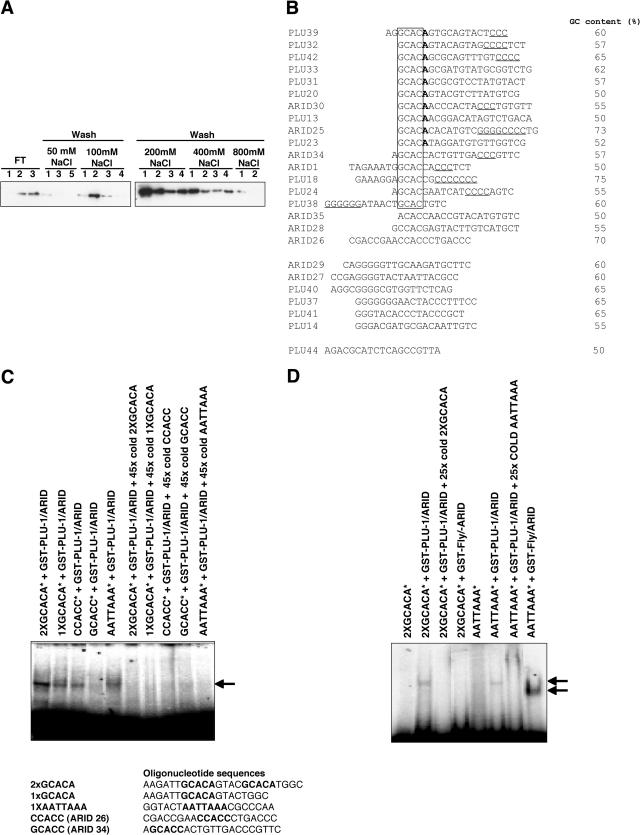
Identification of DNA sequences bound by the PLU-1/JARID1B ARID domain.
The above data suggest that PLU-1/JARID1B could directly bind, via the ARID domain, specific sequences in the metallothionein promoters. To study the capacity of the PLU-1/JARID1B ARID DNA binding domain to bind DNA, a GST fusion protein containing this domain (GST-PLU-1/ARID, amino acid residues 62 to 225) was applied to a native DNA Sepharose column and eluted at increased salt concentrations. The majority of the GST-PLU-1/ARID protein was retained on the DNA at a concentration of 100 mM NaCl, confirming its DNA binding properties (59). A substantial proportion of the protein remained on the column over a wide range of salt concentrations (Fig. 5A), suggesting that the protein can bind different sequences with different affinities.
FIG. 5.
PLU-1/JARID1B binds native DNA and shows a preference for specific sequences. (A) The ARID domain of PLU-1/JARID1B binds to native DNA over a wide range of salt concentrations. A purified GST-ARID fusion protein was applied to a native DNA-cellulose column that was washed with loading buffer containing 50 mM NaCl and then with the increasing salt concentrations shown. Equal aliquots of the flowthrough (FT), wash, and eluted fractions were analyzed by SDS-PAGE before Western blotting using anti-GST as the primary antibody. (B) Sequences of oligonucleotides selected by full-length PLU-1/JARID1B and its specific ARID domain from an initial pool of random nucleotides after eight rounds of PCR/selection. The sequences were analyzed by the MAC vector software. The common motifs are indicated. Sequences were denoted as PLU or ARID, depending on whether they were selected with full-length PLU-1/JARID1B protein or GST-PLU-1/ARID fusion protein, respectively. Underlining indicates three or more flanking C's or G's. (C and D) EMSAs showing binding of selected oligonucleotides by ARID domains. The radiolabeled (*) oligonucleotides containing the motifs indicated are bound with different affinities by the GST-PLU-1/JARID1B ARID fusion protein (GST-PLU-1/ARID) but can be competed off by 45 times excess of specific unlabeled (cold) oligonucleotide. (C) The EMSA showing binding of selected radiolabeled (*) oligonucleotides by GST-PLU-1/ARID and the ARID domain from the Drosophila dead ringer protein (GST-Fly/ARID). (D) The sequences of the oligonucleotides used in the EMSA experiments are shown. The putative binding sequences are shown in bold. Arrows indicate the position of the DNA/GST-PLU-1/ARID complex.
To look for sequence-specific DNA binding activity, full-length PLU-1/JARID1B and the GST-PLU-1-ARID protein were subjected to PCR-assisted DNA binding selection from a random pool of oligonucleotides. Twenty-five independent clones of oligonucleotides, selected by full-length PLU-1/JARID1B and GST-PLU-1/ARID after eight rounds of binding, washing, and amplification, yielded the 25 unique sequences shown in Fig. 5B. The most striking finding is the prevalence of the GCAC motif: 69% of the full-length PLU-1/JARID1B-bound sequences contained them. Moreover, nearly half the GCAC sequences bound by full-length PLU-1/JARID1B and all of the GCAC sequences bound by the GST-PLU-1/ARID contain a run of three or more C's. The GCAC motif was also frequently found to be immediately followed by an A and, in many cases, by both the A and a run of C's. In a few of the selected oligonucleotides, the GCAC motif was directly followed by a C and was associated with a series of C's. All the sequences had a GC content of 50% or more. These data suggest that PLU-1/JARID1B shows a preference for GCAC motifs, followed by an A (and in some cases a C) and a run of C's within 15 bp on either side of this motif.
To confirm that the PLU-1/JARID1B ARID (GST-PLU-1-ARID) binds to the identified DNA motifs, EMSAs were performed using radiolabeled oligonucleotides containing the motifs that had been selected during the PCR-assisted DNA binding selection. The PLU-1/JARID1B ARID domain bound the sequences containing the GCACA motif (1× GCACA) better than the GCACC and CCACC motifs did (Fig. 5C), and it could be competed off by a 25- to 45-fold excess of unlabeled specific oligonucleotide (Fig. 5C), in agreement with the DNA selection binding data. Furthermore, a double GCACA motif (2× GCACA) was bound very strongly. Although the GCACC motif was selected several times from a pool of random oligonucleotides, albeit less frequently than the GCACA motif, the same sequence was not bound effectively by the ARID domain in the EMSA (Fig. 5C, lane GCACC*+GST-PLU-1/ARID). Nevertheless, this motif, when associated with a run of C's, was found frequently within the promoters of genes directly bound by PLU-1/JARID1B (see below).
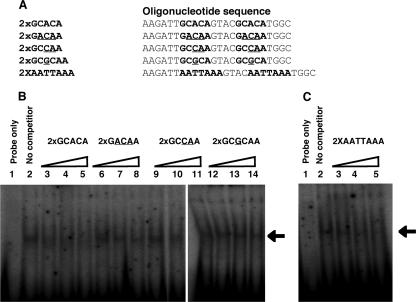
To further confirm the binding consensus of the PLU-1/JARID1B ARID domain, we performed a series of competition assays using the consensus sequences selected in the previous assay and different variants/mutants with a 3-, 2-, or 1-nucleotide change (Fig. 6A, rows 1 to 3). Figure 6B shows that a 5 M excess of unlabeled consensus sequence is sufficient to compete away most of the radiolabeled 2× GCACA oligonucleotide, while all three variants are much less effective at competing with 2× GCACA for binding to the ARID domain.
FIG. 6.
The PLU-1/JARID1B domain specifically binds the GCACA consensus sequence. (A) Sequence details of the oligonucleotide variants used in the EMSAs. The consensus and variant sequences are shown in bold, while the mutated bases in the variant sequences are underlined. (B and C) EMSAs were performed using purified bacterially expressed GST/PLU-1-ARID domain, labeled probe corresponding to PLU-1/JARID1B consensus sequence (2× GCACA), and indicated cold competitor oligonucleotides at 5-, 10-, and 20-fold molar excesses. A fivefold molar excess of cold consensus oligonucleotide is able to compete away most of the consensus oligonucleotide, while the consensus site variants are poor competitors. The AATTAAA sequence is a better competitor than the variant. Arrows indicate the position of DNA/GST-PLU-1/ARID complex.
The ARID domain from the original Drosophila dri protein (GST-Fly/ARID), which selected AT-rich motifs from an oligonucleotide pool (28) and bound the consensus sequence AATTAAA in the EMSA, did not bind the GCACA motif (Fig. 5D). Interestingly, the PLU-1/ARID domain also bound the dri consensus sequence in the EMSA (Fig. 5C), despite the fact that this motif was not selected from a pool of oligonucleotides. To determine whether there was any difference in affinity of the PLU-1/JARID1B ARID domain for the two consensus sequences GCACA and AATTAAA, an EMSA was performed with the radiolabeled PLU-1/JARID1B consensus sequence (2× GCACA) and, as a competitor, the unlabeled AATTAAA sequence (2× AATTAAA). The EMSA experiment showed that the 2× AATTAAA oligonucleotide was a better competitor than the variant oligonucleotides were, confirming that the PLU-1/JARID1B ARID domain can bind the dri AT-rich consensus. However, the EMSA shown in Fig. 6B suggests a lower affinity of the PLU-1/JARID1B ARID domain for the AATTAAA consensus than for the GCACA consensus (compare lanes 3, 4, and 5 of Fig. 6B with lanes 3, 4, and 5 of Fig. 6C). The data suggest that while the purified PLU-1/JARID1B ARID domain is able to bind to an AT-rich motif, it shows preference for a sequence containing a GCACA motif. Considering the selected sequence listed in Fig. 5, the GCACA motif is found most frequently and may be less dependent on the associated run of C's than the GCACC motif is.
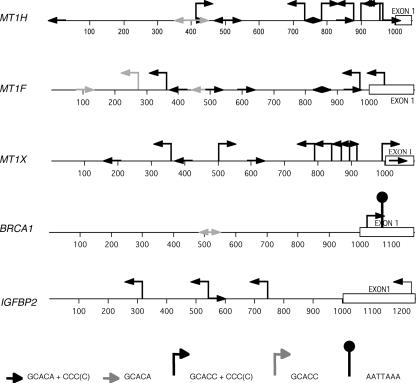
To determine whether the presence of the GCACA/C motif (with or without the following three C's or G's) in promoter regions of regulated genes could relate to a direct regulation of transcription, both strands of the first 1,000 bp upstream of a selection of these genes were examined (Fig. 7 and Table 2). GCACA/C is seen to be prevalent in the promoters of MT1H, MT1F, and MT1X, which we found to bind PLU-1/JARID1B (Fig. 4A). Although the motifs also occur within the region amplified by ChIP PCR, we cannot assume that PLU-1/JARID1B binds between these primers and not to a nearby sequence, because the DNA fragments produced by sonication in the ChIP studies ranged from 500 bp to 2 kb. However, the data support the idea that the GCACA/C motif identified in the selection process, particularly when close to a run of three C's, could be at least one of the consensus binding sites for the PLU-1/JARID1B transcription factor within the human genome. Significantly, the AT-rich consensus sequence is not found in any of the metallothionein promoters. However, a single AATTAAA sequence is found in exon 1 of BRCA1 near a motif consisting of GCACC with three C's, although the BRCA1 promoter is poor in GCACA motifs. By using a different antiserum for ChIP analysis, Yamane et al. (8888) showed the binding of PLU-1/JARID1B to the BRCA1 promoter. It is therefore possible that PLU-1/JARID1B can bind to DNA through either sequence as well as being recruited by other factors.
FIG. 7.
Promoters bound directly by PLU-1/JARID1B contain several putative PLU-1/JARID1B consensus sequences. Schematic representation of the locations and directions of the putative consensus sequence motifs consisting of GCACA/C with or without three or more C's and/or the AATTAA motif.
TABLE 2.
Numbers of motifs found in the proximal sequences and first exons of target genesa
| Gene promoter | No. of indicated motifs found
|
||||
|---|---|---|---|---|---|
| GCACA+ CCC(C) | GCACA | GCACC+ CCC(C) | GCACC | AATTAAA | |
| MTIH | 8 | 2 | 7 | 0 | 0 |
| MT1F | 6 | 2 | 3 | 1 | 0 |
| MT1X | 4 | 0 | 8 | 0 | 0 |
| BRCA1 | 0 | 2 | 1 | 0 | 1 |
| IGFBP2 | 1 | 0 | 4 | 0 | 0 |
For the positions of motifs, see Fig. 7.
Overexpression of PLU-1/JARID1B attenuates the activation of the G2/M block induced by destabilization of the mitotic spindle.
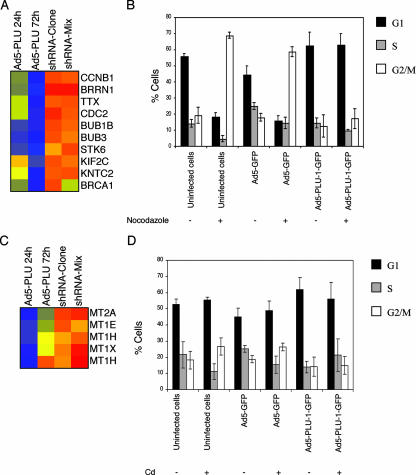
Although changing the levels of PLU-1/JARID1B per se does not change cell cycle progression, some of the genes downregulated by PLU-1/JARID1B do play a role in maintaining the G2/M and spindle cell cycle checkpoints in response to stress (Fig. 8A). We therefore tested the effect of overexpressing PLU-1/JARID1B (and therefore downregulating these genes) on the response of HB2 cells to treatment with the spindle-destabilizing agent nocodazole, which normally results in cells being blocked in G2/M.
FIG. 8.
Overexpression of PLU-1/JARID1B attenuates the G2/M block induced by treatment with either of the microtubule-destabilizing agents nocodazole or cadmium. (A and C) Color-coded TreeView diagrams depicting genes significantly downregulated following the upregulation of PLU-1/JARID1B expression. For details, see the legend for Fig. 2A. (B and D) HB2 cells were infected at an MOI of 500 with no adenovirus (uninfected cells), with the adenovirus GFP vector (Ad5-GFP), or with the adenovirus recombinant vector expressing a GFP-tagged PLU-1/JARID1B gene (Ad5-PLU-1-GFP). After 24 h of infection, cells were treated (+) or not treated (−) with 0.4 μg/ml nocodazole (B) or with 20 μM CdCl2 (D) for a further 24 h. At the end of the incubation, cells were pulse labeled (30 min) with 10 μM BrdU, then harvested, fixed in 1% paraformaldehyde, and stained for DNA content (Hoechst/Triton X-100) and BrdU incorporation (Alexa Fluor 697 intensity) analysis. In uninfected cells, the Hoechst versus BrdU plots were used to assess percentages of cells in G1, S, and G2. In infected cells, profiles were first gated on GFP-positive events before the cell cycle phases were derived. Bars represent the means ± standard errors of the means of three independent experiments.
Cells were infected with Ad5-PLU-1-GFP or Ad5-GFP at an MOI of 500 for 24 h and then treated (or not) for a further 24 h with nocodazole. At the end of the incubation, cells were pulse labeled (30 min) with 10 uM BrdU and then harvested, fixed in 1% paraformaldehyde, and stained for BrdU incorporation (see Materials and Methods) and DNA content (Hoechst staining), followed by flow cytometric analysis. Infected cells expressing high levels of PLU-1/JARID1B-GFP or GFP alone were gated, and the BrdU distribution as well as the DNA content was determined to calculate the percentages of cells in G1, S, and G2/M of the cell cycle. As shown in Fig. 8B, a large proportion (about 60 to 70%) of control noninfected cells and Ad5-GFP-infected cells were arrested by nocodazole in G2/M. The block in G2/M was not seen, however, in the cells expressing high levels of PLU-1/JARID1B, suggesting that the overexpression of PLU-1/JARID1B attenuates the G2/M or mitotic spindle checkpoint.
MT genes, which are also downregulated by PLU-1/JARID1B (Fig. 8C), are induced in response to heavy metals, such as cadmium (31). Cadmium has also been reported to induce DNA damage as well as to destabilize the mitotic spindle (45, 81). We therefore looked at the effects of cadmium on cell cycle progression and checked whether the overexpression of PLU-1/JARID1B modified the effect of cadmium. The treatment of HB2 cells with cadmium showed that cells uninfected or infected with the control Ad-GFP showed a modest increase in the cells in G2/M. While the increase in the number of cells in G2/M was small, it was seen consistently and was not seen in cells infected with the PLU-1/JARID1B recombinant virus (Fig. 8D).
DISCUSSION
The selective increase in expression of the transcriptional repressor PLU-1/JARID1B in breast cancer warranted an investigation into the identification of genes whose expression is regulated by this protein. By using two mammary epithelial cell lines expressing low and high levels of PLU-1/JARID1B, the effects of overexpression and RNAi-induced decrease in expression were investigated by microarray analysis. Eighty-one percent of the 100 genes shown to be inversely regulated in the two systems were transcriptionally repressed when PLU-1/JARID1B expression was upregulated, confirming its role as a transcriptional repressor. In this report, we have studied in more detail the control of expression of MT genes and the tumor suppressor BRCA1 gene, which are transcriptionally repressed by PLU-1/JARID1B. This regulation appears to be operational not only in mammary epithelial cells but also in HeLa cells, where the induction of expression of PLU-1/JARID1B from an inducible promoter also resulted in the downregulation of the expression of these genes. The downregulation of genes involved in the G2/M and spindle checkpoints correlates with the changed response to nocodazole (and to a lesser extent cadmium) when PLU-1/JARID1B is overexpressed.
This is the first study to report a global analysis of the regulation of gene expression by a member of the JARID1 subfamily of the ARID DNA binding proteins. These proteins contain the JmJC domain, which has previously been shown to have demethylase activity (78). Following the recent finding that PLU-1/JARID1B can demethylate tri-, di-, and monomethylated H3K4 (36, 70, 88), we show here, using ChIP analysis, that the level of trimethylated H3K4 on the promoter of MT1H is increased in PLU-1/JARID1B-silenced cells. We have also identified a new unconventional consensus sequence (GCACA) which is bound specifically by the PLU-1/JARID1B ARID domain but not by the ARID domain of the Drosophila dead ringer protein. This result suggests that PLU-1/JARID1B can deliver its H3K4 demethylase activity to the promoters of some downregulated target genes, such as the MT genes, by binding (through the ARID domain) to the GCACA/C consensus sequence. Binding to promoters of other downregulated target genes, which are not rich in the GCACA/C motif, could be through AATTAAA if this sequence is present (as it is in the first exon of BRCA1). EMSA data suggest that the PLU-1/JARID1B ARID domain can bind to this sequence, even though oligonucleotides containing this sequence were not selected from the pool of random oligonucleotides. On the other hand, binding may not be direct but rather may be through other interacting proteins such as c-Myc (70).
ChIP analysis using our α-PLU-1-C antiserum (directed to epitopes in the carboxy terminus) did not precipitate the BRCA1 promoter. In contrast, Yamane et al. showed, by ChIP, that PLU-1/JARID1B is associated with the promoter of tumor suppressor genes, including BRCA1 (88). The antibody used by Yamane et al. was raised against PLU-1/JARID1B N-terminal epitopes, which may suggest that the epitopes in the C-terminal domain of PLU-1/JARID1B are not exposed when bound to the BRCA1 promoter, as they are when bound to the MT promoters.
Transcriptional regulation of MT genes.
MTs are low-molecular-weight, cysteine-rich metal-binding proteins that can be induced to very high levels in response to various stress conditions, including exposure to heavy metals (such as zinc or cadmium), reactive oxygen species, and bacterial or viral infections (10, 18, 30, 31, 40, 53, 54, 68). MTs are known to participate in fundamental cellular processes, such as cell proliferation and apoptosis, and have been involved in multidrug resistance to cancer chemotherapy and free radical scavenging in cells (13, 22, 41, 42, 55, 65, 67). In humans, four isoforms of MT (MT1, MT2, MT3, and MT4) have been described previously, with the MT1 and MT2 isoforms being both basally expressed and highly inducible. The MT1 isoform includes at least 11 genes (including MT1A, -B, -E, -F, -G, -H, -I, and -X), which are clustered within the q13 region on chromosome 16. The MT2, MT3, and MT4 isoforms are located on the same chromosome. Different MT genes in humans may play specific functional roles during development and in various physiological conditions as well as in different organs. While the MTF-1 transcription factor has been found to be responsible for the activation of these genes (33), this is the first report documenting a factor which represses their transcription.
ChIP analysis showed PLU-1/JARID1B to be associated with the promoters of MT genes. Since the sequences selected from a random mixture of nucleotides (by both the full-length PLU-1/JARID1B protein and the ARID domain) contain motifs (GCACA/C, with three C's or three G's) that are abundant in the MT promoters, it is highly likely that the protein binds directly to the promoter sequences. Nevertheless, the data do not exclude the possibility that (i) other domains in PLU-1/JARID1B are involved in direct DNA binding and/or (ii) the protein may be recruited by other DNA binding factors. The mutation of the GCACA/C motifs in the promoters will help resolve this issue.
Data from the literature suggest that there is an inverse correlation between the expression of PLU-1/JARID1B and of both basal and induced expression of MTs. Basal expression of MTs can be detected in the liver of rat, mouse, and human (where PLU-1/JARID1B expression is low [4]), and the proteins are induced severalfold following exposure to heavy metal ions, thus ensuring detoxification and protection from the carcinogenic effects of cadmium (26). In contrast, in rodent testes, where Plu-1/Jarid1B expression is high (48), basal levels of MTs are undetectable and are not induced by heavy metals. This result correlates with the high susceptibility of testes to cadmium-induced carcinogenesis (51, 72, 93), where the low MT level is thought to allow the elimination of cells with DNA damage rather than the mobilization of a repair response which could lead to defective progeny (93). Since the expression of PLU-1/JARID1B is very high in spermatogonia and in specific stages of meiosis (48), it is possible that the low expression of MTs in normal testes relates to the high expression of PLU-1/JARID1B. An inverse relationship between the expression of MTs and PLU-1/JARID1B is also seen in breast cancer cell lines. Thus, high levels of the MTs MT1E and MT2A are found in the breast cancer cell lines Hs578T and MDA-MB-231, which express low levels of PLU-1, while less-invasive estrogen receptor (ER)-positive cell lines, such as MCF7, ZR75, and T47D, show low expression levels of the MT proteins but high levels of PLU-1/JARID1B (3, 4, 46, 74).
The significance of MT-1 gene expression in tumors has not been fully clarified, with their expression varying with tumor/tissue type (38, 39, 77). In breast cancer, an inverse correlation between the expression of MTs and ER staining has been reported (24, 58), as was seen with the breast cancer cell lines. Because both PLU-1/JARID1B and the ER appear to be inversely correlated with the expression of MTs, it is possible that both are involved in the regulation of their expression. We have not been able to demonstrate interaction between the two factors in coimmunoprecipitation experiments with MCF7 cells; however, they could both be operating through different complexes.
PLU-1/JARID1B, BRCA1, and cell cycle checkpoints.
The downregulation of the BRCA1 gene following the overexpression of PLU-1/JARID1B is of great interest. BRCA1 is a tumor suppressor gene whose germ line mutation has been found in almost 45% of hereditary breast cancers and in 80% of families whose members have a high incidence of both breast and ovarian cancer (21, 23, 29, 52). Although gene mutations of BRCA1 are very rare in sporadic breast and ovarian cancer, their expression is frequently reduced or absent in these cases, mediated by epigenetic changes (25, 50). BRCA1 plays a major role in detecting and repairing DNA damage: it is required for effective S-phase progression and is crucial for function of the G2/M-phase checkpoints in the cell cycle, as suggested by the fact that BRCA1-deficient cells exhibit defective G2/M arrest in response to genomic damage (56, 90). The observed overexpression of PLU-1/JARID1B protein in breast cancers may therefore bestow some growth advantage to tumor cells by downregulating the BRCA1 tumor suppressor gene.
Also related to the downregulation of BRCA1 is the downregulation by PLU-1/JARID1B of a specific set of genes that code for homologues of yeast (Saccharomyces cerevisiae) proteins involved in the control of the spindle checkpoint (BUB1B, BUB3, STK6, and TTK), in chromosome condensation (BRRN1 protein kinase), and in the transition from G2 into M (cyclin B1 and CDC2). BUB1B and BUB3 are components of the mitotic checkpoint that delays anaphase until all chromosomes are properly attached to the mitotic spindle; BUB3 localizes BUB1B in the kinetochore (17, 71, 76). BRRN1 is the regulatory subunit of the condensin complex, which is required for the conversion of interphase chromatin into mitotic-like condensed chromosomes (12, 34, 82). Cyclin B1 and CDC2 interact with each other and form a serine/threonine kinase holoenzyme complex, which is essential for control of the cell cycle at the G2/M (mitosis) transition (6, 15, 20, 44, 61, 64). Thus, PLU-1/JARID1B may be involved in the regulation of G2/M checkpoint and late M phase and in regulating the expression of a specific set of genes involved in spindle assembly, chromosomal condensation, and transition through the late stages of mitosis.
A highly relevant observation has been recently reported by Bae and colleagues (2), who identified the genes that are downregulated when BRCA1 is silenced by RNAi. Microarray analysis showed that the downregulation of BRCA1 repressed the transcription of the same group of genes downregulated by PLU-1/JARID1B, i.e., the genes involved in the mitotic spindle checkpoint (e.g., BUB1B, TTK, and STK6) and in the progression into and through mitosis (CDC2). The effects of PLU-1/JARID1B on these genes could therefore be mediated by the downregulation of the expression of BRCA1. An observation of potential relevance to PLU-1/JARID1B and checkpoint control comes from genetic studies of the fission yeast, where the yeast homologue of PLU-1/JARID1B (msc-1) corrects for a defective Chk1 gene (1).
Consistent with the expression profile analysis reported here, cells expressing high levels of PLU-1/JARID1B failed to arrest in G2/M after treatment with the spindle poison nocodazole. Cadmium is also reported to be a spindle poison, although the block in G2/M is, at best, only partial (45, 80, 81). Nevertheless, in cells overexpressing PLU-1/JARID1B, the small increase in the number of cells in G2/M after cadmium treatment was consistently reduced. These combined data suggest that by downregulating the expression of genes whose expression is normally increased in response to stress signals, resulting in a block in G2/M, the PLU-1/JARID1B transcriptional repressor is modulating this response. This function could be highly relevant to any role that this protein plays in the development and progression of breast cancer. Current studies aiming to develop a mouse strain defective in PLU-1/JARID1B function should lead to further clarification of the function of this protein.
Supplementary Material
Acknowledgments
We thank B. Young for enabling us to carry out the Affymetrix Microarrays and the staff at the FACS Laboratory, Cancer Research UK, for excellent support with the fluorescence-activated cell sorter analysis.
This work was funded by a Programme Grant and a fellowship (A.G.S.) from Cancer Research UK and a fellowship from King's College London School of Medicine (S.C.).
Footnotes
Published ahead of print on 20 August 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ahmed, S., C. Palermo, S. Wan, and N. C. Walworth. 2004. A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast. Mol. Cell. Biol. 24:3660-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae, I., J. K. Rih, H. J. Kim, H. J. Kang, B. Haddad, A. Kirilyuk, S. Fan, M. L. Avantaggiati, and E. M. Rosen. 2005. BRCA1 regulates gene expression for orderly mitotic progression. Cell Cycle 4:1641-1666. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, N. L., M. L. Ackland, and E. J. Cornish. 2000. Metallothionein isoform expression by breast cancer cells. Int. J. Biochem. Cell Biol. 32:895-903. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, A., B. Madsen, J. Copier, P. J. Lu, L. Cooper, A. G. Scibetta, J. Burchell, and J. Taylor-Papadimitriou. 2002. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen? Int. J. Cancer 101:581-588. [DOI] [PubMed] [Google Scholar]
- 5.Barrett, A., S. Santangelo, K. Tan, S. Catchpole, K. Roberts, B. Spencer-Dene, D. Hall, A. Scibetta, J. Burchell, E. Verdin, P. Freemont, and J. Taylor-Papadimitriou. 2007. The breast cancer-associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. J. Int. Cancer 121:265-275. [DOI] [PubMed] [Google Scholar]
- 6.Barrett, K. L., D. Demiranda, and K. S. Katula. 2002. Cyclin b1 promoter activity and functional cdk1 complex formation in G1 phase of human breast cancer cells. Cell Biol. Int. 26:19-28. [DOI] [PubMed] [Google Scholar]
- 7.Bartek, J., J. Bartkova, N. Kyprianou, E. N. Lalani, Z. Staskova, M. Shearer, S. Chang, and J. Taylor-Papadimitriou. 1991. Efficient immortalization of luminal epithelial cells from human mammary gland by introduction of simian virus 40 large tumor antigen with a recombinant retrovirus. Proc. Natl. Acad. Sci. USA 88:3520-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bártek, J., J. Bartkova, J. Schneider, J. Taylor-Papadimitriou, J. Kovarik, and A. Rejthar. 1986. Expression of monoclonal antibody-defined epitopes of keratin 19 in human tumours and cultured cells. Eur. J. Cancer Clin. Oncol. 22:1441-1452. [DOI] [PubMed] [Google Scholar]
- 9.Bartek, J., E. M. Durban, R. C. Hallowes, and J. Taylor-Papadimitriou. 1985. A subclass of luminal epithelial cells in the human mammary gland, defined by antibodies to cytokeratins. J. Cell Sci. 75:17-33. [DOI] [PubMed] [Google Scholar]
- 10.Bauman, J. W., J. Liu, Y. P. Liu, and C. D. Klaassen. 1991. Increase in metallothionein produced by chemicals that induce oxidative stress. Toxicol. Appl. Pharmacol. 110:347-354. [DOI] [PubMed] [Google Scholar]
- 11.Berdichevsky, F., C. Gilbert, M. Shearer, and J. Taylor-Papadimitriou. 1992. Collagen-induced rapid morphogenesis of human mammary epithelial cells: the role of the alpha 2 beta 1 integrin. J. Cell Sci. 102:437-446. [DOI] [PubMed] [Google Scholar]
- 12.Cabello, O. A., E. Eliseeva, W. G. He, H. Youssoufian, S. E. Plon, B. R. Brinkley, and J. W. Belmont. 2001. Cell cycle-dependent expression and nucleolar localization of hCAP-H. Mol. Biol. Cell 12:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai, L., G. J. Wang, Z. L. Xu, D. X. Deng, S. Chakrabarti, and M. G. Cherian. 1998. Metallothionein and apoptosis in primary human hepatocellular carcinoma (HCC) from northern China. Anticancer Res. 18:4667-4672. [PubMed] [Google Scholar]
- 14.Chan, S. W., and W. Hong. 2001. Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J. Biol. Chem. 276:28402-28412. [DOI] [PubMed] [Google Scholar]
- 15.Chang, D. C., N. Xu, and K. Q. Luo. 2003. Degradation of cyclin B is required for the onset of anaphase in mammalian cells. J. Biol. Chem. 278:37865-37873. [DOI] [PubMed] [Google Scholar]
- 16.Cloos, P. A., J. Christensen, K. Agger, A. Maiolica, J. Rappsilber, T. Antal, K. H. Hansen, and K. Helin. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307-311. [DOI] [PubMed] [Google Scholar]
- 17.Davenport, J. W., E. R. Fernandes, L. D. Harris, G. A. Neale, and R. Goorha. 1999. The mouse mitotic checkpoint gene bub1b, a novel bub1 family member, is expressed in a cell cycle-dependent manner. Genomics 55:113-117. [DOI] [PubMed] [Google Scholar]
- 18.De, S. K., M. T. McMaster, and G. K. Andrews. 1990. Endotoxin induction of murine metallothionein gene expression. J. Biol. Chem. 265:15267-15274. [PubMed] [Google Scholar]
- 19.Dorn, A., H. Zhao, F. Granberg, M. Hosel, D. Webb, C. Svensson, U. Pettersson, and W. Doerfler. 2005. Identification of specific cellular genes up-regulated late in adenovirus type 12 infection. J. Virol. 79:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ducommun, B., P. Brambilla, M. A. Felix, B. R. Franza, Jr., E. Karsenti, and G. Draetta. 1991. cdc2 phosphorylation is required for its interaction with cyclin. EMBO J. 10:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Easton, D. F., D. Ford, D. T. Bishop, et al. 1995. Breast and ovarian cancer incidence in BRCA1-mutation carriers. Am. J. Hum. Genet. 56:265-271. [PMC free article] [PubMed] [Google Scholar]
- 22.Ebadi, M., M. P. Leuschen, H. el Refaey, F. M. Hamada, and P. Rojas. 1996. The antioxidant properties of zinc and metallothionein. Neurochem. Int. 29:159-166. [DOI] [PubMed] [Google Scholar]
- 23.Ford, D., D. F. Easton, D. T. Bishop, S. A. Narod, D. E. Goldgar, et al. 1994. Risks of cancer in BRCA1-mutation carriers. Lancet 343:692-695. [DOI] [PubMed] [Google Scholar]
- 24.Friedline, J. A., S. H. Garrett, S. Somji, J. H. Todd, and D. A. Sens. 1998. Differential expression of the MT-1E gene in estrogen-receptor-positive and -negative human breast cancer cell lines. Am. J. Pathol. 152:23-27. [PMC free article] [PubMed] [Google Scholar]
- 25.Futreal, P. A., Q. Liu, D. Shattuck-Eidens, C. Cochran, K. Harshman, S. Tavtigian, L. M. Bennett, A. Haugen-Strano, J. Swensen, Y. Miki, et al. 1994. BRCA1 mutations in primary breast and ovarian carcinomas. Science 266:120-122. [DOI] [PubMed] [Google Scholar]
- 26.Goering, P. L., and C. D. Klaassen. 1984. Tolerance to cadmium-induced toxicity depends on presynthesized metallothionein in liver. J. Toxicol. Environ. Health 14:803-812. [DOI] [PubMed] [Google Scholar]
- 27.Granberg, F., C. Svensson, U. Pettersson, and H. Zhao. 2005. Modulation of host cell gene expression during onset of the late phase of an adenovirus infection is focused on growth inhibition and cell architecture. Virology 343:236-245. [DOI] [PubMed] [Google Scholar]
- 28.Gregory, S. L., R. D. Kortschak, B. Kalionis, and R. Saint. 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall, J. M., M. K. Lee, B. Newman, J. E. Morrow, L. A. Anderson, B. Huey, and M. C. King. 1990. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250:1684-1689. [DOI] [PubMed] [Google Scholar]
- 30.Hamer, D. H. 1986. Metallothionein. Annu. Rev. Biochem. 55:913-951. [DOI] [PubMed] [Google Scholar]
- 31.Haq, F., M. Mahoney, and J. Koropatnick. 2003. Signaling events for metallothionein induction. Mutat. Res. 533:211-226. [DOI] [PubMed] [Google Scholar]
- 32.Herrscher, R. F., M. H. Kaplan, D. L. Lelsz, C. Das, R. Scheuermann, and P. W. Tucker. 1995. The immunoglobulin heavy-chain matrix-associating regions are bound by Bright: a B cell-specific trans-activator that describes a new DNA-binding protein family. Genes Dev. 9:3067-3082. [DOI] [PubMed] [Google Scholar]
- 33.Heuchel, R., F. Radtke, O. Georgiev, G. Stark, M. Aguet, and W. Schaffner. 1994. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 13:2870-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirano, T., R. Kobayashi, and M. Hirano. 1997. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E, and a Xenopus homolog of the Drosophila Barren protein. Cell 89:511-521. [DOI] [PubMed] [Google Scholar]
- 35.Huang, T. H., T. Oka, T. Asai, T. Okada, B. W. Merrills, P. N. Gertson, R. H. Whitson, and K. Itakura. 1996. Repression by a differentiation-specific factor of the human cytomegalovirus enhancer. Nucleic Acids Res. 24:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwase, S., F. Lan, P. Bayliss, L. de la Torre-Ubieta, M. Huarte, H. H. Qi, J. R. Whetstine, A. Bonni, T. M. Roberts, and Y. Shi. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128:1077-1088. [DOI] [PubMed] [Google Scholar]
- 37.Jensen, L. R., M. Amende, U. Gurok, B. Moser, V. Gimmel, A. Tzschach, A. R. Janecke, G. Tariverdian, J. Chelly, J. P. Fryns, H. Van Esch, T. Kleefstra, B. Hamel, C. Moraine, J. Gecz, G. Turner, R. Reinhardt, V. M. Kalscheuer, H. H. Ropers, and S. Lenzner. 2005. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin, R., B. H. Bay, V. T. Chow, and P. H. Tan. 2001. Metallothionein 1F mRNA expression correlates with histological grade in breast carcinoma. Breast Cancer Res. Treat. 66:265-272. [DOI] [PubMed] [Google Scholar]
- 39.Jin, R., V. T. Chow, P. H. Tan, S. T. Dheen, W. Duan, and B. H. Bay. 2002. Metallothionein 2A expression is associated with cell proliferation in breast cancer. Carcinogenesis 23:81-86. [DOI] [PubMed] [Google Scholar]
- 40.Kägi, J. H., and A. Schaffer. 1988. Biochemistry of metallothionein. Biochemistry 27:8509-8515. [DOI] [PubMed] [Google Scholar]
- 41.Kelley, S. L., A. Basu, B. A. Teicher, M. P. Hacker, D. H. Hamer, and J. S. Lazo. 1988. Overexpression of metallothionein confers resistance to anticancer drugs. Science 241:1813-1815. [DOI] [PubMed] [Google Scholar]
- 42.Kling, P. G., and P. Olsson. 2000. Involvement of differential metallothionein expression in free radical sensitivity of RTG-2 and CHSE-214 cells. Free Radic. Biol. Med. 28:1628-1637. [DOI] [PubMed] [Google Scholar]
- 43.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312-316. [DOI] [PubMed] [Google Scholar]
- 44.Lee, M. G., and P. Nurse. 1987. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature 327:31-35. [DOI] [PubMed] [Google Scholar]
- 45.Li, W., Y. Zhao, and I. N. Chou. 1993. Alterations in cytoskeletal protein sulfhydryls and cellular glutathione in cultured cells exposed to cadmium and nickel ions. Toxicology 77:65-79. [DOI] [PubMed] [Google Scholar]
- 46.Lu, P. J., K. Sundquist, D. Baeckstrom, R. Poulsom, A. Hanby, S. Meier-Ewert, T. Jones, M. Mitchell, P. Pitha-Rowe, P. Freemont, and J. Taylor-Papadimitriou. 1999. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274:15633-15645. [DOI] [PubMed] [Google Scholar]
- 47.Madsen, B., B. Spencer-Dene, R. Poulsom, D. Hall, P. J. Lu, K. Scott, A. T. Shaw, J. M. Burchell, P. Freemont, and J. Taylor-Papadimitriou. 2002. Characterisation and developmental expression of mouse Plu-1, a homologue of a human nuclear protein (PLU-1) which is specifically up-regulated in breast cancer. Mech. Dev. 119(Suppl. 1):S239-S246. [DOI] [PubMed] [Google Scholar]
- 48.Madsen, B., M. Tarsounas, J. M. Burchell, D. Hall, R. Poulsom, and J. Taylor-Papadimitriou. 2003. PLU-1, a transcriptional repressor and putative testis-cancer antigen, has a specific expression and localisation pattern during meiosis. Chromosoma 112:124-132. [DOI] [PubMed] [Google Scholar]
- 49.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 50.McCoy, M. L., C. R. Mueller, and C. D. Roskelley. 2003. The role of the breast cancer susceptibility gene 1 (BRCA1) in sporadic epithelial ovarian cancer. Reprod. Biol. Endocrinol. 1:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna, I. M., R. M. Bare, and M. P. Waalkes. 1996. Metallothionein gene expression in testicular interstitial cells and liver of rats treated with cadmium. Toxicology 107:121-130. [DOI] [PubMed] [Google Scholar]
- 52.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66-71. [DOI] [PubMed] [Google Scholar]
- 53.Min, K. S., Y. Terano, S. Onosaka, and K. Tanaka. 1991. Induction of hepatic metallothionein by nonmetallic compounds associated with acute-phase response in inflammation. Toxicol. Appl. Pharmacol. 111:152-162. [DOI] [PubMed] [Google Scholar]
- 54.Murata, M., P. Gong, K. Suzuki, and S. Koizumi. 1999. Differential metal response and regulation of human heavy metal-inducible genes. J. Cell. Physiol. 180:105-113. [DOI] [PubMed] [Google Scholar]
- 55.Nagel, W. W., and B. L. Vallee. 1995. Cell cycle regulation of metallothionein in human colonic cancer cells. Proc. Natl. Acad. Sci. USA 92:579-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Narod, S. A., and W. D. Foulkes. 2004. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer 4:665-676. [DOI] [PubMed] [Google Scholar]
- 57.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 58.Oyama, T., H. Take, T. Hikino, Y. Iino, and T. Nakajima. 1996. Immunohistochemical expression of metallothionein in invasive breast cancer in relation to proliferative activity, histology, and prognosis. Oncology 53:112-117. [DOI] [PubMed] [Google Scholar]
- 59.Patsialou, A., D. Wilsker, and E. Moran. 2005. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33:66-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 61.Pines, J., and T. Hunter. 1989. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58:833-846. [DOI] [PubMed] [Google Scholar]
- 62.Quadbeck-Seeger, C., G. Wanner, S. Huber, R. Kahmann, and J. Kamper. 2000. A protein with similarity to the human retinoblastoma binding protein 2 acts specifically as a repressor for genes regulated by the b mating type locus in Ustilago maydis. Mol. Microbiol. 38:154-166. [DOI] [PubMed] [Google Scholar]
- 63.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 64.Riabowol, K., G. Draetta, L. Brizuela, D. Vandre, and D. Beach. 1989. The cdc2 kinase is a nuclear protein that is essential for mitosis in mammalian cells. Cell 57:393-401. [DOI] [PubMed] [Google Scholar]
- 65.Roesijadi, G. 2000. Metal transfer as a mechanism for metallothionein-mediated metal detoxification. Cell. Mol. Biol. (Noisy-le-grand) 46:393-405. [PubMed] [Google Scholar]
- 66.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 67.Satoh, M., M. G. Cherian, N. Imura, and H. Shimizu. 1994. Modulation of resistance to anticancer drugs by inhibition of metallothionein synthesis. Cancer Res. 54:5255-5257. [PubMed] [Google Scholar]
- 68.Schroeder, J. J., and R. J. Cousins. 1990. Interleukin 6 regulates metallothionein gene expression and zinc metabolism in hepatocyte monolayer cultures. Proc. Natl. Acad. Sci. USA 87:3137-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scibetta, A. G., J. Copier, A. Barrett, T. Chaplin, and J. Taylor-Papadimitriou. 2005. Gene expression changes induced by a recombinant E1−/E3− adenovirus type 5 vector in human mammary epithelial cells. Intervirology 48:350-361. [DOI] [PubMed] [Google Scholar]
- 70.Secombe, J., L. Li, L. Carlos, and R. N. Eisenman. 2007. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21:537-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shin, H. J., K. H. Baek, A. H. Jeon, M. T. Park, S. J. Lee, C. M. Kang, H. S. Lee, S. H. Yoo, D. H. Chung, Y. C. Sung, F. McKeon, and C. W. Lee. 2003. Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 4:483-497. [DOI] [PubMed] [Google Scholar]
- 72.Shiraishi, N., J. F. Hochadel, T. P. Coogan, J. Koropatnick, and M. P. Waalkes. 1995. Sensitivity to cadmium-induced genotoxicity in rat testicular cells is associated with minimal expression of the metallothionein gene. Toxicol. Appl. Pharmacol. 130:229-236. [DOI] [PubMed] [Google Scholar]
- 73.Tahiliani, M., P. Mei, R. Fang, T. Leonor, M. Rutenberg, F. Shimizu, J. Li, A. Rao, and Y. Shi. 2007. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 447:601-605. [DOI] [PubMed] [Google Scholar]
- 74.Tai, S. K., O. J. Tan, V. T. Chow, R. Jin, J. L. Jones, P. H. Tan, A. Jayasurya, and B. H. Bay. 2003. Differential expression of metallothionein 1 and 2 isoforms in breast cancer lines with different invasive potential: identification of a novel nonsilent metallothionein-1H mutant variant. Am. J. Pathol. 163:2009-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tan, K., A. L. Shaw, B. Madsen, K. Jensen, J. Taylor-Papadimitriou, and P. S. Freemont. 2003. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 278:20507-20513. [DOI] [PubMed] [Google Scholar]
- 76.Taylor, S. S., E. Ha, and F. McKeon. 1998. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J. Cell Biol. 142:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Theocharis, S. E., A. P. Margeli, J. T. Klijanienko, and G. P. Kouraklis. 2004. Metallothionein expression in human neoplasia. Histopathology 45:103-118. [DOI] [PubMed] [Google Scholar]
- 78.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 79.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19:381-391. [DOI] [PubMed] [Google Scholar]
- 80.Wang, Z., T. A. Chin, and D. M. Templeton. 1996. Calcium-independent effects of cadmium on actin assembly in mesangial and vascular smooth muscle cells. Cell Motil. Cytoskeleton 33:208-222. [DOI] [PubMed] [Google Scholar]
- 81.Wang, Z., and D. M. Templeton. 1996. Cellular factors mediate cadmium-dependent actin depolymerization. Toxicol. Appl. Pharmacol. 139:115-121. [DOI] [PubMed] [Google Scholar]
- 82.Watrin, E., and V. Legagneux. 2005. Contribution of hCAP-D2, a non-SMC subunit of condensin I, to chromosome and chromosomal protein dynamics during mitosis. Mol. Cell. Biol. 25:740-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 84.Whitson, R. H., T. Huang, and K. Itakura. 1999. The novel Mrf-2 DNA-binding domain recognizes a five-base core sequence through major and minor-groove contacts. Biochem. Biophys. Res. Commun. 258:326-331. [DOI] [PubMed] [Google Scholar]
- 85.Wilsker, D., A. Patsialou, P. B. Dallas, and E. Moran. 2002. ARID proteins: a diverse family of DNA binding proteins implicated in the control of cell growth, differentiation, and development. Cell Growth Differ. 13:95-106. [PubMed] [Google Scholar]
- 86.Wilsker, D., L. Probst, H. M. Wain, L. Maltais, P. W. Tucker, and E. Moran. 2005. Nomenclature of the ARID family of DNA-binding proteins. Genomics 86:242-251. [DOI] [PubMed] [Google Scholar]
- 87.Winer, J., C. K. Jung, I. Shackel, and P. M. Williams. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 270:41-49. [DOI] [PubMed] [Google Scholar]
- 88.Yamane, K., K. Tateishi, R. J. Klose, J. Fang, L. A. Fabrizio, H. Erdjument-Bromage, J. Taylor-Papadimitriou, P. Tempst, and Y. Zhang. 2007. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25:801-812. [DOI] [PubMed] [Google Scholar]
- 89.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]
- 90.Zhang, J., and S. N. Powell. 2005. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol. Cancer Res. 3:531-539. [DOI] [PubMed] [Google Scholar]
- 91.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress.’ EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao, H., F. Granberg, L. Elfineh, U. Pettersson, and C. Svensson. 2003. Strategic attack on host cell gene expression during adenovirus infection. J. Virol. 77:11006-11015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou, T., G. Zhou, W. Song, N. Eguchi, W. Lu, E. Lundin, T. Jin, and G. Nordberg. 1999. Cadmium-induced apoptosis and changes in expression of p53, c-jun and MT-I genes in testes and ventral prostate of rats. Toxicology 142:1-13. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.