Abstract
RNAs 33 nucleotides in length can direct accurate initiation of subgenomic RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase (RdRp), provided that the native sequences are maintained at five positions: −17, −14, −13, −11, and the +1 initiation site. The functional groups in the bases of these essential nucleotides required to interact with RdRp were examined by using chemically synthesized RNAs containing base analogs at each of the five positions. Analysis using a template competition assay revealed that the mode of recognition for the initiation nucleotide (+1) is distinct from that of the other essential nucleotides in the promoter. Competition experiments also determined that three template nucleotides are sufficient for stable interaction with RdRp. These results identify base moieties in the brome mosaic virus subgenomic promoter required for efficient RNA synthesis and support the hypothesis that the recognition of a RNA promoter by a viral RdRp is analogous to the recognition of DNA promoters by DNA-dependent RNA polymerases.
Replication of viral RNA genomes requires specific recognition of RNA features by proteins. The viral and cellular proteins that comprise the RNA-dependent RNA polymerase (RdRp) complex are responsible for directing viral RNA synthesis from the infecting RNA templates. Unlike other polymerases, RdRps can initiate RNA synthesis from either the end of the RNA template or from an internal promoter (1). The recognition elements that determine template specificity, ensuring amplification of the appropriate viral RNA species, are not well characterized. An in-depth biochemical knowledge of the features that determine specificity of viral RNA replication is directly relevant to viral pathogenesis and is important for elucidating the principles for protein-RNA interaction.
To investigate the mechanism of RNA-directed RNA synthesis, we studied brome mosaic virus (BMV), the type member of the bromovirus group of plant viruses in the alphavirus-like superfamily of (+)-strand RNA viruses (2). Three RNAs designated 1, 2, and 3, and a subgenomic RNA4 that is initiated from (−)-strand RNA3 comprise the BMV genome. In a highly specific manner, enriched BMV RdRp preparations from infected barley can synthesize (−)-strand RNA from (+)-strand templates and subgenomic (+)-strand products from (−)-strand templates (3–5).
We have developed a system to study BMV subgenomic RNA initiation from minimal templates, designated proscripts, because they contain both the promoter and template for (+)-strand RNA synthesis. The 20 nts 3′ of the subgenomic initiation nucleotide are recognized as the subgenomic core promoter (6, 7). In vitro, these 20 nts efficiently and accurately direct (+)-strand RNA synthesis (5, 8). By using this functional assay, we demonstrated that nucleotides −17, −14, −13, and −11 relative to the subgenomic initiation site must be maintained for RNA synthesis and are recognized in a sequence-specific fashion (8). RdRp interaction with the BMV core promoter represents an unusual class of protein-RNA interaction in that binding to RNA is not initially determined by the structure of the RNA.
In this paper, we have systematically replaced each of the base moieties predicted to interact with the BMV RdRp by using chemically synthesized base analogs and examined the effects on RNA synthesis. Our predictions were confirmed and extended. We also observed the importance of the 2′-hydroxyl (OH) group of the ribose at position −11 relative to the initiation site. We also have found that the mode of recognition for the initiation nucleotide is distinct from those at positions −17, −14, −13, and −11. A template competition analysis revealed the minimal RNA sequence needed to stably interact with RdRp. These results elucidate the main features in the subgenomic promoter required to interact with RdRp.
MATERIALS AND METHODS
Synthesis of Proscripts.
PCR was used to generate cDNA copies of the (−)-strand BMV RNA3 encompassing the subgenomic promoter from the cDNA clone of RNA pB3TP8 (9). Pairs of primers, one of which contained a T7 promoter, allowed proscript RNAs to be generated by using T7 RNA polymerase (Ampliscribe, Epicentre Technologies, Madison, WI) as described previously (5). Transcription reactions that incorporate inosines were preformed with 2 mM of the primer GpG, which allows initiation to take place and ITP in place of GTP in the reaction. RNAs were purified with Qiagen columns (Chatsworth, CA) using the manufacturer’s protocol to remove NTPs and proteins remaining from the T7 transcription reaction. RNAs were visually inspected by denaturing PAGE and quantified by UV absorbance.
Synthesis of 2′-O-TBDMSi-3′-O-phosphoramidites of purine riboside (10), 2-amino purine riboside (11), and pyrimidine-2-one riboside (12) were performed as previously described. 3′-O-phosphoramidite of 7-deaza-2′-deoxyguanosine was purchased from Glen Research (Sterling, VA). Chemical synthesis of the proscripts containing these base analogs were performed on an ABI 394 automated DNA synthesizer (Applied Biosystems) using conventional phosphoramidite elongation cycles according to Wincott et al. (13). After subsequent ethanolic ammonium hydroxide and triethylamine trihydrofluoride treatment to cleave the exocyclic amino and 2′-OH protecting groups, the proscripts were purified and analyzed by anion-exchange HPLC (13). Mass spectral analysis of each chemically synthesized proscript was performed on a Voyager-DE matrix-associated laser desorption ionization-time of flight spectrometer (Perseptive Biosystem, Framingham, MA), and all were within 0.1% of the expected mass (Table 1).
Table 1.
Matrix-associated laser desorption ionization-time of flight atomic mass determination of the proscripts containing base analogs
| Proscript name | Mass, daltons
|
Δ, amu | |
|---|---|---|---|
| Calc. | Obs. | ||
| −17 G/2′-dG | 10571.2 | 10566.1 | 5.1 |
| −17 G/2-Aminopurine | 10570.2 | 10568.8 | 1.4 |
| −17 G/N7-deaza-2′-dG | 10569.2 | 10559.6 | 9.6 |
| −14 A/Purine | 10572.2 | 10577.8 | 5.6 |
| −13 C/Pyrimidine-2-one | 10571.2 | 10564.6 | 7.3 |
| −11 G/2′-dG | 10571.2 | 10563.9 | 7.3 |
| −11 G/N7-deaza-2′-dG | 10569.2 | 10569.5 | 0.3 |
| −11 G/2-Aminopurine | 10570.2 | 10571.2 | 1.0 |
| −1 G/2-Aminopurine | 10570.2 | 10569.3 | 0.9 |
| +1 C/Pyrimidine-2-one | 10571.2 | 10566.4 | 4.8 |
| +2 A/Purine | 10572.2 | 10569.3 | 2.9 |
Calc., calculated; Obs., observed.
RdRp Activity Assay and Product Analysis.
BMV RdRp was prepared from infected barley as described previously (14). Standard assays consisted of 25 nM of template RNA (unless stated otherwise) with 10 μl of RdRp in a 40-μl reaction containing 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12.5 mM DTT, 0.5% (vol/vol) Triton X-100, 2 mM MnCl2, 200 μM ATP and UTP, 500 μM GTP, and 250 nM [α-32P]CTP (Amersham). Reactions were incubated at 30°C for 90 min and stopped by phenol/chloroform extraction followed by ethanol precipitation in the presence of 5 μg of glycogen and 0.4 M ammonium acetate. Products were separated by electrophoresis on 20% denaturing (8 M urea) polyacrylamide gels. Gels were wrapped in plastic and exposed to film at −80°C. Product bands were quantified by using a PhosphorImager (Molecular Dynamics), and values were compared to the amount of product generated from the wild-type (WT) template (−20/13) to derive the relative percent activity of mutant templates. All values shown represent the mean of at least three independent experiments with SD shown. WT −20/13 contains sequence complementary to viral (+)-strand RNA3 from positions 1222 to 1252.
Template competition assays were performed under the reaction conditions stated above except that 25 nM of proscript −20/15, directing synthesis of a 15-nt product, was incubated with increasing concentrations of various competitors. The amount of RdRp used in these reactions is limiting as previously demonstrated (8). The product generated from the −20/15 proscript was quantitated as above and plotted against the concentration of competitor to determine the concentration of competitor needed to reduce the 15-nt product by 50%, designated as the I50 value.
RESULTS
Functional Groups Required for RNA Synthesis.
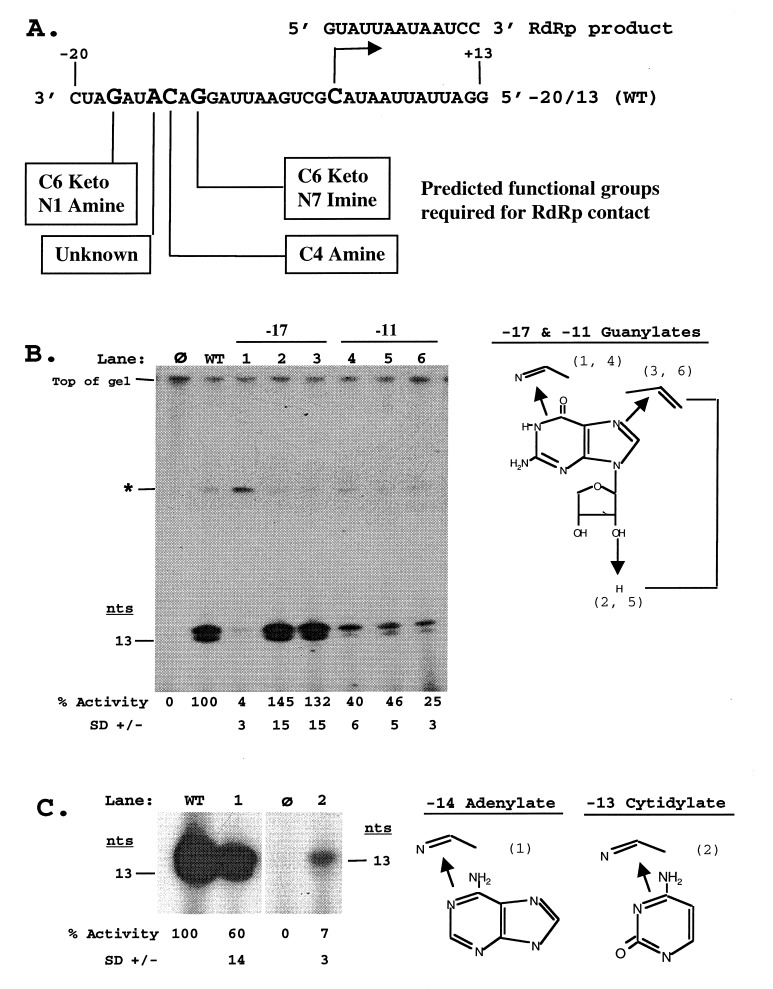
Nucleotides located at positions −17, −14, −13, and −11 relative to the subgenomic initiation site (+1) were required for RNA synthesis, and predictions were made regarding the functional moieties mediating this effect (Fig. 1A) (8). RNAs containing base analogs at each of the four critical positions were made to determine the importance of these functional groups (Fig. 1). As the WT control, a 33-nt proscript (designated −20/13) was constructed, which contains the WT promoter sequence 20 nts 3′ of the subgenomic initiation start site in (−)-strand RNA3. This proscript directs the synthesis of a 13-nt product, the first 11 nts of which are BMV sequence followed by two guanylates that allow labeling of RdRp products with [α-32P]CTP (Fig. 1 B and C, lane WT).
Figure 1.
Functional moieties in the subgenomic promoter required for initiation of RNA synthesis. (A) Predicted functional groups required for interaction with the BMV RdRp. The sequence shown is proscript −20/13 containing the WT BMV subgenomic promoter directing synthesis of a 13-nt product and serves as the WT control. The subgenomic initiation site is denoted by an arrow with the sequence of the RdRp product shown above. The base moieties predicted to interact with the RdRp by previous mutational studies (8) are indicated below the four nucleotides essential for RNA synthesis. (B) Recognition of the guanylate residue at positions −17 and −11. The bands denoted by ∗ represent terminal transferase labeling of the input template. (C) Recognition of the adenylate at position −14 and the cytidylate at position −13. The structures of the particular base for each position and of the nucleosides in the case of the guanylate at positions −17 and −11 are shown with arrows indicating defined changes in the functional groups mediated by the insertion of various base analogs. Numbers in parenthesis indicate the lane in the autoradiograph containing the RdRp reaction products from proscripts containing the indicated base analog. Lane WT represents the products directed by the −20/13 proscript, and lane ø represents the products of a control reaction with no added template. The reaction products were separated by denaturing PAGE and visualized by autoradiography with their sizes denoted on the side. The predominant RdRp product was 14 nts because of the nontemplated addition of one residue at the 3′ end of the RNA product (8). Accurate initiation was verified by described enzymatic manipulations (8) and comparison to T7-generated size markers. Values listed below the gels represent the percent activity from promoters containing each base analog compared to that from the WT promoter sequence.
We first examined the recognition of the guanylate at position −17 (Fig. 1B). The C6 keto (and possibly the N1) group was predicted to interact with the BMV RdRp. A proscript RNA containing the base analog 2-aminopurine, which removes the C6 keto group and forms a double bond with the N1 moiety, decreased RNA synthesis to background levels (Fig. 1B, lane 1). The mutational analysis did not implicate roles for the C2 amine and the N7 imine (8). The more limited role of the C2 amine in interaction with RdRp can be surmised by the severity of the 2-aminopurine substitution, which indicates that the C6 keto and possibly the N1 groups are the determining moieties. As negative control, a change of the −17 ribose to a deoxyribose was unaffected in RdRp recognition (Fig. 1B, lane 2). Finally, a direct examination of the N7 imine by a replacement with a carbon along with a change of the ribose to deoxyribose had no effect on the ability of the proscript to direct RNA synthesis by RdRp (Fig. 1B, lane 3).
The guanylate at position −11 was predicted to be recognized in a bidentate fashion mediated by both the C6 keto (along with the N1 amine) and the N7 imine. Incorporation of the 2-aminopurine base analog (11) at this position significantly decreased the ability to direct RNA synthesis to 40% (Fig. 1B, lane 4). However, RNA synthesis was not reduced to background levels, consistent with the prediction that the N7 imine was also important. The N7 deaza base analog was available only in a deoxyribose form; therefore, a control RNA with deoxyguanosine at position −11 was first tested. A deoxyribose at position −11 reduced RNA synthesis to 46% of WT (Fig. 1B, lane 5), implicating that the RNA backbone does mediate some aspect of subgenomic promoter recognition. Removal of both the N7 imine and the 2′-OH further reduced synthesis to 25% of WT, indicating recognition of the predicted N7 imine (Fig. 1B, lane 6). Again RNA synthesis was not abolished because this base analog retained the C6 keto group.
A likely candidate for RdRp recognition of the adenylate at position −14 is the exocyclic C6 amine group. To test the importance of this group, a purine riboside analog (10) was substituted for the adenylate at this position. An RNA with this change retained synthesis at 60% of WT, suggesting some other features of the adenylate are important for recognition by RdRp (Fig. 1C, lane 1). In contrast, the −13 cytidylate was predicted to interact with RdRp by the exocyclic C4 amine group. A proscript containing the base analog pyrimidine-2-one (12), which specifically removes this functional group, reduced the level of RNA synthesis to 7% (Fig. 1C, lane 2).
RdRp Contacts Within the Subgenomic Promoter.
A template competition assay was used to test whether various promoter mutations affected the ability of the RNA to interact with the BMV RdRp (Table 2). The amount of synthesis from a WT promoter directing the production of a 15-nt product (proscript −20/15) was determined in the absence and presence of various competitor templates. If a mutant proscript used as a competitor had lost its ability to be recognized by RdRp, then its presence in a reaction should not adversely affect the amount of synthesis from the WT −20/15 proscript.
Table 2.
Mutations in the subgenomic promoter that affect the ability to interact with RdRp
| Competitor, molar ratio | % Activity from proscript −20/15 |
|---|---|
| −20/13 WT | |
| 1:1 | 51% |
| −14 A/U | |
| 1:1 | 110% |
| 5:1 | 100% |
| −13 C/G | |
| 1:1 | 130% |
| 5:1 | 110% |
| −11 G/C | |
| 1:1 | 135% |
| 5:1 | 95% |
| +1 C/G | |
| 1:1 | 110% |
| 5:1 | 58% |
As a positive control, the WT proscript −20/13 (generating a 13-nt product) was tested for its ability to inhibit synthesis of the 15-nt product from −20/15. When this proscript was present in the same molar amount as proscript −20/15, the level of 15-nt synthesis was reduced by half. Mutations at positions −14, −13, and −11, which abolished the ability to direct synthesis, did not inhibit the BMV RdRp from productively interacting with a WT promoter even when present in molar excess (Table 2), the same result as was obtained with mutations at position −17 (8). In contrast, a change of the initiation cytidylate to a guanylate (abolishing its ability to direct RNA synthesis) reduced synthesis from proscript −20/15 by more than 40% when present in 5-fold molar excess (Table 2). Thus, even though mutations in the core promoter and in the initiation nucleotide all were drastically reduced in the ability to direct RNA synthesis, these results suggested that the nucleotides in the subgenomic core promoter and the initiation site are recognized in different fashions.
Minimal Template Needed for RdRp to Interact with the Subgenomic Promoter.
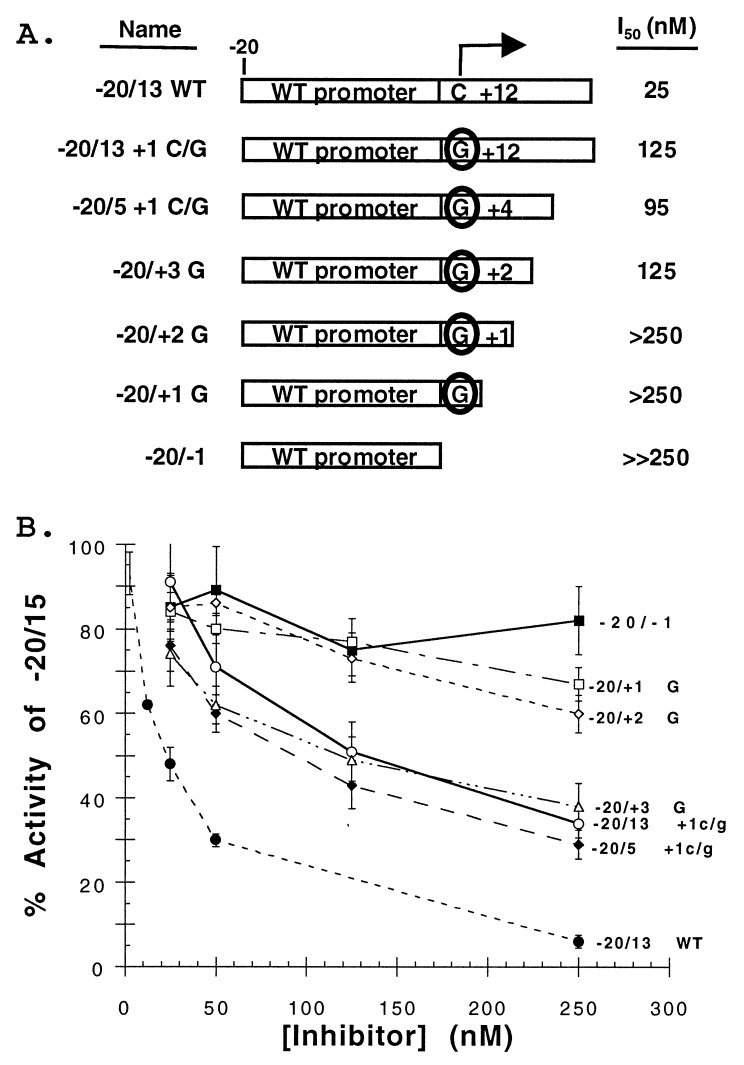
To delineate the length of template required for stable interaction with RdRp, proscripts containing template truncations were constructed and tested for their ability to inhibit synthesis of the 15-nt product from proscript −20/15. The concentration of the competitor proscript required to reduce the activity from −20/15 by 50% was termed the I50 value. This refinement of the template competition assay allowed us to compare various proscripts’ ability to interact with RdRp. Better competitors (able to interact more strongly with RdRp and, therefore, reduce synthesis from proscript −20/15) will have lower I50 values. These truncated proscripts all contained WT promoter sequences but varied in the length of their templates from 13, 5, 3, 2, 1, and 0 nts (Fig. 2A). To uncouple the effects of RNA synthesis from binding, these truncated proscripts all contained a mutated initiation site (+1c/g). Comparison of the I50 value for proscripts −20/13 +1c/g and −20/13 WT revealed a 5-fold difference in the ability to inhibit synthesis of the 15-nt product (Fig. 2B), consistent with the results observed in Table 2.
Figure 2.
Template requirements for stable interaction with RdRp. (A) Schematic of proscripts containing 5′ truncations of the template. The initiation site is denoted with an arrow. Proscripts containing the +1 c/g mutation (circled) are unable to direct RNA synthesis. The names and I50 values for each construct are listed to the sides. (B) Determination of I50 values. The amount of activity from the −20/15 proscript directing synthesis of a 15-nt product was measured in the presence of increasing amounts (up to 10-fold molar excess, 250 nM) of competitor templates. The I50 value was determined as the concentration of competitor needed to reduce the 15-nt product from 25 nM of the −20//15 proscript by 50%. The identities of the various proscript competitors are labeled to the right. Data points represent the mean of at least three independent experiments with the SD expressed as error bars.
Decreasing the lengths of the template portion of the competitor RNAs from 13 to 3 nts had no adverse effect on their ability to interact with RdRp (I50 values ranging from 95 to 125 nM, Fig. 2B). However, a template of 2 nts in length failed to reduce 15-nt synthesis from proscript −20/15 by 50% in the concentration range tested (up to 10-fold molar excess) (Fig. 2B). As would be expected, this defect also was observed in proscripts having 1 or 0 nts in the template (Fig. 2B). These results indicated that a minimum length of the proscript required for stable interaction with RdRp is 3 nts. This requirement does not depend on the identities of the nucleotides at positions +2 and +3 because proscripts containing different nucleotides at these positions had similar I50 values (R.W.S. and C.C.K. data not shown).
Recognition of the Nucleotides Surrounding the Subgenomic Initiation Site.
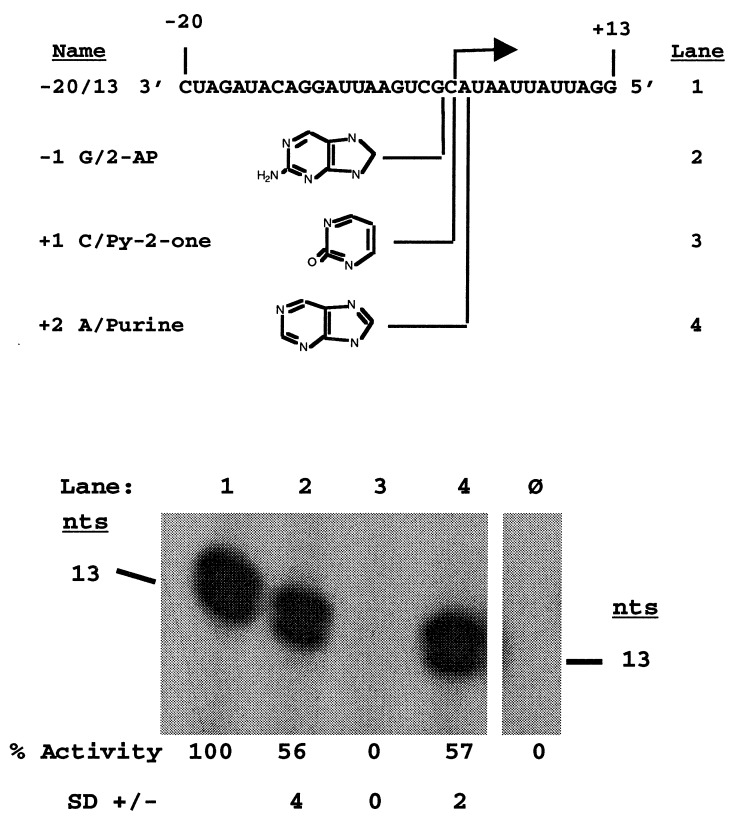
We next wanted to determine how RdRp recognized the correct initiation site. Proscripts containing base analogs at positions −1, +1, and +2 were constructed. Change of the −1 guanylate to 2-aminopurine and the +2 adenylate to a purine with no exocyclic groups both resulted in proscripts that are decreased in RNA synthesis by only 40-45% (Fig. 3, lanes 2 and 4). Conversely, removal of the exocyclic C4 amine group from the +1 cytidylate reduced RNA synthesis to undetectable levels (Fig. 3, lane 3). These results are consistent with findings from mutational analyses of the nucleotides in this same region (15). Recognition of the +1 nucleotide by BMV RdRp will be discussed below.
Figure 3.
Recognition of the subgenomic initiation site. The sequence of the WT −20/13 proscript is shown with the initiation site denoted. The structure of the base analogs incorporated at positions −1, +1, and +2 are listed below with the names of the various proscripts shown on the left. The lanes containing the RdRp reaction products from the proscripts containing the base analogs are indicated on the right. Lane ø represents the products of a control reaction with no added template. The reaction products were separated by denaturing PAGE and visualized by autoradiography with their sizes denoted on the side. Values listed below the gels represent the percent activity from promoters containing each base analog compared to that from the WT promoter sequence.
DISCUSSION
Although initiation of subgenomic RNA synthesis has been studied in other RNA viruses (16-20), a biochemical understanding of the promoters responsible for directing synthesis is lacking for all RNA viruses. For BMV, the in vitro synthesis of subgenomic RNA4 using (−)-strand RNA 3 template with its 3′ end 20 nts from the initiating cytidylate was first demonstrated by Miller and colleagues (4). Using minimal RNA templates directing the production of 13-nt RNA products, we previously have identified four critical positions (−17, −14, −13, and −11 relative to the initiation site) required for RNA synthesis and, at least for position −17, required for recognition by the BMV RdRp (8).
The current work uses chemically synthesized RNAs containing base analogs to further define how these critical nucleotides in the subgenomic promoter contribute to recognition by the BMV RdRp. The functional groups predicted to be important by mutational analysis (8) were confirmed by these data. The guanylate at position −17 interacts with RdRp through the C6 keto group and possibly the N1 amine, which also was affected by the replacement of the guanine with a 2-aminopurine. BMV RdRp also was shown to recognize the cytidylate at position −13 through the exocyclic C4 amine group. However, the recognition of the other two nucleotides appears to be more complex than originally postulated.
The guanylate at position −11 contains at least three moieties recognized by RdRp: C6 keto and N7 imine in the base, and the 2′ hydroxyl of the ribose. Both of the base functional groups were predicted from previous mutational data, but the ribose moiety was not. Protein recognition of a 2′-hydroxyl is not unexpected because it can function as both a hydrogen bond donor or acceptor, thereby potentially interacting with a variety of amino acid side chains. Alternatively, a change in the sugar conformation enacted by the deoxyribose replacement may affect the local geometry of the surrounding nucleotides (for example, positions −14 and −13) (21). However, the 2′-hydroxyl at position −17 is not important for RNA synthesis (Fig. 1A, lanes 2 and 5), signifying that not every 2′-hydroxyl group is required for recognition. Specific recognition of a limited number of 2′-hydroxyls has been observed for the MS2 coat protein and Escherichia coli alanine-tRNA synthetase (22, 23). These results suggest that the BMV RdRp may be able to tolerate deoxyriboses in at least a subset of positions within the subgenomic promoter and even perhaps the template residues.
Although no predictions were made for the recognition of the −14 adenylate, the removal of the exocyclic C6 amine did not dramatically affect the promoter’s ability to direct RNA synthesis (Fig. 1C, lane 1). A closer examination of the nucleotide substitution data (8) revealed that, regardless of sequence context, a bulky C2 substitution had a consistent deleterious effect on RNA synthesis at this position. Perhaps the sequence identity at position −14 is essential not only for interaction with the BMV RdRp, but also the prevention of steric clashes as has been postulated for the recognition of the branch-site adenylate in a group II intron (10). However, the specific features of the adenylate at position −14 recognized by RdRp remain to be elucidated.
Our results support a model of the direct readout of the nucleotides in the BMV subgenomic core promoter in a manner comparable to promoter recognition by the bacteriophage DNA-dependent RNA polymerases. The phage T7 and T3 RNA polymerases contact specific nucleotides within a 17-nt consensus promoter primarily at nucleotides −11 to −9 relative to the start site (24, 25). This comparison implies that the BMV RdRp makes hydrogen bond contacts with the functional groups defined in this study. Alternatively, the functional groups may affect the presentation of structure(s) within the core promoter required for recognition by RdRp. Results from template competition assays were consistent with positions −17, −14, −13, and −11 being required for stable interaction with RdRp (Table 2 and ref. 8). Contact sites in the protein subunit(s) of RdRp remain to be determined, but the analysis of the RNA will guide this process.
The stable interaction between the BMV RdRp and the subgenomic promoter requires at least three template residues (Fig. 2). However, the template nucleotides at positions +2 and +3 can be replaced with other nucleotides, suggesting that contacts with BMV RdRp are not specific immediately following the subgenomic initiation site. The results obtained with base analog inserted at the +2 position of the subgenomic promoter support this view (Fig. 3). Only the exocyclic C4 amine group of the +1 cytidylate was essential for RNA synthesis (Fig 3, lane 3). It can be envisioned that 3 nts of the template are required to provide a platform for stable interaction with the RdRp.
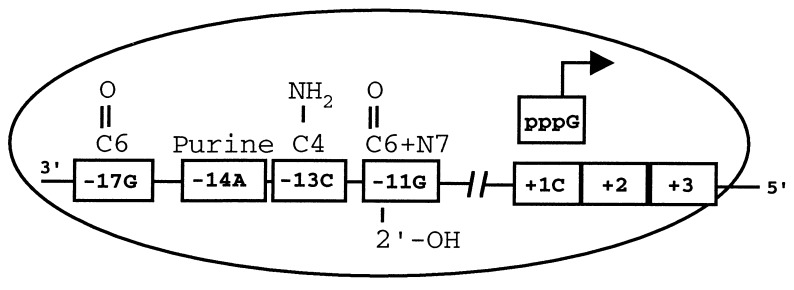
Kao and Sun (26) have demonstrated that the BMV RdRp possesses an initiating nucleotide binding site that is specific for GTP, but GTP can be replaced by specific oligonucleotide primers. The importance of the +1 C4 amine functional group may be mediated by interaction with GTP already bound by RdRp, and not directly with the RdRp complex. This hypothesis dictates that selection of the proper initiation site occurs by hydrogen bonding, or structural constraints (27), between an GTP “primer” located in the BMV RdRp and the +1 nucleotide in the template. This trimolecular recognition of the proper initiation site depends on proper orientation of the RdRp on the template, presumably through contacts with the four essential nucleotides within the promoter sequence (Fig. 4). Changes in the sequence identity of the initiation site then would abolish the ability to initiate RNA synthesis, but only reduce the binding affinity with the functional groups contained in an otherwise WT subgenomic promoter.
Figure 4.
A model for the interaction between the BMV RdRp and the subgenomic promoter elements needed to initiate RNA synthesis. Essential nucleotides are boxed with the key features putatively required for hydrogen bond formation with amino acid residues in RdRp shown above. The 2′-hydroxyl at position −11 also contributes to RNA synthesis. Recognition of the initiating nucleotide may occur by the rGTP primer bound by the RdRp. The oval represents a low-resolution structure of the RdRp complex.
Although we have identified several moieties in the BMV core promoter important for RNA synthesis, we cannot eliminate the possibility that additional moieties may modulate recognition by RdRp. Examination of the effects of multiple changes in the RNA have provided results consistent with this view. Using proscripts that have inosines replacing all guanines (i.e., removing the exocyclic C2 amine in the various guanines), the level of RNA synthesis was at 8% of WT −20/13 (B. Higgs and C.C.K., data not shown). This result suggests that some properties affected by the C2 amines in one or more of the five substituted guanines are important for RNA synthesis either by interacting with RdRp or perturbing some aspect of RNA structure. More biophysical examination of the interaction between RdRp and the subgenomic core promoter is necessary.
Protein-RNA interaction is fundamental for viral replication and to many cellular activities (28). Delineation of the principles viral RdRps use to recognize their correct template in the cellular milieu offers key insights into a complex step in the viral life cycle and into the generalities governing the dynamics of protein-RNA interaction. The production of subgenomic RNAs is a common scheme used by many different (+)-strand RNA viruses (1, 15). Therefore, the base-specific recognition of the subgenomic core promoter we have described for BMV is likely to occur in other (+)-strand RNA viruses.
Acknowledgments
We thank our colleagues at Ribozyme Pharmaceuticals, P. Haeberli for preparation of purine and 2-aminopurine phosphoramidites, A. Karpeisky for preparation of pyrimidine-2-one phoshoramidite, and V. Mokler and L. Maloney for the synthesis and purification of the different modified −20/13 proscripts. We also thank M. Chapman, S. Adkins, J. Richardson, J. Drummond, and T. Widlanski for many helpful discussions. Funding was provided by U.S. Department of Agriculture Grant 9702126 and National Science Foundation Grant MCB9507344. R.W.S. is supported by a National Institutes of Health Genetics Training Grant to the Indiana University Biology Department.
ABBREVIATIONS
- RdRp
RNA-dependent RNA polymerase
- BMV
brome mosaic virus
- WT
wild type
References
- 1. Buck K W. Adv Virus Res. 1996;47:159–251. doi: 10.1016/S0065-3527(08)60736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldbach R, LeGall O, Welink J. Semin Virol. 1991;2:19–25. [Google Scholar]
- 3.Hardy S F, German T L, Loesch-Fries L S, Hall T C. Proc Natl Acad Sci USA. 1979;76:4956–4960. doi: 10.1073/pnas.76.10.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller W A, Dreher T W, Hall T C. Nature (London) 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 5.Adkins S, Siegel R W, Sun J-H, Kao C C. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 6.Marsh L E, Dreher T W, Hall T C. Nucleic Acids Res. 1988;16:981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.French R, Ahlquist P. J Virol. 1988;62:2411–2420. doi: 10.1128/jvi.62.7.2411-2420.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel R W, Adkins S, Kao C C. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janda M, French R, Ahlquist P. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 10.Liu Q, Green J B, Khodadai A, Haeberli P, Beigelman L, Pyle A M. J Mol Biol. 1997;267:163–171. doi: 10.1006/jmbi.1996.0845. [DOI] [PubMed] [Google Scholar]
- 11.Konforti B, Abramovitz D, Duarte C, Karpeisky A, Beigelman L, Pyle A M. J Mol Cell. 1998;1:433–441. doi: 10.1016/s1097-2765(00)80043-x. [DOI] [PubMed] [Google Scholar]
- 12.Murray J B, Adams C J, Arnold J R P, Stockley P G. Biochem J. 1995;311:487–494. doi: 10.1042/bj3110487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun J-H, Adkins S, Faurote G, Kao C C. Virology. 1996;226:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 15.Adkins S, Stawicki S S, Faurote G, Siegel R W, Kao C C. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 16.Ou J-H, Rice C M, Dalgarno L, Strauss E G, Strauss J H. Proc Natl Acad Sci USA. 1982;79:5235–5239. doi: 10.1073/pnas.79.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levis R, Schlesinger S, Huang H V. J Virol. 1990;64:1726–1733. doi: 10.1128/jvi.64.4.1726-1733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Der Vossen E A G, Notenboom T, Bol J F. Virology. 1995;212:663–672. doi: 10.1006/viro.1995.1524. [DOI] [PubMed] [Google Scholar]
- 19.Johnston J C, Rochon D M. Virology. 1995;214:100–109. doi: 10.1006/viro.1995.9950. [DOI] [PubMed] [Google Scholar]
- 20.Kim K-H, Hemenway C. Virology. 1997;232:187–197. doi: 10.1006/viro.1997.8565. [DOI] [PubMed] [Google Scholar]
- 21.Auffinger P, Westhof E. J Mol Biol. 1997;274:54–63. doi: 10.1006/jmbi.1997.1370. [DOI] [PubMed] [Google Scholar]
- 22.Baidya N, Uhlenbeck O C. Biochemistry. 1995;34:12363–12368. doi: 10.1021/bi00038a033. [DOI] [PubMed] [Google Scholar]
- 23.Musier-Forsyth K, Schimmel P. Nature London. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 24.Schick C, Martin C T. Biochemistry. 1995;34:666–672. doi: 10.1021/bi00002a034. [DOI] [PubMed] [Google Scholar]
- 25.Li T, Ho H H, Maslak M, Schick C, Martin C T. Biochemistry. 1996;35:3722–3727. doi: 10.1021/bi9524373. [DOI] [PubMed] [Google Scholar]
- 26.Kao C C, Sun J-H. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran S, Kool E T. Proc Natl Acad Sci USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Draper D E. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]