Abstract
The Cdc34 E2 ubiquitin (Ub) conjugating enzyme catalyzes polyubiquitination of a substrate recruited by the Skp1-Cullin 1-F-box protein-ROC1 E3 Ub ligase. Using mutagenesis studies, we now show that human Cdc34 employs distinct sites to coordinate the transfer of Ub to a substrate and the assembly of polyubiquitin chains. Mutational disruption of the conserved charged stretch (residues 143 to 153) or the acidic loop residues D102 and D103 led to accumulation of monoubiquitinated IκBα while failing to yield polyubiquitin chains, due to a catalytic defect in Ub-Ub ligation. These results suggest an ability of human Cdc34 to position the attacking Ub for assembly of polyubiquitin chains. Analysis of Cdc34N85Q and Cdc34S138A revealed severe defects of these mutants in both poly- and monoubiquitination of IκBα, supporting a role for N85 in stabilizing the oxyanion and in coordinating, along with S138, the attacking lysine for catalysis. Finally, Cdc34S95D and Cdc34E108A/E112A abolished both poly- and monoubiquitination of IκBα. Unexpectedly, the catalytic defects of these mutants in di-Ub synthesis can be rescued by fusion of a glutathione S-transferase moiety at E2's N terminus. These findings support the hypothesis that human Cdc34 S95 and E108/E112 are required to position the donor Ub optimally for catalysis, in a manner that might depend on E2 dimerization.
By eliminating proteins in a selective and rapid fashion, ubiquitin (Ub)-dependent proteolysis imposes profound alteration over a wide range of biological processes that include cell growth and death, development, signal transduction, transcriptional control, genomic integrity, membrane trafficking, and tumor suppression (7, 22). Of central importance is the unique modification step, termed ubiquitination, through which a target protein is tagged with lysine 48-linked polyubiquitin chains. The polyubiquitinated protein is recognized by the 26S proteasome, which catalyzes degradation of the substrate.
Ubiquitination requires activation of the monomeric Ub through formation of an initial thiol-ester bond with the E1 activating enzyme, followed by transfer of the thiol-ester-linked Ub to an E2 conjugating enzyme (7). Assembly of signaling Ub chains attached to a substrate is typically mediated by an E3 Ub ligase, owing to its capability to recognize both the substrate and an E2 enzyme (22). Two main classes of E3 are defined by the presence of either a HECT domain or a RING finger motif. While HECT E3s utilize a conserved catalytic cysteine residue forming a thiol-ester intermediate with Ub, RING E3s recruit the E2 enzyme via the RING domain and promote the transfer of Ub from E2 to a bound substrate (22).
The cullin (CUL)-based RING family of E3 Ub ligases (17, 19) is characterized by two signature components, namely, a CUL family molecule containing a conserved CUL domain (10, 36, 38) and its RING finger partner, ROC1/Rbx1/Hrt1 (9, 16, 25, 27, 29) (or APC11 in APC/C [16]). The prototypical SCF (Skp1-CUL1-F-box protein-ROC1) E3 utilizes CUL1 to assemble both an Skp1-F-box protein heterodimeric substrate-targeting module and a ROC1 RING-based Ub core ligase (18, 34, 39). The multiple interactions among SCF components place the F-box protein (a member of a large family of substrate-targeting molecules [8]) within the proximity of ROC1, which recruits an E2 conjugating enzyme (1, 25, 27). Consequently, a substrate, once bound to the F-box protein, is positioned optimally for accepting a Ub moiety in an E2-catalyzed transfer reaction. Ub transfer and the assembly of polyubiquitin chains are stimulated by Nedd8, which is conjugated to CUL1 at K720 (reviewed in reference 17).
Cdc34 is the class II E2 conjugating enzyme capable of working with SCF and the VHL E3 for ubiquitination in vitro (3, 15, 28, 29, 34). Saccharomyces cerevisiae Cdc34 (ScCdc34), encoded by an essential gene, promotes the G1-to-S-phase transition by mediating the Ub-dependent degradation of the Sic1 CDK inhibitor in a fashion depending on the SCFCdc4 E3 (5, 11). The human Cdc34 cDNA can functionally substitute for ScCdc34 (24), underscoring a functional conservation between the two orthologs.
Previous studies have identified multiple sites required for the function of Cdc34. Work with yeast (12, 26), subsequently confirmed for humans (33), has established the indispensable role of the C-terminal tail of Cdc34 in mediating interactions with SCF. In addition, conserved ScCdc34 S97 (S95 in human Cdc34) is essential for cell viability (14) and is required for supporting E2 monomer interactions or polyubiquitin chain assembly on the E2 protein (32). Other critical ScCdc34 sites include a conserved acidic insertion loop (residues 103 to 114 [residues 102 to 113 in humans]) (14, 23) and the charged cluster (residues D144, D148, R150, K151, and E154 [residues D143, R149, K150, and E153 in humans]) (23).
How Cdc34 assembles a Ub chain onto a substrate remains poorly understood. In an attempt to understand how an SCF substrate engages a Ub-charged Cdc34 (Cdc34∼S∼Ub) for nucleophilic attack, a diffusion-driven collision model was proposed, based on experimental data suggesting a requirement for prompt release of Ub-charged Cdc34 from SCF (2). However, an alternative view was presented in a more recent study, arguing that the productive interactions between SCF and Cdc34∼S∼Ub drive the transfer of Ub to a bound substrate (21). In addition, Petroski and Deshaies (20) recently proposed that attachment of the first Ub to the Sic1 substrate is a rate-limiting event. In their study, it was revealed that while the conserved ScCdc34 acidic loop is required for assembly of extensive polyubiquitin chains, it is dispensable for the attachment of the first Ub to Sic1.
One intriguing issue is whether Cdc34 functions in a dimeric form, which was originally proposed by Ellison and colleagues, who suggested that conjugation of a thiol-linked Ub promoted dimerization of ScCdc34 (32). We have reported that when placed into proximity by forced dimerization, human Cdc34 is evidently more active to catalyze Ub-Ub ligation (4). It was then proposed that juxtaposed human Cdc34 molecules might dimerize, creating a new binding site to properly position the acceptor Ub, reminiscent of the MMS2-Ubc13 heterodimer model (31). However, it is also possible that forced E2 dimerization may drive high local concentrations of Cdc34∼S∼Ub. This proximity effect could explain increased di-Ub synthesis, provided that one thiol-linked Ub could serve as the donor and the other could serve as the acceptor. Indeed, it was demonstrated recently that Ube2g2, an E2 conjugating enzyme with the acidic loop, requires two Ub-charged E2 molecules to catalyze di-Ub formation (13).
To further our understanding of Cdc34-catalyzed polyubiquitination, we have constructed and characterized a panel of mutant human Cdc34 enzymes. Our results have revealed intriguing properties of human Cdc34 in coordinating the attacking lysine, the donor, and the acceptor Ub for catalysis.
MATERIALS AND METHODS
Plasmids.
Cdc34 point mutants were created by site-directed mutagenesis with a QuikChange kit (Stratagene), using pET3a-HA-His-Cdc34 (33) as the template and the following primers: for Cdc34N85Q, CCAAGATGTGGCACCCTCAAATCTACGAGACGGGGGACG (5′) and CGTCCCCCGTCTCGTAGATTTGAGGGTGCCACATCTTGG (3′); for Cdc34Y87A, GGCACCCTAACATCGCCGAGACGGGGGACG (5′) and CGTCCCCCGTCTCGGCGATGTTAGGGTGCC (3′); for Cdc34S95D, CGGGGGACGTGTGTATCGACATCCTCCACCCGCCGG (5′) and GCCCCCTGCACACATAGCTGTAGGAGGTGGGCGGCC (3′); for Cdc34D102A/D103A, CCGCCGGTGGCCGCCCCCCAGAG (5′) and CTCTGGGGGGCGGCCACCGGCGG (3′); for Cdc34E108A/E112A, CAGAGCGGGGCGCTGCCCTCAGCGAGGTGGAAC (5′) and GTTCCACCTCGCTGAGGGCAGCGCCCCGCTCTG (3′); and for Cdc34S138A, CGAGCCCAACACCTTCGCGCCCGCAAACG (5′) and CGTTTGCGGGCGCGAAGGTGTTGGGCTCG (3′). Cdc34D143A/M147A/R149A/K150A/E153A (Cdc345A) was generated by two rounds of mutagenesis. In the first step, Cdc34D143A/M147A/K150A was created using the primers GCAAACGTGGCCGCCTCCGTGGCGTACAGGGCGTGGAAAGAGAGC (5′) and GCTCTCTTTCCACGCCCTGTACGCCACGGAGGCGGCCACGTTTGC (3′). The resulting DNA was then used as a template to construct Cdc345A by employing primers CCGTGGCGTACGCGGCGTGGAAAGCGAGCAAGGGGAAGG (5′) and CCTTCCCCTTGCTCGCTTTCCACGCCGCGTACGCCACGG (3′). Glutathione S-transferase (GST)-Cdc34 mutants were constructed using pGEX-GST-Cdc34 (4) as the template and the PCR primers described above. To create pFast-FKBP-L-Flag-Cdc34E108A/E112A, pFast-L-Flag-Cdc34 (4) was used as the template and the mutagenesis primers were as described above.
To generate Ub74, a stop codon was introduced at Ub Gly75 by PCR, using pET15-His-HA-PK-Ub (29) as the template and primers GGTCCTGCGTCTGAGATGAGGTTAAGGATCCGGCTGC (5′) and CCAGGACGCAGACTCTACTCCAATTCCTAGGCCGACG (3′).
To construct pFast-Flag-IKKβS177E/S181E, pRC-β-actin-HA-IKKβS177E/S181E (see reference 29 and references therein) was used as a template for PCR with the primers AAAAAGCTTGATTATAAAGATGATGATGATAAAATGAGCTGGTCACCTTCCC (5′) and AAAAAGCTTTCATGAGGCCTGCTCCAGG (3′). The resulting fragment, deleted of the hemagglutinin (HA) tag but containing the Flag epitope at the N terminus of IKKβS177E/S181E, was then subcloned into the pFast-Bac1 vector (Bac-to-Bac baculovirus expression system; Invitrogen), creating pFast-Flag-IKKβS177E/S181E.
To construct pGEX-2TK-IκBα(1-54), the DNA sequence containing the first 54 amino acids of IκBα was amplified by PCR (Expand High Fidelity PCR system; Roche) from pGEX-IκBα(1-54) (see reference 29 and references therein), using a BamHI restriction site-containing 5′ primer, ACGGGATCCCGTATGTTCCAGGCGGC, and an EcoRI restriction site-containing 3′ primer, GGAATTCCTCAGAGGCGGATCTCCTG, per the manufacturer's protocols (restriction sites are shown in bold). The ends were digested with the BamHI and EcoRI enzymes overnight and ligated into a similarly digested pGEX-2TK vector (Amersham Biosciences).
All constructs were verified by DNA sequencing.
Baculoviruses.
To construct baculoviruses expressing Flag-IKKβS177E/S181E and FKBP-L-Flag-Cdc34E108A/E112A, pFast-Flag-IKKβS177E/S181E and pFast-FKBP-L-Flag-Cdc34E108A/E112A, respectively, were transformed into competent DH10Bac cells. The bacmid DNAs were isolated following the manufacturer's instructions. Baculoviruses were prepared as described previously (4).
Proteins.
The wild-type and mutant forms of human Cdc34, encoded in the pET3a-HA-His-Cdc34 vector, were expressed in bacteria and purified as previously described (33), with modifications. Following affinity purification with Ni-nitrilotriacetic acid (QIAGEN), imidazole-eluted Cdc34 was dialyzed against buffer A (25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.01% Nonidet P-40, 10% glycerol, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg/ml of antipain and leupeptin) plus 100 mM NaCl. The resulting material was chromatographed on a Mono Q HR 10/10 column (Amersham Pharmacia Bioscience), and bound proteins were eluted with a gradient of 100 to 500 mM NaCl in buffer A. Cdc34 peaked at about 360 mM NaCl, as determined by Coomassie staining following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After concentration with a centrifugal filter device (Millipore), Cdc34 was filtrated by fast-performance liquid chromatography, using a Superose 6 tricorn column (Amersham Pharmacia Bioscience) preequilibrated with buffer A plus 150 mM NaCl. Cdc34 migrated at a position similar to that of bovine serum albumin (BSA). All Cdc34 point mutants behaved identically to the wild type. The yield for wild-type and mutant Cdc34 was between 0.5 and 1 mg per 2 liters of culture.
GST-Cdc34 and mutants, encoded in pGEX-GST-Cdc34, were expressed in bacteria and purified as previously described (4). All of the mutants behaved identically to the wild-type enzyme and possessed Stokes radii consistent with dimers (4). The yield for GST-Cdc34 and mutants was 0.5 to 1 mg per 1 liter of culture.
For expression of His-HA-PK-UbK48R or His-HA-PK-Ub74, pET15-His-HA-PK-UbK48R or pET15-His-HA-PK-Ub74 was transformed into pJY2 (Affiniti)-containing Escherichia coli BL21(DE3) cells. Induction and purification (through Ni-nitrilotriacetic acid) were identical to those described for the preparation of Cdc34 (33). Purified His-HA-PK-Ub74 was kept in phosphate-buffered saline plus 1 mM DTT. The yield for Ub74 was 20 mg (5 mg/ml) per 3 liters of culture.
FKBP-L-Flag-Cdc34E108A/E112A was expressed in infected High Five cells (two T150 flasks) treated with or without AP20187. Both the free and AP20187-bound dimeric FKBP-L-Flag-Cdc34E108A/E112A proteins were purified through an M2 affinity matrix. Detailed procedures were described previously (4). The yields of monomeric and dimeric FKBP-L-Flag-Cdc34E108A/E112A were 75 μg (0.75 mg/ml) and 100 μg (1 mg/ml), respectively.
For large-scale purification of Flag-IKKβS177E/S181E, 3 × 107 Sf9 cells were infected (multiplicity of infection [MOI] = 10) with baculovirus for 48 h at 27°C. Cells were harvested, extract was prepared, and the protein was purified as described previously (4).
SCFβTrCP2, ROC1-CUL1324-776, PK-Ub, Nedd8, and Ubc12 were prepared using procedures described previously (29, 33). GST-2TK-IκBα(1-54) was expressed in bacteria and purified by affinity chromatography using a glutathione matrix followed by gel filtration using a Superose 6 tricorn column (Amersham Pharmacia Bioscience).
Preparation of 32P-GST-IκBα(1-54) for ubiquitination.
To prepare IKKβ-phosphorylated 32P-GST-2TK-IκBα(1-54), we used a two-step procedure. Initially, GST-2TK-IκBα(1-54) was phosphorylated by IKKβ in a reaction mixture (100 μl) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 0.5 mM DTT, 1 mM ATP, 0.1 mg/ml of BSA, GST-2TK-IκBα (23 μg), and Flag-IKKβS177E/S181E (0.1 μg). The reaction mix was incubated for 15 min at 37°C. A second aliquot of kinase (0.1 μg) was added, and the incubation was continued for 15 min at 37°C. EDTA was then added to a final concentration of 10 mM. The IKKβ-phosphorylated GST-2TK-IκBα was adsorbed to glutathione beads (30 μl), and the resulting matrix was washed to remove IKKβ and ATP. The bound IKKβ-phosphorylated GST-2TK-IκBα was then 32P labeled by cyclic AMP kinase in a reaction mixture (100 μl) that contained 20 mM Tris-HCl (pH 7.4), 12 mM MgCl2, 50 mM NaCl, 25 μM ATP, 0.1 mg/ml of BSA, 2 mM NaF, [γ-32P]ATP (50 μCi), and cyclic AMP kinase (10 units; Sigma). The reaction mix was incubated at 37°C for 30 min on a thermomixer (1,300 rpm; Eppendorf). Following washing of the beads, the bound 32P-labeled substrate was eluted with buffer A plus 50 mM NaCl and 20 mM glutathione. The eluate was adjusted by buffer exchange to contain 25 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.01% NP-40, 10% glycerol, and 50 mM NaCl. After concentration, the 32P-GST-2TK-IκBα concentration was estimated to be 0.5 pmol per μl. The yield for this procedure is about 20%.
Neddylation.
SCFβTrCP2 (∼0.5 pmol) bound to anti-HA beads (20 μl) or ROC1-CUL1324-776 was neddylated in a reaction mixture (final volume of 30 μl) that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 0.1 mg/ml BSA, Nedd8 (0.5 μM), APP-BP1/Uba3 (3.3 nM), and Ubc12 (1.5 μM). The mixture was incubated for 5 min at room temperature.
Ubiquitination.
Neddylated SCFβTrCP2 (∼25 nM or indicated amount) bound to anti-HA beads (20 μl) was incubated in a mixture that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 0.1 mg/ml BSA, 32P-GST-2TK-IκBα (20 nM), Ub (8 μM), E1 (10 nM), and Cdc34 or mutant (in the indicated amounts). The reaction mix was incubated at 37°C for 60 min or specified times on a thermomixer (1,300 rpm; Eppendorf). The reaction products were visualized by autoradiography after separation by 4 to 20% SDS-PAGE.
Di-Ub synthesis. (i) Multiple-turnover assay.
The reaction mixture (30 μl) contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 0.1 mg/ml BSA, 32P-UbK48R (50 pmol), bovine Ub (bUb; 150 pmol), E1 (0.6 pmol), and the indicated amount of Cdc34 or mutant Cdc34. For reactions carried out with the neddylated ROC1-CUL1 complex, purified ROC1-CUL1324-776 (1 μg) was preincubated with Nedd8 (75 pmol), APP-BP1/Uba3 (0.15 pmol), and Ubc12 (25 pmol) in the above reaction mixture lacking 32P-Ub, E1, and Cdc34 for 5 min at room temperature. In all cases, ubiquitination was initiated by the addition of Cdc34 and the reaction was incubated for the indicated times at the specified temperature. Where indicated, DTT was added to a final concentration of 0.1 M prior to the addition of SDS loading buffer (10 μl). Aliquots were separated by 4 to 20% SDS-PAGE followed by autoradiography. Quantitation of the ubiquitination products was performed using a phosphorimager (Bio-Rad).
(ii) Single-turnover pulse-chase analysis.
Pulse-chase was carried out in two steps. Initially, an E2 charging reaction was assembled in a mixture (5 μl) that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM NaF, 10 nM okadaic acid, 2 mM ATP, 0.6 mM DTT, 0.1 mg/ml BSA, 32P-UbK48R (12 μM), E1 (0.1 μM), and human Cdc34 or mutant Cdc34 (4 μM). The reaction mix was incubated for 5 min at 37°C. EDTA (1 μl at 0.5 M) was added to a final concentration of 50 mM (in 10 μl). The reaction (in a final volume of 10 μl) was then chased with Ub74 or bUb (125 μM) for the indicated times at 37°C.
Ub conjugation assay.
To measure the conjugation of Ub to GST-Cdc34 or mutants, the reaction was carried out in a mixture (30 μl) that contained 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 0.6 mM DTT, 2 mM ATP, 2 mM NaF, 10 nM okadaic acid, 0.1 mg/ml BSA, 32P-Ub (200 pmol), E1 (0.5 pmol), and E2 (20 pmol). The reaction mix was incubated at 16°C for 30 min. Where indicated, DTT (0.1 M) was added. One-half of the reaction mixture was then separated by 4 to 20% SDS-PAGE, and the product was visualized by autoradiography.
RESULTS
Construction of a panel of human Cdc34 mutants.
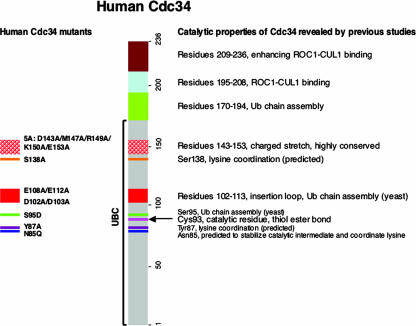
In order to execute catalysis, human Cdc34 must possess several functional sites that mediate multifaceted interactions to accommodate SCF, and hence the substrate, as well as the thiol-linked donor Ub and the “attacking” acceptor Ub. Previous mutagenesis studies, carried out predominantly with budding yeast, have revealed a significant role for the Cdc34 C terminus in interactions with SCF (12, 26, 32) and have identified multiple sites within the UBC domain required for the Cdc34 function (14, 20, 23, 32), as summarized in Fig. 1. In a previous study by Pickart and colleagues (35), a conserved asparagine residue (N85 in human Cdc34) was proposed to play a role in contributing to formation of an oxyanion hole that stabilizes the negatively charged transition state during conjugation. In a recently published elegant study by Yunus and Lima (37), the SUMO E2 conjugating enzyme Ubc9 uses N85, Y87, and D127 to coordinate the attacking lysine residue for catalysis, yielding an isopeptide bond linkage between SUMO and a target protein, such as p53 or RanGap1. Yunus and Lima suggested that this lysine coordination may be conserved and that in human Cdc34, N85, Y87, and S138 may carry out such an activity.
FIG. 1.
Functional domains of human Cdc34. A schematic representation of domains within human Cdc34 which are responsible for catalysis, multi-Ub chain assembly, and interactions with ROC1-CUL1 for the efficient synthesis of Ub polymers is shown. Mutants made and analyzed in this study are indicated on the left, with their positions marked.
These studies prompted us to construct and characterize a panel of human Cdc34 mutants (N85Q, Y87A, S95D, D102A/D103A, E108A/E112A, S138A, and 5A [D143A/M147A/R149A/K150A/E153A] mutants) (Fig. 1). All mutant human Cdc34 proteins were expressed in bacteria and purified to apparent homogeneity by affinity chromatography, Mono Q ion exchange, and gel filtration (see Materials and Methods).
Inhibition of ligation of Ub to the IκBα substrate lysine by mutations at human Cdc34 S95 or E108/E112.
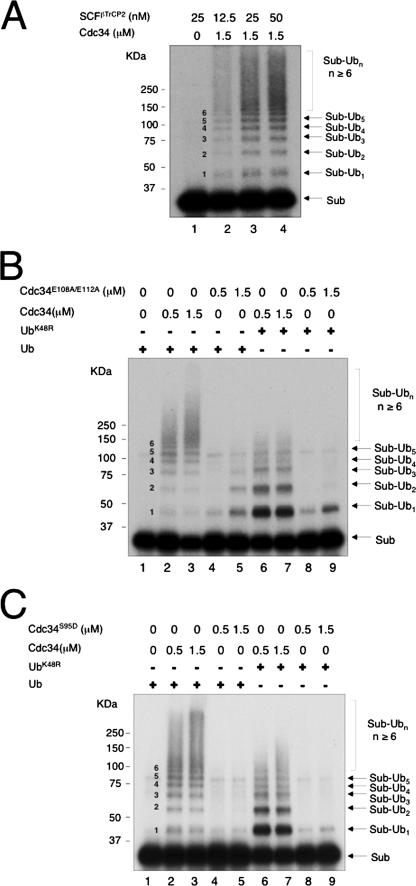
We first measured and compared the effects of purified human Cdc34 and mutants on ubiquitination of IκBα by using a reconstituted system as described previously (29, 34). This assay employed 32P-labeled, IKKβ-phosphorylated IκBα as a substrate, immunopurified SCFβTrCP2 (comprised of Skp1, neddylated CUL1, βTrCP2/HOS, and ROC1) as E3, human Cdc34 or a mutant as E2, a saturating amount of E1, and Ub. Autoradiographic analysis revealed that when wild-type Cdc34 was used, substrate-Ub conjugates formed a laddered pattern in an SCFβTrCP2 (Fig. 2A) and E2 (Fig. 2B, lanes 2 and 3) concentration-dependent fashion. In contrast, Cdc34E108A/E112A poorly supported the polyubiquitination reaction (Fig. 2B, lanes 4 and 5). To evaluate the ability of wild-type and mutant E2 to mediate the ligation of the substrate lysine to Ub, we used UbK48R in place of wild-type Ub to inhibit polyubiquitin chain assembly. The results showed that Cdc34E108A/E112A was much less effective than the wild type in supporting mono-ubiquitination (Fig. 2B, compare lanes 6 and 7 with lanes 8 and 9). These data demonstrated that the E108A/E112A substitution in human Cdc34 inhibited both poly- and monoubiquitination. Notably, while wild-type Cdc34 formed polyubiquitinated chains as early as 5 min after incubation (Fig. 3C and D and 4C and D), Cdc34E108A/E112A yielded no detectable high-molecular-weight Ub conjugates after 60 min (Fig. 2B, lane 5), further underscoring the pronounced catalytic defects of this mutation. Similarly, replacement of S95 with Asp rendered human Cdc34 inactive in the poly- and monoubiquitination of IκBα (Fig. 2C). Altogether, these results established that the integrity of S95 and E108/E112 is required for the ability of human Cdc34 to catalyze the ligation of the attacking substrate lysine to Ub.
FIG. 2.
Ligation of Ub to IκBα by SCFβTrCP2 requires human Cdc34 S95 and E108/E112. The ubiquitination reaction was carried out as described in Materials and Methods, with titration of SCFβTrCP2 (A) and comparison of wild-type Cdc34 with Cdc34E108A/E112A (B) or Cdc34S95D (C) in the poly- and monoubiquitination of IκBα by SCFβTrCP2. The reaction products are indicated, with the number of Ub moieties attached to the substrate marked in the middle.
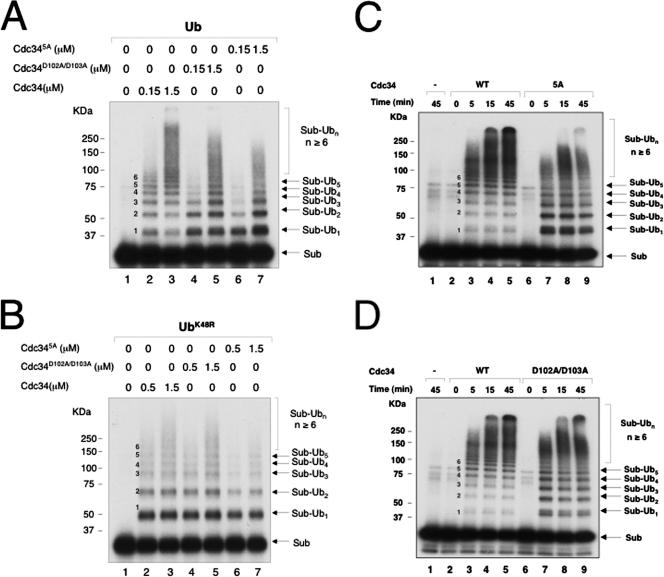
FIG. 3.
Human Cdc34 D102/D103 and D143/M147/R149/K150/E153 are required for polyubiquitination of IκBα by SCFβTrCP2. Reactions were carried out as described in Materials and Methods. The effects of alanine substitutions at human Cdc34 D102/D103 or D143/M147/R149/K150/E153 were analyzed by E2 titration experiments with wild-type Ub (A) or UbK48R (B) and by time course analyses (C and D) with 1.5 μM of E2. The reaction products are indicated, with the number of Ub moieties attached to the substrate marked in the middle.
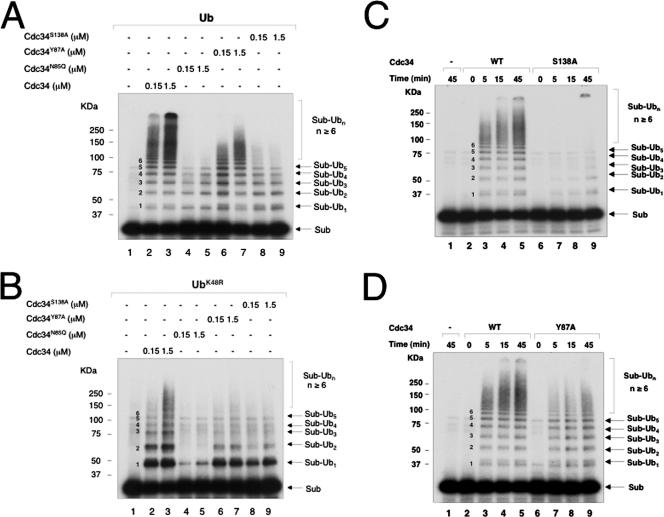
FIG. 4.
Inhibitory effects of Cdc34N85Q, Cdc34Y87A, and Cdc34S138A in the polyubiquitination of IκBα by SCFβTrCP2. The reactions were carried out as described in Materials and Methods. The effects of mutations at human Cdc34 N85, Y87, and S138 were analyzed by E2 titration with wild-type Ub (A) or UbK48R (B) and by kinetic experiments (C and D) with 1.5 μM of E2. The reaction products are indicated, with the number of Ub moieties attached to the substrate marked in the middle.
We previously observed that human Cdc34 formed polyubiquitinated IκBα through Ub lysine residues other than K48 with a very slow kinetics (34). In keeping with this observation, human Cdc34 was found to support only low levels of polyubiquitination of IκBα (Fig. 2B and C, compare lanes 1 and 2 with lanes 6 and 7). Note that the UbK48R preparation used in this study was derived from the BL21(DE3)/pJY2 strain, which eliminates misincorporation of lysine for arginine. In addition, the inefficient use of UbK48R for Ub chain assembly was also observed with yeast Cdc34 in the polyubiquitination of Sic1 (30).
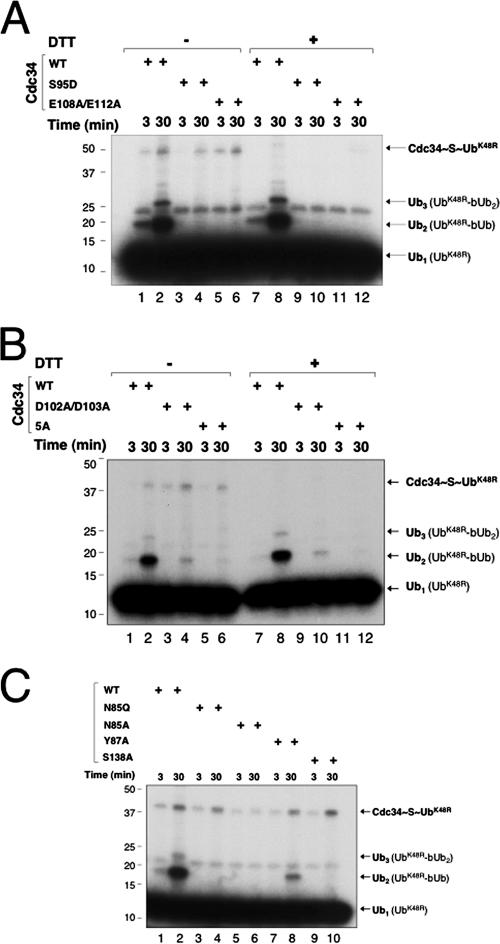
Mutations at D102/D103 or D143/M147/R149/K150/E153 inhibit the ability of human Cdc34 to form a polyubiquitin chain on IκBα.
Mutations at D102/D103 or D143/M147/R149/K150/E153 in human Cdc34, however, yielded phenotypes that substantially differed from those observed with S95D and E108A/E112A mutants. Cdc34D102A/D103A or Cdc34D143A/M147A/R149A/K150A/E153A (Cdc345A) accumulated mono-ubiquitinated IκBα while inhibiting the formation of polyubiquitin chain formation (Fig. 3A). When UbK48R was used in place of wild-type Ub, these mutants supported mono-ubiquitination of IκBα with an efficiency comparable to that of wild-type E2 (Fig. 3B). Further kinetic analyses confirmed the accumulation of monoubiquitinated IκBα by Cdc345A or Cdc34D102A/D103A, albeit Cdc345A appeared to increase mono-ubiquitinated IκBα to higher levels than did Cdc34D102A/D103A (Fig. 3C and D). Collectively, these results indicated that mutations at D102/D103 or D143/M147AR149/K150/E153 retained the ability of human Cdc34 to catalyze the ligation of a substrate lysine to Ub while severely compromising the E2's capability to support the formation of polyubiquitin chains.
Cdc34 N85Q, Y87A, and S138A mutants inhibit IκBα ubiquitination to various degrees.
We next analyzed human Cdc34N85Q, Cdc34Y87A, and Cdc34S138A by examining their effects in supporting IκBα polyubiquitination (Fig. 4A, C, and D) or monoubiquitination (Fig. 4B) in both E2 titration (Fig. 4A and B) and kinetics (Fig. 4C and D) experiments. Cdc34N85Q appeared to resemble Cdc34E108A/E112A and Cdc34S95D in abolishing both polyubiquitination (Fig. 4A, compare lanes 2 and 3 with lanes 4 and 5) and monoubiquitination (Fig. 4B, compare lanes 2 and 3 with lanes 4 and 5). Cdc34S138A still supported monoubiquitination, albeit at significantly lower levels than those of the wild type (Fig. 4B, compare lanes 2 and 3 with lanes 8 and 9). However, this mutation severely impaired Cdc34's ability to catalyze polyubiquitin chain formation (Fig. 4C). Mutation at Y87 exhibited a lesser inhibitory effect, as Cdc34Y87A was able to catalyze polyubiquitination, although with slower kinetics than that of the wild type (Fig. 4D). Note that unlike Cdc345A or Cdc34D102A/D103A, neither Cdc34N85Q, Cdc34Y87A, nor Cdc34S138A accumulated monoubiquitinated IκBα (compare Fig. 4 with Fig. 3C and D).
Cdc34N85Q, Cdc34Y87A, Cdc34S95D, Cdc34D102A/D103A, Cdc34E108A/E112A, Cdc34S138A, and Cdc345A are all defective in Ub-Ub ligation.
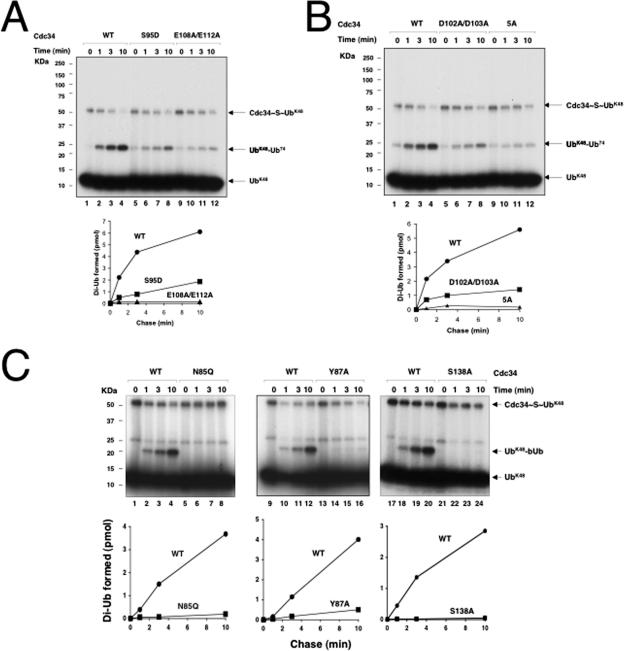
We determined whether the above-mentioned mutations influenced the ability of E2 to catalyze Ub-Ub ligation by first employing the multiple-turnover di-Ub synthesis assay. In this system, 32P-UbK48R was used as the donor and bUb was used as the acceptor in the presence of ROC1-CUL1-Nedd8, which stimulated di-Ub synthesis by a factor of 20 (4). As shown, all of the Cdc34 mutants (Cdc34S95D, Cdc34E108A/E112A, Cdc34D102A/D103A, Cdc345A, Cdc34N85Q, Cdc34Y87A, and Cdc34S138A) retained the ability to form E2∼S∼Ub (Fig. 5A, lanes 1 to 6, B, lanes 1 to 6, and C). However, no detectable di-Ub synthesis was observed with Cdc34S95D (Fig. 5A, lanes 3, 4, 9, and 10), Cdc34E108A/E112A (Fig. 5A, lanes 5, 6, 11, and 12), Cdc345A (Fig. 5B, lanes 5, 6, 11, and 12), Cdc34N85Q or Cdc34N85A (Fig. 5C, lanes 3 to 6), and Cdc34S138A (Fig. 5C, lanes 9 and 10). Cdc34D102A/D103A (Fig. 5B, lanes 3, 4, 9, and 10) and Cdc34Y87A (Fig. 5C, lanes 7 and 8) supported di-Ub formation with much lower efficiencies than that of the wild type.
FIG. 5.
Effects of human Cdc34 mutants in Ub-Ub ligation, as measured by multiple-turnover di-Ub synthesis assay. Time course experiments were carried out to measure the ligation of 32P-UbK48R to bUb, as described in Materials and Methods, with 4 μM of wild-type human Cdc34 or mutant Cdc34, as specified. Where indicated, DTT was added (0.1 M) after the incubation to disrupt the thiol-ester. The reaction products are indicated.
We next developed a single-round pulse-chase assay in which Cdc34∼S∼32P-UbK48 was initially formed and Ub charging was then terminated by the addition of EDTA (50 mM; at this concentration, no Cdc34∼S∼Ub was detected [data not shown]). When chased with Ub74 (can only serve as an acceptor Ub), di-Ub was produced (Fig. 6A, lanes 2 to 4). As shown, Cdc34S95D (Fig. 6A, lanes 6 to 8), Cdc34E108A/E112A (Fig. 6A, lanes 10 to 12), Cdc34D102A/D103A (Fig. 6B, lanes 6 to 8), and Cdc345A (Fig. 6B, lanes 10 to 12) were all defective in supporting the synthesis of di-Ub, with Cdc34E108A/E112A and Cdc345A having the most pronounced defects (see graphs for a quantitative comparison).
FIG. 6.
Effects of human Cdc34 mutants in Ub-Ub ligation, as measured by a single-round pulse-chase assay. Human Cdc34 or mutant Cdc34 (4 μM), as specified, was charged with 32P-UbK48R and chased with either Ub74 (A and B) or bUb (C), using a protocol described in detail in Materials and Methods. The reaction products are indicated. Formation of di-Ub by wild-type or mutant Cdc34 was quantitated and expressed graphically.
A similar single-round pulse-chase assay was carried out to analyze Cdc34N85Q, Cdc34Y87A, and Cdc34S138A, with the exception that bUb was used in place of Ub74. As shown, the synthesized 32P-UbK48R-bUb dimer can easily be distinguished from the intrinsic radioactive UbK48R dimeric material, given their different gel mobilities (Fig. 6C, lanes 1 to 4). The results showed that all three mutants were inactive (Fig. 6C, lanes 6 to 8, 14 to 16, and 22 to 24; see graphs for quantitative comparisons).
Taken together, these results demonstrated that mutations at N85, Y87, S95, E108/E112, D102/D103, S138, and D143/M147AR149/K150/E153 inhibited the ability of human Cdc34 to catalyze the ligation of Ub molecules.
GST fusion reverses catalytic defects caused by mutation at human Cdc34 S95 or E108/E112.
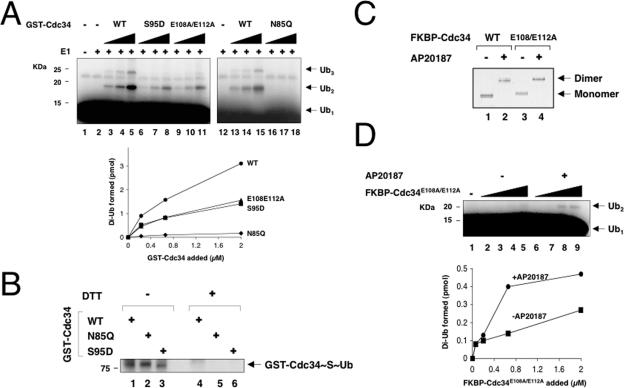
To further our understanding of the catalytic properties of mutants at human Cdc34 N85, S95, and E108/E112, we asked whether the enzymatic defects of these mutants can be overcome through GST fusion, which activates wild-type human Cdc34, presumably by dimerization (4). Purified GST-fused Cdc34, Cdc34S95D, Cdc34E108A/E112A, and Cdc34N85Q possessed identical Stokes radius values, consistent with their existence as dimers (4). Interestingly, both GST-Cdc34S95D and GST-Cdc34E108A/E112A supported the synthesis of di-Ub with an efficiency of 50% that of the wild type (Fig. 7A, compare lanes 3 to 5 with lanes 6 to 8 as well as lanes 9 to 11; see the graph for quantitation). In contrast, GST-Cdc34N85Q was inactive (Fig. 7A, compare lanes 13 to 15 and lanes 16 to 18), despite the fact that this fusion enzyme supported the formation of E2∼S∼Ub as efficiently as the wild type did (Fig. 7B, compare lanes 1 and 2). Thus, GST fusion selectively reversed the catalytic defects of Cdc34S95D and Cdc34E108A/E112A.
FIG. 7.
Effects of E2 dimerization in reversing the catalytic defects of Cdc34N85Q, Cdc34S95D, and Cdc34E108A/E112A. (A) GST fusion restored the ability of Cdc34S95D and Cdc34E108A/E112A, but not Cdc34N85Q, to catalyze di-Ub synthesis. The di-Ub synthesis reaction was measured by the multiple-turnover assay as described in Materials and Methods, with increasing amounts of wild-type or mutant GST-Cdc34 as indicated. The reaction mix was incubated at 37°C for 60 min. Formation of di-Ub by wild-type or mutant GST-Cdc34 was quantitated and expressed graphically. (B) GST-Cdc34N85Q forms thiol-ester conjugates with Ub. A Ub conjugation assay was performed as described in Materials and Methods. Where indicated, DTT was added (0.1 M) after the incubation to disrupt the thiol-ester. (C) Dimerization of FKBP-Cdc34E108A/E112A in the presence of AP20187. Native gel electrophoresis was performed with purified FKBP-Cdc34 and FKBP-Cdc34E108A/E112A (1 μg) isolated from infected High Five cells treated with or without AP20187. (D) FKBP-based dimerization stimulates the ability of Cdc34E108A/E112A to synthesize di-Ub. The di-Ub synthesis reaction was measured by the multiple-turnover assay as described in Materials and Methods, with increasing amounts of the monomeric or dimeric (AP20187-bound) form of FKBP-Cdc34E108A/E112A. The reaction mix was incubated at 37°C for 60 min. Formation of di-Ub by monomeric or dimeric FKBP-Cdc34E108A/E112A was quantitated and expressed graphically.
Next, we employed the FKBP homodimerization system, which has been used previously to demonstrate that dimerized FKBP-Cdc34 (induced by the AP20187 chemical dimerizer) is more active in di-Ub synthesis than the monomeric form (4). For this purpose, we constructed baculoviruses expressing FKBP-Flag-Cdc34E108A/E112A and FKBP-Flag-Cdc34S95D. However, for reasons that are unclear to us, FKBP-Flag-Cdc34S95D was expressed poorly, precluding our assessment of its catalytic properties. FKBP-Flag-Cdc34E108A/E112A was expressed in infected insect cells treated with or without AP20187 and then affinity purified. Native gel electrophoresis experiments showed that AP20187-treated FKBP-Flag-Cdc34 or FKBP-Flag-Cdc34E108A/E112A migrated significantly slower than the untreated protein (Fig. 7C), confirming that FKBP-Flag-Cdc34E108A/E112A bound with AP20187 is a dimer.
When assayed for di-Ub synthesis, the AP20187-treated, dimeric form of FKBP-Flag-Cdc34E108A/E112A was more active than the monomeric form (Fig. 7D). However, unlike GST-Cdc34E108A/E112A, which possessed 50% of the wild-type activity (Fig. 7A), dimeric FKBP-Flag-Cdc34E108A/E112A was only 10% as active as the wild type (data not shown). The basis for the difference between the GST- and FKBP-based dimerization systems in the extents of activation of Cdc34E108A/E112A in di-Ub synthesis is presently unclear. Possibly, in the case of GST-Cdc34E108A/E112A or GST-Cdc34S95D, the GST dimer may place the two Cdc34 mutant molecules sufficiently close to form a dimer, thus largely bypassing the requirement for S95 and/or E108/E112. However, in the case of the AP20817-bound FKBP dimer, the orientation of the E2 enzyme might be less favorable to form a dimer without the assistance of E108/E112. In all, these results indicate that mutations at S95 and E108/E112 can be reactivated by GST fusion and that FKBP-based dimerization also activates Cdc34E108A/E112A, albeit to a limited degree.
DISCUSSION
Coordination of the acceptor Ub for assembly of polyubiquitin chains.
Our mutagenesis studies have revealed an intriguing role played by the highly conserved charged stretch (residues 143 to 153) within the UBC domain of human Cdc34 (Fig. 1). Disruption of this site by quintuple mutations (5A) resulted in the accumulation of monoubiquitinated IκBα while yielding far fewer polyubiquitin chains than did wild-type Cdc34 (Fig. 3A and C). This defect is most likely due to a catalytic deficiency in the assembly of polyubiquitin chains, as Cdc345A failed to catalyze Ub-Ub ligation measured in both multiple (Fig. 5B, lanes 5 and 6)- and single (Fig. 6B, lanes 9 to 12)-turnover di-Ub synthesis assays. Cdc34D102A/D103A exhibited defects similar to those of Cdc345A in accumulating mono-ubiquitinated IκBα, albeit to a lesser extent (Fig. 3A, lanes 4 and 5, and D, lanes 7 to 9). These data suggest that Cdc345A and Cdc34D102A/D103A are fully, or near fully, capable of accommodating the attacking lysine from IκBα and aligning the Cd34∼S∼Ub thiol-ester to orchestrate chemical catalysis, resulting in an isopeptide bond linking Ub to the substrate. However, these mutants were inadequate in coordinating the attack by the acceptor Ub on the Cd34∼S∼Ub thiol-ester and hence ineffective at producing a Ub chain with (Fig. 3) or without (Fig. 5B and 6B) attachment to a substrate. Whether human Cdc34 D102/D103 and the charged stretch (residues 143 to 153) represent elements of an E2 functional site that anchors/positions the acceptor Ub awaits future structural and biochemical work. Consistent with our findings, it was previously observed that a yeast Cdc34 mutant (D144A/D148A/R150A/K151A/E154A) was able to catalyze the ligation of Ub to histones but failed to support the assembly of polyubiquitin chains (23). Notably, this mutant exhibited only a slight growth defect at low temperatures (23). Possibly, multiple Cdc34-mediated contacts are involved in accommodating the acceptor Ub and a single site defect can be tolerated in vivo.
Whatever the precise mechanism, our findings are consistent with a view that coordination of the acceptor Ub is a major determinant for polyubiquitination catalyzed by Cdc34. This coordination must depend on formation of a pocket/surface for accommodating the acceptor Ub, which allows the precise positioning of K48 in the vicinity of the carbonyl oxygen of Gly76 of the donor Ub thiol bonded to Cdc34. Cdc34 alone is capable of catalysis to produce di-Ub (Fig. 6). However, the binding of a neddylated SCF-substrate complex may trigger a set of interactions that influence Cdc34's positioning of the acceptor Ub, enhancing both the efficiency and length of polyubiquitin chain assembly. Indeed, it has been shown that neddylated SCF markedly stimulates human Cdc34's ability to catalyze di-Ub synthesis and polyubiquitin chain assembly (4). Moreover, SCF was shown to accelerate the discharge of Ub from yeast Cdc34 to synthesize di-Ub (20). Altogether, these findings have led us to speculate that the decision for mono-ubiquitination versus polyubiquitination, at least in RING E3-mediated reactions, may be determined predominantly by the multifaceted interactions between E2, E3, and an E3-bound substrate, which either permit or disallow accommodation of the acceptor Ub.
Accommodation of the attacking lysine and the donor Ub for catalysis.
As revealed by this study, side chains from human Cdc34 N85, S95, E108/E112, and S138 are all involved in the chemical attack of the striking lysine residue on Cdc34∼S∼Ub. Mutational substitution at these sites inhibited mono-ubiquitination of IκBα (Fig. 2B and C and 4A to C) and blocked Ub-Ub ligation (Fig. 5A and C and 6A and C, lanes 5 to 8).
The attachment of the first Ub to IκBα requires optimal orientation of Cdc34∼S∼Ub proximal to the attacking lysine (K21 or K22). Human Cdc34 N85, a highly conserved residue in all E2 enzymes, may have a dual role in catalysis. On the one hand, this residue has been proposed to stabilize the oxyanion formed during the attack of lysine on E2∼S∼Ub (35). Consistent with this view, Ubc9 N85 is found proximal to the carbonyl oxygen of the terminal glycine in a putative product complex, Ubc9-Nup358/RANBP2-RanGAP1-SUMO (37). On the other hand, human Cdc34 N85, along with Y87 and S138, was implicated as having a role in positioning the attacking lysine within the active site and activating the nucleophile (37). Our mutagenesis studies are in agreement with the proposed role for human Cdc34 N85 and S138 in catalysis. However, human Cdc34Y87A was still active in monoubiquitination of IκBα (Fig. 4B, compare lanes 2 and 3 with lanes 6 and 7) but was defective in producing polyubiquitin chains (Fig. 4A and D) and in di-Ub synthesis (Fig. 5C and 6C), suggesting a function for the Y87 side chain in Ub chain assembly. It should be mentioned that the yeast Cdc34 S139A mutant (equivalent to the human Cdc34 S138A mutant) exhibited no growth defects (14). In contrast, the biological significance of the conserved asparagine has been demonstrated for Ubc13 (35) and Ubc9 (37). In all, these findings argue strongly for the essential role of the conserved asparagine residue in E2 catalysis.
Human S95 and E108/E112 may have distinct roles in the positioning of the donor Ub for catalysis. One possibility is that human Cdc34 S95, positioned near the catalytic residue C93, participates in stabilizing/positioning the C-terminal tail of the donor Ub. In the nuclear magnetic resonance structural model of Ubc1(1-150)∼S∼Ub (6), the Ub C-terminal tail, L71 to G76, is positioned in a shallow cleft formed by two Ubc1 segments, namely, L89 to I91, immediately upstream of the catalytic residue C88, and N119 to P121. Moreover, this E2-Ub interface is supported by hydrogen bonding interactions involving side chains from Ubc1 E117 and Ub R72. In addition, Ubc9 S95, adjacent to the catalytic residue C93, is hydrogen bonded to SUMO T95, presumably stabilizing the SUMO Gly96-Gly97 bond for catalysis (37). Cdc34S95D was shown to possess 50% of the wild-type activity in conjugating Ub (Fig. 5A, compare lanes 2 and 4), which is consistent with that observed with the equivalent ScCdc34 mutant (32). However, Cdc34S95D exhibited <10% of the wild-type activity in supporting IκBα mono-ubiquitination (Fig. 2C) and in Ub-Ub ligation (Fig. 5A and 6A). Taking these results together, it can be speculated that human Cdc34 S95 contacts the Ub's C-terminal tail, thereby contributing to the formation of E2∼S∼Ub with maximal efficiency and to the positioning of the donor Ub's Gly76 optimally for catalysis. It remains possible that the side chains from human Cdc34 E108/E112 engage in hydrogen bonding/electrostatic interactions with the C-terminal tail of the donor Ub (e.g., R74 and/or R72).
However, a direct role played by human S95 and/or E108/E112 in catalysis, as proposed above, appears at odds with the results of our GST fusion experiments and with previous studies on ScCdc34. Surprisingly, the catalytic defects of Cdc34E108/E112A and Cdc34S95D in di-Ub synthesis can be rescued by fusion of GST at the N terminus of the enzyme (Fig. 7A). It was shown previously that in yeast, a Cdc34 S97D mutation (equivalent to human Cdc34 S95D mutation) resulted in a nonviable phenotype (14) and the mutant E2 was defective in self-association and in multi-Ub chain assembly (32). In addition, mutant K (E109A/D111A/E113A [23]; similar to human Cdc34E108A/E112A used in this study) and an acidic loop deletion mutant (14) were devoid of Cdc34 function. Interestingly, however, overexpression of mutant K rescued Cdc34 activity (23). Even more strikingly, the defects of the loop deletion mutant can be overcome by combined S97D and S73K mutations (14). The observed intragenic suppression of Cdc34 mutants argues for a functional interaction between these positions. In all, these findings appear to favor a crucial, but indirect, role for human Cdc34 S95 and E108/E112 in catalysis. It may be that these residues are responsible for inducing E2 dimerization, which, as we have shown previously, stimulates Ub-Ub ligation (4). Forced Cdc34 dimerization might help to create a new binding surface critical for proper positioning of the donor Ub. Clearly, the merit of this hypothesis awaits future investigation at both the biochemical and structural levels.
Notably, the human Cdc34 acidic loop appears to be associated with two activities. It utilizes residues including D102/D103 to assist in polyubiquitin chain assembly, a function conserved in ScCdc34. In addition, the human Cdc34 acidic loop contributes to the transfer of the thiol-linked Ub to the attacking lysine via interactions mediated by side chains from E108/E112.
Acknowledgments
We thank Victor M. Rivera of ARIAD Pharmaceuticals, Inc., for providing the pC4-Fv1E plasmid and the AP20187 chemical dimerizer. We thank Inger Tappin for assistance in preparation of baculovirus. We are grateful to J. Hurwitz for his insightful discussions during the course of this study. We appreciate Raymond Deshaires for his comments on stimulation of di-Ub synthesis by FKBP-based dimerization of human Cdc34.
K.W. was supported by a postdoctoral training grant from NCI. This study was supported by Public Health Service grants GM61051, CA095634, and CA111515 to Z.-Q.P.
Footnotes
Published ahead of print on 13 August 2007.
REFERENCES
- 1.Chen, A., K. Wu, S. Y. Fuchs, P. Tan, C. Gomez, and Z.-Q. Pan. 2000. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J. Biol. Chem. 275:15432-15439. [DOI] [PubMed] [Google Scholar]
- 2.Deffenbaugh, A. E., K. M. Scaglione, L. Zhang, J. M. Moore, T. Buranda, L. A. Sklar, and D. Skowyra. 2003. Release of ubiquitin-charged Cdc34-S-Ub from the RING domain is essential for ubiquitination of the SCF(Cdc4)-bound substrate Sic1. Cell 114:611-622. [DOI] [PubMed] [Google Scholar]
- 3.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 4.Gazdoiu, S., K. Yamoah, K. Wu, C. R. Escalante, I. Tappin, V. Bermudez, A. K. Aggarwal, J. Hurwitz, and Z. Q. Pan. 2005. Proximity-induced activation of human Cdc34 through heterologous dimerization. Proc. Natl. Acad. Sci. USA 102:15053-15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goebl, M. G., J. Yochem, S. Jentsch, J. P. McGrath, A. Varshavsky, and B. Byers. 1988. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science 241:1331-1335. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton, K. S., M. J. Ellison, K. R. Barber, R. S. Williams, J. T. Huzil, S. McKenna, C. Ptak, M. Glover, and G. S. Shaw. 2001. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure 9:897-904. [DOI] [PubMed] [Google Scholar]
- 7.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 8.Jin, J., T. Cardozo, R. C. Lovering, S. J. Elledge, M. Pagano, and J. W. Harper. 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18:2573-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamura, T., D. M. Koepp, M. N. Conrad, D. Skowyra, R. J. Moreland, O. Iliopoulos, W. S. Lane, W. G. Kaelin, Jr., S. J. Elledge, R. C. Conaway, J. W. Harper, and J. W. Conaway. 1999. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284:657-661. [DOI] [PubMed] [Google Scholar]
- 10.Kipreos, E. T., L. E. Lander, J. P. Wing, W. W. He, and E. M. Hedgecock. 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85:829-839. [DOI] [PubMed] [Google Scholar]
- 11.Koepp, D. M., J. W. Harper, and S. J. Elledge. 1999. How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97:431-434. [DOI] [PubMed] [Google Scholar]
- 12.Kolman, C. J., J. Toth, and D. K. Gonda. 1992. Identification of a portable determinant of cell cycle function within the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 11:3081-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, W., D. Tu, A. T. Brunger, and Y. Ye. 2007. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446:333-337. [DOI] [PubMed] [Google Scholar]
- 14.Liu, Y., N. Mathias, C. N. Steussy, and M. G. Goebl. 1995. Intragenic suppression among CDC34 (UBC3) mutations defines a class of ubiquitin-conjugating catalytic domains. Mol. Cell. Biol. 15:5635-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, F., A. Kentsis, R. Osman, and Z. Q. Pan. 2005. Inactivation of VHL by tumorigenic mutations that disrupt dynamic coupling of the pVHL:HIF-1α complex. J. Biol. Chem. 280:7985-7996. [DOI] [PubMed] [Google Scholar]
- 16.Ohta, T., J. J. Michel, A. J. Schottelius, and Y. Xiong. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3:535-541. [DOI] [PubMed] [Google Scholar]
- 17.Pan, Z. Q., A. Kentsis, D. C. Dias, K. Yamoah, and K. Wu. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23:1985-1997. [DOI] [PubMed] [Google Scholar]
- 18.Patton, E. E., A. Willems, D. Sa, L. Kuras, D. Thomas, K. L. Craig, and M. Tyers. 1998. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12:692-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell. Biol. 6:9-20. [DOI] [PubMed] [Google Scholar]
- 20.Petroski, M. D., and R. J. Deshaies. 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123:1107-1120. [DOI] [PubMed] [Google Scholar]
- 21.Petroski, M. D., G. Kleiger, and R. J. Deshaies. 2006. Evaluation of a diffusion-driven mechanism for substrate ubiquitination by the SCF-Cdc34 ubiquitin ligase complex. Mol. Cell 24:523-534. [DOI] [PubMed] [Google Scholar]
- 22.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 23.Pitluk, Z. W., M. McDonough, P. Sangan, and D. K. Gonda. 1995. Novel CDC34 (UBC3) ubiquitin-conjugating enzyme mutants obtained by charge-to-alanine scanning mutagenesis. Mol. Cell. Biol. 15:1210-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plon, S. E, K. A. Leppig, H. N. Do, and M. Groudine. 1993. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc. Natl. Acad. Sci. USA 90:10484-10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seol, J. H., R. M. Feldman, W. Zachariae, A. Shevchenko, C. C. Correll, S. Lyapina, Y. Chi, M. Galova, J. Claypool, S. Sandmeyer, K. Nasmyth, and R. J. Deshaies. 1999. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13:1614-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silver, E. T., T. J. Gwozd, C. Ptak, M. Goebl, and M. J. Ellison. 1992. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 11:3091-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skowyra, D., D. M. Koepp, T. Kamura, M. N. Conrad, R. C. Conaway, J. W. Conaway, S. J. Elledge, and J. W. Harper. 1999. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and rbx1. Science 284:662-665. [DOI] [PubMed] [Google Scholar]
- 28.Skowyra, D., K. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 29.Tan, P., S. Y. Fuchs, A. Chen, K. Wu, C. Gomez, Z. Ronai, and Z. Q. Pan. 1999. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell 3:527-533. [DOI] [PubMed] [Google Scholar]
- 30.Tang, X., S. Orlicky, Z. Lin, A. Willems, D. Neculai, D. Ceccarelli, F. Mercurio, B. H. Shilton, F. Sicheri, and M. Tyers. 2007. Suprafacial orientation of the SCF(Cdc4) dimer accommodates multiple geometries for substrate ubiquitination. Cell 129:1165-1176. [DOI] [PubMed] [Google Scholar]
- 31.VanDemark, A. P., R. M. Hofmann, C. Tsui, C. M. Pickart, and C. Wolberger. 2001. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105:711-720. [DOI] [PubMed] [Google Scholar]
- 32.Varelas, X., C. Ptak, and M. J. Ellison. 2003. Cdc34 self-association is facilitated by ubiquitin thiolester formation and is required for its catalytic activity. Mol. Cell. Biol. 23:5388-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, K., A. Chen, P. Tan, and Z.-Q. Pan. 2002. The Nedd8-conjugated ROC1-CUL1 core ubiquitin ligase utilizes Nedd8 charged surface residues for efficient polyubiquitin chain assembly catalyzed by Cdc34. J. Biol. Chem. 277:516-527. [DOI] [PubMed] [Google Scholar]
- 34.Wu, K., S. Y. Fuchs, A. Chen, P. Tan, C. Gomez, Z. Ronai, and Z.-Q. Pan. 2000. The SCFHos/β-TRCP-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 20:1382-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, P. Y., M. Hanlon, M. Eddins, C. Tsui, R. S. Rogers, J. P. Jensen, M. J. Matunis, A. M. Weisman, C. P. Wolberger, and C. M. Pickart. 2003. A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J. 22:5241-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, H., J.-M. Peters, R. W. King, A. M. Page, P. Hieter, and M. W. Kirschner. 1998. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science 279:1219-1222. [DOI] [PubMed] [Google Scholar]
- 37.Yunus, A. A., and C. D. Lima. 2006. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat. Struct. Mol. Biol. 13:491-499. [DOI] [PubMed] [Google Scholar]
- 38.Zachariae, W., A. Shevchenko, P. D. Andrews, R. Ciosk, M. Galova, M. J. R. Stark, M. Mann, and K. Nasmyth. 1998. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279:1216-1219. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, N., B. A. Schulman, L. Song, J. J. Miller, P. D. Jeffrey, P. Wang, C. Chu, D. M. Koepp, S. J. Elledge, M. Pagano, R. C. Conaway, J. W. Conaway, J. W. Harper, and N. P. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]