Abstract
The size of an organ must be tightly controlled so that it fits within an organism. The mammalian lens is a relatively simple organ composed of terminally differentiated, amitotic lens fiber cells capped on the anterior surface by a layer of immature, mitotic epithelial cells. The proliferation of lens epithelial cells fuels the growth of the lens, thus controling the size of the lens. We report that the Notch signaling pathway defines the boundary between proliferation and differentiation in the developing lens. The loss of Notch signaling results in the loss of epithelial cells to differentiation and a much smaller lens. We found that the Notch effector Herp2 is expressed in lens epithelium and directly suppresses p57Kip2 expression, providing a molecular link between Notch signaling and the cell cycle control machinery during lens development.
The Notch signaling cascade, a well-conserved, cell-cell communication pathway, determines cell fate during animal development (13, 17). Notch receptors and their ligands (Delta, Serrate/Jagged, F3/Cortactin, and NB3/DNER) are transmembrane proteins with large extracellular domains (4). When Notch receptors are engaged by their ligands, the receptors undergo proteolytic cleavage, leading to the release of the Notch intracellular domain. The Notch intracellular domain translocates to the nucleus, where it forms a trimeric complex with the DNA-binding protein RBP-Jκ [or RBP-J, also known as CSL for CBF1/Su(H)/Lag-1] and the coactivator Mastermind (Mam) and, at the same time, the domain releases RBP-J from a corepressor complex. The trimeric complex further recruits histone acetyltransferases (p300 and/or PCAF/GCN5) and chromatin-remodeling complexes (BRM, TRA1/TRRAP, and Dom) to form a transcriptional activator (9, 11, 14, 15). Genes activated by Notch signaling include the Hes family of transcription repressors, homologues of Drosophila Hairy/Enhancer of split (2, 6, 18). More recently, another family of transcription repressors, Herp (Hes-related repressor protein; also known as Hesr/Hey/Hrt/Chf/gridlock), was identified and shown to be activated by Notch (10, 26). However, few genes are known to be regulated by either the Hes or the Herp family of transcription factors.
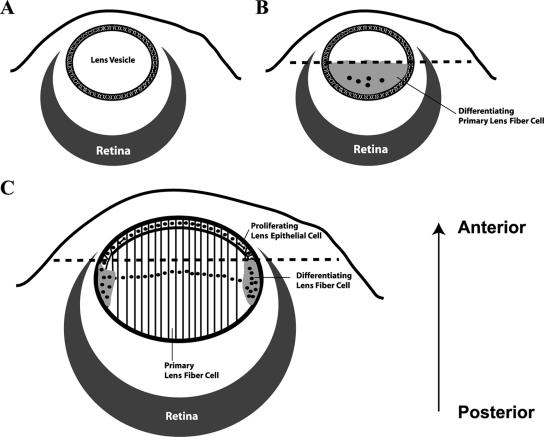
Ocular lens development can be divided into two stages (Fig. 1). The first stage results in the formation of a lens vesicle (Fig. 1A) and the primary lens. The lens vesicle is derived from the lens placode region of the head ectoderm. The optic vesicle (future retina) induces invagination of the lens placode, which eventually pinches off the head ectoderm to form a hollow sphere, the lens vesicle (Fig. 1A). Cells in the posterior portion of the lens vesicle, facing the optic vesicle or retina, differentiate into primary lens fiber cells (Fig. 1B) under the induction of a putative signal emitted from the retina. The anterior portion of the lens vesicle remains undifferentiated. During the second stage of lens development, the anterior epithelial cells continue to proliferate and their progeny differentiate into secondary lens fiber cells in the transition zone (or bow region), which is located around the lens equator where the epithelium terminates (Fig. 1C). Thus, the growth of the lens is realized through the addition of secondary fiber cells and is fueled by mitotic activities in the epithelium. The mechanism(s) that determines the boundary of differentiation during the lens vesicle stage and during the formation of secondary lens fiber cells remains unknown.
FIG. 1.
Diagrams showing different developmental stages of the ocular lens. (A) Lens vesicle. (B) Formation of primary lens fiber cells. (C) Formation of secondary lens fiber cells. Dashed lines in panels B and C represent the boundary of differentiation, epithelial cells anterior to which are not induced to differentiate. The arrow (posterior to anterior) represents the orientation of all lens images throughout this paper.
Here we report that the Notch signaling pathway controls the size of the lens epithelium by defining the boundary between proliferation and differentiation during lens development. A loss of Notch signaling causes the lens epithelium to shrink because the epithelial cells are lost to differentiation. As a result, the lens and the eye are much smaller than normal. We show that the Notch effector Herp2 is expressed in lens epithelium and directly suppresses p57Kip2 expression, providing a molecular link between the Notch signaling pathway and the cell cycle control machinery. This link likely explains, at least in part, the ability of the Notch signaling pathway to maintain the proliferation potential of progenitor cells in a large number of developing systems.
MATERIALS AND METHODS
Mice.
Rbp-J conditional knockout mice (30) were obtained from T. Honjo at Kyoto University. Le-Pax6-Cre transgenic mice were produced by P. Overbeek at Baylor College of Medicine. We obtained the R26R Cre reporter line from the Jackson Laboratory. All mice were genotyped by PCR.
For LacZ staining, embryos were dissected in cold phosphate-buffered saline (PBS) and washed in cold 0.1 M phosphate buffer before being fixed in 0.24% glutaraldehyde, 5 mM EDTA, 2 mM MgCl2, and 10 mM phosphate buffer at 4°C while rocking for 45 min. After fixation, embryos were stained overnight in 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, and 1 mg/ml 5-bromo-4-chloro-3-indolyl-galactoside at room temperature while rocking. Stained embryos were rinsed with PBS and photographed. Histological analyses of embryos were carried out as described previously (33).
Yeast one-hybrid screening.
We used the yeast one-hybrid kit from Clontech (Palo Alto, CA). Conserved element E-I or E-II from the mouse p57Kip2 gene was subcloned into pHis1-1 and pLacZi-1 and integrated into the genome of the YM4271 yeast strain to generate YE-I and YE-II. An embryonic day 17.5 (E17.5) mouse cDNA library in pACT2 (Clontech) was screened in YE-I and YE-II. Approximately 2 × 106 clones were screened. Clones that survived 3-amino-1,2,3-triazole (3-AT) selection were tested for LacZ expression. Those without LacZ expression were eliminated. The clones that expressed LacZ were used to extract the library plasmid DNA. Extracted DNA was transformed into Escherichia coli and then transformed back into YE-I, YE-II, and YE-p53 (provided with the kit) to test for specificity.
Cell culture and CAT and chromatin immunoprecipitation (ChIP) assays.
Cos7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The chloramphenicol acetyltransferase (CAT) assay was carried out 24 h after transfection using the CAT enzyme-linked immunosorbent assay kit (Roche). CAT expression levels were normalized to cotransfected β-galactosidase.
The ChIP assay was carried out according to the protocol suggested by the manufacturer of ChIP-grade anti-His6 antibodies (catalog no. ab9108; Abcam). His6-tagged cyclin D1 and Herp2 were subcloned into pCDNA3.1 (Invitrogen). The expression vectors were transfected into NIH 3T3 cells by using Lipofectamine 2000 (Invitrogen). Twenty-four hours after the transfection, formaldehyde was added to medium to cross-link proteins with DNA and the cells were lysed in 50 mM HEPES, pH 7.5-140 mM NaCl-1 mM EDTA-1% Triton X-100-0.1% sodium deoxycholate-0.1% sodium dodecyl sulfate. Cell lysates were sonicated using a Microson ultrasonic cell disruptor (four cycles of 30 s on and 30 s off) to shear genomic DNA to an average fragment size of 500 to 1,000 bp and then immunoprecipitated with anti-His6 antibodies. The precipitated DNA was purified with a QIAquick PCR purification kit, followed by PCR amplification of two regions of the mouse p57Kip2 promoter. The primers were 5′-TACAAGGCAGGCCCTGTAATCGGA-3′ and 5′-CCCCGCCGCCCCAGCAGTAAGCAG-3′ for region a and 5′-CTCTGCAGGGCCTTTCAAGTATGT-3′ and 5′-TTGGCTGGAAGTAGTTATGCTAGA-3′ for region b.
Immunofluorescence analysis and in situ hybridization.
Mouse embryos at the desired stages of development were harvested after a 2-h pulse of bromodeoxyuridine (BrdU) and were fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin wax. Sections (5 mm) were cut and collected.
Immunofluorescence staining was performed according to the instructions provided by the antibody suppliers. In brief, dewaxed and rehydrated sections were heated in 10 mM sodium citrate (pH 6.0) in a microwave oven (700 W, 10 min) to retrieve the antigen. Slides were washed three times for 5 min in PBS. Primary antibodies were diluted in blocking buffer (PBS, 2% bovine serum albumin, 0.1% Triton X-100). We used 1:50 rabbit anti-p57 (catalog no. ab4058; Abcam Inc., Cambridge, MA) and 1:50 goat anti-Jagged 1 (C-20; Santa Cruz Biotechnology), 1:100 rabbit anti-cleaved caspase-3 (Cell Signaling), 1:100 mouse anti-E-cadherin (BD Bioscience), and 1:10 rabbit anti-β-crystallin (a gift from P. Overbeek, Baylor College of Medicine). Tissue sections were incubated with antibody overnight at 4°C. After being washed with PBS three times, sections were incubated with the following secondary antibodies for 1 h at room temperature: Alexa 594-conjugated donkey anti-goat immunoglobulin G (IgG) (Invitrogen), fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG (Santa Cruz Biotechnology), and FITC-conjugated rabbit anti-mouse IgG (Sigma). Nuclei were counterstained with 100 ng/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma) in PBS. Slides were viewed with a Zeiss LSM-510 META confocal microscope by using LSM 510 (version 3.2) software to acquire images, and the lens region was exported as the region of interest. BrdU incorporation was visualized with a BrdU staining kit (Amersham Biosciences).
In situ hybridization was performed as described previously (23, 27). The probe for Herp2 was a PCR clone of its open reading frame obtained from Deepak Srivastava (University of Texas Southwestern Medical Center). The probe for Notch3 was the SacII-to-XbaI fragment of the cDNA (a gift from U. Lendahl, Karolinska Institute) cloned into pBS.
RESULTS
Generation of lens-specific Rbp-J deletion mice.
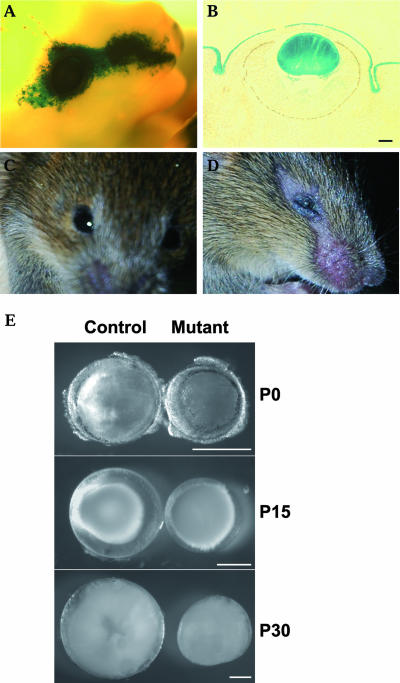
It was previously reported that Notch2 and -3 are expressed in lens epithelium and that Jag1 is expressed in differentiating lens fiber cells (3, 12, 20). These expression patterns suggest that Notch signaling may be involved in ocular lens development. To determine whether the function of Notch signaling is important for lens development, we decided to delete Rbp-J in the lens since it is required for all Notch receptors. In order to inactivate Rbp-J specifically in the lens, we employed a transgenic mouse line (Le-Pax6-Cre) in which the Cre recombinase expression is driven by a lens-specific enhancer of Pax6 (1). This Pax6 enhancer is active at the beginning of lens induction (1). Using a Cre reporter strain (29), we found that Cre recombinase activity in the Le-Pax6-Cre transgenic line is confined to the lens and the ectoderm covering the eye (Fig. 2A and B). Mice carrying a floxed Rbp-J allele (Rbp-Jflox) (30) were crossed with Le-Pax6-Cre mice. Extensive breeding indicated that the Cre transgene resides on the same chromosome as Rbp-J does. To bring the transgene and Rbp-Jflox together meiotically, we crossed Rbp-J+/flox/Le-Pax6-Cre mice with wild-type mice. A few recombinant animals in which Rbp-Jflox and the Cre transgene were linked were obtained from over 250 offspring. The Cre transgene is apparently active in the female germ line, because the Rbp-Jflox allele in the recombinants was converted to Rbp-Jd (the deleted allele) by passing through the female germ line. We then crossed Rbp-J+/d/Le-Pax6-Cre mice with Rbp-Jflox/flox mice to obtain animals with the Rbp-Jflox/d/Le-Pax6-Cre genotype. In these animals with the Rbp-Jflox/d/Le-Pax6-Cre genotype, the floxed Rbp-J allele should be inactivated in both the lens and skin covering the eye, producing Rbp-J-null cells in these areas.
FIG. 2.
Loss of Notch signaling causes small-lens phenotype in mice. (A) LacZ reporter expression in Le-Pax6-Cre/R26R mice. (B) A section through the stained region shown in panel A. (C) A P14 Rbp-J+/Flox mouse. (D) A P14 Rbp-Jflox/d/Le-Pax6-Cre mouse. (E) Postnatal lenses in control and Rbp-J mutant mice at P0, P15, and P30. Scale bars, 250 μm.
Eyes are smaller in lens-specific Rbp-J deletion mice.
At birth, Rbp-Jflox/d/Le-Pax6-Cre mice were not different from their Rbp-Jflox/+ littermates, which were used as controls in our experiments. Eyelids opened in control mice at about 2 weeks after birth (Fig. 2C). In mutant mice, eyes were smaller than normal and eyelids had barely opened by 2 weeks of age and remained that way into adulthood (Fig. 2D). The hairless stripe from the temporal to the nasal side of the eye in the mutants was due to the loss of Notch signaling in skin cells (31), indicating that the Cre transgene worked as expected. We dissected lenses from control and mutant mice between postnatal day 0 (P0) and P30, the time point when lens growth stops. As shown in Fig. 2E, mutant lenses were much smaller than control lenses. All Rbp-J mutants showed similar reductions in lens size, indicating a strong penetrance of the mutant phenotype.
Herp2 is expressed in lens epithelium.
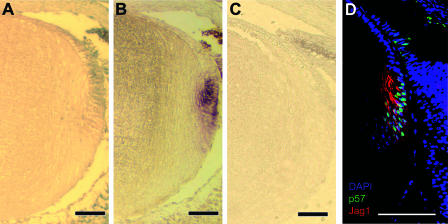
To understand the molecular mechanism(s) underlying the small-lens phenotype in Rbp-J mutants, we first determined which of the Notch effectors is expressed in the lens. To that end, we used in situ hybridization to survey the expression of the Hes and Herp families of transcription repressors. At E17.5, Herp2 was expressed in lens epithelium (Fig. 3A) in a domain right above (but also overlapping with) that of p57Kip2 (Fig. 3B), a cyclin-dependent kinase (Cdk) inhibitor (19, 22) that is required for cell cycle withdrawal during lens fiber cell differentiation (32, 33). Herp2 expression was lost in Rbp-J mutants (Fig. 3C; also see Fig. 6C), demonstrating that Notch signaling is required for the expression of this transcriptional repressor. We also examined Jag1 expression by immunofluorescence. This Notch ligand was expressed in the differentiating fiber cells that express p57Kip2 (Fig. 3D). The expression of Jag1 is likely induced by the same signal that induces p57Kip2 expression. The Jag1 expression pattern places it in the right position to activate Notch signaling (and hence Herp2 expression) in adjacent epithelial cells. Jag1 expression was not detected in more mature, center-localized lens fiber cells.
FIG. 3.
Notch signaling in the ocular lens. (A) In situ hybridization of Herp2 in a section of an E17.5 wild-type lens. Note the orientation of the sections: anterior up and posterior down, as shown in Fig. 1. (B) In situ hybridization of p57Kip2 in an adjacent section of panel A. (C) In situ hybridization of Herp2 in a section of an E17.5 Rbp-J mutant lens. (D) Immunofluorescent staining of Jag1 and p57Kip2 in sections of an E17.5 wild-type lens. The sections were counterstained for DNA with DAPI. Scale bars, 50 μm.
FIG. 6.
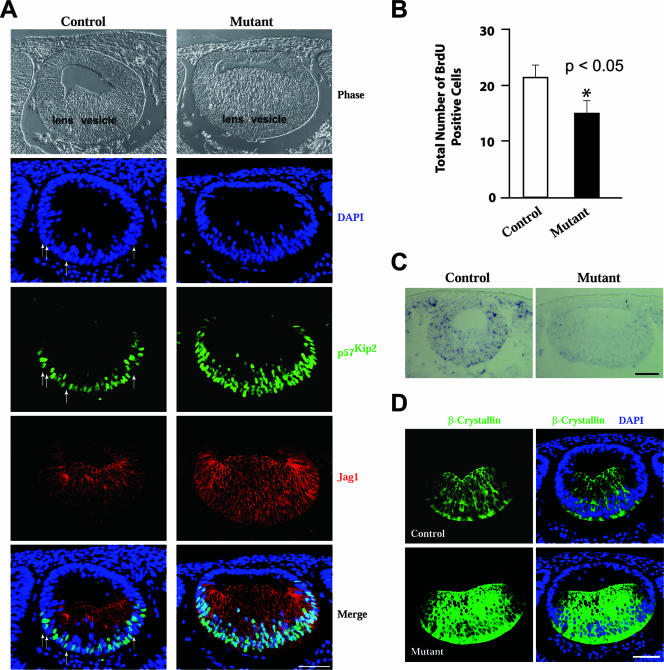
Notch signaling suppresses the differentiation of primary lens fiber cells. (A) Sections across the eye region in E11.5 control and Rbp-J mutant embryos were immunostained for p57Kip2 and Jag1. Arrows indicate some of the control cells that do not express p57Kip2. (B) Quantification of BrdU incorporation in control and mutant lens vesicles. Error bars indicate standard deviations. *, P value was <0.05. (C) In situ hybridization of Herp2 in sections of E11.5 control and mutant eyes. (D) Immunostaining of β-crystallin in sections of E11.5 control and mutant eyes. Scale bars, 50 μm.
Herp2 can interact with a conserved element in the promoter of p57Kip2.
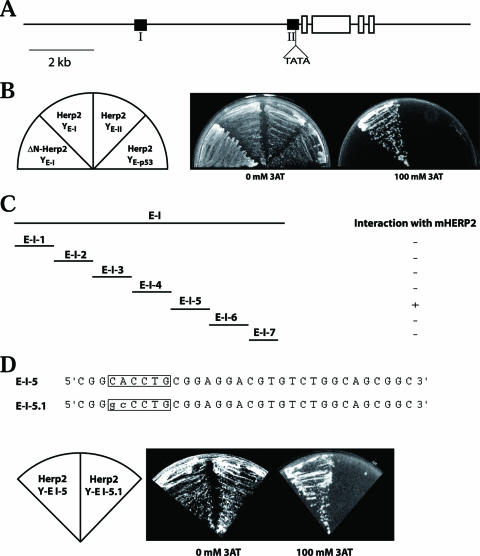
The Herp2 and p57Kip2 expression patterns suggest that Herp2 may suppress p57Kip2 expression, directly or indirectly, to prevent lens epithelial cells from exiting the cell cycle. To gain insight into the regulation of the expression of p57Kip2, we compared a 7-kb region 5′ to the first exon of the mouse gene with the equivalent region of the human gene and identified two significantly conserved elements, E-I (214 bp, with a P value of 8e−55) and E-II (199 bp, with a P value of 1e−24) (Fig. 4A). Sequence analysis indicated that the conserved regions E-I and E-II do not contain any repetitive sequences. The distance between E-I and E-II as well as the distance from these two elements to the transcription start is also conserved between humans and mice, suggesting that these two elements might play a role in regulating the expression of p57Kip2. To identify transcription factors that may interact with E-I or E-II, we performed yeast one-hybrid screens. We constructed two yeast (Saccharomyces cerevisiae) strains in which the expression of His3 was under the control of a minimal yeast promoter and one copy of E-I or E-II (only the region of position numbers −199 to −70 of E-II was used to avoid the TATA box). Both strains (YE-I and YE-II) could survive on His dropout plates due to the basal level expression of His3, but neither could survive on His dropout plates containing 60 mM 3-AT (an inhibitor of His3). We screened a mouse E17.5 cDNA library (fused with the Gal4 transactivation domain at the 5′ end of the cDNAs) by using His dropout and 60 mM 3-AT for selection. About 2 × 106 clones were screened. After the elimination of false positive clones, one clone was identified for E-I but none were identified for E-II (Fig. 4B). The clone identified for E-I encodes the full-length Herp2. The Gal4 activation domain (Gal4AD) is fused in frame 108 nucleotides 5′ to the starting ATG of Herp2. Gal4AD-Herp2 conferred YE-I, but not YE-II, resistance up to 100 mM 3-AT (Fig. 4B). An N-terminal deletion that destroyed the DNA-binding domain of Herp2 resulted in the inability to survive on 3-AT-containing medium (Fig. 4B). As a further test for specificity, we found that Gal4AD-Herp2 could not activate His3 expression via the p53-binding site (Fig. 4B).
FIG. 4.
Herp2 interacts with the E-I element of p57Kip2 promoter in yeast. (A) Diagram of the mouse p57Kip2 gene showing the conserved elements E-I and E-II (black boxes). E-I and E-II were identified through sequence comparison among human, rat, and mouse p57KIP2 genes. Empty boxes represent exons. (B) Herp2 interacts with only E-I. Neither E-II nor the p53-binding site interacts with Herp2. The interaction was abolished by the deletion of the N-terminal region of Herp2, which contains the DNA-binding domain. (C) Mapping of the Herp2-binding site within E-I. (D) The E box in E-I-5 is essential for interaction with Herp2. Mutating the E box destroyed the ability of E-I-5 to interact with Herp2.
We mapped the binding site of Herp2 in E-I by dividing E-I into seven smaller elements (32 bp each for the first six elements and 22 for the last element). We found that only E-I-5 could drive His3 expression through Gal4AD-Herp2 (Fig. 4C). As a member of the helix-loop-helix family of transcription factors, Herp2 should bind to the E box (CAXXTG). Indeed, there is an E box in E-I-5, CACCTG (Fig. 4C). When it was mutated to GCCCTG, the mutant E-I-5 (E-I-5.1) was no longer able to drive His3 expression through Gal4AD-Herp2 (Fig. 4D). These results indicate that Herp2 interacts with the E box CACCTG in E-I.
Herp2 can suppress the expression of p57Kip2.
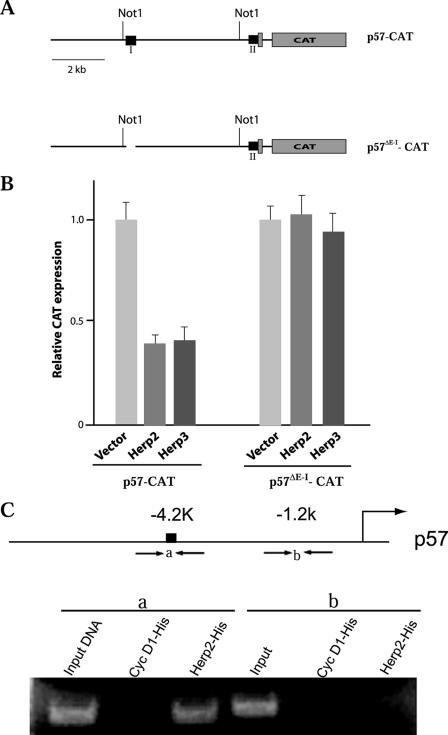
The interaction of Herp2 with E-I in yeast suggests that this Notch effector can suppress p57 expression directly. To test this possibility, we generated two constructs, p57-CAT and p57ΔE-I-CAT, by using the 7-kb mouse p57Kip2 promoter region and its E-I-deleted version to drive CAT reporter expression (Fig. 5A). The CAT constructs were transfected into Cos7 cells, together with a Herp2 expression vector. As shown in Fig. 5B, Herp2 suppressed CAT expression in an E-I-dependent fashion. Herp3, another member of the Herp family of repressors, also suppressed the p57Kip2 promoter (Fig. 5B).
FIG. 5.
Suppression of p57Kip2 promoter in mammalian cells. (A) Diagrams showing the CAT constructs. (B) Herp2 and Herp3 suppress CAT reporter expression driven by the p57Kip2 promoter. CAT expression in empty vector-transfected cells was set to 1.0. Error bars indicate standard deviations. (C) ChIP assay results demonstrating the interaction of Herp2 with the E-I region of p57Kip2 in NIH 3T3 cells.
Furthermore, we carried out ChIP assays to assess whether E-I is a target of Herp2. His6-tagged Herp2 as well as His6-cyclin D1 (as a control) expression vectors were transfected into NIH 3T3 cells. After cross-linking with formaldehyde, the transfected cells were lysed and sonicated for ChIP with anti-His6 antibodies. The precipitates were PCR amplified using two primer sets, one for E-I (containing the E box identified in Fig. 4C) and the other for a region located at kb −1.2 of mouse p57Kip2. As shown in Fig. 5C, only the E-I region was present in the immunoprecipitates brought down by His6-Herp2. Taken together, these data indicate that Herp2 can directly suppress the expression of p57Kip2.
Increased number of p57Kip2-expressing cells in Rbp-J mutants at primary lens stage.
The ability of Herp2 to suppress p57Kip2 expression prompted us to ask whether there is an increase in the number of p57Kip2-expressing (thus nondividing) cells in the Rbp-J mutant that might account for the smaller-lens phenotype. We first examined the process of primary lens fiber cell differentiation in Rbp-J mutants. At this stage of development, the epithelial cells in the posterior half of the lens vesicle have begun to differentiate, expressing p57Kip2 and Jag1 (Fig. 6A), while the anterior half of the lens vesicle remains undifferentiated. The size and the cell number (see Fig. S2A in the supplemental material) of the lens vesicle in Rbp-J mutants are comparable to those in the control, indicating that the small lens size in the mutants is not a result of defective lens vesicle formation. However, the number of p57Kip2-positive cells in the mutants was much greater: p57Kip2 expression was detected not only in almost every cell in the posterior region but also in some cells in the more anterior region. More p57Kip2 expression in the posterior region indicates that Notch signaling also functions in the posterior part of the lens vesicle, and the expression in the anterior region indicates that the p57Kip2 expression domain in the mutants expands into a region that does not normally express p57Kip2. In support of the function of Notch signaling in the posterior region, Herp2 expression was detected by in situ hybridization in the posterior portion of control lens vesicles, but not in lens vesicles from Rbp-J mutants at E11.5 (Fig. 6C). Furthermore, Notch3 is expressed throughout the lens vesicle (and in other eye structures as well) as detected by in situ hybridization (see Fig. S1 in the supplemental material).
As a result of the increase of p57Kip2-expressing cells both anteriorly and posteriorly, the number of cells undergoing active DNA synthesis decreases in the mutant lens vesicle relative to control (Fig. 6B and see Fig. S3 in the supplemental material), indicating a decline in the proliferation potential of the mutant lens vesicle.
Enhanced differentiation of primary lens fiber cells in Rbp-J mutants.
The overall differentiation of primary lens fiber cells also proceeded faster in the absence of Notch signaling. Primary fiber cell elongation was greater in the Rbp-J mutant than in the control (Fig. 6A, compare images for the control and mutant phases and Jag1 immunostaining; see Fig. S2B in the supplemental material), as was the expression of β-crystallin, a marker of lens fiber cell differentiation (Fig. 6D). Furthermore, the number of p27Kip1-positive cells also increased slightly in the mutant (see Fig. S4 in the supplemental material). This result is consistent with the notion that Notch signaling could also suppress the expression of p27Kip1 (8, 24). p27Kip1 plays a minor role in lens fiber cell differentiation (33). Its protein levels (25), but not its message levels (33), are increased in differentiating fiber cells. Taken together, these results indicate that Notch signaling suppresses the differentiation of primary lens fiber cells.
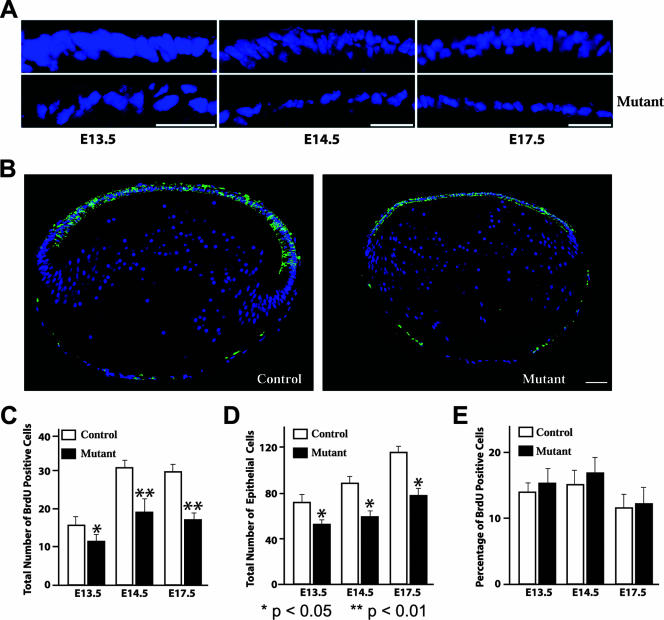
Thinning of the lens epithelium in Rbp-J mutants.
Next, we examined the process of secondary lens fiber cell formation, which lasts until adulthood in mice (16). During this stage of lens development, secondary fiber cells are generated in a region called the transition zone which is located where the epithelium terminates at the equator of the lens. Continued proliferation of anterior lens epithelial cells generates secondary fiber cells. In Rbp-J mutant mice, fewer epithelial cells are left due to the enhanced differentiation of primary lens fiber cells, which predicts that the mutant lens epithelium will be smaller and contain fewer cells. Indeed, we found that the epithelium in the mutant lens was thinner and contained fewer cells than the epithelium in the control did (Fig. 7A). No disruption of the epithelial structure was revealed by E-cadherin immunostaining (Fig. 7B). However, it is apparent from the E-cadherin staining that the size of the mutant lens epithelium is smaller than that in the control, as is the lens itself (Fig. 7B). To rule out the possibility that Notch signaling is required for the proliferation of lens epithelial cells, we analyzed BrdU incorporation in control and mutant lenses. The total number of BrdU-positive cells was significantly reduced in the mutants compared to that in the controls (Fig. 7C and see Fig. S5 in the supplemental material), and so was the total number of epithelial cells (Fig. 7D), resulting in similar BrdU indices in mutants and controls (Fig. 7E). We also asked whether an increase in the rate of apoptotic cell death could account for the apparent loss of cells in mutant lens epithelium. Activated caspase-3 was used as an indicator of apoptosis. No differences in activated caspase-3 staining were observed. In fact, no apoptotic cells were detected in either control or mutant lenses (data not shown). These data indicate that the Notch signaling pathway is not essential for the proliferation of lens epithelial cells and that the loss of Rbp-J does not induce apoptosis. Thus, the thinning of the mutant epithelium is not the result of an inability to proliferate or an increased tendency to die, but rather, it results from the early loss of epithelial cells to differentiation (Fig. 6A).
FIG. 7.
The size of lens epithelia is reduced in Rbp-J mutants. (A) Close-up images of DAPI-stained lens sections. (B) Immunofluorescence staining of E-cadherin in sections derived from E17.5 embryos. Sections were counterstained for DNA with DAPI. (C to E) Quantification of BrdU incorporation and cell numbers in lens epithelia. BrdU incorporation was analyzed by immunostaining. Error bars indicate standard deviations. Scale bars, 50 μm.
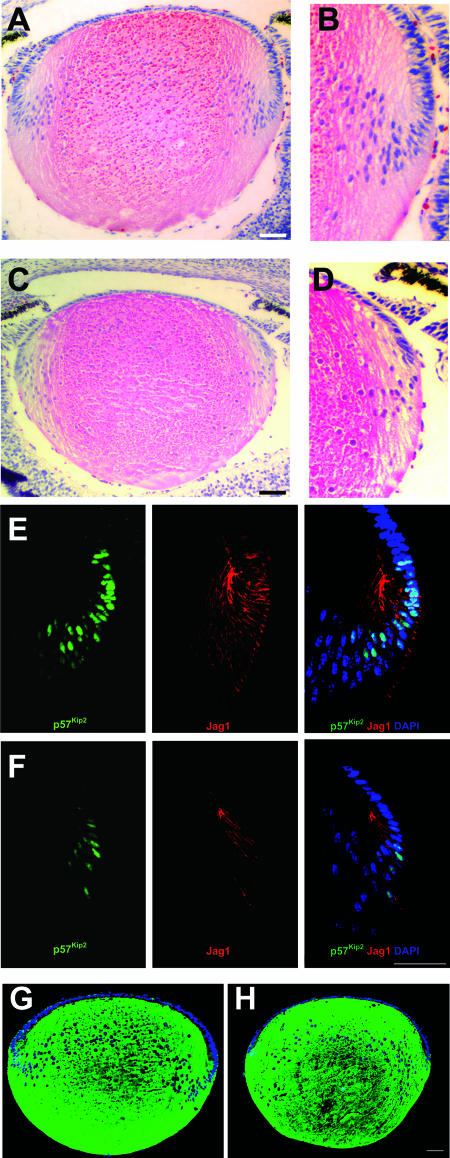
Impaired production of secondary lens fiber cells in Rbp-J mutants.
The smaller-than-normal size of the mutant lens epithelium and the reduced production of epithelial cells as indicated by the total number of BrdU-positive cells (Fig. 7C) predict that the generation of secondary fiber cells is less active in Rbp-J mutants than in controls. Therefore, we analyzed secondary lens fiber cell formation at E17.5. The mutant transition zone was more anterior and contained fewer cells than the transition zone of the control (Fig. 8A to D). Immunofluorescence staining of p57Kip2 confirmed that there were fewer cells undergoing cell cycle withdrawal and differentiation in the mutant than in the control (Fig. 8E and F). Further, the number of cells expressing Jag1 also decreased dramatically in the mutant (Fig. 8E and F).
FIG. 8.
Formation of secondary lens fiber cells is decreased in Notch signaling mutants. (A) Microphotograph of a hematoxylin-and-eosin-stained E17.5 control lens section. (B) Higher magnification of the transition zone shown in panel A. (C) Microphotograph of an hematoxylin-and-eosin-stained E17.5 Rbp-J mutant lens section. (D) Higher magnification of the transition zone shown in panel C. (E and F) Immunofluorescent Jag1 and p57Kip2 staining in section of control (E) and Rbp-J mutant (F) lenses. (G and H) Immunofluorescent β-crystallin staining in sections of control (G) and Rbp-J mutant (H) lenses. Sections in panels E to H were counterstained for DNA with DAPI. Scale bars, 50 μm.
Given the apparent reduction in the generation of secondary fiber cells, we determined whether differentiation itself is impaired in Rbp-J mutants by analyzing β-crystallin expression. As shown in Fig. 8G and H, the expression of this lens fiber cell marker did not differ between the control and the mutant, indicating that the loss of Notch signaling does not affect the differentiation process per se. However, the less-stained core, which is formed by primary lens fiber cells, is located further posterior in the mutant (Fig. 8H), a likely result of more anterior formation of secondary fiber cells due to anterior-ward shifting of the transition zone in mutants. Taking these data together, we conclude that the loss of Notch signaling reduces the production of secondary lens fiber cells, leading to a smaller-than-normal ocular lens. This reduction may become even greater postnatally, as the difference in size between the wild type and the mutant is much larger at P30 than at P15 (Fig. 2E).
The small-lens phenotype in Rbp-J mutants is rescued by simultaneous inactivation of p57Kip2.
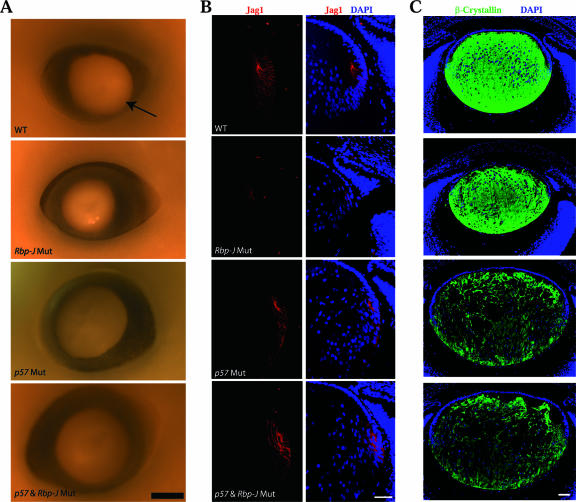
If the aberrant p57Kip2 expression pattern underlies the small-lens phenotype, we should be able to restore the lens size in Rbp-J mutants by genetically removing this Cdk inhibitor. To that end, we generated double-mutant embryos lacking p57Kip2 and Rbp-J in the lens. We previously showed that deleting p57Kip2 increases the lens size and reduces the levels of β-crystalline expression (33). As expected, p57Kip2 single- and p57Kip2/Rbp-J double-mutant lenses were slightly bigger than wild-type lenses at E16.5 (Fig. 9A). Importantly, the double-mutant lens was as big as the p57Kip2 single-mutant lens and bigger than the Rbp-J mutant lens, demonstrating that the deletion of p57Kip2 restored the lens size in Rbp-J mutants. Furthermore, the removal of p57Kip2 normalized the number of Jag1-expressing lens fiber cells undergoing differentiation in the transition zone, as indicated by increased Jag1 expression in the double mutant relative to its expression in the Rbp-J single mutant (Fig. 9B). In p57Kip2/Rbp-J double-mutant lenses, the β-crystallin expression is also reduced to the same level as that in the p57Kip2 single-mutant lens (Fig. 9C). These results strongly support the notion that the Notch signaling pathway protects lens epithelial cells by suppressing p57Kip2 expression.
FIG. 9.
Removal of p57Kip2 normalizes ocular lens sizes in Rbp-J mutants (Rbp-J Mut). (A) Microphotographs of whole-mount lenses at E16.5. (B) Immunofluorescent staining of Jag1 at E16.5. (C) Immunofluorescent staining of β-crystallin at E16.5. Sections were counterstained for DNA with DAPI. WT, wild type. Scale bars, 50 μm.
DISCUSSION
Little is known about the signaling mechanisms and the interactions between them that specify the size of an organ so that it fits within an organism. The ocular lens is a simple organ composed of two types of cells: the mitotically active epithelial cells and the amitotic fiber cells. The former cells are the precursors of the latter cells. Therefore, mechanisms that control the size of this epithelial precursor pool and the mitotic activities of the epithelial cells within the pool determine the size of the lens. Lens epithelial cells are induced to form fiber cells by a differentiation signal(s) emitted from the retina. The nature of the signal remains unknown, but fibroblast growth factors are prime candidates (21). It is believed that the signal forms a gradient from posterior to anterior in the optic cup (21). This gradient may therefore determine the boundary of differentiation (Fig. 1) and, hence, the size of the epithelial pool, if the differentiation of lens fiber cells requires a certain strength of the signal.
Our results indicate that the Notch signaling pathway also plays a role in defining the differentiation boundary, in addition to the proposed differentiation signal gradient. In the absence of Notch signaling, some more anteriorly localized, would-be epithelial cells in the lens vesicle prematurely start to differentiate, resulting in the shrinkage of the future lens epithelium, the precursor pool of lens fiber cells. Consequently, the production of secondary lens fiber cells is reduced and smaller lenses are generated in mutants with defective Notch signaling.
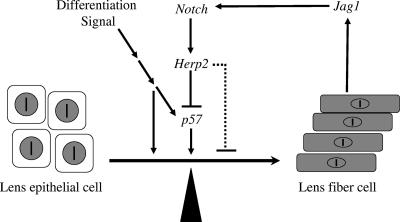
The expression of Jag1 by differentiating fiber cells suggests that Jag1 may be induced by the differentiation signal to activate Notch receptors on the adjacent epithelial cells, preventing them from differentiating. Thus, the lens fiber cell differentiation signal sets up a negative feedback loop at cellular level through the Notch signaling pathway to limit the number of epithelial cells undergoing differentiation (Fig. 10). The disruption of this feedback mechanism results in the apparent strengthening of the differentiation signal and premature differentiation.
FIG. 10.
A model describing the role of the Notch signaling in keeping the balance between proliferation and differentiation during lens development.
The feedback loop may also function in the posterior region of the lens vesicle where the first few differentiated cells (perhaps those at the posterior tip of the lens vesicle that are exposed to the strongest differentiation signal first) express Jag1, activate Notch receptors on neighboring cells, and prevent them from differentiating. Only when the differentiation signal gets strong enough can those cells overcome Notch-mediated suppression and start to differentiate. In support of that possibility, Notch3 and Herp2 expression was also found in the posterior part of the lens vesicle, and the differentiation of primary lens fiber cell in Rbp-J mutants was stronger than that in controls. Furthermore, this feedback mechanism could also function to protect the lens epithelium during the formation of secondary fiber cells. The loss of this protection could shift the transition zone in the anterior direction. However, we do not have direct evidence to support that possibility at present. To test that likelihood, one would need to inactivate the Notch signaling after the primary lens stage.
How does the Notch signaling suppress lens fiber cell differentiation? We found that the transcriptional repressor, Herp2, is expressed in epithelial cells in a Notch signaling-dependent way. We also found that p57Kip2 is a direct target of Herp2. Given the importance of this Cdk inhibitor in the differentiation of lens fiber cells (32, 33), by blocking its expression, Herp2 (and hence the Notch signaling) may indirectly inhibit lens fiber cell differentiation. However, Herp2 could suppress other genes directly involved in the differentiation (Fig. 10, dashed line), which will require further investigation. Since p57Kip2 is a Cdk inhibitor, the suppression of its expression by Notch signaling also helps maintain the proliferation potential of lens epithelial cells.
Notch signaling has been implicated in the development of various structures of the eye. A hypomorphic Notch2 allele results in an aberrant bulbous structure, retrolenticular hyperplasia, and microphthalmia (22a). The mild small-eye phenotype may be caused in part by the effect on lens development described here. Jag1-deficient mice have been generated (30a). Although heterozygous mice show defects in irises and corneas, no impairment in the lens was reported and Jag1-null mice died too early (before E10.5) for the analysis of the role of Jag1 on lens development. By deleting Rbp-J specifically in the lens, our work revealed a critical role for the Notch signaling pathway in balancing proliferation and differentiation during ocular lens development. Notch signaling performs this balance by controlling the number of cells undergoing differentiation without affecting the proliferation or differentiation processes themselves. Directly repressing the expression of p27Kip1 (8, 24) and p57Kip2 (this study and reference 7) or inducing their degradation (5, 28) is perhaps the general mechanism used by the Notch signaling to control tissue growth in a large number of developing systems.
Supplementary Material
Acknowledgments
We are indebted to P. Overbeek of Baylor College of Medicine for sharing Le-Pax6-Cre transgenic mice with us and to T. Honjo of Kyoto University in Japan for providing us with Rbp-J conditional knockout mice. We thank D. Srivastava of University of Texas Southwestern Medical Center for providing us with the cDNAs for Herp1, -2, and -3 and S. Hamilton and Z. Songyang of Baylor College of Medicine for their technical support.
This work was supported in part by NIH grant EY12825 and in part by research grant 6-FY00-812 from the March of Dimes Birth Defects Foundation.
Footnotes
Published ahead of print on 20 August 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ashery-Padan, R., T. Marquardt, X. Zhou, and P. Gruss. 2000. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 14:2701-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, A. M., and J. W. Posakony. 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9:2609-2622. [DOI] [PubMed] [Google Scholar]
- 3.Bao, Z. Z., and C. L. Cepko. 1997. The expression and function of Notch pathway genes in the developing rat eye. J. Neurosci. 17:1425-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 5.Dohda, T., A. Maljukova, L. Liu, M. Heyman, D. Grander, D. Brodin, O. Sangfelt, and U. Lendahl. 2007. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp. Cell Res. 313:3141-3152. [DOI] [PubMed] [Google Scholar]
- 6.Egan, S. E., B. St-Pierre, and C. C. Leow. 1998. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr. Top. Microbiol. Immunol. 228:273-324. [DOI] [PubMed] [Google Scholar]
- 7.Georgia, S., R. Soliz, M. Li, P. Zhang, and A. Bhushan. 2006. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev. Biol. 298:22-31. [DOI] [PubMed] [Google Scholar]
- 8.Havrda, M. C., M. J. Johnson, F. O'Neill, C., and L. Liaw. 2006. A novel mechanism of transcriptional repression of p27(kip1) through Notch/HRT2 signaling in vascular smooth muscle cells. Thromb. Haemostasis 96:361-370. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 96:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iso, T., L. Kedes, and Y. Hamamori. 2003. HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell Physiol. 194:237-255. [DOI] [PubMed] [Google Scholar]
- 11.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 12.Jones, E. A., M. Clement-Jones, and D. I. Wilson. 2000. JAGGED1 expression in human embryos: correlation with the Alagille syndrome phenotype. J. Med. Genet. 37:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadesch, T. 2004. Notch signaling: the demise of elegant simplicity. Curr. Opin. Genet. Dev. 14:506-512. [DOI] [PubMed] [Google Scholar]
- 14.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 12:2269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, H., Y. Taniguchi, H. Kurooka, S. Minoguchi, T. Sakai, S. Nomura-Okazaki, K. Tamura, and T. Honjo. 1997. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development 124:4133-4141. [DOI] [PubMed] [Google Scholar]
- 16.Kuszak, J. R., and M. J. Costello. 2004. Structure of the vertebrate lens, p. 91-105. In F. J. Lovicu and M. L. Robinson (ed.), Development of the ocular lens. Cambridge University Press, Cambridge, United Kingdom.
- 17.Lai, E. C. 2004. Notch signaling: control of cell communication and cell fate. Development 131:965-973. [DOI] [PubMed] [Google Scholar]
- 18.Lecourtois, M., and F. Schweisguth. 1995. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 9:2598-2608. [DOI] [PubMed] [Google Scholar]
- 19.Lee, M. H., I. Reynisdottir, and J. Massague. 1995. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 9:639-649. [DOI] [PubMed] [Google Scholar]
- 20.Lindsell, C. E., J. Boulter, G. diSibio, A. Gossler, and G. Weinmaster. 1996. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol. Cell. Neurosci. 8:14-27. [DOI] [PubMed] [Google Scholar]
- 21.Lovicu, F. J., and J. W. McAvoy. 2005. Growth factor regulation of lens development. Dev. Biol. 280:1-14. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka, S., M. C. Edwards, C. Bai, S. Parker, P. Zhang, A. Baldini, J. W. Harper, and S. J. Elledge. 1995. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 9:650-662. [DOI] [PubMed] [Google Scholar]
- 22a.McCright, B., X. Gao, L. Shen, J. Lozier, Y. Lan, M. Maguire, D. Herzlinger, G. Weinmaster, R. Jiang, and T. Gridley. 2001. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491-502. [DOI] [PubMed] [Google Scholar]
- 23.Moorman, A. F., A. C. Houweling, P. A. de Boer, and V. M. Christoffels. 2001. Sensitive nonradioactive detection of mRNA in tissue sections: novel application of the whole-mount in situ hybridization protocol. J. Histochem. Cytochem. 49:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Murata, K., M. Hattori, N. Hirai, Y. Shinozuka, H. Hirata, R. Kageyama, T. Sakai, and N. Minato. 2005. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol. Cell. Biol. 25:4262-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagahama, H., S. Hatakeyama, K. Nakayama, M. Nagata, K. Tomita, and K. Nakayama. 2001. Spatial and temporal expression patterns of the cyclin-dependent kinase (CDK) inhibitors p27Kip1 and p57Kip2 during mouse development. Anat. Embryol. 203:77-87. [DOI] [PubMed] [Google Scholar]
- 26.Nakagawa, O., D. G. McFadden, M. Nakagawa, H. Yanagisawa, T. Hu, D. Srivastava, and E. N. Olson. 2000. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc. Natl. Acad. Sci. USA 97:13655-13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 28.Sarmento, L. M., H. Huang, A. Limon, W. Gordon, J. Fernandes, M. J. Tavares, L. Miele, A. A. Cardoso, M. Classon, and N. Carlesso. 2005. Notch1 modulates timing of G1-S progression by inducing SKP2 transcription and p27 Kip1 degradation. J. Exp. Med. 202:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 30.Tanigaki, K., H. Han, N. Yamamoto, K. Tashiro, M. Ikegawa, K. Kuroda, A. Suzuki, T. Nakano, and T. Honjo. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443-450. [DOI] [PubMed] [Google Scholar]
- 30a.Xue, Y., X. Gao, C. E. Lindsell, C. R. Norton, B. Chang, C. Hicks, M. Gendron-Maguire, E. B. Rand, G. Weinmaster, and T. Gridley. 1999. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 8:723-730. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto, N., K. Tanigaki, H. Han, H. Hiai, and T. Honjo. 2003. Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr. Biol. 13:333-338. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, P., N. J. Liegeois, C. Wong, M. Finegold, H. Hou, J. C. Thompson, A. Silverman, J. W. Harper, R. A. DePinho, and S. J. Elledge. 1997. Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature 387:151-158. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, P., C. Wong, R. A. DePinho, J. W. Harper, and S. J. Elledge. 1998. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 12:3162-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.